Abstract
Thoracic aortic aneurysm (TAA) is a genetic disease predisposing to aortic dissection. It is important to identify the genetic modifiers controlling penetrance and expressivity to improve clinical prognostication. Exome sequencing was performed in 27 subjects with syndromic or familial TAA presenting with extreme phenotypes (15 with severe TAA; 12 with mild or absent TAA). Family-based analysis of a subset of the cohort identified variants, genes, and pathways segregating with TAA severity among three families. A rare missense variant in ADCK4 (p.Arg63Trp) segregated with mild TAA in each family. Genes and pathways identified in families were further investigated in the entire cohort using the optimal unified sequence kernel association test, finding significance for the gene COL15A1 (p=0.025) and the retina homeostasis pathway (p=0.035). Thus, we identified candidate genetic modifiers of TAA severity by exome-based study of extreme phenotypes, which may lead to improved risk stratification and development of new medical therapies.
Keywords: Aorta, Aneurysm, Genetics, Exome, Modifier, Marfan
Introduction
Thoracic aortic aneurysm (TAA) is an aortopathy characterized by dilation of the proximal aorta and risk of life-threatening complications such as aortic dissection and sudden cardiac death. TAA accounts for at least 10,000 deaths per year in the United States.[1] When a diagnosis of TAA is made, the ability to predict complications of disease is limited. Guidelines for surgery incorporate aortic diameter, genetic diagnosis, and family history, but clinical prediction models are imperfect.[2,3] There is a critical need to improve methods for stratifying a patient's risk in order to optimize clinical decisions such as indications for aortic replacement surgery (ARS), medical therapy, activity restrictions, and frequency of follow up. Broadening the use of individual genotype data is one promising strategy to improve risk stratification.
TAA is clinically and genetically heterogeneous. TAA is associated with autosomal dominant connective tissue disorders (CTDs) including Marfan syndrome (MFS) (FBN1 mutations), Loeys-Dietz syndrome (TGFBR1/TGFBR2), and vascular type Ehlers-Danlos syndrome (COL3A1).[4-6] Nonsyndromic TAA is also frequently genetic and may occur as autosomal dominant familial TAA, which in 15-20% of cases is associated with mutations in genes encoding vascular smooth muscle cell proteins such as ACTA2 and MYH11.[7] Thus existing knowledge of the genetic basis of TAA indicates that pathogenesis is mediated by extracellular matrix (ECM), TGFβ signaling, and the smooth muscle contraction.
Disease severity and the risk for complications are variable not only between individuals with different mutations in the same gene, but also between relatives with the same mutation, making prognosis challenging.[8,9] The severity of non-cardiovascular phenotypic features is also variable and may correlate with TAA severity.[5,10] The genetic mechanisms impacting the degree of TAA severity have not been established. For this study, we hypothesized that whole exome sequencing (WES) in subjects with extreme TAA phenotypes would identify genetic variants that modify TAA severity. We tested this hypothesis by complementary analyses using family-based and case-control approaches.
Methods
Detailed methods are available in the Online Resource Methods.
Study cohort and classification of TAA phenotypes
Subjects were prospectively enrolled from the Cincinnati Children's Hospital Medical Center Cardiovascular Genetics clinic from January 2012 to December 2014. This study was approved by the local Institutional Review Board, and all subjects gave informed consent. Subjects eligible for this study had syndromic or familial TAA or carried the same mutation as a first degree relative with confirmed TAA. Genetic rare variant analyses in this study were restricted to Caucasian subjects. Clinical data, including clinical genetic testing results, were collected by review of the electronic medical record. Subjects were defined as mutation positive if they had a pathogenic or likely pathogenic variant identified by clinical testing in one of 24 genes associated with TAA (Online Resource Table 1). Cardiovascular imaging was performed clinically and included echocardiography, magnetic resonance imaging (MRI), or computed tomography. Published nomograms were used to calculate z-scores for subjects age 18 years or younger.[11] To divide the cohort into extreme phenotype groups, we identified two groups: severe and no/mild TAA. Severe TAA was defined by a history of aortic dissection, need for ARS according to guidelines [2], aortic diameter ≥ 5 cm, or z-score ≥ +6. No/mild TAA was defined by a maximum aortic root or ascending aorta diameter ≤ 4.5 cm or z-score ≤ +4 and no history of dissection or ARS. The no/mild TAA group included mutation positive subjects without TAA (aortic diameter < 4 cm or z-score ≤ +2).[2,12]
WES and variant inclusion criteria
Sequencing was performed in the Cincinnati Children's Genetic Variation and Gene Discovery Core. After quality filtering, there were in total 176,885 variant calls (average 6551 per subject). Rare variants were defined as variants with minor allele frequency <0.01 or absent in public databases from the 1000 Genomes Project and the National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project. Variants were filtered for those that were predicted to result in a protein change.
Family-based analysis for divergent extreme TAA phenotypes
Pairs of first degree relatives within the study cohort who were mutation positive by clinical genetic testing but demonstrated divergent extreme phenotypes were identified from 3 families. For each pedigree the rare protein-changing variants unshared between first degree relatives with divergent phenotypes were selected. These unshared variants and genes were then analyzed across the pedigrees. To aid interpretation of the unshared rare variants within each pedigree, genes were ranked according to functional similarity to known TAA-causing genes using the prioritization algorithm of ToppGene (http://toppgene.cchmc.org).[13] Four bioinformatics damage prediction programs were also used to annotate missense variants. To investigate pathway-level associations, unshared gene lists for each pedigree were analyzed for functional enrichment of Gene Ontology terms and pathways using ToppGene.
