Abstract
The liver plays a crucial role in a variety of physiological processes. Sexual dimorphism is markedly defined in liver disorders, such as fatty liver diseases and liver cancer, but barely addressed in the normal liver. Distinct sex hormone signaling between male and female livers is the major driving factor for hepatic sexual dimorphism. Over 6,000 genes are differently expressed between male and female livers in mice. Here we address how sex hormone receptors, estrogen receptor alpha (ERα) and androgen receptor (AR), mediate sexually dimorphic gene expression in mouse livers. We identified 5,192 ERα target genes and 4,154 AR target genes using ChIP-Seq. Using liver-specific ERα or AR knockout mice, we further identified direct and functional target genes of ERα (123 genes) and AR (151 genes) that contribute to hepatic sexual dimorphism. We also found that the most significant sexually dimorphic gene expression was initiated at birth by comparing hepatic gene expression data from the embryonic stage E10.5 to the postnatal stage P60 during liver development. Overall, our study indicates that sex hormone receptor signaling drives sexual dimorphism of hepatic gene expression throughout liver development.
Keywords: sexual dimorphism, mouse liver, androgen receptor (AR), estrogen receptor alpha (ERα)
1. Introduction
The liver provides many essential functions, including regulation of metabolism, control of cholesterol and serum protein biosynthesis and transport, detoxification of drugs, immune response, and so on (1). Male and female livers show significant sex-dependent differences in physiological and pathological processes, such as cholesterol metabolism, detoxification function, and the prevalence of liver diseases, including fatty liver diseases, hepatitis, and hepatocellular carcinoma (2–7). Many studies have shown that phenotypic differences between male and female livers are derived from sexual dimorphism of hepatic gene expression (8–10). Distinct sex hormone signaling between male and female livers is the major driving factor for hepatic sexual dimorphism (11).
Sex hormones, estrogen in females and androgen in males, are responsible for crucial cellular functions, including cell proliferation, differentiation, and homeostasis, by binding to its receptors to regulate the receptor activity. In mouse liver, estrogen receptor alpha (ERα) and androgen receptor (AR) are the major sex hormone receptors that mediate hepatic sex hormone signaling in females and males, respectively (12). ERα and AR have been shown to govern the expression of sexually dimorphic genes in many hepatic processes and disorders. For example, ERα regulated the prevention of tumorigenesis, anti-inflammatory pathway, body weight dynamics, and lipolysis, while AR controlled the susceptibility of tumorigenesis, DNA damage from oxidative stress, energy homeostasis, and epigenetic regulation (13–21). These studies suggest that ERα- and AR-mediated sex hormone signaling plays important roles in the progression of liver diseases, such as hepatocellular carcinoma, steatosis, and hepatitis (2–7), however, how ERα and AR regulate sexual dimorphic gene expression in normal livers has never been addressed in a genome-wide manner.
Hence, in this study, we performed a whole-genome gene expression analysis to identify differently expressed genes in male and female livers using microarrays. Next, we identified ERα or AR binding sites-associated target genes using ChIP-Seq and we further identified ERα or AR direct target genes using liver-specific ERα or AR knockout mice to authenticate the key functional genes that are mediated by sex hormone receptor signaling and contributing to hepatic sexual dimorphism. Meanwhile, we also applied the pathway analysis to address the functions of these sexual dimorphic genes. Lastly, we explored the possible time when these sexually dimorphic genes were initiated during liver development by comparing hepatic gene expression data from the embryonic stage E10.5 to the postnatal stage P60. Here, we provide a comprehensive genomic profiling of sex hormone receptor signaling in sexually dimorphic gene expression in the normal liver.
2. Materials and methods
2.1 Animals
ARloxP/loxP mice were purchased from EMMA. AlfpCre mice were a gift from Dr. Klaus Kaestner at the University of Pennsylvania. We crossed ARloxP/loxP mice (control) with AlfpCre mice to obtain liver-specific AR ablation, ARloxP/loxP;AlfpCre mice (mutant). All animals were fed ad libitum and had free access to water. Livers were collected from ~12 week-old mice and stored at −80°C. All animal experiments performed were in compliance with ethical regulations and were approved by the Mayo Clinic Institutional Animal Care and Use Committee.
