Abstract
The androgen receptor (AR) for the male hormone androgen plays an important role in regulation of cell survival or death depending on the nature of cellular context and extracellular stimuli. The pro-survival function of AR is mediated mainly by transcriptional regulation of its target genes. By contrast, the prodeath function of AR can be transcription-dependent or -independent, although the underlying mechanism of the latter is incompletely understood. Here we report that, in androgen-independent prostate cancer cells, AR promotes UV-induced apoptosis through down-regulation of basal expression of p21 independently of its transcriptional activity. Down-regulation of basal p21 expression depends on AR N-terminal interacting protein PIRH2, an E3 ligase for proteasomal degradation of p53. Silencing of PIRH2 up-regulates p53, which in turn activates p21 transcription. Consistent with this, knockdown of PIRH2 suppresses UV-induced AR-dependent apoptosis. Our data suggest that AR primes androgen-independent prostate cancer cells to DNA damage-induced apoptosis through the PIRH2-p53-p21 axis.
Keywords: Androgen receptor, Prostate cancer, p21, Apoptosis, PIRH2, p53
1. Introduction
Androgen receptor (AR) is a member of the steroid hormone receptor superfamily [1,2]. Like other steroid hormone receptors, AR has three known functional domains: the N-terminal transactivation domain, the C-terminal DNA-binding domain (DBD), and the ligand-binding domain (LBD) [2]. There are several regulatory elements in the N-terminal transactivation domain, including AF1 (activation function 1) and post-translational modification sites for phosphorylation and ubiquitination [3–5]. The AR N-terminal transactivation domain also interacts with dozens of proteins [6] including PIRH2 (p53-induced RING-H2 protein; also known as AR-NIP, AR N-terminal interacting protein) [7,8]. However, the biological function of the interaction between AR and PIRH2 is incompletely understood.
The androgen-AR signaling pathway plays a critical role in regulation of cell death [9–11]. While AR exerts its pro-survival and pro-growth functions through transcriptional regulation of its target genes [12,13], it also has pro-death activity, especially in late stage androgen-independent prostate cancer [14,15]. Previously, we have shown that AR can augment UV-induced cell death through promotion of Bax mitochondrial translocation or generation of apoptotic AR proteolytic fragments [14,16], demonstrating that AR is able to regulate death independent of its transcriptional activity.
The AR N-terminal interacting protein PIRH2 interacts with more than 20 proteins and exerts its functions through various mechanisms [8,17]. It has been reported that PIRH2 can inhibit AR N- and C-terminal interactions [8]. PIRH2 also enhances recruitment of AR to the promoter of the prostate specific antigen (PSA) gene, or promotes degradation of the AR co-repressor, HDAC1 [18,19]. Furthermore, PIRH2 functions as an E3 ligase for a group of signaling molecules involved in DNA damage response, including p53, p73, and p27 [7,17,20,21].
The cell cycle inhibitor p21 is a target gene of p53 [22–24], and its level of expression often determines the sensitivity of cells to genotoxic stresses such as UV irradiation [25–27]. Different E3 ligases, such as SCF and CRL4, have been shown to be involved in p21 degradation, [28,29]. Here we show that AR down-regulates basal p21 expression through the PIRH2-p53-p21 axis in androgen-independent prostate cancer cells, thereby priming the cells to UV-induced apoptosis.
2. Materials and methods
2.1. Reagents and cell culture
Antibodies against p21 (H164) and AR (H280) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody against β-actin was from Millipore (Billerica, MA). Antibody against PIRH2 was from Abcam (Cambridge, MA). The fluorogenic caspase-3 substrate DEVD-AFC (carbobenzoxy-Asp-Glu-Val-Asp-7-amino-4- trifluoromethyl coumarin), protein synthesis inhibitor cycloheximide (CHX), and proteasomal inhibitor MG-132 were from Calbiochem (La Jolla, CA). Human prostate cancer cell LNCaP subline 104-R cells and AR knockdown 104-R (AR siRNA) cells were generated and maintained as described previously [14]. Both cell lines were cultured in Dulbecco’s Modified Eagle medium (DMEM) with 10% dextran-coated, charcoal-stripped fetal bovine serum [9].
