Abstract
Fetal exposure to synthetic glucocorticoids reprograms distinct neural circuits in the developing brain, often in a sex-specific manner, via mechanisms that remain poorly understood. To reveal whether such reprogramming is associated with select molecular signatures, we characterized the transcriptome of primary, embryonic mouse cerebral cortical and hypothalamic neural progenitor/stem cells derived from individual male and female embryos exposed to the synthetic glucocorticoid, dexamethasone. Gene expression profiling by RNA-Seq identified differential expression of common and unique genes based upon brain region, sex, and/or dexamethasone exposure. These gene expression datasets provide a unique resource that will inform future studies examining the molecular mechanisms responsible for region- and sex-specific reprogramming of the fetal brain brought about by in utero exposure to excess glucocorticoids.
Keywords: glucocorticoids, development, cortex, hypothalamus, neural stem cells, sex differences
Introduction
The rise in endogenous glucocorticoids during late gestation is critical for maturation of fetal organ systems including the lungs, liver, and brain (Moisiadia and Matthews, 2014; Peffer et al., 2015). Some of the adverse events associated with preterm birth result from insufficient time for glucocorticoids to promote sufficient organ development for viability outside the uterus. Therefore, mothers at risk for premature labor are administered synthetic glucocorticoids (e.g. dexamethasone (Dex) or betamethasone) because this treatment results in reduced respiratory distress, intraventricular hemorrhage and necrotizing enterocholitis in the infant (Crowley, 1995; Roberts and Dalziel, 2006) and decreases infant morbidity and mortality. However, long term follow-up studies, particularly in term-born children exposed to antenatal glucocorticoids, have uncovered some cases of adverse consequences on brain development associated with fetal exposure to synthetic glucocorticoids. For example, excess glucocorticoids during fetal development may increase risk for various neuropsychiatric disorders with presumed developmental origins including, but not limited to, schizophrenia, anxiety, depression, autism spectrum disorders, attention deficit hyperactivity disorder, as well as disruptions in hypothalamic-pituitary-adrenal axis function (Karemaker et al., 2008; Wyrwoll and Homes, 2012). These results highlight the potential adverse effects of antenatal glucocorticoids on various brain regions during development particularly the cortex and hypothalamus (reviewed in Peffer et al., 2015; Schmitt et al., 2014).
The cortex is part of the telencephalon and regulates higher order abilities, emotion, and cognition (Rubenstein et al., 2011). During embryonic development, cortical layers arise from progenitor cells within the ventricular zone of the lateral ventricles (Rakic, 1988). In comparison, the hypothalamus is a part of the diencephalon and integrates inputs from various central and peripheral sources to maintain homeostasis of various organ systems and whole body metabolism. During development, distinct hypothalamic nuclei originate from the proliferative zone of the third ventricle (Altman and Bayer, 1986). Furthermore, the hypothalamus develops with earlier formed neurons migrating farther than later formed neurons, which is in contrast to the pattern seen in cortical development where newer formed neurons must migrate past older cells in deeper layers towards their final destination in superficial layers (Molyneaux et al., 2007; Rakic and Lombroso, 1998). Even though the development of these two distinct brain regions are by most comparisons opposite, cells that encompass the hypothalamus and cortex originate from the proliferation and differentiation of neural progenitor/stem cells (NPSCs).
We have previously examined global gene expression of Dex-dependent genes in cortical NPSCs by microarray (Peffer et al., 2014) and in hypothalamic NPSCs by RNA-Seq (Frahm et al., 2016). To provide a resource and genome-wide information that could aid in understanding the selective effects of antenatal glucocorticoids on cortical versus hypothalamic development, we present a detailed bioinformatics analysis of RNA-Seq data sets, combining the previously published expression data of hypothalamic NPSCs exposed to Dex with a newly generated RNA-Seq analysis of male and female embryonic cerebral cortex NPSCs exposed to Dex.
Materials and Methods
Animals
Separate cortical and hypothalamic NPSC cultures were generated from individual C57BL/6 mice on embryonic day(E) 14.5, dissociated into single cells, and grown as three-dimensional neurosphere cultures as previously described (Frahm et al., 2016; Peffer et al., 2014; Salvi et al., 2009; Samarasinghe et al., 2011). NPSCs were maintained separate for individual embryos. These cultures were passaged to expand and enrich for transcriptional analysis, which utilized RNA prepared from cells at the third passage (P3). Fetal sex was determined for each sample by digesting tail tissue overnight in 200μl of nonionic detergent buffer with 1.2μl of proteinase K overnight at 56°C. The next day, the samples were heat inactivated at 95°C for 10 minutes and isolated DNA subjected to PCR analysis to detect the Y chromosome Sry gene
RNA-Seq Analysis
Cortical and hypothalamic NPSCs from 3 biological replicates (individual embryos) were each treated with 100 nM Dex (Sigma Chemicals, MO) or vehicle (ethanol [EtOH]) for 4h and harvested in TRIzol® (Invitrogen, 15596-026). For hypothalamic NPSCs (Frahm et al., 2016), RNAs were isolated using the Machery-Nagel Nucelospin RNA II kit while cortical RNAs were isolated from Trizol (Life Technologies) lysates following organic extraction and ethanol precipitation. An RNA-Seq library was prepared using the TruSeq Stranded Total RNA kit (RS-122-2201; Illumina) and then subjected to next generation sequencing by the Tufts University Genomics Core. After sequencing, cortical NPSC reads and raw fastq reads from the previously published hypothalamic NPSCs (Frahm et al., 2016) were processed together for further bioinformatics analysis. The hypothalamic RNA-Seq data was re-processed for this study to ensure the use of identical bioinformatics pipelines for both the cortex and hypothalamic data. Hypothalamic reads were aligned to the Ensembl GRCm38 Mouse genome with Tophat v2.0.9 while cortical reads were aligned with Tophat2 v2.0.13, using default parameters. Reads were aligned to the Ensembl GRCm38 Mouse genome with Tophat2 v2.0.13 using default parameters. Read quantification was performed with HTSeq using the union method. Differential expression analysis for hypothalamus and cortex was performed using edgeR with a gene expression filter of >5 counts per million (CPM) in at least three samples as expression above background. Additional statistical analysis was performed using R (Bioconductor). Common Dex-dependent genes were obtained by taking the intersection of the significant (FDR 5%) Dex-dependent genes significant in both male and female Dex vs. EtOH gene lists. Comparisons to published studies were done using NextBio (Base Illumina, Carlsbad, CA). The raw and processed data was submitted to GEO (accession #GSE95363).