2.2 Gene expression profiling
Total RNA was purified from both control and mutant livers using the TRIzol reagent (Invitrogen, Carlsbad, CA) and the RNeasy Mini kits (QIAGEN, Valencia, CA) and measured for gene expression profiles using mRNA-Seq with the HiSeq 2000 (Illumina). Sequencing reads were aligned and analyzed using Tophat and Cufflinks to identify differentially expressed genes between control and mutant livers. Meanwhile, we also re-analyzed differential gene expression in livers between control and liver-specific ERα knockout mice from microarray data (GSE36514) (22). Additionally, we also analyzed differential gene expression in normal mouse livers between males and females using GSE32244 data (5). Genes showing at least 2-fold changes with P values of less than 0.05 between control and mutant livers were considered as differentially expressed genes. We also collected gene expression data in embryonic and postnatal male mouse livers from the embryonic day 10.5 (E10.5), E11.5, E12.5, E13.5, E14.5, E16.5, the postnatal day 0 (P0, new born) P7, P30, and P60 (two-month old) mice in the GEO database, including GSE6998 and GSE21224 (23, 24), and re-analyzed these data together using the Affy program in the Bioconductor package.
2.3 The analysis of ERa and AR target genes
In addition to obtain ERα- or AR-regulated genes from gene expression profiling in livers of liver-specific ERα or AR knockout mice, we also analyzed genome-wide ERα and AR binding of ChIP-Seq data from E-MTAB-805 (5). By intersecting ERα- or AR-regulated genes with ERα- or AR-associated target genes from ChIP-Seq data, we obtained directly regulated ERα or AR target genes. Next, we performed the Ingenuity pathway analysis to identify the functions of these ERα or AR target genes. The default settings were used for our Ingenuity pathway analysis (version 2016). The P-values were calculated by comparing the number of queried genes with the number of all genes in each functional pathway using Fisher’s exact test.