Case-control design for rare variant association testing between extreme TAA phenotype groups
We utilized the optimal unified sequence kernel association test (SKAT-O) to perform burden and non-burden association tests using rare variants.[14] The two phenotype groups were defined as a dichotomous variable. Due to the limited power to test rare variants individually in the study cohort, collapsing (i.e. grouping) variants onto genetic regions is necessary. To investigate family-based findings within the overall cohort, each gene that segregated with TAA severity across two or more families was individually tested using SKAT-O in all 27 samples. The significant pathways that were identified in family-based ToppGene enrichment analyses were also tested in the overall cohort, excluding family members in which the enrichment was identified. These samples were excluded because SKAT-O is a non-burden test and these samples, which are by definition enriched for the tested pathways, would skew the model and limit the effects of the other samples in the cohort. In separate exploratory global analysis, all genes containing at least two variants in the overall cohort were tested using gene-level SKAT-O. Pathway-level associations were also globally investigated by collapsing variants within Gene Ontology Biological Process (GO BP) terms.
Statistical analysis
Demographic information, clinical genetic data, additional cardiovascular characteristics, and overall number of autosomal rare variants, were compared between TAA extreme phenotype groups with 2x2 Fisher's exact test (categorical variables) or student's t-test (continuous variables). A p-value < 0.05 was used to define statistical significance.
In family-based variant segregation analysis, simulation was used to estimate the significance of observing any variant overlapping all 3 pedigrees. A detailed description of the simulation method is in the Online Resource Methods. For family-based functional enrichment analysis using ToppGene, a p-value < 0.05 after Bonferroni correction was used to define statistical significance.
SKAT-O analyses were performed using the statistical program R (version 3.2.1) and package ‘SKAT’ (version 1.21). The uniform variant weighting scheme was used. Age and gender were included as covariates. When investigating the targeted subset of genes and pathways identified in family-based analyses, a p-value < 0.05 was used to define statistical significance.[15] For exome wide gene-level SKAT-O, simulation was used to estimate significance. Details are provided in the Online Resource Methods.
Results
Genetic and cardiovascular features of cohort with extreme TAA phenotypes
Genetics evaluation
Demographic and clinical data for the 27 subjects in this study are shown in Table 1. Clinical genetic testing for TAA was previously performed in 26 of 27 subjects. Overall, 20 subjects were positive for TAA-causing mutations in FBN1 (n=11), TGFBR1 (3), TGFBR2 (3), and ACTA2 (3) (Online Resource Table 2). A likely pathogenic variant in TGFB2 was incidentally identified with WES in one subject. The remaining 6 subjects had TAA and autosomal dominant family history of TAA but no identified disease-causing mutation through clinical testing or WES. Four of these 6 genotype-negative subjects were examined by a geneticist experienced in CTD evaluation while two were referred exclusively for genetic counseling. All 4 subjects examined were found to have signs of CTD, including one who met clinical criteria for MFS based on systemic features and TAA (Online Resource Table 3).[16] One subject without a formal examination by a geneticist has a brother with TAA and clinically documented signs of CTD.
Table 1. Cohort description and TAA severity groups.
| Entire cohort, N (%) | Severe TAA (n=15) | No/mild TAA (n=12) | P-value | |
|---|---|---|---|---|
| Age (y)† | 29 ± 16 | 25 ± 15 | 35 ± 16 | 0.10 |
|
| ||||
| Sex | 0.13 | |||
|
| ||||
| Male | 18 (67) | 12 (80) | 6 (50) | |
| Female | 9 (33) | 3 (20) | 6 (50) | |
| Clinical TAA genetic testing performed | 26 (96) | 15 (100) | 11 (92) | 0.44 |
|
| ||||
| Pathogenic mutation | ||||
|
| ||||
| Any TAA gene | 21 (78) | 11 (73) | 10 (83) | 0.66 |
| FBN1 | 11 (41) | 5 (33) | 6 (50) | 0.45 |
| TGFBR1 | 3 (11) | 2 (13) | 1 (8) | |
| TGFBR2 | 3 (11) | 2 (13) | 1 (8) | |
| TGFB2* | 1 (4) | 0 (0) | 1 (8) | |
| ACTA2 | 3 (11) | 2 (13) | 1 (8) | |
| Unknown | 6 (22) | 4 (27) | 2 (17) | |
|
| ||||
| Mitral valve prolapse | 1.0 | |||
|
| ||||
| Present | 8 (30) | 4 (27) | 4 (33) | |
| Absent | 19 (70) | 11 (73) | 8 (67) | |
|
| ||||
| Bicuspid aortic valve | 0.6 | |||
|
| ||||
| Present | 4 (15) | 3 (20) | 1 (8) | |
| Absent | 22 (81) | 11 (73) | 11 (92) | |
| Not reported | 1 (4) | 1 (7) | 0 (0) | |
|
| ||||
| Variant counts† | ||||
|
| ||||
| All variants | 6551 ± 148 | 6560 ± 157 | 6541 ± 141 | 0.75 |
| Rare variants | 331 ± 27 | 334 ± 18 | 328 ± 35 | 0.55 |
Identified with whole exome sequencing
Reported as mean ± standard deviation
Upon WES, there was no significant difference in the overall number of protein-changing rare variants between the severe TAA phenotype (334 ± 18 variants/subject) and no/mild TAA phenotype (328 ± 35) groups (P=0.55). In addition to the variants in TAA genes that were interpreted as pathogenic or likely pathogenic through clinical genetic testing, 13 other rare missense variants in TAA genes that were not classified as pathogenic or likely pathogenic were identified (Table 2). Population allele frequency data, bioinformatics predictions, and previous submissions to ClinVar were used to classify these variants in accordance with the 2015 guidelines set forth by the American College of Medical Genetics and Genomics.[17] Most of these additional non-disease causing variants in TAA genes were found in mutation positive subjects. The MFAP5 or MYLK variant identified in a genotype-negative subject with mild TAA may be disease-causing based on strong damage predictions, as we have previously described.[18]
Table 2. Heterozygous rare variants in TAA genes within study cohort not classified as disease-causing.