2.2. Immunoblot analysis
Cells were harvested and lysed in NP-40 buffer (145 mM NaCl, 5 mM MgCl2, 20 mM HEPES, 1 mM EGTA, 1 mM EDTA, 10 mM DL-dithothreitol (DTT), 0.2% Nonidet P40) with a protease inhibitor cocktail (100 μM PMSF, 10 μg/ml Leupeptin, 10 μg/ml Aprotinin, 1 mM Benzamidine). Equal amounts of cell lysates (60 μg) were subjected to SDS–PAGE and transferred onto a PVDF membrane. The membrane was blocked in 3% fat-free milk or 3% BSA for 1 h and incubated sequentially with the primary antibody overnight at 4 °C. The target proteins were detected using Super-Signal chemiluminescence reagents (Pierce Biotechnology, Inc., Rockford, IL).
2.3. Caspase-3 activity assay
Assays of Caspase-3 activity were performed as previously described [30]. Briefly, cell lysate (40 μg) in buffer A (25 mM HEPES, pH 7.4, 5 mM EDTA, 2 mM DTT, 0.1% Chaps) was incubated at 37 °C for 1 h with fluorogenic caspase-3 substrate DEVD-AFC in the dark. Fluorogenic intensities were then monitored using a Gemini EM microplate spectrofluorometer (Molecular Devices) at 360 nm excitation and 445 nm emission.
2.4. RNA interference
All siRNA duplexes were designed and obtained from Dharmacon Research. The siRNA targeting sequence for AR is 5′-CTTCGACCATTTCTGACAA-3′; for PIRH2, 5′-CCAACAGACTTGTGAAGAA-3′; for p53, 5′-AGACCTATGGAAACTACTT-3′; and that for luciferase, 5′-CGUACGCGGAAUACUUCGA-3′. The siRNAs were transfected into cells using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions; after 48 h, cells were harvested for further experimentation.
2.5. Lentivirus encoding siRNA
The lentiviral RNA interference expression vector pLKO.1, VSVG envelope protein vector pCMV-VSV-G, and lentiviral packaging vector pCMV-dR8.2 dvpr were from Addgene [31]. For each target gene, a pair of complementary 64-base hairpin oligonucleotides, each containing a palindromic 19-base pair of targeted siRNA sequences in inverted repeat orientation, were synthesized (Integrated DNA Technologies). After annealing, the 64-mer duplex was inserted into AgeI- and EcoRI-digested pLKO.1. The siRNA/pLKO.1 constructs were then transfected into HEK293T cells with pCMV-VSV-G and pCMV-dR8.2 dvpr, and viral supernatants were collected 48 h post transfection. These supernatants were used immediately to infect target cells, which were selected for three weeks with puromycin to remove uninfected cells prior to experimentation.
2.6. Analytical tools and statistics
The relative densities of p21 protein signals in immunoblots were measured and analyzed using the Image Processing and Analysis in Java (Image J) program (http://rsbweb.nih.gov/ij/). Statistical analysis for comparisons of caspase-3 activities and the relative densities of p21 protein were performed using the two-tailed Student’s t test. p-values less than 0.05 were considered statistically significant.