Comparisons with Published E14.5 Mouse Cortical Zones Dataset
The in-house dataset was compared to the reads per kilobase of transcript per million mapped (RPKM) values for the samples belonging to the different embryonic zones in the mouse cortex, as downloaded from the author's website (http://rakiclab.med.yale.edu/transcriptome/index.aspx) (Ayoub et al., 2011). The dataset and in-house datasets were filtered to remove genes that were not common to both. It is important to note that published cortical zone data (Ayoub et al., 2011) were analyzed using the University of California Santa Cruz mm9 reference genome, whereas the in-house data were analyzed using the Ensembl GRCm38 reference genome.
Results
Characteristics of the Mouse Fetal Cortical NPSC Transcriptome
A transcriptome of significantly expressed genes was obtained from a newly generated RNA-Seq data set using a counts based method and setting a cutoff of >5 CPM for expression above background (see Methods section). We first combined both male and female cortical vehicle or Dex-treated NPSC gene lists in pie charts shown in Figure 1, which display identified expression of 10,250 protein coding genes, 155 long noncoding RNAs, 31 short noncoding RNAs, and 48 pseudogenes (Figure 1a). Additional analysis of the categories of long noncoding RNAs expressed in both male and female NPSCs include 61 lincRNAs, 57 processed transcripts, 32 antisense, 3 sense intronic, 1 sense overlapping, and 1 bidirectional promoter lncRNA (Figure 1b) while the short noncoding RNAs included 12 snRNA, 7 scaRNA, 7 miscellaneous RNA, 3 miRNA, and 2 mitochondrial ribosomal RNA (Figure 1c). These data expand our previous microarray analysis of E14.5-derived primary cerebral cortical NPSCs, which used an Affymetrix genome array that contained nearly 14,000 protein-coding genes but did not assess non-protein coding genes.
Figure 1.
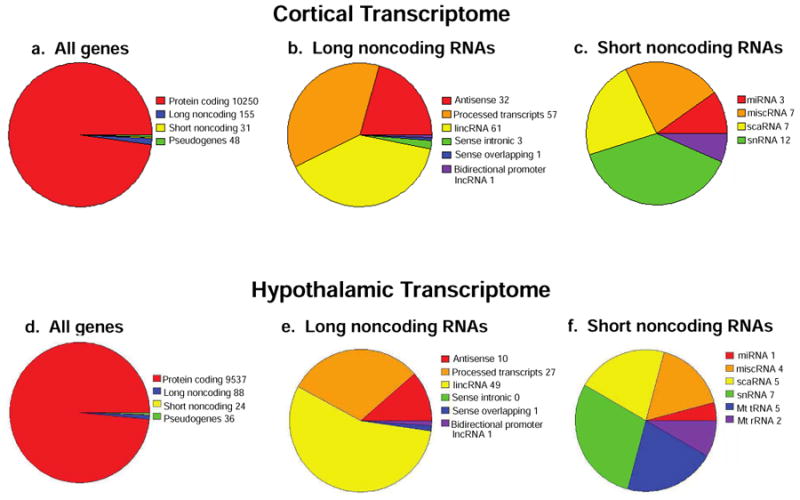
RNA-Seq profiling of cerebral cortical and hypothalamic NPSCs. Pie charts detail the distribution of total genes identified (a), long noncoding RNAs (b), and short noncoding RNAs (c) expressed > 5 mean CPM following ethanol vehicle or Dex treatment in both male and female cortical NPSCs. Distribution of total genes identified (d) and long-noncoding RNAs (e), and short noncoding RNAs (f) expressed > 5 mean CPM following ethanol vehicle or Dex treatment in both male and female hypothalamic NPSCs.
Comparison of the Mouse Fetal Cortical and Hypothalamic NPSC Transcriptomes
To ensure appropriate statistical comparisons of embryonic cerebral cortical versus hypothalamic NPSCs, we re-evaluated an RNA-Seq data set from hypothalamic NPSCs using a count-based method rather than FPKM as utilized previously (Frahm et al., 2016). For combined male and female vehicle-treated hypothalamic NPSCs, we detected RNA for 9,537 protein coding, 88 long noncoding, 24 short noncoding, and 36 pseudogenes (Figure 1d). For long noncoding RNAs, there were 10 antisense, 27 processed transcripts, 49 lincRNAs, 1 sense overlapping, and 1 bidirectional promoter lncRNA (Figure 1e). When examining short noncoding RNAs our data identified 1 miRNA, 4 miscellaneous RNA, 5 scaRNA, 7snRNA, 5 mitochondrial tRNA, and 2 mitochondrial ribosomal RNA (Figure 1f). This new analysis of the hypothalamic NPSC derived transcripts identified 1,405 fewer protein coding, 599 fewer long noncoding, and 1,206 fewer short noncoding genes reflecting differences in gene assignments generated from FPKM versus count-based analysis of RNA-Seq data. Importantly, high levels of progenitor/stem cell expressed genes Nes (654 CPM), Olig2 (277 CPM), Sox2 (241 CPM) were still among the top genes identified using both methods. In addition, hypothalamic mature neuron markers Sim1, Trh, Pomc, Npy showed no significant counts above background (> 5) in the count-based method as previously reported from FPKM analysis of the hypothalamic NPSC transcriptome (Frahm et al., 2016). Finally, we observe a similar pattern of expression of nuclear receptors, which included Coup-TF1, Coup-TF2, Ear2, Erra, GR, Lxrb, Nur77, Pparg, Rara, Rev-ErbA-a, Rev-ErbA-b, Rora, Rorb, Rxra, Rxrb, Tlx, Tr2, Tr4, Tra, Trb, in male and female cortical and hypothalamic NPSCs (Supplemental File 1) as revealed in previous findings using FPKM (Frahm et al., 2016).