| TAA gene in exome | Nucleotide variant | Amino acid change | Allele frequency* | Bio-informatics predictions† | ClinVar classification | Subject ID | TAA gene with mutation | TAA severity |
|---|---|---|---|---|---|---|---|---|
| CBS | c.146C>T | p.Pro49Leu | 0.00018 | +/+/+/+ | Pathogenic‡ | 13TAA1028-1b | ACTA2 | Severe |
| COL3A1 | c.1804C>A | p.Pro602Thr | 0.00440 | -/+/+/+ | Benign or likely benign | 12TAA1018-1 | TGFBR2 | Mild |
| COL3A1 | c.2035G>A | p.Ala679Thr | 0.01369 | -/-/-/- | Benign or likely benign | 14TAA1132-1 | Unknown | Severe |
| COL5A1 | c.2852A>G | p.Asn951Ser | 0.01023 | +/-/-/- | Benign or likely benign | 12TAA1037-2 | Unknown | Severe |
| COL5A1 | c.341C>A | p.Ala114Asp | 0.00061 | +/+/+/+ | Benign or likely benign | 13TAA1084-1 | FBN1 | Mild |
| COL5A2 | c.4240G>A | p.Asp1414Asn | 0.00099 | +/+/+/- | Benign or likely benign | 12TAA1019-1 | TGFBR1 | Severe |
| FBN1 | c.6043G>A | p.Asp2015Asn | NR | -/-/+/- | Benign or likely benign | 13TAA1042-3 | TGFBR1 | Mild |
| FBN2 | c.1592G>C | p.Gly531Ala | 0.00128 | +/+/+/+ | Benign or likely benign | 13TAA1042-3 | TGFBR1 | Mild |
| FBN2 | c.1592G>C | p.Gly531Ala | 0.00128 | +/+/+/+ | Benign or likely benign | 14TAA1042-4 | TGFBR1 | Severe |
| FBN2 | c.1040G>A | p.Arg347His | 0.00451 | -/+/+/- | Benign or likely benign | 13TAA1098-1 | TGFB2 | Mild |
| LOX | c.476C>A | p.Pro159Gln | 0.00555 | +/-/-/- | NR | 12TAA1037-2 | Unknown | Severe |
| MFAP5 | c.341G>A | p.Arg114Gln | NR | +/+/-/+ | NR | 14TAA1142-1 | Unknown | Mild |
| MYH11 | c.5821A>T | p.Thr1941Ser | 0.00124 | -/-/+/- | Benign or likely benign | 12TAA1039-1 | FBN1 | Severe |
| MYLK | c.2693G>A | p.Arg898Gln | 0.00021 | +/+/+/- | NR | 14TAA1142-1 | Unknown | Mild |
Frequency data from Exome Aggregation Consortium (ExAC)
Bioinformatics prediction programs: SIFT/Polyphen2 HDIV/MutationTaster/PROVEAN
CBS mutations cause disease via autosomal recessive mechanism
NR: Not reported
Extreme TAA phenotype groups
The severe TAA group (n=15 subjects) included 13 with history of ARS, which was performed emergently for type A aortic dissection (4) or electively (9). The aortic diameter was documented to be least 5.0 cm in 7 of 8 adults undergoing elective ARS. Pre-operative imaging data was not available for one adult, but ARS was performed soon after TAA diagnosis. The one pediatric subject undergoing elective ARS had aortic root z-score of +9.5. The no/mild TAA group (n=12) included 6 subjects with mild TAA and 6 who had no clinical evidence of TAA but are at risk because they are mutation positive and have at least one first degree relative with TAA who carries the same mutation. All mutation-negative subjects had at least mild TAA. None in the mild TAA group who carried a TGFβ receptor mutation had aortic diameter greater than 4.0 cm. Only one severe TAA subject and one no/mild TAA subject had clinical hypertension.
Family-based analyses of pedigrees with extreme TAA phenotypes
Identification of unshared rare variants within pedigrees
To compare subjects who have the same pathogenic mutation but clearly different TAA severity, 3 pairs of first degree relatives from each pedigree were studied (Figure 1). Pedigree I includes a 9 year old boy with MFS (maternally inherited FBN1 mutation; p.Cys1806Tyr) and severe aortic root dilation (z-score = +6.4), whose mother has no evidence of TAA at age 26. This boy's brother reportedly died secondary to a neonatal aortic dissection. Pedigree II includes a subject with a TGFBR1 mutation (p.Asn478Ser) who underwent elective ARS at age 59 years for ascending aorta diameter of 5.1 cm. In contrast, his sister carries the mutation but has no evidence of TAA at age 55. Neither has findings of Loeys-Dietz syndrome or other CTD, leading to the diagnosis of familial TAA due to the TGFBR1 mutation. Finally, Pedigree III includes a subject with Loeys-Dietz syndrome (TGFBR2 mutation; p.Arg229Pro) resulting in aortic dissection at age 27, whose mutation positive sister only has evidence of mild TAA (aortic root 4 × 3.4 × 3.3 cm on cardiac MRI) at age 32. In each of these families the subject with severe TAA was younger or very close in age to the included relative with no/mild TAA.
Fig. 1.