3. Results
3.1. UV induces p21 degradation in an AR-dependent but androgen-independent manner
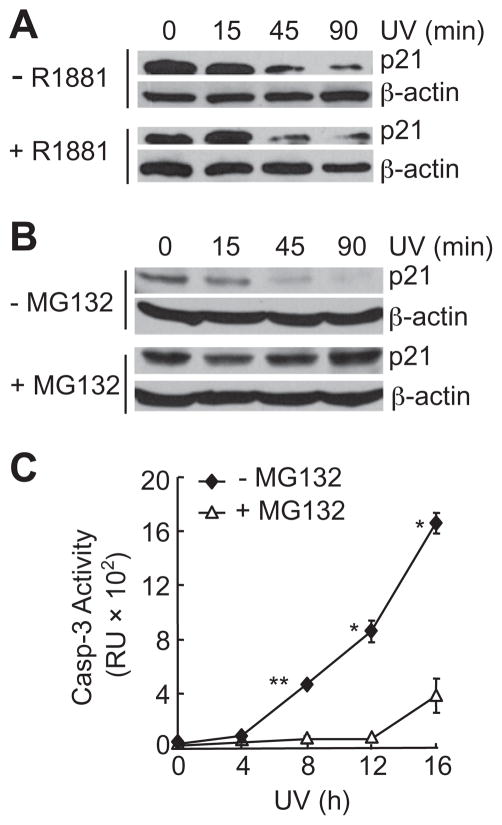
Previously, we have showed that AR can sensitize UV-induced AR positive prostate cancer cell death in an androgen-independent manner, by promoting Bax mitochondrial translocation or generation of pro-apoptotic AR N-terminal fragments [14,16]. In searching for additional downstream effector(s) that may mediate androgen-independent pro-death activity of AR, we noticed that the protein level of p21, which is an AR target gene, was significantly reduced in androgen-independent AR-positive 104-R cells 90 min after UV stimulation (Fig. 1A). The reduction of p21 proteins was inhibited by the proteasomal inhibitor MG-132 (Fig. 1B), but not influenced by androgen (Fig. 1A). These results demonstrated that p21 underwent proteasomal degradation upon UV stimulation, consistent with the previous reports [27]. Interestingly, UV-induced apoptotic cell death was also inhibited by MG-132 but not by androgen (Fig. 1C) [13]. The degradation of p21 was not a consequence of cell death, as it occurred within 1 h following UV irradiation, while the onset of the cell death was detected only after 4–8 h (Fig. 1C). Thus, proteasomal degradation of p21 proteins may contribute to UV-induced apoptosis in AR-positive prostate cancer cells.
Fig. 1.
UV-induced degradation of p21 protein in AR-positive 104-R cells. (A) 104-R cells were treated with or without a synthetic androgen, R1881 (2 nM) for 2 h followed by UV irradiation (100 J/m2). The p21 protein levels were analyzed by immunobloting with anti-p21 antibody. (B and C) 104-R cells were treated with or without proteasome inhibitor MG-132 (10 μM) for 2 h. The p21 protein levels were analyzed by immunoblotting (B), as described in (A). Caspase activity was measured using fluorogenic substrate Ac-DEVD-AFC (C). *p < 0.05, **p < 0.01.
3.2. AR regulates basal p21 expression independently of its transcription activity
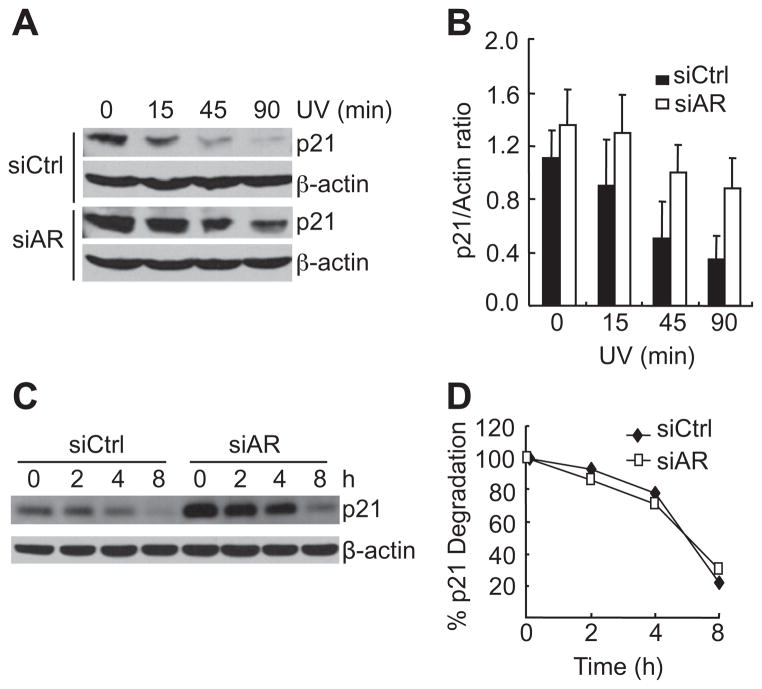