We then compared the transcriptome from E14.5-derived primary cerebral cortical and hypothalamic NPSCs. For genes that were selectively expressed in cortical NPSCs but did not reach our cutoff for expression above background in hypothalamic NPSCs, there were 631 protein coding, 75 long noncoding, and 11 short noncoding RNAs (Figure 2a). Long noncoding genes selective expressed in cortical NPSCs included 23 antisense, 23 lincRNA, 26 processed transcripts, and 3 sense intronic (Figure 2b). Short noncoding transcripts selectively expressed in cortical NPSCs included 5 snRNAs, 2 scaRNAs, 3 miscellaneous RNAs, and 1 miRNA (Figure 2c). Conversely for genes selectively expressed only in hypothalamic NPSCs, there were only 39 protein coding (Figure 2d) and 6 long noncoding RNAs that included 1 antisense and 5 lincRNAs (Figure 2e). Since our RNA prep did not enrich for small RNAs, the short noncoding transcripts identified in our analysis are likely to underrepresent the small RNA transcriptome in these cells.
Figure 2.
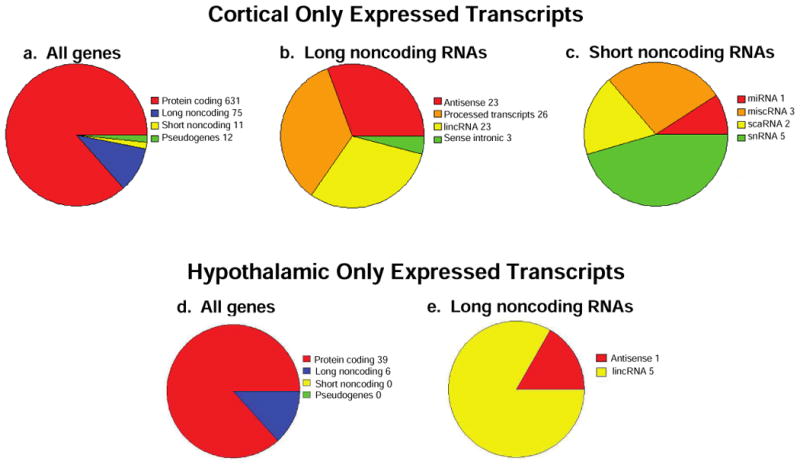
RNA-Seq profiling of unique transcripts in cerebral cortical and hypothalamic NPSCs. Pie charts detail the distribution of total genes identified (a), long noncoding RNAs (b), and short noncoding RNAs (c) expressed above a threshold of 5 mean CPM in both male and female cortical NPSCs following ethanol vehicle or Dex treatment but below that threshold in hypothalamic NPSCs. Distribution of all genes (d) and long-non coding RNAs (e) expressed with the above threshold in hypothalamic NSPCs but not cortical NPSCs following ethanol vehicle or Dex treatment.
Comparison of Gene Expression in Male and Female Cortical- NPSCs with a Published Dataset
Our in-house cerebral cortical NPSC dataset was compared to a previously published RNA-Seq analysis of the transcriptome in different embryonic zones from the E14.5 mouse cortex (Ayoub et al., 2011). A comparison of the hypothalamic NPSC transcriptome obtained using FPKM analysis to relevant published data sets was reported previously by our group (Frahm et al., 2016). During cortical development, the layer of cells adjacent to the ventricle is the location where the first neurons are produced and referred to as the ventricular zone (VZ, Rakic and Lombroso, 1998). As neurogenesis proceeds, another layer of proliferative cells arise within the subventricular zone (SVZ), which is located above the VZ (Molyneaux et al., 2007). Additionally, the cortical plate (CP) is formed by neurons that have migrated out to the pial surface of the cortex after they have undergone their last mitotic division (Rakic and Lombroso, 1998). As shown in Figure 3, the highest correlation between both our cerebral cortical and hypothalamic NPSC transcriptome derived from males and females was with the transcriptome derived from the VZ and SVZ of the E14.5 cerebral cortex. The lowest correlation in this analysis using the primary E14.5 derived NPSCs from both tissues was with transcripts expressed in the E14.5 CP. These results corroborate the stem and progenitor cell properties of our cultures as both of these cell types would be present in the VZ and SVZ to different extents at E14.5 but not highly represented in the CP, which is populated predominantly by newborn neurons (Molyneaux et al., 2007).
Figure 3.
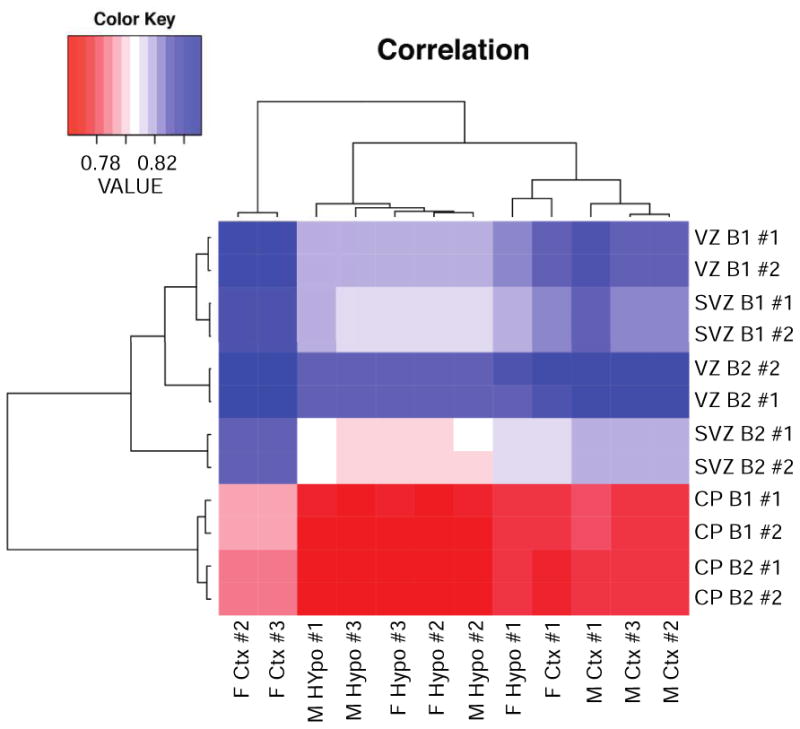
Comparison of RNA expression in male and female vehicle-treated cortical and hypothalamic NPSCs with a published dataset of RNA expression in different mouse embryonic cortical zones (SVZ, VZ, CP) using biological replicates (designated as B1, B2) and 2 technical replicates (#1, #2) of each zone.
