Abstract
Twitter, as a social media platform, has become an increasingly useful data source for health surveillance studies, and personal health experiences shared on Twitter provide valuable information to the surveillance. Twitter data are known for their irregular usages of languages and informal short texts due to the 140 character limit, and for their noisiness such that majority of the posts are irrelevant to any particular health surveillance. These factors pose challenges in identifying personal health experience tweets from the Twitter data. In this study, we designed deep neural networks with 3 different architectural configurations, and after training them with a corpus of 8,770 annotated tweets, we used them to predict personal experience tweets from a set of 821 annotate tweets. Our results demonstrated a significant amount of improvement in predicting personal health experience tweets by deep neural networks over that by conventional classifiers: 37.5% in accuracy, 31.1% in precision, and 53.6% in recall. We believe that our method can be utilized in various health surveillance studies using Twitter as a data source.
I. Introduction
Twitter, as a social media platform, has become an increasingly useful data source for a wide range of health surveillance studies. They include investigations of influenza pandemics [1–10], Haitian cholera outbreak [11], Ebola outbreak [12], non-medical use of a psychostimulant drug (Ad-derall) [13], drug abuse [14], smoking [15], suicide risks [16], migraine headaches [17], pharmaceutical product safety [18–22], disease outbreaks during festivals [23], detection of Schizophrenia [24], food-borne illness [25, 26], and even dental pains [27].
In most of these studies, Twitter data were collected by keyword search, which can still leave a significant amount of irrelevant tweets in the study data. For example, Freifeld et al. [18] showed that only 4,401 (7.2%) tweets relevant to the study were discovered from a random sample of 61,402 tweets which were from the collected 6.9 million tweets for 23 medicine products from November 1, 2012 through May 31, 2013. This suggests that a large amount of noisy irrelevant tweets exists in the Twitter data collected.
Various manual and simple methods were used to select samples for research, making the research outcomes difficult to compare and to reproduce. With the sheer volume of Twitter posts, manual approaches will not work well and an automated method is needed. In addition, for the long term ongoing activities for health surveillance, an automatic method capable of correctly identifying study Twitter data is needed.
Personal health experiences shared on Twitter play an important role in health surveillance. Personal experience tweets (PETs) are tweets that describe a person’s encounters, observations, and important events related to his or her life. In studying health related activities, such experiences pertain to changes of a person’s health, due to an illness, a disease, or a treatment. Personal experience tweets contain patient generated information related to their health and such information is an important source of information for study of health related issues. Below are examples of personal health experience tweets:
Feeling dizzy every time I took pregabalin so I google-d the side effects of it
Just starting lyrica, tho it reduced the pain, i cant sleep at the night
Twitter data possess unique characteristics which are not found in many other sources of data. First, each tweet is limited to 140 characters, making users quite creative in coming up with various short texts which do not follow the spelling and grammar of the languages used in order to include the needed information within the limit. Furthermore, emotional expressions in the forms of emoticons and emoji’s are commonly seen in the Twitter posts. Most challengingly to health surveillance, Twitter data are noisy and contain a significant amount of tweets irrelevant to the health issues being studied. The irrelevant, noisy tweets can be those for promoting products, news, and even spamming.
For health related studies, data collected from Twitter require human annotation to confirm and discover what was posted by the users. Annotation is a laborious, time consuming process requiring a significant amount of effort from domain experts, which can be attributed to the slow progress in scaling up the many developed methods to the continuous and ongoing process of health activity surveillance.
For health surveillance, it is important to have an effective and efficient method to identify personal health experience tweets. In this paper, we present our work of developing a deep neural network-based approach to identify such tweets, and compare and discuss the performance of our approach with that of the conventional methods.
II. Related Work
Jiang and colleague introduced the concept of personal experience tweets in discovering drug effects by mining Twitter data [21]. Authors trained three conventional classifiers (naïve Bayes, SVM, and maximum entropy) with a corpus of 600 tweets (300 PETs and 300 non-PETs), and used the trained models to classify 285 tweets. Data sets used in their study seem to be small and the performance may not be generalized to the population of Twitter data.