3. Results
3.1. Sex dimorphism in hepatic gene expression and development
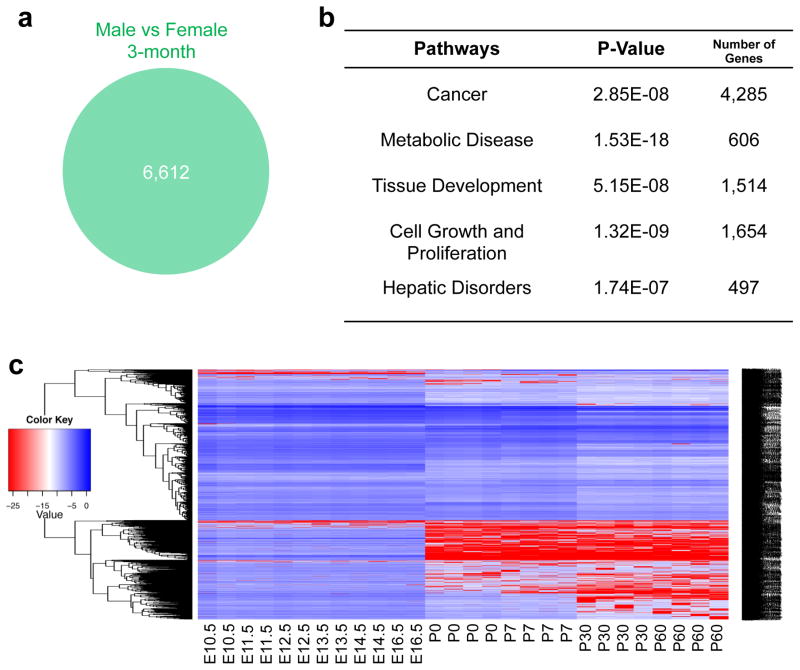
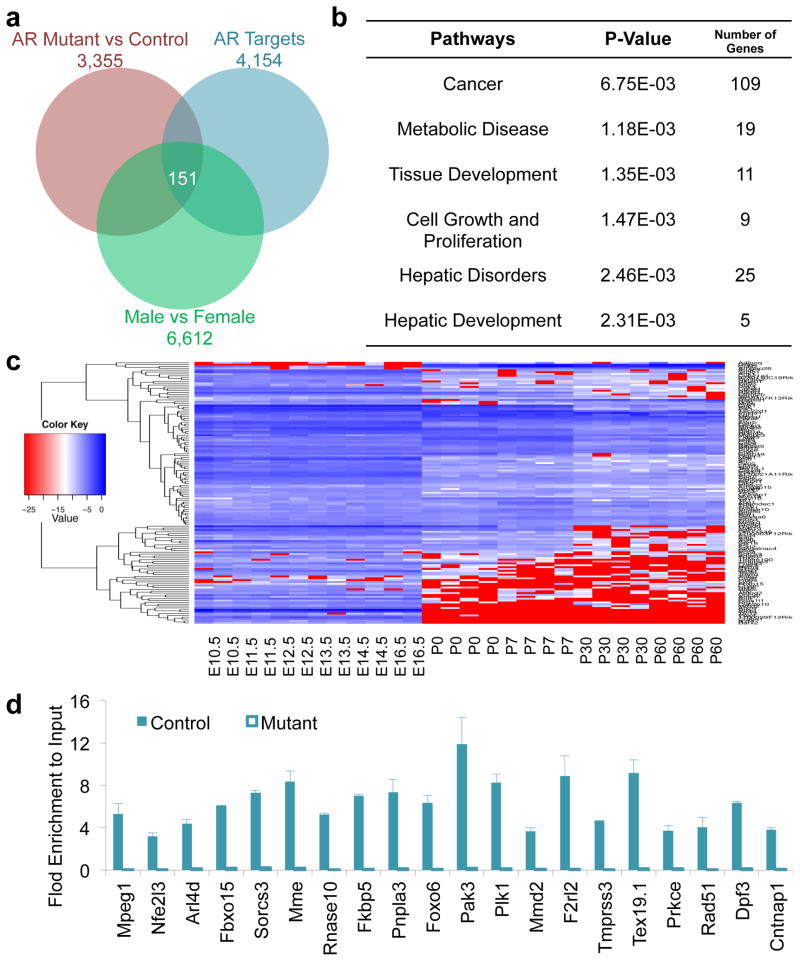
To investigate sexual dimorphism of global gene expression differences in the mouse liver, we analyzed the ratio of normalized intensities from gene expression microarrays in paired samples (male and female livers of three-month old mice). A total of 6,612 differentially expressed genes with at least 1.5 fold changes were identified between male and female mouse livers (Fig. 1a). To understand the functions of these genes, we performed the Ingenuity pathway analysis to identify the most significantly enriched pathways related to gene functions. The pathway enrichment analysis showed that differentially expressed genes between sexes were involved in five major pathways (Fig. 1b). Among them, the most enriched one, composed of 4,285 differentially expressed genes, was related to cancer, and others were related to cell growth and proliferation, tissue development, metabolic diseases, and hepatic disorders (Fig. 1b). These enriched pathways for sexual differentially expressed genes will pave a new way for an in-depth study on the underlying of sex dimorphism in hepatic gene expression and functions during pathological processes, such as hepatic tumorigenesis.
Figure 1.
Sexual dimorphism in hepatic gene expression and development. (a) Total 6,612 genes differentially expressed in male and female livers of 3-month old mice (1.5-fold). (b) The Ingenuity pathway analysis of sexually dimorphic genes in the mouse liver. (c) Expression profiles of sexually dimorphic genes during liver development. E10.5-E16.5, embryonic days 10.5 to 16.5. P0-P60, postnatal days 0 to 60.