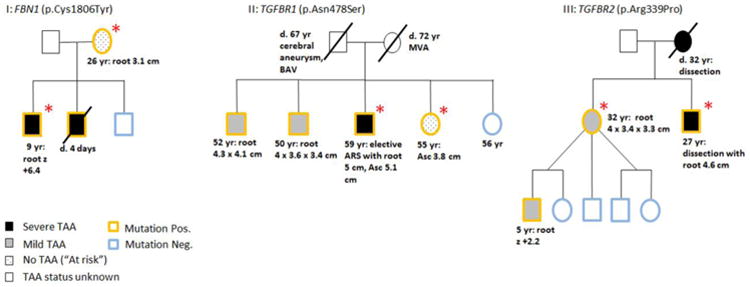
Three pedigrees containing first degree relatives with divergent extreme TAA phenotypes. - Red asterisks indicate subjects sequenced for family-based variant analyses. ARS: aortic replacement surgery, Asc: ascending aorta, BAV: bicuspid aortic valve, MVA: motor vehicle accident
Genes and variants associate with TAA severity across pedigrees
For each pedigree, a list of the rare variants that were unshared between first degree relatives was tabulated. The number of unshared rare variants in each pedigree were similar (Figure 2). In each pedigree, the subject with severe TAA had more rare variants overall and more with strong bioinformatics damage predictions (Online Resource Table 4). Lists of unshared rare variants and genes were compared between pedigrees. Those overlapping across at least 2 pedigrees were selected for further analysis.
Fig. 2.
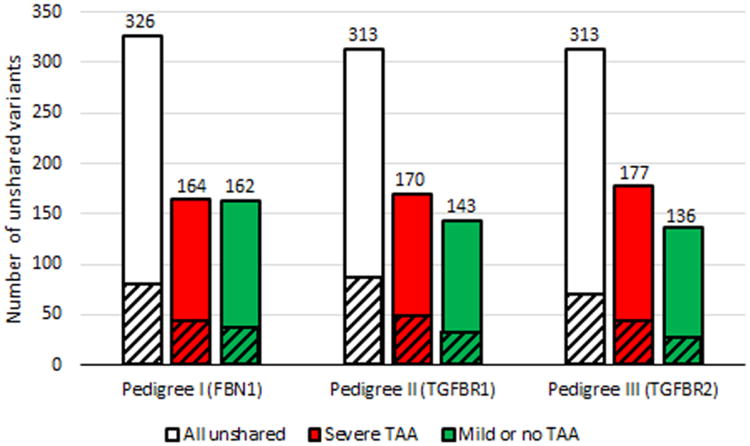
Distribution of unshared rare variants between first degree relatives with divergent TAA phenotypes. - Cross-hatched areas represent the number of variants predicted to be damaging by 4 of 4 bioinformatics programs or frameshift, nonsense, stop loss, splice site, or initiation codon variants
In total there were 50 genes with unshared rare variants overlapping across pedigrees. These are organized in Figure 3 to convey variant- and gene-level annotation data, including damage predictions and functional similarity to TAA genes based on ToppGene ranking. To our knowledge, none of these genes is reported to cause TAA independently. For most genes, the rare variants were family specific. However, there were 6 genes (marked by asterisks in Figure 3) for which the identical variant was identified in at least 2 pedigrees. This includes a heterozygous variant in ADCK4 (c.187C>T, p.Arg63Trp) identified in individuals with mild TAA in all 3 pedigrees. This variant is predicted to affect function by all 4 bioinformatics programs. Based upon the reported minor allele frequency for this variant in control populations (0.005987), simulations utilizing 5000 sequencing data sets predict the likelihood for this occurring by chance as 0.009, strongly supporting the significance of this observation. Meanwhile, variants in GATA2, KLC4, THSD7B, MCL1, and ZNF98 overlapped at least two pedigrees. Each was identified in subjects with concordant TAA phenotypes (Figure 3). None of these 6 variants, including the ADCK4 variant, was otherwise identified in the overall cohort.
Fig. 3.
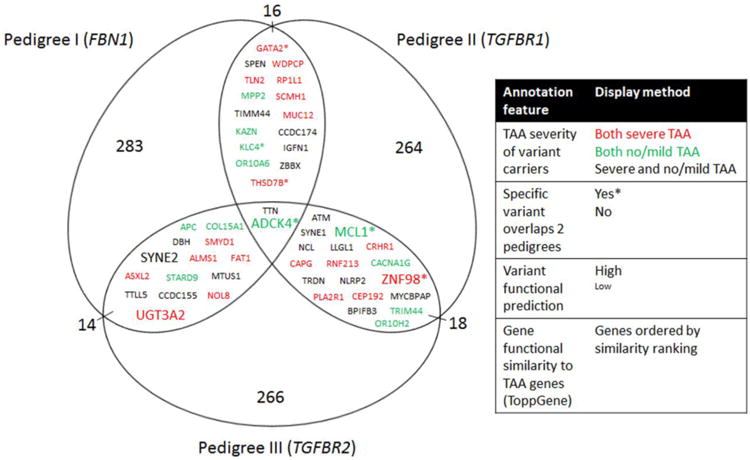
Genes and specific variants overlap across pedigrees. - Variants that were unshared between first degree relatives were collected for each pedigree. Two genes overlapped all 3 pedigrees, and 14 to 18 genes overlapped any two pedigrees. These included ADCK4 for which the same variant was identified in subjects with mild TAA in all 3 pedigrees. Five other specific variants overlapped at least 2 of the 3 pedigrees (asterisks). The predicted functional impact of variants was stratified as high (missense variants predicted damaging in at least 4 of 4 programs or frameshift/stop gain/splice variant) or low (missense variants predicted damaging in 0-3 programs or inframe insertions or deletions) and displayed by font size. The genes are ordered based on ToppGene ranking for similarity to TAA genes, reading from top to bottom and left to right within each overlap segment
In order to further investigate the significance of the 50 overlapping genes, each was tested for association with TAA severity in the overall cohort using SKAT-O. Among these prioritized genes, COL15A1 was most significantly associated with TAA severity (p=0.025). The COL15A1 variants (p.Phe851Leu, p.Ile1304Met) segregating with mild TAA in pedigrees are each predicted damaging by at least two prediction programs. The COL15A1 variant (p.Phe851Leu) identified in Pedigree III was also identified within the overall cohort in a subject with mild TAA who carries a TGFB2 mutation. Thus, in total three subjects with mild TAA carried a rare coding variant in COL15A1.