The Mouse Fetal Cortical and Hypothalamic NPSCs Dexamethasone Transcriptomes
RNA-Seq analysis identified 1054 protein coding, 30 long noncoding, 3 short noncoding, and 3 pseudogenes in male and female cortical NPSCs (Figure 5a). The long noncoding genes in male and female cortical NPSCs included 11 processed transcripts, 10 lincRNA, and 9 antisense (figure 5b). The short noncoding RNAs included 2 snRNAs and 1 miRNAs. The hypothalamic NPSCs dexamethasone transcriptome identified 101 protein coding RNAs and 1 long noncoding RNA (Figure 5c) which consisted of an antisense RNA. The top 25 induced and 25 repressed Dex-dependent protein-coding genes in both male and female cortical NPSCs are presented in Figure 4. The highly Dex-induced genes Fam107a, Cftr, Pln4, Ptk2b were previously identified in cortical NPSCs using microarray analysis (Peffer et al., 2014), which did not identify the top Dex-repressed genes identified in this RNA-Seq analysis (Figure 4). It is unclear why putative Dex-repressed genes were underrepresented in the previous microarray analysis of cerebral cortical NPSCs. To provide a visual comparison of Dex-dependent genes in cerebral cortical versus hypothalamic NPSCs derived from both males and females E14.5 embryos, volcano plots were generated. As shown in Figure 6, the most highly significant and most robustly upregulated genes (i.e. upper right quadrant) are similar in all groups. Specific genes in this category include Hif3a and Fam107a, which have been previously been validated by quantitative RT-PCR as highly induced genes in cerebral cortical (Peffer et al., 2014) and hypothalamic (Frahm et al., 2016) NPSCs. However, hypothalamic male and female NPSCs are largely devoid of robustly down-regulated genes in comparison to the cortical NPSCs (lower left quadrant). Two members of the Fos/Jun family (i.e. Fosb and Junb) are uniquely downregulated by Dex in cortical but not hypothalamic NPSCs (Figure 6, Supplemental File 2).
Figure 5.
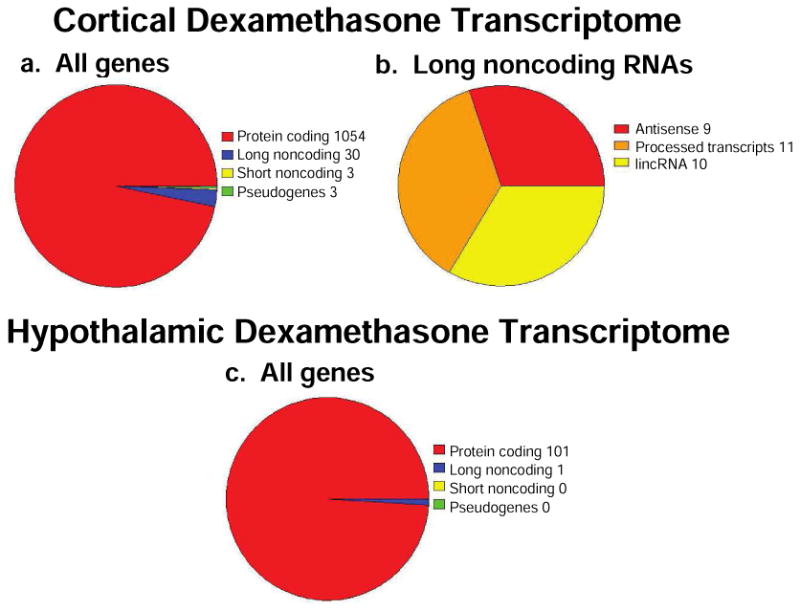
RNA-Seq profiling of Dex-dependent genes in cortical and hypothalamic NPSCs. Pie charts detail the distribution Dex-dependent total genes identified (a) and long noncoding RNAs (b) from both male and female cortical NPSCs that display mean CPM > 5. Distribution of Dex-dependent total genes identified (c) from both male and female hypothalamic NPSCs that display significant CPM (>5).
Figure 4.
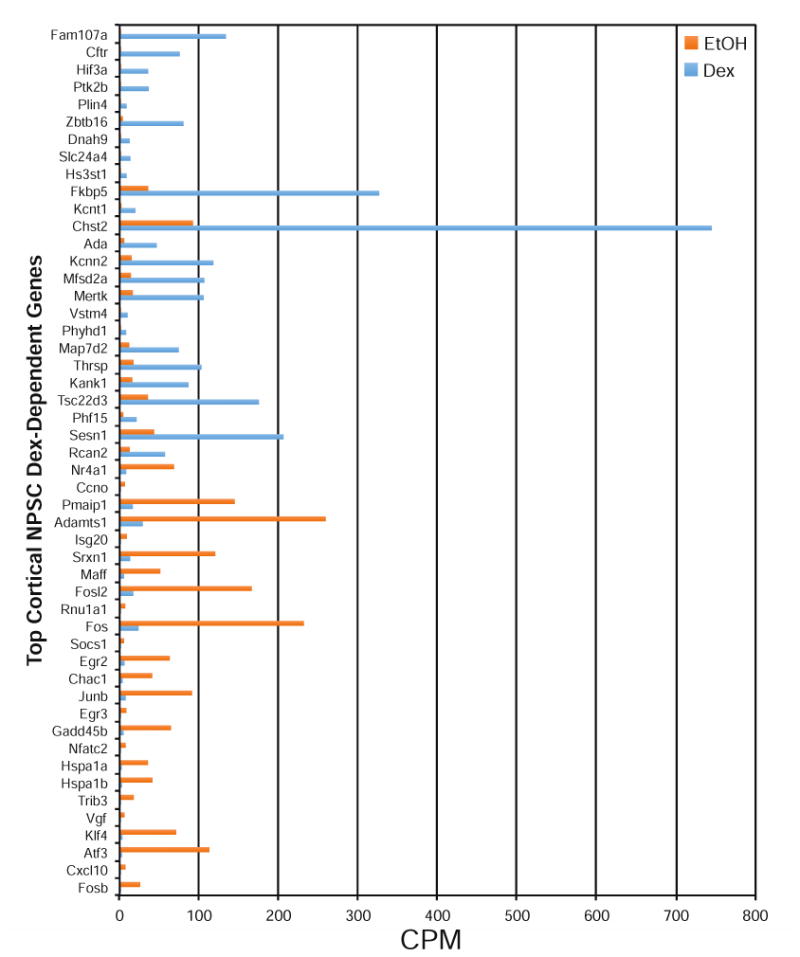
Top 25 Induced and Repressed Dex-Dependent Genes Identified in Combined Male and Female Cortical NPSCs using RNA-Seq analysis (p < 0.05).
Figure 6.