To determine whether the degradation of p21 is regulated by AR, 104-R cells were stably transfected with control siRNA (siCtrl) or siRNA against AR (siAR), which significantly knocked down AR [14]. Upon UV-irradiation, p21 was rapidly degraded in cells transfected with siCtrl: a decrease of 80% in p21 level occurred 45 min after UV stimulation (Fig. 2A). By contrast, p21 degradation was significantly reduced in cells transfected with siAR (Fig. 2A). Basal p21 expression, however, was much higher in AR knockdown cells than that in the control cells (Fig. 2A). Quantitative immunoblot analysis revealed that the rate of p21 degradation was similar between AR-positive and AR-negative cells (Fig. 2B). Consistent with this result, the half-life of p21 proteins was similar between AR-positive and -negative cells (Fig. 2C and D). These results suggest that AR negatively regulates basal p21 expression, rather than its degradation rate.
Fig. 2.
AR controls basal p21 expression, but not its degradation rate. (A) 104-R cells stably transfected with either control siRNA (siCtrl) or AR siRNA (siAR) [14]. Cells were irradiated by UV (100 J/m2) for various periods of time, as indicated. p21 protein levels were analyzed by immunoblotting with anti-p21 antibody. (B) Quantitation of p21 protein levels using image J software. The data are from three independent experiments (C) 104-R cells were treated with MG-132 (10 μM) for 12 h, completely washed, and then treated with CHX (50 μg/ml) for various periods of time, as indicated. The p21 protein levels were analyzed by immunoblotting with anti-p21 antibody. (D) Quantitation of p21 protein levels using image J software.
3.3. Basal p21 level is controlled by the AR binding protein PIRH2 pathway
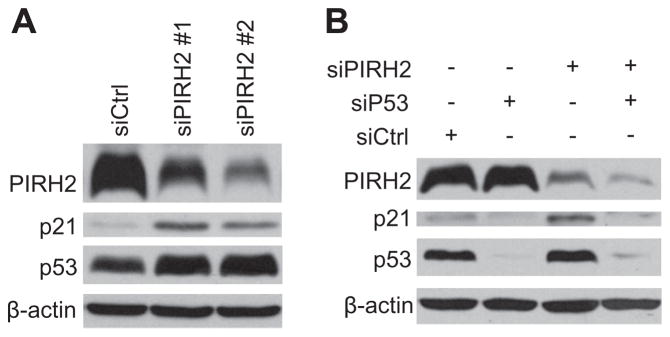
PIRH2 was initially identified as an AR N-terminal interacting protein that acts as an E3 ligase to ubiquitinate p53, thereby triggering p53 degradation [7]. Since p21 is a p53 target gene in cells stimulated by various stress signals including genotoxic stress [22,32], we were curious about whether AR may control basal p21 expression through the PIRH2-p53 axis. To test this hypothesis, we used the siRNA approach. AR-positive 104-R cells were transfected with control siRNA (siCtrl) or two different siRNAs against PIRH2 (siPIRH2). Immunoblotting analysis revealed that PIRH2 expression was significantly reduced by siPIRH2 (Fig. 3A). Under the same conditions, p53 and p21 protein levels are upregulated (Fig. 3B). Furthermore, up-regulation of p21 expression by siPIRH2 was diminished when p53 was simultaneously knocked down (Fig. 3B). These data demonstrate that PIRH2, via p53, controls basal p21 expression.