Previous studies have shown that liver-specific gene expression was initiated during liver development. However, when sex-related gene expression occurs during liver development is barely understood, though early studies in 1970s suggest that sex-specific expression of a few selected genes might be initiated in two major phases of liver development, in the early postnatal period and at puberty. Thus, we collected and analyzed gene expression profiles in the mouse livers from embryonic day E10.5 to E16.5 and from the postnatal day 0 (P0, new born) to P7, P30, and P60 (two-month old) mice. The most significant changes in the expression of sexually dimorphic genes occurred at birth (Fig. 1c). These results suggest that the major sexually dimorphic genes may be initiated at birth or in the early postnatal period during liver development.
3.2. Sexually dimorphic gene expression potentially mediated by ERa in the female mouse liver
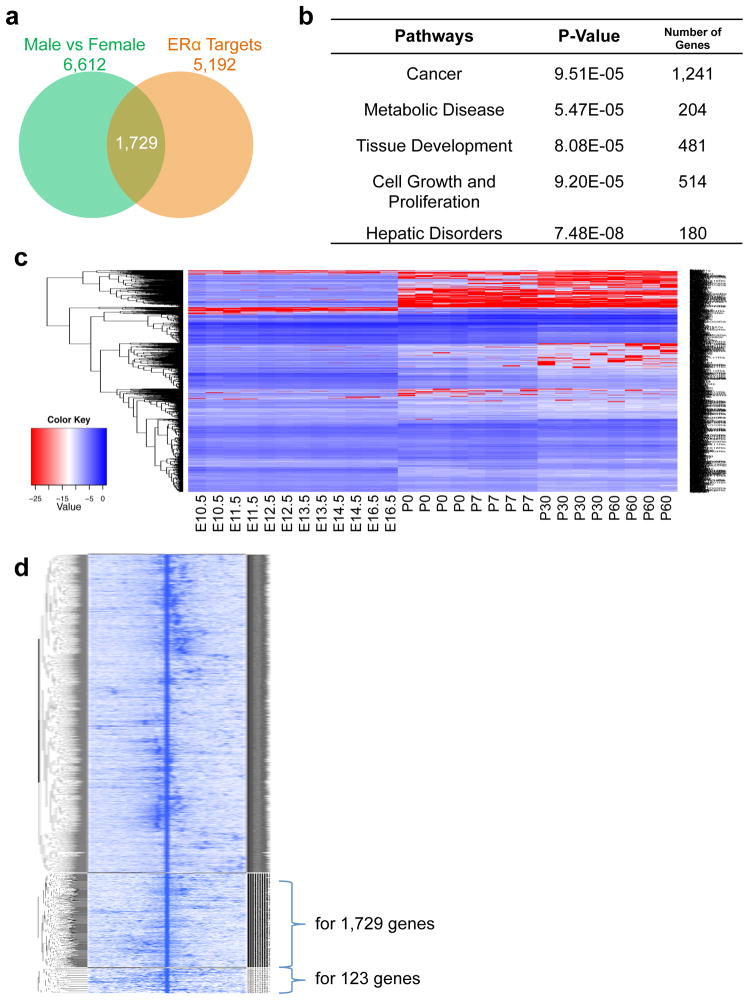
To examine how many of these sexually dimorphic genes were regulated by ERα in the female mouse liver, we analyzed global ERα binding sites and associated target genes from ERα ChIP-Seq data and identified 5,192 ERα associated target genes (Fig. 2a). By intersecting them with 6,612 sexually differentially expressed genes, we found that 1,729 sexually differentially expressed genes were potential ERα target genes (Fig. 2a). These data suggest that ERα may regulate sexually dimorphic gene expression in liver. The Ingenuity pathway analysis showed that about 72% of these genes (1,241 genes out of 1,729 genes) were enriched in the cancer pathway, indicating that the majority of these sexually differentially expressed and also potential ERα target genes were highly correlated to cancer (Fig. 2b). By tracing the expression of these genes during liver development, we found that the most significant changes of these genes primarily occurred in the liver of newly born mice and hepatic expression of these genes were markedly decreased after birth (Fig. 2c). Varied expression of potential ERα target genes was in parallel with the changes of sexually dimorphic genes during liver development, suggesting that ERα could mediate sexually dimorphic gene expression in the mouse liver.