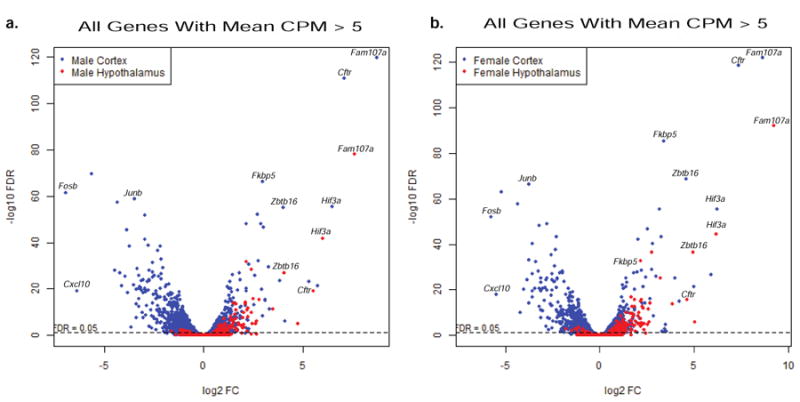
Volcano Plots of Comparison of Male (a) and Female (b) Cortical and Hypothalamic NPSC Dex-dependent Genes.
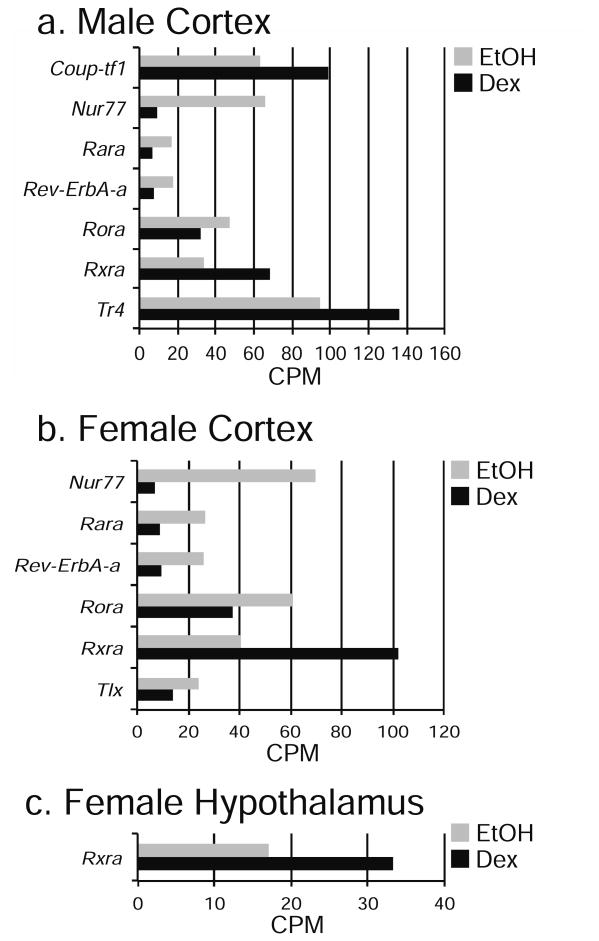
When comparing Dex-dependent genes in both cortical and hypothalamic NPSCs, there were 67 genes identified as either induced or repressed in combined male and female NPSCs from both tissues (Table 1, Supplemental File 2). Not surprisingly, top known GR-target genes such as Fam107a, Cftr, Fkbp5 and the novel GR-target gene Hif3a recently identified in hypothalamic NPSCs (Frahm et al., 2016) were included in this list. The 67 Dex-dependent genes that were regulated in both male and female cortical and hypothalamic NPSCs showed similar magnitudes of change, particularly for highly induced genes (Figure 6). Interestingly, we also identified sex-specific Dex-dependent changes in the nuclear receptors in male and female cortical NPSCs for Nur77, Rara, Rev-ErbA-b, Rora, Rxra, Coup-tf1 and Tr4, in only male cortical NPSCs Coup-TF1, and for hypothalamus only Rxra in females (Figure 7) suggesting the possibility of region specific crosstalk between distinct nuclear receptors in the fetal brain.
Table 1.
Common and Distinct Dex-Dependent Genes Identified in Male and/or Female Cortical and/or Hypothalamic NPSCs using RNA-Seq analysis.
| Comparisons | Genes |
|---|---|
| Male & Female Cortex & Hypothalamus Common Dex-Regulated | 67 |
| Male & Female Cortex Unique Dex-Regulated | 1000 |
| Male & Female Hypothalamus Unique Dex-Regulated | 14 |
| Male Cortex Unique Dex-Regulated | 206 |
| Female Cortex Unique Dex-Regulated | 145 |
| Male Hypothalamus Unique Dex-Regulated | 8 |
| Female Hypothalamus Unique Dex-Regulated | 21 |
Figure 7.
Dex-Dependent Differences in Nuclear Receptor Expression in Cerebral Cortical Male NPSCs, Cerebral Cortical Female NPSCs, and Female Hypothalamic NPSCs identified using RNA-Seq analysis (p < 0.05).
Further comparisons of the RNA-Seq data set identified 1,000 genes uniquely Dex-dependent in male and female cerebral cortical NPSCs, while only 14 genes were uniquely Dex-dependent in hypothalamic NPSCs (Table 1). Furthermore, 206 genes were uniquely Dex-dependent in male cortical NPSCs and 145 uniquely Dex-dependent in female cortical NPSCs. As evident from the Figure 4, Table 1, and Volcano plots in Figure 6, hypothalamic NPSCs have far fewer unique Dex-dependent genes than cortical NPSCs. However, for hypothalamic NPSCs, there were 8 genes uniquely Dex-dependent in males and 21 genes uniquely Dex-dependent in females. Overall, these results provide the dexamethasone transcriptome in male and female cerebral cortical- and hypothalamic-derived embryonic neural stem cells.
Conclusions
Our genome-wide analysis examined the transcriptome in E14.5-dervived primary NPSCs maintained in vitro from male and female cerebral cortex and hypothalamus and their response to glucocorticoid exposure (4h, 100 nM Dex). For direct comparison with cortical NPSCs, we chose to remap our previously published hypothalamic NPSCs transcriptome using a counts-based method. When comparing gene expression profiles between cortical and hypothalamic NPSCs, many genes were expressed within both regions, although we did identify 729 cortical-specific and 45 hypothalamic-specific genes. Therefore, while the transcriptome of E14.5 cerebral cortical and hypothalamic NPSCs may be largely devoted to maintaining the proliferative capacity of these cells, unique gene expression profiles exist within these cells that reflect their distinct programming and responses to differentiation cues. At E14.5 when we isolated our NPSCs neurogenesis is almost complete in the hypothalamus (Shimagori et al. 2010) and just underway in the cortex (reviewed in Carson et al., 2016); therefore, differences in their gene expression profiles may be a result of previous programing of these cells in vivo and represent unique networks that impact specific programming of stem and progenitor cells from these regions and provide new targets to study region specific brain development.