Fig. 3.
Basal p21 protein levels are controlled by the PIRH2-p53 pathway. (A) 104-R cells were transfected with either control siRNA (siCtrl) or two different PIRH2 siRNAs (siPIRH2 #1 and #2) for 48 h. Expression levels of p21, p53, and actin were analyzed by immunoblotting using corresponding antibodies. (B) 104-R cells were transfected with siCtrl, siPIRH2, or p53 siRNA (siP53). Expression levels of p21 and p53 were analyzed by immunoblotting, as described in (A).
3.4. AR sensitizes UV-induced cell death through the PIRH2-p53-p21 axis
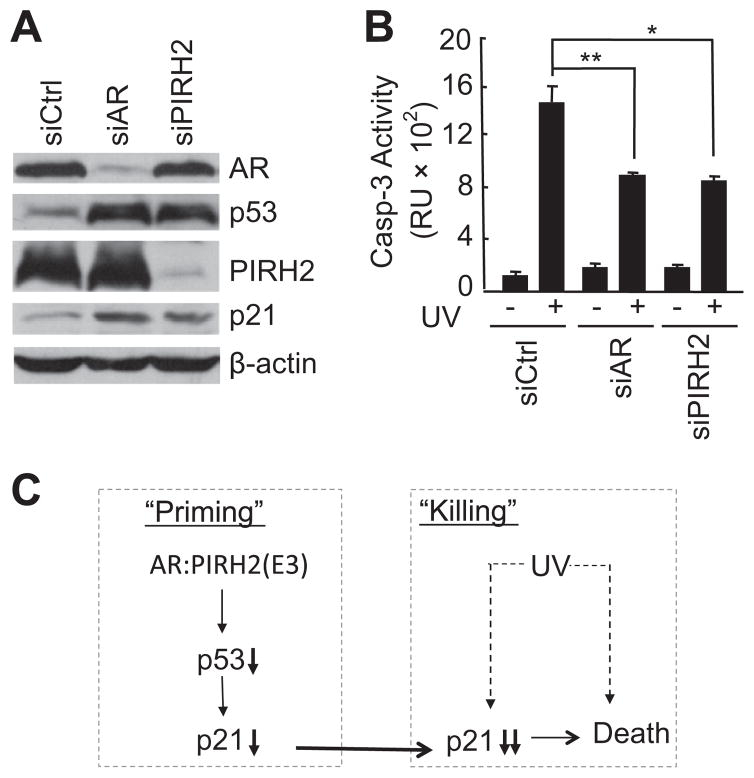
To determine whether AR sensitizes UV-induced cell death through the PIRH2-p53-p21 axis, 104-R cells were infected with lentiviruses encoding siCtrl, siAR, or siPIRH2. Immunoblotting analysis revealed that levels of AR and PIRH2 were significantly reduced by their corresponding siRNAs (Fig. 4A). As expected, knockdown of either AR or PIRH2 resulted in up-regulation of p21 expression (Fig. 4A). Knockdown of AR also impaired negative regulation of p53 by PIRH2, resulting in the enhancement of p53 expression (Fig. 4A). Importantly, UV-induced AR-mediated apoptosis was significantly inhibited in PIRH2-knocked down cells (Fig. 4B). Thus, the PIRH2-p53-p21 axis contributes to AR-mediated sensitization of prostate cancer cells to UV-induced apoptosis (Fig. 4C).
Fig. 4.
AR via the PIRH2-p53-p21 axis primes androgen-independent 104-R cells to UV-induced apoptosis. (A) and (B) 104-R cells were infected with lentiviruses encoding either AR shRNA or PIRH2 shRNA. Expression of PIRH2, AR, and p21 was analyzed by immunoblotting with corresponding antibodies (A). Apoptotic caspase assay (B). (C) Schematic illustration of how AR via the PIRH2-p53-p21 axis sensitizes androgen-independent prostate 104-R cancer cells to UV irradiation. AR promotes PIRH2-mediated p53 degradation, thereby reducing basal p21 expression. This will reduce the apoptotic threshold to UV-induced apoptosis.