Recently, in developing an efficient and effective method of constructing a corpus of personal experience tweets, Jiang and colleagues [28] iteratively trained three conventional classifiers (IB1 – nearest neighbors, J48 – decision tree, and MLP – multilayer perceptron) with annotated tweets to derive a corpus of 8,770 annotated tweets (2,067 PETs and 6,703 non-PETs). While their prediction performance on the training data looked strong, but in each iteration, the predictions on the unannotated data did not perform well as on the training data. This is because that authors wanted to reduce the annotation cost by only annotating the predicted positive tweets from which only the prediction precision could be measured, and it ranged from 0.28 to 0.49.
III. Method
Identifying personal health experience tweets is a binary classification problem. Due to the uniqueness of Twitter data, commonly used linear classifiers do not perform well. Deep neural networks are known for their ability to perform well for situations where linear solutions fail. In this project, we designed deep neural networks with three different architectural configurations, and trained and tested them with annotated personal experience tweets related to the use of 4 dietary supplements.
A. Data Sets
From May 30, 2014 to December 8, 2014, we collected 108,528 number of tweets related to 4 dietary supplements (Echinacea, Melatonin, St. John’s Wort, and Valerian.) through the use of Twitter REST APIs1. Names of the dietary supplements were used as keywords in retrieval of data. Any retweets and non-English tweets were discarded. Of the collected tweets, a corpus of 8,770 annotated tweets was constructed [28], and it was used as the training set in this project. We also randomly identified and annotated 821 tweets (485 PETs and 336 non-PETs) from the collected data to form the test set. Tweets in the test dataset were randomly chosen from all four dietary supplements and across the timespan of the data collection.
B. Features
The retrieved tweets not only include the textual data but also the metadata. Upon experimenting and observation, we identified 19 features that can be useful in this study.
Count of frequent terms. They are the textual terms (tokens) frequently appearing in one class but not in the opposite class. Four features related to frequent terms were extracted after scanning the training data, and they are for the positive class and negative class in the tweet text and the Twitter user name which can be phrases.
Count of URLs. Irrelevant tweets tend to include URLs in the tweet text, and a small number of relevant tweets contain URLs to provide additional information.
Count of emotion words. To some extent, the sentiment of an individual tweet expresses the type of a Twitter user’s experience. For instance, a pleasant experience may be indicated by a happy expression.
Twitter client application. Commercial purpose and spam tweets tend to use client applications which can automatically post to Twitter – for instance, twitterfeed.com, whereas individual Twitter users tend to use a different set of Twitter clients such as Twitter mobile apps and the official Twitter Website [29].
Counts of personal pronouns, first person pronouns and second person pronouns. To distinguish personal from non-personal tweets, the usage of personal pronouns can be valuable information because personal tweets tend to use personal pronouns more frequently than non-personal tweets as studied by Elgersma et al. on personal blogs [30].
In addition, we also include in our features the counts of unique words and total number of words, as well as Twitter user id.