Functional enrichment analysis identified significant pathways in families
Enrichment analysis of each pedigree's list of unshared rare variants using ToppGene identified 7 significant GO terms or pathways (Figure 4). Similar to our gene-level, targeted investigation of family-based findings in the overall cohort, we tested whether the gene sets identified by enrichment analysis in families could be validated in the remainder of the cohort using SKAT-O. Interestingly, the retina homeostasis genes that were enriched in Pedigree I were also significantly associated with TAA severity (p=0.035) (Online Resource Table 5). In contrast, variants in known TAA genes were not collectively associated with TAA severity using SKAT-O (p=0.54). Replication of a family-based finding within the overall cohort strongly supports these retina genes as candidate modifiers. Thus, our tiered approach identified significant candidate modifiers at the variant, gene, and pathway levels, as summarized in Figure 5.
Fig. 4.
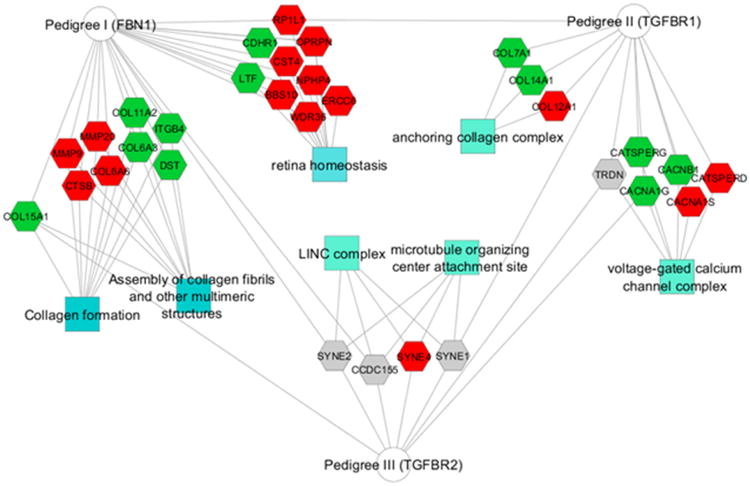
Rare variants unshared between first degree relatives were enriched for GO and pathway annotations. - Pedigrees (circles) are connected to significantly enriched annotations (squares) by edges that pass through the genes contributing to the enrichment (hexagons). Genes found in severe TAA subjects are red, genes in mild TAA subjects are green, and genes in both severe and mild TAA subjects are gray. None of the enrichments overlapped multiple pedigrees, but some genes within the significant enrichments overlapped pedigrees as indicated by edges connecting a gene to more than one pedigree (e.g. COL15A1)
Fig. 5.
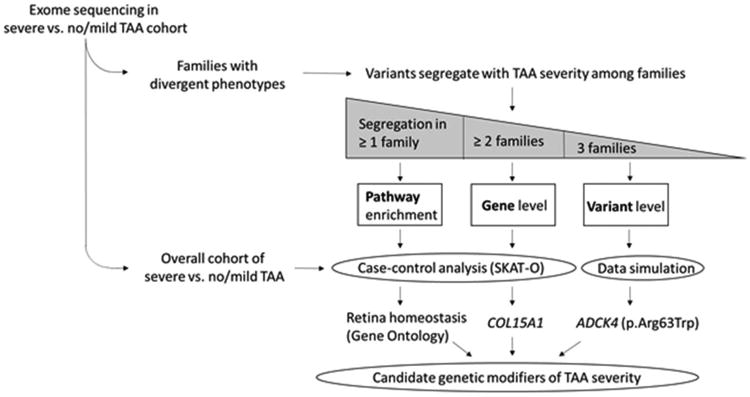
Identifying candidate modifiers of TAA severity: summary of study design and primary results. -Rare coding variants among subjects with extreme TAA phenotypes (severe vs no/mild) were analyzed using a tiered approach. First, family-based analyses were performed using a subset of the cohort in families with divergent extreme phenotypes to identify segregation with TAA severity at the level of variant, gene, and pathway. Second, case-control analyses were performed in the entire cohort for the family-based gene and pathway level findings and data simulation for variant level findings. Strong candidate modifiers of TAA severity were identified at each level, including a variant in ADCK4 (p.Arg63Trp), the gene COL15A1, and a pathway important for retina homeostasis. SKAT-O: optimal unified sequence kernel association test.
Exome wide case-control analysis of extreme TAA phenotype groups
Exome wide SKAT-O identifies a candidate modifier gene and pathways
In addition to the targeted investigation of family-based findings, we performed exome wide rare variant analysis. Based on simulated data sets, the only individual gene to achieve exome wide empirical significance was PADI3 (p=7.5×10-4) (Online Resource Table 6). Simulations found 243 out of 5000 data sets (4.9%) that contained genes with p-values less than 7.5×10-4, supporting the significance of this finding. Three mild TAA subjects carried a missense PADI3 variant (rs142129409, rs144080386, rs144944758), each predicted damaging by all 4 programs. After collapsing variants within annotated GO BPs, the most significant SKAT-O pathways associated with TAA severity were endosome transport, lipoprotein metabolism, apoptosis, and photoreceptor homeostasis (Table 3). Thus, exome wide SKAT-O analyses identify additional modifier candidate genes and pathways that may indirectly support family-based findings.