4. Discussion
The male hormone receptor AR plays a critical role in regulation of prostate cancer survival or death, depending on the nature of extracellular stimuli [9,10]. While the pro-survival function of AR is ligand-dependent, the pro-death function of AR can be ligand-independent, through promotion of Bax mitochondria translocation or generation of a pro-apoptotic AR N-terminal fragment [14,16]. It has not been known whether AR can promote cell death through other mechanisms. In this report, we demonstrate that AR can promote UV-induced apoptosis in androgen-independent prostate cancer cells through the PIRH2-p53-p21 axis. Loss of AR led to an increase in p21 protein level without affecting the rate of UV-induced p21 degradation (Fig. 2). Silencing of AR-associated protein PIRH2 resulted in accumulation of p53, up-regulation of p21, and consequent resistance to UV-induced apoptosis (Fig. 3). Finally, knockdown of PIRH2 abrogated UV-induced, AR-mediated apoptosis (Fig. 4). Thus, in addition to promotion of Bax mitochondrial translocation and generation of an apoptotic AR proteolytic fragment, AR utilizes the PIRH2-p53-p21 axis to inhibit basal p21 expression, thereby reducing the apoptotic threshold in prostate cancer cells.
Down-regulation of basal p21 expression by AR primes androgen-independent prostate cancer 104-R1 cells to UV-induced apoptosis. The p21 gene is a target of AR-mediated transcriptional control [33]. Ironically, our results show that AR negatively controls p21 protein level independently of AR transcriptional activity (Fig. 1A). Interestingly, although UV-irradiation induced p21 degradation in 104-R1 cells, the rate of p21 degradation was not controlled by AR (Fig. 2). Instead, AR down-regulated basal p21 expression (Figs. 2 and 3). Importantly, down-regulation of basal p21 expression by AR sensitized 104-R1 cells to UV-irradiation (Figs. 1C and 4B). This observation is also consistent with the fact that the level of p21 is extremely low and unstable in AR-positive 104-R1 cells compared with other AR negative prostate cancer cells, such as PC3 (data not shown). Although the rate of UV-induced degradation of p21 was similar between AR-positive and AR-negative 104-R1 cells (Fig. 2B), down-regulation of p21 basal expression by AR allows UV to more rapidly reduce the amount of p21 to the level that it is no longer able to inhibit UV-induced apoptosis.
The regulation of basal p21 expression by AR in prostate cancer cells depends on the cell context. Our results show that AR promoted UV-induced apoptosis in p53-positive androgen-independent 104-R cells by down-regulating basal p21 expression (Figs. 2 and 3). However, we have also shown that AR up-regulated p21 in a ligand-dependent manner, resulting in the inhibition of JNK activation and apoptosis in TNFα-treated 104-R cells [13]. Thus, AR may control the threshold of cell death by controlling p21 level through both transcription dependent and independent mechanisms in cell type and stimulus dependent manners.
The PIRH2-p53 axis mediates the effect of AR on basal p21 expression. Previously, it has been shown that PIRH2 negatively regulates p53 through ubiquitin-mediated proteasomal degradation [7,34]. Our results show that knockdown of PIRH2, an E3 ligase of p53 [7], abrogated the down-regulation of p21 by AR in unstimulated 104-R cells (Fig. 3). This suggests that PIRH2 acts downstream of AR in regulation of p21 expression and UV-induced apoptosis. Future studies are needed to determine whether PIRH2 directly serves as the E3 ligase for p21. How the interaction between AR and PRIH2 regulates the PRIH2 E3 ligase activity remains unknown. Since knockdown of AR impaired PIRH2 E3 ligase activity toward p53 (Fig. 4A), it is possible that AR may stabilize or promote PIRH2 E3 ligase activity so that it can catalyze p53 proteasomal degradation. Future studies are needed to explore this idea.