C. Deep Neural Networks
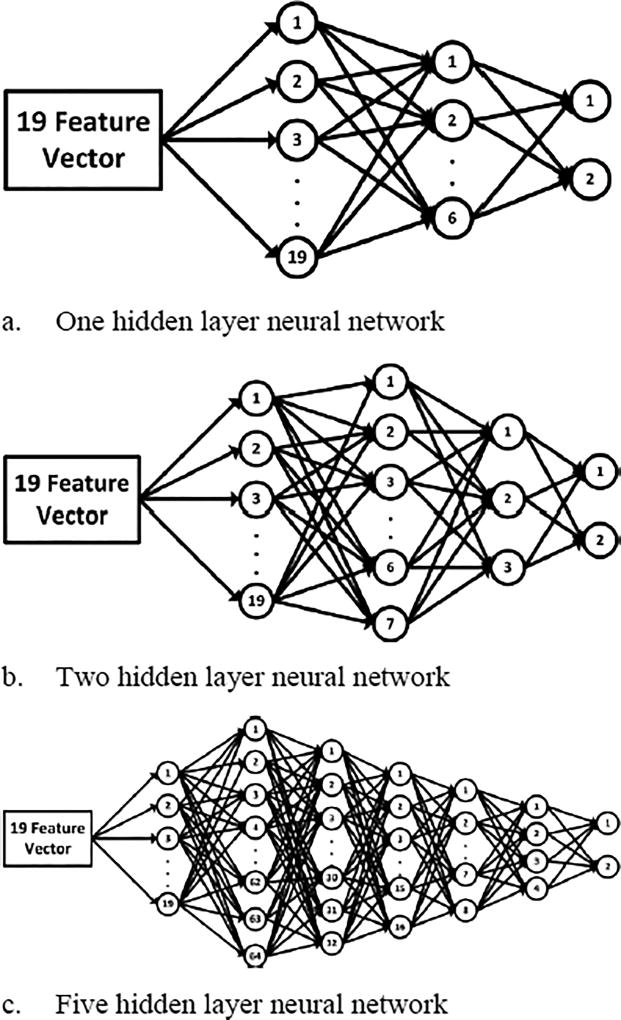
Neural networks with three different architectural configurations were chosen in this study as shown in Figure 1. These configurations consisted of 1-hidden-layer neural network with 19 inputs mapped to 6-neuron hidden layer producing a 2-class output as positive (PET) or negative (non-PET). The second configuration consisted of 2-hidden layer neural network with 19 inputs connecting first hidden layer with 7 neurons followed by another with 3 neurons connecting to the 2-class output layer. And the 3rd configuration is a 5-hidden layer neural network with 19-neuron input connecting to first hidden layer which consists of 64 neurons followed by second, third, fourth and fifth hidden layers consisting of 32, 16, 8 and 4 neurons respectively, passing final output to the 2-class output layer.
Figure 1.
Deep neural network (DNN) architectures used in this study.
The neural networks were implemented using the Google’s TensorFlow platform2 along with scikit-learn libraries3. For each of the three configurations, a training set consisting of 8,770 annotated tweets were used and iterated over 2,000 epochs with a batch size of 128 – we chose a sufficiently large enough number of epochs to ensure that each individual configuration will reach to a stable state. All three models were tested with a test set of 821 annotated tweets. For calculating the cost, gradient descent optimizer with value 0.001 was used. Softmax with cross entropy with logit was used for loss calculation.
D. Baseline Classifiers
To benchmark the prediction performance of the deep neural networks, we chose the following commonly used classifiers: 1) IB1 – k nearest neighbor, 2) J48 – decision tree, 3) LR – logistic regression, and 4) SVM – support vector machine. The performance of these classifiers served as the baseline in comparison. Weka4, which includes the implementation of all these classifiers, was used to gather the performance data on the same data sets.
IV. Results
We used the same training data (8,770 tweets) to train all the classifiers and later used the trained models to classify the positive tweets (PETs) and negative tweets (non-PETs) on the same set of test data (821 tweets). Listed in Table I are the results of the classifiers we tested. In the table, precision, recall and F1 are only for the positive class (PET), and accuracy and ROC (which is the area under curve) are for both positive and negative classes. LR stands for logistic regression, and DNN1, DNN2, and DNN5 represent 1-hidden layer, 2-hidden layer and 5-hidden layer neural networks respectively. For each performance measure, the highest (best) value is in boldface.
TABLE I.