Although we did not enrich for small RNAs in previous RNA-Seq studies of hypothalamic NPSCs, we did detect a number of long noncoding RNAs (687) and short noncoding RNAs (1,230; Frahm et al., 2016). This was largely due to our prior mapping using FPKMs that will be biased towards expression of small RNAs. Therefore, we only identified 88 long noncoding and 24 short noncoding RNAs for hypothalamic NPSCs and in cortical NPSCs 155 long noncoding and 31 short noncoding RNAs using a counts-based method. Due to the assumptions and normalizations used for FPKMs, we remapped our hypothalamic NPSCs along with cortical NPSCs for comparison and to allow for future additional comparisons with other published datasets. For example, Lv et al., 2013 collected 17 datasets generated from embryonic mouse brains and filtered to identify 29,837 long noncoding RNA candidates. This vast difference in identified long noncoding RNAs originated from difference in starting material (whole brain versus neural stem cells in two select brain regions) and their analysis (enrichment for identifying long noncoding RNAs). Nonetheless, our analysis identified coding and noncoding gene lists for both cortical and hypothalamic NPSCs and generated data sets for subsequent analysis. Future work may reveal whether the long noncoding RNAs we identified in primary NPSC cultures have an impact in vivo on stem or progenitor cell proliferation or differentiation.
An examination of the Dex-dependent transcriptome identified 67 Dex-dependent genes as either induced or repressed by Dex in both male and female cortical and hypothalamic NPSCs. Further bioinformatic analysis revealed unique Dex-dependent genes for region (cortical versus hypothalamic) and sex (male versus female). We did find it striking that our analysis identified over 1000 cortical-specific Dex-dependent genes but only 14 hypothalamic-specific Dex-dependent genes. Interestingly, as volcano plots illustrated, the majority of these cortical-specific Dex-dependent genes are robustly repressed genes. It seems unlikely that differences in Dex-dependent repression of target genes in cortical versus hypothalamic NPSCs result from cell-type specific engagement of the major GR mRNA decay pathway as mRNAs of the critical components of this pathway (i.e. Dcp1a, Hrsp12, Pnrc2, Upf1, Upf1; Park et al., 2016) are expressed to similar extents (i.e. from RNA-seq data) in cortical and hypothalamic NPSCs. Finally, the difference in Dex-dependent genes in hypothalamic versus cortical NPSCs could result from experimental or biological variations in the primary NPSCs from these two regions. For example, our cortical NPSCs s may contain more GRs (127.8 CPM ± SEM 5.3) than hypothalamic NPSCs (88.7 CPM ± SEM 2.0), which would be consistent with higher levels of GR present in the adult mouse cortex compared to the hypothalamus (Mahfouz et al., 2016).
Our genome-wide analysis approach of measuring the sexually dimorphic Dex transcriptome found unique genes for region and sex. Importantly, Klf4 was previously identified and validated as a male Dex-dependent gene in hypothalamic NPSCs (Frahm et al., 2016) and in the current dataset was also identified as hypothalamic-specific male NPSCs Dex-dependent gene. Recently, global analysis for highly expressed genes that correlated with GR expression revealed a high degree of similarity among brain regions such as the cortex, striatum, thalamus, and midbrain but not the hypothalamus (Mahfouz et al., 2016). These findings support the notion the adult hypothalamus may have a unique global transcriptional response to glucocorticoids distinct from many other brain regions, which despite their enormous differences in cellular composition share a closely related GR transcriptional signature. Our results suggest that such unique responses to glucocorticoids may be established early in development and evident in hypothalamic stem cells.
Queries using the Transcriptomine® tool (Becnel et al., 2017) confirmed that some of the most robust Dex-dependent (induced) genes in male and female cortical or hypothalamic NPSCs (e.g. Fkbp5, Per1, Tsc22d3) are GR targets common to many different cell types and tissues. In contrast, Hif3a and Cftr, both highly induced in cortical and hypothalamic NPSCs of both sexes, have only been detected as GR targets in a handful of cell types. For example, Hif3a was identified as a GR target in primary astrocytes, an oligodendrocyte cell line and a pituitary corticotroph cell line suggesting that this member of the Hif family may function outside of an angiogenic response to influence brain function. The response of Cftr is much more varied with induction being noted in gastrocnemius and the Oli-neu oligodendrocyte cell line but repression in the A549 cell line and whole liver. The phosphodiesterase Pde1b is Dex repressed in cortical and hypothalamic NSPCs of both males and females but only found as a GR target (i.e. repressed) in one other data set (derived from primary astrocytes). Pde1b is induced by chronic and acute stress in adult mice (Hufgard et al., 2017) and may impact depressive and anxiety like behavior in mice (Hufgard et al., 2017; Siuciak et al., 2007).
One of the genes identified in our study as Dex-dependent in specific cell types or sexes, such as Pde3a (Dex repressed in male and female hypothalamic but not cortical NSPCs) was only identified as a GR target gene in T-cell leukemic cells. Pde3a is a cGMP-inhibited cyclic phosphodiesterase that is highly expressed in the developing brain (Reinhardt and Bondy, 1996) but has not been examined for its impact on neural stem cells. The calcineurin regulator Rcan2, was uniquely Dex-dependent in female hypothalamic NSPCs in our data set and only found to be a GR target in primary fibroblasts and the A549 lung epithelial cell line. Rcan2 is expressed in select nuclei of the adult hypothalamus and regulates energy balance (Sun et al., 2011) but its function in the developing hypothalamus has not been investigated. Finally, the catenin family member Arvcf gene, was only found to be Dex-dependent (induced) in male hypothalamic NSPCs of our data set and not identified as a GR target in any data set contained within Transcriptomine (Becnel et al., 2017). While Arvcf is a target of other nuclear receptors, its only response in brain was found in whole hypothalamic tissue from neonatal female mice treated with 17β-estradiol. Arvcf is expressed within a specific region of the developing brain (i.e. the ganglionic eminence) (Ulfig and Chan 2004) and has been suggested to be a human risk allele with neurodevelopmental origins for schizophrenia (Sim et al., 2012) as well as white matter injury following premature birth (Boardman et al., 2014).