Acknowledgments
This work was supported by a National Institutes of Health grant to JX (CA128114).
References
- 1.Chang C, Kokontis J, Swift S, Liao ST. Molecular cloning and structural analysis of complementary DNA of human and rat androgen receptors. Prog Clin Biol Res. 1990;322:53–63. [PubMed] [Google Scholar]
- 2.Chang CS, Kokontis J, Liao ST. Structural analysis of complementary DNA and amino acid sequences of human and rat androgen receptors. Proc Natl Acad Sci USA. 1988;85:7211–7215. doi: 10.1073/pnas.85.19.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J. 2005;391:449–464. doi: 10.1042/BJ20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gioeli D, Ficarro SB, Kwiek JJ, Aaronson D, Hancock M, Catling AD, White FM, Christian RE, Settlage RE, Shabanowitz J, Hunt DF, Weber MJ. Androgen receptor phosphorylation. Regulation and identification of the phosphorylation sites. J Biol Chem. 2002;277:29304–29314. doi: 10.1074/jbc.M204131200. [DOI] [PubMed] [Google Scholar]
- 5.Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc Natl Acad Sci USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beitel L. Androgen Receptor Interacting Proteins and Coregulators Table, 2010. < http://androgendb.mcgill.ca/ARinteract.pdf>.
- 7.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 8.Beitel LK, Elhaji YA, Lumbroso R, Wing SS, Panet-Raymond V, Gottlieb B, Pinsky L, Trifiro MA. Cloning and characterization of an androgen receptor N-terminal-interacting protein with ubiquitin-protein ligase activity. J Mol Endocrinol. 2002;29:41–60. doi: 10.1677/jme.0.0290041. [DOI] [PubMed] [Google Scholar]
- 9.Kokontis JM, Hsu S, Chuu CP, Dang M, Fukuchi J, Hiipakka RA, Liao S. Role of androgen receptor in the progression of human prostate tumor cells to androgen independence and insensitivity. Prostate. 2005;65:287–298. doi: 10.1002/pros.20285. [DOI] [PubMed] [Google Scholar]
- 10.Isaacs JT. Apoptosis: translating theory to therapy for prostate cancer. J Natl Cancer Inst. 2000;92:1367–1369. doi: 10.1093/jnci/92.17.1367. [DOI] [PubMed] [Google Scholar]
- 11.Heisler LE, Evangelou A, Lew AM, Trachtenberg J, Elsholtz HP, Brown TJ. Androgen-dependent cell cycle arrest and apoptotic death in PC-3 prostatic cell cultures expressing a full-length human androgen receptor. Mol Cell Endocrinol. 1997;126:59–73. doi: 10.1016/s0303-7207(96)03970-6. [DOI] [PubMed] [Google Scholar]
- 12.Debes JD, Tindall DJ. The role of androgens and the androgen receptor in prostate cancer. Cancer Lett. 2002;187:1–7. doi: 10.1016/s0304-3835(02)00413-5. [DOI] [PubMed] [Google Scholar]
- 13.Tang F, Kokontis J, Lin Y, Liao S, Lin A, Xiang J. Androgen via p21 inhibits tumor necrosis factor alpha-induced JNK activation and apoptosis. J Biol Chem. 2009;284:32353–32358. doi: 10.1074/jbc.M109.042994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y, Kokontis J, Tang F, Godfrey B, Liao S, Lin A, Chen Y, Xiang J. Androgen and its receptor promote Bax-mediated apoptosis. Mol Cell Biol. 2006;26:1908–1916. doi: 10.1128/MCB.26.5.1908-1916.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh S, Hu YC, Rahman M, Lin HK, Hsu CL, Ting HJ, Kang HY, Chang C. Increase of androgen-induced cell death and androgen receptor transactivation by BRCA1 in prostate cancer cells. Proc Natl Acad Sci USA. 2000;97:11256–11261. doi: 10.1073/pnas.190353897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godfrey B, Lin Y, Larson J, Haferkamp B, Xiang J. Proteasomal degradation unleashes the pro-death activity of androgen receptor. Cell Res. 2010;20:1138–1147. doi: 10.1038/cr.