Prediction Results
| Classifier | Accuracy | Precision (PET) |
Recall (PET) |
F1 (PET) |
ROC |
|---|---|---|---|---|---|
| IB1 | 0.479 | 0.689 | 0.214 | 0.327 | 0.537 |
| J48 | 0.565 | 0.721 | 0.431 | 0.539 | 0.618 |
| LR | 0.493 | 0.714 | 0.237 | 0.356 | 0.649 |
| SVM | 0.440 | 0.654 | 0.109 | 0.187 | 0.513 |
| DNN1 | 0.777 | 0.945 | 0.662 | 0.778 | 0.802 |
| DNN2 | 0.772 | 0.953 | 0.647 | 0.770 | 0.803 |
| DNN5 | 0.765 | 0.945 | 0.638 | 0.762 | 0.795 |
V. Discussions
As shown in Table I, all three DNN classifiers outperform all conventional classifiers tested (IB1, J48, LR, and SVM) by a noticeable margin, with DNN1 and DNN2 being the best for the deep neural networks and J48 for the conventional classifiers. Summarized in Table II are the significant performance improvements of predicting PETs with DNN classifiers over that with the best conventional classifier (J48).
TABLE II.
Prediction Performance Improvements
| Accuracy | Precision (PET) |
Recall (PET) |
F1 (PET) |
ROC | |
|---|---|---|---|---|---|
| J48 | 0.565 | 0.721 | 0.431 | 0.539 | 0.618 |
| Best of DNN1 & DNN2 | 0.777 | 0.953 | 0.662 | 0.778 | 0.803 |
| Change | 37.5% | 31.1% | 53.6% | 44.3% | 29.8% |
These significant improvements in performance can help with health surveillance tasks. The improved accuracy will help predict both true positive tweets (PETs) and true negative tweets (non-PETs) more accurately. The higher precision of predicting positive tweets (PETs) will help include more positive tweets (PETs) in the result, effectively reducing the number of irrelevant, noisy tweets and the annotation effort. The increased recall will help minimize the number of actual positive tweets (PETs) missed by the imperfect classifiers – in other words, the result will miss fewer number of actual positive tweets (PETs).
Another observation of our results is that more number of hidden layers in the neural network does not seem to help improve the prediction performance significantly on the test dataset we used. The single hidden layer architecture performed the best in our study, and the 5-hidden layer neural network performed the worst among the 3 configurations tested. This may indicate that 1) the single hidden layer architecture could suffice for predicting positive tweets (PETs), or 2) a larger data size may be needed to train deep neural networks for more accurate performance measure.
Although the simplest neural network performed (nearly) best, the cost of training the model was higher than that of training the more sophisticated neural network models. This is because it takes longer time (more epochs) for the single hidden layer model to reach to a stable state - we observed that it took about 1,500 epochs for the single hidden layer model but roughly 100 epochs for the 5-hidden layer neural network to reach the stable state. This suggests that if the training time is essence and slightly poorer performance is acceptable, the 5-layer model can be the choice, but if the abundant computational power is available and a simple architecture is preferred, the single hidden layer architecture can be the choice.
In this study, a small set of annotated tweets were used to test the algorithms. As we realize, the data set, which was chosen randomly, may not be representative to the tweet population. In our future research, we plan to continue collecting and annotating personal health experience tweets and investigate if our method will be applicable to the larger sets of Twitter data.
VI. Conclusion
In this research, we demonstrated that deep neural networks performed significantly better in classifying personal health experience tweets (PETs) from non-personal health experience tweets (non-PETs) than the conventional classifiers did, indicating the effectiveness of deep neural networks for health surveillance tasks. We believe that our method can be utilized to automate health surveillance activities that use Twitter as the data source.
Acknowledgments
Authors wish to thank Yongbing Tang for collecting the Twitter data, and Cecelia Lai for annotating the tweets.
Footnotes
Research supported in part by the National Institutes of Health grant 1R15LM011999-01.
Contributor Information
Keyuan Jiang, Department of Computer Information Technology and Graphics, Purdue University Northwest, Hammond, IN 46321 USA.
Ravish Gupta, Department of Computer Information Technology and Graphics, Purdue University Northwest, Hammond, IN 46321 USA.
Matrika Gupta, Department of Computer Information Technology and Graphics, Purdue University Northwest, Hammond, IN 46321 USA.