RNA-Seq analysis is a powerful tool to assess how endogenous (e.g. circulating glucocorticoids) and exogenous (e.g. Dex treatment) factors impact gene transcription and subsequent brain development but still requires careful validations. For example, our stringent statistical analysis excluded the well-known-GR target gene Gjb6 as significantly Dex-dependent in cortical and hypothalamic NPSCs. Gjb6 was identified as Dex-induced and validated in our previous studies using microarray in cortical NPSCs (Peffer et al., 2014) and previous mapping of this hypothalamic NPSCs dataset using RNA-Seq analysis (Frahm et al., 2016). It was also identified as Dex regulated in a primary astrocyte data set contained within Transcriptomine. During this recent analysis, Gjb6 did not pass the >5 CPM cutoff and as a result was not included in our Dex-dependent gene list. In addition, when examining common Dex-dependent genes in both male and female cerebral cortical and hypothalamic NPSCs, the Arl4d gene was identified as repressed in cerebral cortical NPSCs but induced in hypothalamic NPSCs. Arl4d was identified as a Dex-inducible gene in previous microarray analysis of cerebral cortical NPSCs published by our group and validated in that report by qRT-PCR analysis from independent biological samples (Peffer et al., 2014). For that reason we omitted it from Supplemental File 2. It should also be noted that Arl4d has consistently been induced and never repressed by Dex in various data sets contained within Transcriptomine (e.g. whole mouse and rat liver, primary mouse astrocytes, the mouse AtT-20 corticotrophic pituitary cell line, the human A549 lung epithelial cell line, and the 7438 mouse mammary carcinoma cell line). These examples are important to note and reinforce the need to carefully examine gene expression profiles from large available datasets, but do not detract from the utility of RNA-Seq to identify region- and sex-specific GR-target genes.
In conclusion, we have generated a data resource of the Dex transcriptome in E14.5-derived mouse male and female cortical and hypothalamic NPSCs. Using RNA-Seq analysis, we identified common and unique genes due to region, sex, and/or treatment that can serve as a resource to investigate the responses of the developing cerebral cortex and hypothalamus to excess endogenous glucocorticoids that might arise as a result of maternal stress or due to therapeutic treatments with synthetic glucocorticoids.
Supplementary Material
Supplemental File 1. Nuclear receptor expression (CPM) in cerebral cortical and hypothalamic male and female NPSCs identified using RNA-Seq analysis.
Supplemental File 2. Common and unique Dex-Dependent Genes in cerebral cortical and hypothalamic male and female NPSCs identified using RNA-Seq analysis.
-
-
Fetal exposure to excess glucocorticoids disrupts brain development differentially in males and females in regions such as the cerebral cortex and hypothalamus.
-
-
To determine potential mechanisms, the dexamethasone transcriptome in male and female cerebral cortical and hypothalamic embryonic neural stem cells was determined utilizing RNA-Seq analysis.
-
-
Unique region- and sex-specific Dex-dependent genes were identified providing a resource for examining the mechanisms by which excess glucocorticoids alter fetal brain development.
Acknowledgments
We would like to thank Ross Carson for his technical assistance in these studies. This project used the UPCI Cancer Bioinformatics Services, which is supported in part by the National Cancer Institute award P30CA047904. This study also was supported a NURSA Data Resource Project from an U24 grant DK097748 (DBD, APM, URC), a T32 training grant T32DK007052 from the National Institute of Diabetes and Digestive and Kidney Diseases (KAF) an ACS grant (RSG-09-054-0 1-GMC, APM), G. Harold and Leila Y. Mathers Charitable Foundation Award (APM), NIH T32-HD071834-01A1 (AR), NIH K12 HD052892-09 (AR), Magee-Womens Clinical Trainee Research Award (AR), and the University of Pittsburgh Medical Center Department of Pediatrics Endowed Instructorship (AR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman J, Bayer SA. The development of the rat hypothalamus. Adv Anat Embryol Cell Biol. 1986;100:1–178. [PubMed] [Google Scholar]
- Ayoub AE, Oh S, Xie Y, Leng J, Cotney J, Dominguez MH, Noonan JP, Rakic P. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci USA. 2011;108:14950–5. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel LB, Ochsner SA, Darlington YF, McOwiti A, Kankanamge WH, Dehart M, Naumov A, McKenna NJ. Discovering relationships between nuclear receptor signaling pathways, genes, and tissues in Transcriptomine. Sci Signal. 2017;10:476. doi: 10.1126/scisignal.aah6275. [DOI] [PubMed] [Google Scholar]
- Boardman JP, Walley A, Ball G, Takousis P, Krishnan ML, Hughes-Carre L, Aljabar P, Serag A, King C, Merchant N, Srinivasan L, Froguel P, Hajnal J, Rueckert D, Counsell S, Edwards AD. Common genetic variants and risk of brain injury after preterm birth. Pediatrics. 2014;133:e1655–63. doi: 10.1542/peds.2013-3011. [DOI] [PubMed] [Google Scholar]
- Carson R, Monaghan-Nichols AP, DeFranco DB, Rudine AC. Effects of antenatal glucocorticoids on the developing brain. Steroids. 2016;114:25–32. doi: 10.1016/j.steroids.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley PA. Antenatal corticosteroid therapy: a meta-analysis of the randomized trials, 1972 to 1994. Am J Obstet Gynecol. 1995;173:322–335. doi: 10.1016/0002-9378(95)90222-8. [DOI] [PubMed] [Google Scholar]
- Frahm KA, Peffer ME, Zhang JY, Luthra S, Chakka AB, Couger MB, Chandran UR, Monaghan AP, DeFranco DB. Research Resource: The dexamethasone transcriptome in hypothalamic embryonic neural stem cells. Mol Endocrinol. 2016;30:144–54. doi: 10.1210/me.2015-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufgard JR, Williams MT, Skelton MR, Grubisha O, Ferreira FM, Sanger H, Wright ME, Reed-Kessler TM, Rasmussen K, Duman RS, Vorhees CV. Phosphodiesterase-1b (Pde1b) knockout mice are resistant to forced swim and tail suspension induced immobility and show upregulation of Pde10a. Psychopharmacology. 2017 doi: 10.1007/s00213-017-4587-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Karemaker R, Kavelaars A, ter Wolbeek M, Tersteeg-Kamperman M, Baerts W, Veen S, Samsom JF, Visser GH, van Bel F, Heijnen CJ. Neonatal dexamethasone treatment for chronic lung disease of prematurity alters the hypothalamus-pituitary-adrenal axis and immune system activity at school age. Pediatrics. 2008;121:e870–8. doi: 10.1542/peds.2007-2454. [DOI] [PubMed] [Google Scholar]