Figure 2.
ERα-mediated sexually dimorphic gene expression in the mouse liver. (a) Expression of 1,729 ERα targets is sexually dimorphic. (b) The Ingenuity pathway analysis of sexually dimorphic ERα target genes in the mouse liver. (c) Expression profiles of sexually dimorphic ERα target genes during liver development. E10.5-E16.5, embryonic days 10.5 to 16.5. P0-P60, postnatal days 0 to 60. (d) Heatmap of all ERα binding sites including those associated with 1,729 genes from Fig. 2a and 123 genes from Fig. 4a.
3.3. Sexually dimorphic gene expression potentially mediated by AR in the male mouse liver
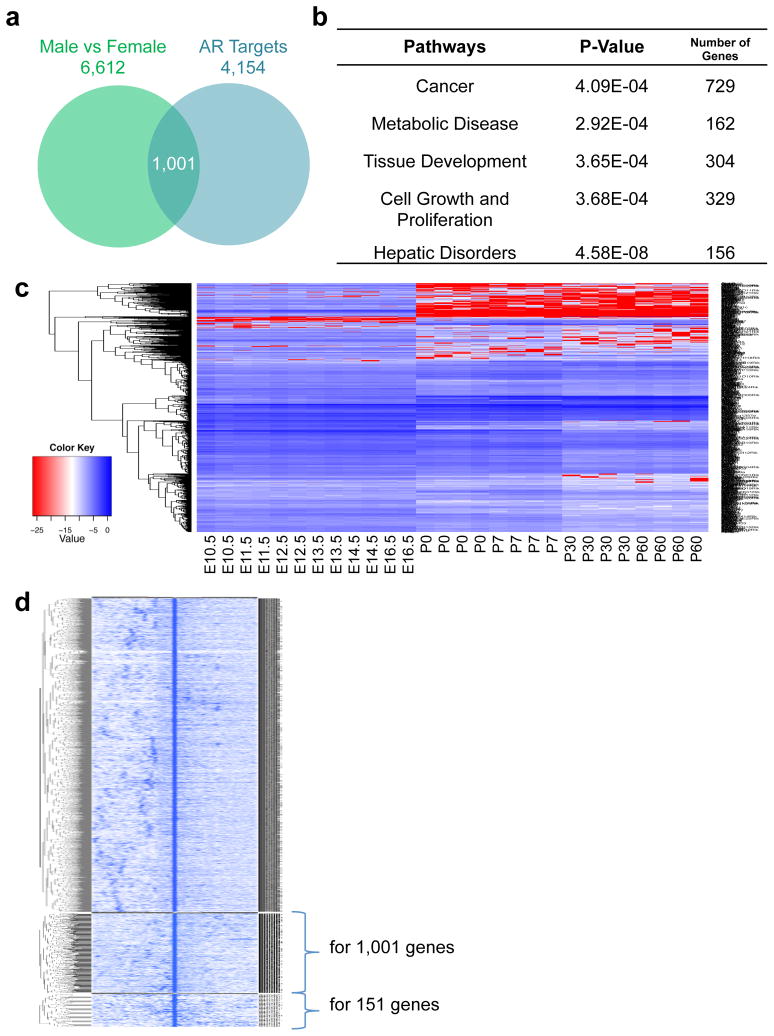
To examine whether AR participates in the regulation of sexually dimorphic gene expression in the mouse liver, we conducted AR ChIP-Seq analysis to reveal genome-wide AR binding sites and their associated target genes. We found 4,154 potential AR target genes in the male mouse liver, which regulated about 1,001 sexually differentially expressed genes after intersecting with the total 6,612 sexually dimorphic genes (Fig. 3a). The Ingenuity pathway analysis showed that these 1,001 genes were highly involved in cancer (73%), cell growth and proliferation, and tissue development (Fig. 3b). Further analysis also showed that the expression of most of these genes significantly changed at birth during liver development (Fig. 3c). Varied expression of potential AR target genes was paralleled with the changes of sexually dimorphic genes during liver development, suggesting that AR could also participate in modulating sexually dimorphic gene expression in the mouse liver.