Table 3. Gene Ontology biological processes most associated with TAA severity using SKAT-O.
| Gene set | No. Markers | P-value | Genes with variants in TAA cohort |
|---|---|---|---|
| Endosome transport | 17 | 1.1×10-3 | ABCA1, ANKRD27, DOPEY1, DOPEY2, EEA1, LYST, MON2, TOM1, YKT6, ZFYVE16 |
| Regulation of neuron apoptosis | 2 | 1.5×10-3 | PCSK9, PPT1 |
| Lipoprotein metabolic process | 11 | 1.7×10-3 | ANGPTL3, APOA4, CETP, FNTA, GPAA1, PIGV, PIGZ, PPT1, SCARF1 |
| Photoreceptor cell maintenance | 21 | 3.5×10-3 | CDH23, CLRN1, GPR98, PCDH15, USH1C, USH2A |
| Peptide metabolic process | 2 | 4.1×10-3 | FURIN |
Discussion
Reduced penetrance and variable expression complicate the clinical management of syndromic and familial TAA. To test the hypothesis that genetic modifiers contribute to phenotype severity, we have studied subjects with extreme phenotypes using WES. TAA provides an opportunity to study genetic modifiers because syndromic and familial TAA are usually autosomal dominant conditions with reduced penetrance and reproductive fitness is adequate to study family members. The identification of TAA modifiers will improve methods for predicting the risk of progression to severe TAA or dissection and identify novel targets for medical therapy. Comprehensive prediction models will facilitate precise family-specific and individual-based clinical recommendations, such as frequency of surveillance, activity restriction, medical therapy, and timing for ARS.
TAA features histopathological findings of collagen fibril dysregulation in the context of different disease-causing mutations.[19-21] Therefore, the collagen genes and pathways identified in this study are plausible candidate modifiers across different genetic causes of heritable TAA. Among these candidates, COL15A1 presented the strongest independent evidence of association, hypothetically related to its role in basement membrane structure or control of angiogenesis.[22] It is also notable that variants in COL3A1, COL5A1, and COL5A2 comprised 5 of the 13 variants identified in TAA genes that were not independently causative mutations (Table 2). Dysregulated TGFβ signaling is clearly associated with TAA, but, somewhat unexpectedly, annotated TGFβ signaling pathway gene sets were not associated with TAA severity in these analyses. This does not contradict established mechanisms because TGFβ signaling intersects with other signaling pathways and has diverse pleiotropic downstream effects that are incompletely defined but include ECM homeostasis. We speculate that the effects of certain heterozygous variants in collagen-related genes are subclinical in isolation but impact pathogenesis when combined with a TAA-causative mutation. This may contrast with the essential TGFβ signaling pathway that may be more likely to cause disease independently.
We observed compelling convergence between family-based findings and SKAT-O analyses for genes important for retina homeostasis. There is indirect evidence to suggest common mechanisms between ocular and aortic homeostasis. We have previously reported that 1) ocular findings may precede progression of TAA and 2) ocular findings correlate with TAA severity in pediatric patients.[23,10] It is interesting that two top candidate genes identified in this study, ADCK4 and COL15A1, are also associated with retinal disorders.[24,25] However, the possible association between different types of ocular abnormalities and TAA severity remains unclear. Detailed ocular phenotyping in heritable TAA is indicated and may identify clinical features useful for TAA risk prediction. Functional studies of these retina homeostasis genes (e.g. RP1L1) within the aorta are needed.
Our findings also implicate genes important for energy metabolism, oxidative stress, and apoptosis as candidate modifiers. There is prior evidence that these pathways are associated with the development of TAA in diverse genetic contexts.[26-31] The protein product of ADCK4 is AarF Domain Containing Kinase 4, which translocates to the inner mitochondrial membrane to participate in the synthesis of coenzyme Q10 and electron transport.[32] ADCK4 is associated with autosomal recessive nephrotic syndrome but is also expressed in the thoracic aorta (www.gtexportal.org) and in cultured vascular smooth muscle cells derived directly from human ascending aorta (data not shown). Tissues and cells that have high energy requirements are known to be affected by mitochondrial dysfunction. Thus, the ADCK4 variant identified in this study may mediate aortic smooth muscle function or survival through mitochondrial energy metabolism and oxidative stress pathways. Identification of apoptosis and lipoprotein metabolic pathways in our exome wide SKAT-O analyses further supports a role for apoptosis and lipid metabolism pathways in TAA pathogenesis. Together these findings strongly warrant further investigation.
Throughout this study, we identified candidate modifiers in subjects with severe TAA (i.e. modifiers that worsen disease) and mild TAA (i.e. modifiers that lessen disease). For instance, the variant in ADCK4 segregated with mild TAA, suggesting a protective effect. The role of protective rare variants is relatively understudied but essential for the development of novel therapies.[33] Genomic analyses and functional studies must increasingly focus on protective genetic mechanisms. It is likely that combinations of rare variants interact to affect phenotype.[33] Variants within the same pathway may have opposing biological effects. Our observation of collagen-related variants in both severe and mild TAA demonstrates this complexity. These challenges are partially addressed by existing statistical methods such as SKAT-O, which has non-burden and epistatic variant-variant interaction functions. However, there remains a need to develop novel statistical and experimental platforms to define how specific variants interact to influence phenotype. This is particularly challenging for autosomal dominant diseases and will require low and high throughput experimental approaches.[34,35] Ultimately, these methods will define mechanisms by which aggregated variants correlate with specific phenotypes and clinical outcomes.