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung YS, Qian Y, Chen X. Pirh2 RING-finger E3 ubiquitin ligase: its role in tumorigenesis and cancer therapy. FEBS Lett. 2012;586:1397–1402. doi: 10.1016/j.febslet.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung YS, Hakem A, Hakem R, Chen X. Pirh2 E3 ubiquitin ligase monoubiquitinates DNA polymerase eta to suppress translesion DNA synthesis. Mol Cell Biol. 2012;31:3997–4006. doi: 10.1128/MCB.05808-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logan IR, Gaughan L, McCracken SR, Sapountzi V, Leung HY, Robson CN. Human PIRH2 enhances androgen receptor signaling through inhibition of histone deacetylase 1 and is overexpressed in prostate cancer. Mol Cell Biol. 2006;26:6502–6510. doi: 10.1128/MCB.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hattori T, Isobe T, Abe K, Kikuchi H, Kitagawa K, Oda T, Uchida C, Kitagawa M. Pirh2 promotes ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. Cancer Res. 2007;67:10789–10795. doi: 10.1158/0008-5472.CAN-07-2033. [DOI] [PubMed] [Google Scholar]
- 21.Jung YS, Qian Y, Chen X. The p73 tumor suppressor is targeted by Pirh2 RING finger E3 ubiquitin ligase for the proteasome-dependent degradation. J Biol Chem. 2011;286:35388–35395. doi: 10.1074/jbc.M111.261537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gartel AL, Tyner AL. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp Cell Res. 1999;246:280–289. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- 23.Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther. 2002;1:639–649. [PubMed] [Google Scholar]
- 24.Reinke V, Lozano G. Differential activation of p53 targets in cells treated with ultraviolet radiation that undergo both apoptosis and growth arrest. Radiat Res. 1997;148:115–122. [PubMed] [Google Scholar]
- 25.Murray SA, Zheng H, Gu L, Jim Xiao ZX. IGF-1 activates p21 to inhibit UV-induced cell death. Oncogene. 2003;22:1703–1711. doi: 10.1038/sj.onc.1206327. [DOI] [PubMed] [Google Scholar]
- 26.Rieber M, Strasberg Rieber M. Apoptosis-inducing levels of UV radiation and proteasome inhibitors produce opposite effects on p21(WAF1) in human melanoma cells. Int J Cancer. 2000;86:462–467. doi: 10.1002/(sici)1097-0215(20000515)86:4<462::aid-ijc3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 27.Soria G, Podhajcer O, Prives C, Gottifredi V. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene. 2006;25:2829–2838. doi: 10.1038/sj.onc.1209315. [DOI] [PubMed] [Google Scholar]
- 28.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- 30.Xiang J, Chao DT, Korsmeyer SJ. BAX-induced cell death may not require interleukin 1 beta-converting enzyme-like proteases. Proc Natl Acad Sci USA. 1996;93:14559–14563. doi: 10.1073/pnas.93.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, Weinberg RA, Novina CD. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 33.Lu S, Liu M, Epner DE, Tsai SY, Tsai MJ. Androgen regulation of the cyclin-dependent kinase inhibitor p21 gene through an androgen response element in the proximal promoter. Mol Endocrinol. 1999;13:376–384. doi: 10.1210/mend.13.3.0254. [DOI] [PubMed] [Google Scholar]
- 34.Corcoran CA, Montalbano J, Sun H, He Q, Huang Y, Sheikh MS. Identification and characterization of two novel isoforms of Pirh2 ubiquitin ligase that negatively regulate p53 independent of RING finger domains. J Biol Chem. 2009;284:21955–21970. doi: 10.1074/jbc.M109.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]