- Lv J, Cui W, Liu H, He H, Xiu Y, Guo J, Liu H, Liu Q, Zeng T, Chen Y, Zhang Y, Wu Q. Identification and Characterization of Long Non-Coding RNAs Related to Mouse Embryonic Brain Development from Available Transcriptomic Data. PLoS ONE. 2013;8:e71152. doi: 10.1371/journal.pone.0071152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz A, Lelieveldt BP, Grefhorst A, van Weert LT, Mol IM, Sips HC, van den Heuvel JK, Datson NA, Visser JA, Reinders MJ, Meijer OC. Genome-wide coexpression of steroid receptors in the mouse brain: Identifying signaling pathways and functionally coordinated regions. Proc Natl Acad Sci USA. 2016;113:2738–43. doi: 10.1073/pnas.1520376113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisiadia VG, Matthews SG. Glucocorticoids and fetal programming part 1: Outcomes. Nat Rev Endocrinol. 2014;10:391–402. doi: 10.1038/nrendo.2014.73. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal sub-type specification in the cerebral cortex. Nature Reviews Neuroscience. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Park OH, Park J, Yu M, An HT, Ko J, Kim YK. Identification and molecular characterization of cellular factors required for glucocorticoid receptor-mediated mRNA decay. Genes Dev. 2016;30:2093–2105. doi: 10.1101/gad.286484.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peffer ME, Chandran UR, Luthra S, Volonte D, Garabedian MJ, Monaghan AP, DeFranco DB. Caveolin-1 regulates genomic action of the glucocorticoid receptor in neural stem cells. Mol Cell Biol. 2014;34:2611–23. doi: 10.1128/MCB.01121-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peffer ME, Zhang JY, Umfrey L, Rudine AC, Monaghan AP, DeFranco DB. Minireview: The impact of antenatal therapeutic synthetic glucocorticoids on the developing fetal brain. Mol Endocrinol. 2015;29:658–66. doi: 10.1210/me.2015-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–6. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Rakic P, Lombroso PJ. Development of the cerebral cortex: 1. Forming the cortical structure. J Am Acad Adolesc Psychiatry. 1998;37:116–7. doi: 10.1097/00004583-199801000-00026. [DOI] [PubMed] [Google Scholar]
- Reinhardt RR, Bondy CA. Differential cellular pattern of gene expression for two distinct cGMP-inhibited cyclic nucleotide phosphodiesterases in developing and mature rat brain. Neuroscience. 1996;72:567–78. doi: 10.1016/0306-4522(95)00520-x. [DOI] [PubMed] [Google Scholar]
- Roberts D, Dalziel S. Antenatal corticosteroids for accelerating fetal lung maturation for women at risk of preterm birth. Cochrane Database Syst Rev. 2006;19 doi: 10.1002/14651858.CD004454.pub2. CD004454. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL. Annual Research Review: Development of the cerebral cortex: implications for neurodevelopmental disorders. J Child Psychol Psychiatry. 2011;52:339–55. doi: 10.1111/j.1469-7610.2010.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi R, Arsenijevic Y, Giacomini M, Rey JP, Voirol MJ, Gaillard RC, Risold PY, Pralong F. The fetal hypothalamus has the potential to generate cells with a gonadotropin releasing hormone (GnRH) phenotype. PLoS One. 2009;4:e4392. doi: 10.1371/journal.pone.0004392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarasinghe RA, Di Maio R, Volonte D, Galbiati F, Lewis M, Romero G, DeFranco DB. Nongenomic glucocorticoid receptor action regulates gap junction intercellular communication and neural progenitor cell proliferation. Proc Natl Acad Sci USA. 2011;108:16657–62. doi: 10.1073/pnas.1102821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A, Malchow B, Hasan A, Falkai P. The impact of environmental factors in severe psychiatric disorders. Front Neurosci. 2014;8:19. doi: 10.3389/fnins.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, Qi L, Qian J, Blackshaw S. A genomic atlas of mouse hypothalamic development. Nat Neurosci. 2010;13:767–75. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim K, Chan WY, Woon PS, Low HQ, Lim L, Yang GL, Lee J, Chong SA, Sitoh YY, Chan YH, Liu J, Tan EC, Williams H, Nowinski WL. ARVCF genetic influences on neurocognitive and neuroanatomical intermediate phenotypes in Chinese patients with schizophrenia. J Clin Psychiatry. 2012;73:320–6. doi: 10.4088/JCP.10m06491. [DOI] [PubMed] [Google Scholar]
- Siuciak JA, McCarthy SA, Chapin DS, Reed TM, Vorhees CV, Repaske DR. Behavioral and neurochemical characterization of mice deficient in the phosphodiesterase-1B (DE1B) enzyme. Neuropharmacology. 2007;53:113–24. doi: 10.1016/j.neuropharm.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Sun XY, Hayashi Y, Xu S, Kanou Y, Takagishi Y, Tang YP, Murata Y. Inactivation of the Rcan2 gene in mice ameliorates the age- and diet-induced obesity by causing a reduction in food intake. PLoS One. 2011;6:e14605. doi: 10.1371/journal.pone.0014605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulfig N, Chan WY. Expression of ARVCF in the human ganglionic eminence during fetal development. Dev Neurosci. 2004;26:38–44. doi: 10.1159/000080710. [DOI] [PubMed] [Google Scholar]
- Wyrwoll CS, Homes MC. Prenatal excess glucocorticoids exposure and adult affective disorders: A role for serotonergic and catecholamine pathways. Neuroendocrinology. 2012;95:47–55. doi: 10.1159/000331345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File 1. Nuclear receptor expression (CPM) in cerebral cortical and hypothalamic male and female NPSCs identified using RNA-Seq analysis.
Supplemental File 2. Common and unique Dex-Dependent Genes in cerebral cortical and hypothalamic male and female NPSCs identified using RNA-Seq analysis.