Limitations to this study include the small sample size. We aimed to optimize the power of rare variant analysis by studying subjects with extreme phenotypes and employing complementary family-based and case-control designs. Pathway-based analysis may overcome sample size limitations by reducing dimensionality while optimizing biological plausibility. However, this approach is dependent on existing knowledge and curation methods. This study cohort is genetically heterogeneous. Certain modifier genes may depend on the primary disease-causing genotype, but shared mechanisms, and therefore shared modifiers, likely exist. Our replication of family-based analysis findings in the overall cohort supports the latter. Nevertheless, validation of all identified candidates is needed in a larger cohort. Common variants and non-coding variants in regulatory regions were not studied but also may impact phenotype. For example, differential expression of mutant versus non-mutant alleles in TAA genes due to genetic variation in their regulatory regions could affect disease penetrance. The determinants of TAA severity are likely multifactorial, but the study of genetic modifiers is a first step to understanding complex multifactorial etiologies, including factors impacting possible differences in TAA severity between men and women. Because patients were recruited in a subspecialty Cardiovascular Genetics clinic, there may be ascertainment bias in the cohort. Finally, TAA is a progressive condition and therefore phenotype assignment may change as the patient ages. We limited this theoretical risk by primarily studying adult-aged subjects and by comparing extreme phenotypes, recognizing that the study of young patients is necessary to predict early disease progression.
In conclusion, we have identified strong candidates for modifying TAA severity. This study identifies candidates consistent with existing knowledge of TAA pathogenesis as well as new genes and pathways suggesting novel mechanisms. Together, these hypothesis generating findings initiate a path toward risk stratification through genetic testing at an early stage of disease and identifying novel therapeutic targets.
Supplementary Material
Acknowledgments
We thank the Heart Institute Research Core (recruitment), the Genetic Variation and Gene Discovery Core (sequencing), Phil Dexheimer, Osniel Ramos-Gonzales, and Amy Opoka for their assistance.
Sources of Funding: This study was supported by a National Institutes of Health (Bethesda, MD) K12HD068371 (BJL), a Burroughs Wellcome Fund (Research Triangle Park, NC) Clinical Scientist Award in Translational Research #1008496 (SMW), and an American Heart Association (Dallas, TX) Innovative Research Grant 14IRG18830027 (RBH).
Abbreviations
- ARS
aortic replacement surgery
- CTD
connective tissue disorder
- ECM
extracellular matrix
- GO BP
Gene Ontology Biological Process
- MFS
Marfan syndrome
- MRI
magnetic resonance imaging
- SKAT-O
optimal unified sequence kernel association test
- TAA
thoracic aortic aneurysm
- TGFβ
transforming growth factor beta
- WES
whole exome sequencing
Footnotes
Disclosures: The authors declare that they have no conflict of interest.
Human subjects/informed consent statement: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
References
- 1.Kochanek KD, Murphy SL, Xu J. Deaths: Final Data for 2011. Natl Vital Stat Rep. 2015;63(3):1–120. [PubMed] [Google Scholar]
- 2.Hiratzka LF, Bakris GL, Beckman JA, Bersin RM, Carr VF, Casey DE, Jr, et al. ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with Thoracic Aortic Disease: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology, American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons, and Society for Vascular Medicine. Circulation. 2010;121(13):e266–369. doi: 10.1161/CIR.0b013e3181d4739e. 2010. [DOI] [PubMed] [Google Scholar]
- 3.Pape LA, Tsai TT, Isselbacher EM, Oh JK, O'Gara PT, Evangelista A, et al. Aortic diameter >or = 5.5 cm is not a good predictor of type A aortic dissection: observations from the International Registry of Acute Aortic Dissection (IRAD) Circulation. 2007;116(10):1120–1127. doi: 10.1161/circulationaha.107.702720. [DOI] [PubMed] [Google Scholar]
- 4.Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352(6333):337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 5.Loeys BL, Schwarze U, Holm T, Callewaert BL, Thomas GH, Pannu H, et al. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med. 2006;355(8):788–798. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- 6.Superti-Furga A, Steinmann B, Ramirez F, Byers PH. Molecular defects of type III procollagen in Ehlers-Danlos syndrome type IV. Hum Genet. 1989;82(2):104–108. doi: 10.1007/BF00284038. [DOI] [PubMed] [Google Scholar]
- 7.Milewicz DM, Trybus KM, Guo DC, Sweeney HL, Regalado E, Kamm K, et al. Altered Smooth Muscle Cell Force Generation as a Driver of Thoracic Aortic Aneurysms and Dissections. Arterioscler Thromb Vasc Biol. 2017;37(1):26–34. doi: 10.1161/atvbaha.116.303229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faivre L, Collod-Beroud G, Loeys BL, Child A, Binquet C, Gautier E, et al. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am J Hum Genet. 2007;81(3):454–466. doi: 10.1086/520125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milewicz DM, Regalado ES. Use of genetics for personalized management of heritable thoracic aortic disease: how do we get there? J Thorac Cardiovasc Surg. 2015;149(2 Suppl):S3–5. doi: 10.1016/j.jtcvs.2014.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landis BJ, Ware SM, James J, Shikany AR, Martin LJ, Hinton RB. Clinical Stratification of Pediatric Patients with Idiopathic Thoracic Aortic Aneurysm. J Pediatr. 2015;167(1):131–137. e131–135. doi: 10.1016/j.jpeds.2015.02.042. [DOI] [PubMed] [Google Scholar]