Ricardo A. Calix, Department of Computer Information Technology and Graphics, Purdue University Northwest, Hammond, IN 46321 USA
Gordon R. Bernard, Department of Medicine, Vanderbilt University, Nashville, TN 37232 USA
References
- 1.Chew C, Eysenbach G. "Pandemics in the age of Twitter: content analysis of Tweets during the 2009 H1N1 outbreak," (in eng) PLoS One. 2010;5(11):e14118. doi: 10.1371/journal.pone.0014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Signorini A, Segre AM, Polgreen PM. "The use of Twitter to track levels of disease activity and public concern in the U.S. during the influenza A H1N1 pandemic," (in eng) PLoS One. 2011;6(5):e19467. doi: 10.1371/journal.pone.0019467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collier N, Son NT, Nguyen NM. "OMG U got flu? Analysis of shared health messages for bio-surveillance," (in eng) J Biomed Semantics. 2011 Oct;2(Suppl 5):S9. doi: 10.1186/2041-1480-2-S5-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilge U, Bozkurt S, Yolcular BO, Ozel D. "Can social web help to detect influenza related illnesses in Turkey?" (in eng) Stud Health Technol Inform. 2012;174:100–4. [PubMed] [Google Scholar]
- 5.Nagel AC, Tsou MH, An L, Gawron JM, Gupta DK, Spitzberg B, Yang J, Han S, Peddecord KM, Sawyer MH, Lindsay S. "The complex relationship of realspace events and messages in cyberspace: case study of influenza and pertussis using tweets," (in eng) J Med Internet Res. 2013 Oct;15(10):e237. doi: 10.2196/jmir.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gesualdo F, et al. "Can Twitter Be a Source of Information on Allergy? Correlation of Pollen Counts with Tweets Reporting Symptoms of Allergic Rhinoconjunctivitis and Names of Antihistamine Drugs," (in eng) PLoS One. 2015;10(7):e0133706. doi: 10.1371/journal.pone.0133706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broniatowski DA, Paul MJ, Dredze M. "National and local influenza surveillance through Twitter: an analysis of the 2012–2013 influenza epidemic," (in eng) PLoS One. 2013;8(12):e83672. doi: 10.1371/journal.pone.0083672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fung IC, et al. "Chinese social media reaction to the MERS-CoV and avian influenza A(H7N9) outbreaks," (in eng) Infect Dis Poverty. 2013 Dec;2(1):31. doi: 10.1186/2049-9957-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagar R, Yuan Q, Freifeld CC, Santillana M, Nojima A, Chunara R, Brownstein JS. "A case study of the New York City 2012–2013 influenza season with daily geocoded Twitter data from temporal and spatiotemporal perspectives," (in eng) J Med Internet Res. 2014 Oct;16(10):e236. doi: 10.2196/jmir.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen C, Tsou MH, Aslam A, Nagel A, Gawron JM. "Applying GIS and Machine Learning Methods to Twitter Data for Multiscale Surveillance of Influenza," (in eng) PLoS One. 2016;11(7):e0157734. doi: 10.1371/journal.pone.0157734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chunara R, Andrews JR, Brownstein JS. "Social and news media enable estimation of epidemiological patterns early in the 2010 Haitian cholera outbreak," (in eng) Am J Trop Med Hyg. 2012 Jan;86(1):39–45. doi: 10.4269/ajtmh.2012.11-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odlum M, Yoon S. "What can we learn about the Ebola outbreak from tweets?," (in eng) Am J Infect Control. 2015 Jun;43(6):563–71. doi: 10.1016/j.ajic.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanson CL, Burton SH, Giraud-Carrier C, West JH, Barnes MD, Hansen B. Tweaking and tweeting: exploring Twitter for nonmedical use of a psychostimulant drug (Adderall) among college students. J Med Internet Res. 2013 Apr;15(4):e62. doi: 10.2196/jmir.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chary M, Genes N, McKenzie A, Manini AF. "Leveraging social networks for toxicovigilance," (in eng) J Med Toxicol. 2013 Jun;9(2):184–91. doi: 10.1007/s13181-013-0299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sofean M, Smith M. "Sentiment analysis on smoking in social networks," (in eng) Stud Health Technol Inform. 2013;192:1118. [PubMed] [Google Scholar]