- 11.Colan SD, McElhinney DB, Crawford EC, Keane JF, Lock JE. Validation and re-evaluation of a discriminant model predicting anatomic suitability for biventricular repair in neonates with aortic stenosis. J Am Coll Cardiol. 2006;47(9):1858–1865. doi: 10.1016/j.jacc.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Lopez L, Colan SD, Frommelt PC, Ensing GJ, Kendall K, Younoszai AK, et al. Recommendations for quantification methods during the performance of a pediatric echocardiogram: a report from the Pediatric Measurements Writing Group of the American Society of Echocardiography Pediatric and Congenital Heart Disease Council. J Am Soc Echocardiogr. 2010;23(5):465–495. doi: 10.1016/j.echo.2010.03.019. quiz 576-467. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91(2):224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farlow JL, Robak LA, Hetrick K, Bowling K, Boerwinkle E, Coban-Akdemir ZH, et al. Whole-Exome Sequencing in Familial Parkinson Disease. JAMA Neurol. 2016;73(1):68–75. doi: 10.1001/jamaneurol.2015.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loeys BL, Dietz HC, Braverman AC, Callewaert BL, De Backer J, Devereux RB, et al. The revised Ghent nosology for the Marfan syndrome. J Med Genet. 2010;47(7):476–485. doi: 10.1136/jmg.2009.072785. [DOI] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schubert JA, Landis BJ, Shikany AR, Hinton RB, Ware SM. Clinically relevant variants identified in thoracic aortic aneurysm patients by research exome sequencing. Am J Med Genet A. 2016;170(5):1288–1294. doi: 10.1002/ajmg.a.37568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halme T, Savunen T, Aho H, Vihersaari T, Penttinen R. Elastin and collagen in the aortic wall: changes in the Marfan syndrome and annuloaortic ectasia. Exp Mol Pathol. 1985;43(1):1–12. doi: 10.1016/0014-4800(85)90050-4. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, LeMaire SA, Chen L, Shen YH, Gan Y, Bartsch H, et al. Increased collagen deposition and elevated expression of connective tissue growth factor in human thoracic aortic dissection. Circulation. 2006;114(1 Suppl):I200–205. doi: 10.1161/circulationaha.105.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maleszewski JJ, Miller DV, Lu J, Dietz HC, Halushka MK. Histopathologic findings in ascending aortas from individuals with Loeys-Dietz syndrome (LDS) Am J Surg Pathol. 2009;33(2):194–201. doi: 10.1097/PAS.0b013e31817f3661. [DOI] [PubMed] [Google Scholar]
- 22.Marneros AG, Olsen BR. The role of collagen-derived proteolytic fragments in angiogenesis. Matrix Biol. 2001;20(5-6):337–345. doi: 10.1016/s0945-053x(01)00151-2. [DOI] [PubMed] [Google Scholar]
- 23.Ware SM, Shikany A, Landis BJ, James JF, Hinton RB. Twins with progressive thoracic aortic aneurysm, recurrent dissection and ACTA2 mutation. Pediatrics. 2014;134(4):e1218–1223. doi: 10.1542/peds.2013-2503. [DOI] [PubMed] [Google Scholar]
- 24.Korkmaz E, Lipska-Zietkiewicz BS, Boyer O, Gribouval O, Fourrage C, Tabatabaei M, et al. ADCK4-Associated Glomerulopathy Causes Adolescence-Onset FSGS. J Am Soc Nephrol. 2016;27(1):63–68. doi: 10.1681/asn.2014121240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duvvari MR, van de Ven JP, Geerlings MJ, Saksens NT, Bakker B, Henkes A, et al. Whole Exome Sequencing in Patients with the Cuticular Drusen Subtype of Age-Related Macular Degeneration. PLoS One. 2016;11(3):e0152047. doi: 10.1371/journal.pone.0152047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunetti-Pierri N, Pignatelli R, Fouladi N, Towbin JA, Belmont JW, Craigen WJ, et al. Dilation of the aortic root in mitochondrial disease patients. Mol Genet Metab. 2011;103(2):167–170. doi: 10.1016/j.ymgme.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Emrich FC, Okamura H, Dalal AR, Penov K, Merk DR, Raaz U, et al. Enhanced caspase activity contributes to aortic wall remodeling and early aneurysm development in a murine model of Marfan syndrome. Arterioscler Thromb Vasc Biol. 2015;35(1):146–154. doi: 10.1161/atvbaha.114.304364. [DOI] [PubMed] [Google Scholar]
- 28.Kuang SQ, Medina-Martinez O, Guo DC, Gong L, Regalado ES, Reynolds CL, et al. FOXE3 mutations predispose to thoracic aortic aneurysms and dissections. J Clin Invest. 2016;126(3):948–961. doi: 10.1172/jci83778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang HH, van Breemen C, Chung AW. Vasomotor dysfunction in the thoracic aorta of Marfan syndrome is associated with accumulation of oxidative stress. Vascul Pharmacol. 2010;52(12):37–45. doi: 10.1016/j.vph.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Branchetti E, Poggio P, Sainger R, Shang E, Grau JB, Jackson BM, et al. Oxidative stress modulates vascular smooth muscle cell phenotype via CTGF in thoracic aortic aneurysm. Cardiovasc Res. 2013;100(2):316–324. doi: 10.1093/cvr/cvt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YC, Huang HY, Chang CJ, Cheng CH, Chen YT. Mitochondrial GLUT10 facilitates dehydroascorbic acid import and protects cells against oxidative stress: mechanistic insight into arterial tortuosity syndrome. Hum Mol Genet. 2010;19(19):3721–3733. doi: 10.1093/hmg/ddq286. [DOI] [PubMed] [Google Scholar]
- 32.Ashraf S, Gee HY, Woerner S, Xie LX, Vega-Warner V, Lovric S, et al. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest. 2013;123(12):5179–5189. doi: 10.1172/jci69000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harper AR, Nayee S, Topol EJ. Protective alleles and modifier variants in human health and disease. Nat Rev Genet. 2015;16(12):689–701. doi: 10.1038/nrg4017. [DOI] [PubMed] [Google Scholar]
- 34.Zilberberg L, Phoon CK, Robertson I, Dabovic B, Ramirez F, Rifkin DB. Genetic analysis of the contribution of LTBP-3 to thoracic aneurysm in Marfan syndrome. Proc Natl Acad Sci U S A. 2015;112(45):14012–14017. doi: 10.1073/pnas.1507652112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miles LA, Garippa RJ, Poirier JT. Design, execution, and analysis of pooled in vitro CRISPR/Cas9 screens. Febs j. 2016;283(17):3170–3180. doi: 10.1111/febs.13770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