- 16.Jashinsky J, et al. "Tracking suicide risk factors through Twitter in the US," (in eng) Crisis. 2014;35(1):51–9. doi: 10.1027/0227-5910/a000234. [DOI] [PubMed] [Google Scholar]
- 17.Nascimento TD, et al. "Real-time sharing and expression of migraine headache suffering on Twitter: a cross-sectional infodemiology study," (in eng) J Med Internet Res. 2014 Apr;16(4):e96. doi: 10.2196/jmir.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freifeld CC, Brownstein JS, Menone CM, Bao W, Filice R, Kass-Hout T, Dasgupta N. "Digital drug safety surveillance: monitoring pharmaceutical products in twitter," (in eng) Drug Saf. 2014 May;37(5):343–50. doi: 10.1007/s40264-014-0155-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coloma PM, Becker B, Sturkenboom MC, van Mulligen EM, Kors JA. Evaluating Social Media Networks in Medicines Safety Surveillance: Two Case Studies. Drug Saf. 2015 Aug; doi: 10.1007/s40264-015-0333-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginn R, Pimpalkhute P, Nikfarjam A, Patki A, O'Connor K, Sarker A, Smith K, Gonzalez G. Mining Twitter for adverse drug reaction mentions: a corpus and classification benchmark. Proceedings of the fourth workshop on building and evaluating resources for health and biomedical text processing. 2014 [Google Scholar]
- 21.Jiang K, Zheng Y. The 9th International Conference on Advanced Data Mining and Applications (ADMA 2013) Hangzhou, China, 2013: Springer-Verlag; Mining Twitter Data for Potential Drug Effects; pp. 434–443. [Google Scholar]
- 22.O'Connor K, Pimpalkhute P, Nikfarjam A, Ginn R, Smith KL, Gonzalez G. "Pharmacovigilance on twitter? Mining tweets for adverse drug reactions," (in ENG) AMIA Annu Symp Proc. 2014;2014:924–33. [PMC free article] [PubMed] [Google Scholar]
- 23.Yom-Tov E, Borsa D, Cox IJ, McKendry RA. "Detecting disease outbreaks in mass gatherings using Internet data," (in eng) J Med Internet Res. 2014 Jun;16(6):e154. doi: 10.2196/jmir.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McManus K, Mallory EK, Goldfeder RL, Haynes WA, Tatum JD. "Mining Twitter Data to Improve Detection of Schizophrenia," (in eng) AMIA Jt Summits Transl Sci Proc. 2015;2015:122–6. [PMC free article] [PubMed] [Google Scholar]
- 25.Harris JK, et al. "Health department use of social media to identify foodborne illness - Chicago, Illinois, 2013–2014," (in eng) MMWR Morb Mortal Wkly Rep. 2014 Aug;63(32):681–5. [PMC free article] [PubMed] [Google Scholar]
- 26.Harris JK, et al. "Using Twitter to Identify and Respond to Food Poisoning: The Food Safety STL Project," (in eng) J Public Health Manag Pract. 2017 Feb; doi: 10.1097/PHH.0000000000000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heaivilin N, Gerbert B, Page JE, Gibbs JL. "Public health surveillance of dental pain via Twitter," (in eng) J Dent Res. 2011 Sep;90(9):1047–51. doi: 10.1177/0022034511415273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang K, Calix RA, Gupta M. Construction of a Personal Experience Tweet Corpus for Health Surveillance. ACL 2016. 2016:128. [Google Scholar]
- 29.Westman S, Freund L. Information interaction in 140 characters or less: genres on twitter; Proceedings of the third symposium on Information interaction in context; 2010. pp. 323–328. [Google Scholar]
- 30.Elgersma E, de Rijke M. Personal vs non-personal blogs: initial classification experiments; Proceedings of the 31st annual international ACM SIGIR conference on Research and development in information retrieval; 2008. pp. 723–724. [Google Scholar]