Figure 3.
AR-mediated sexually dimorphic gene expression in the mouse liver. (a) Expression of 1,001 AR targets is sexually dimorphic. (b) The Ingenuity pathway analysis of sexually dimorphic AR target genes in the mouse liver. (c) Expression profiles of sexually dimorphic AR target genes during liver development. E10.5-E16.5, embryonic days 10.5 to 16.5. P0-P60, postnatal days 0 to 60. (d) Heatmap of all AR binding sites including those associated with 1,001 genes from Fig. 3a and 151 genes from Fig. 5a.
3.4. Direct and functional ERa target genes in the female mouse liver
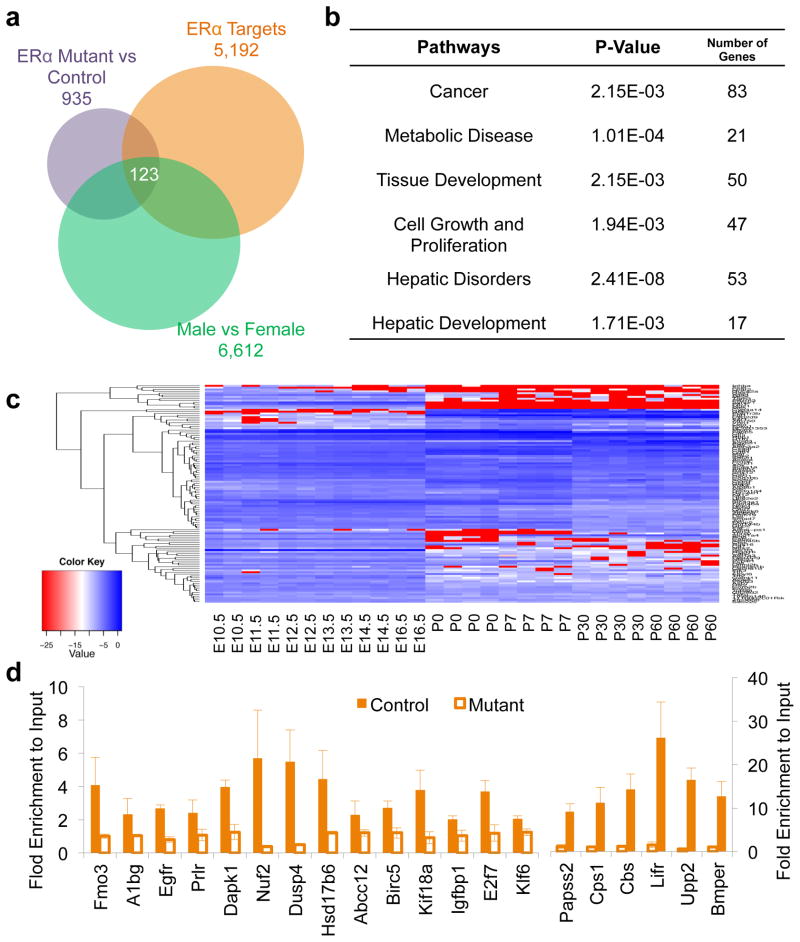
Not all ERα-associated target genes from ChIP-Seq data are regulated by ERα. Thus, to identify direct and functional ERα target genes, we also need to identify ERα-regulated genes. “Loss-of-function” analysis is usually used to identify the functionally regulated target genes of a transcription factor. Therefore, we analyzed differentially expressed genes in livers between female control and liver-specific ERα knockout mice and found about 953 ERα-regulated genes in the female livers, which could be regulated directly or indirectly by ERα (Fig. 4a). By intersecting them with 5,193 ERα-associated target genes from ChIP-Seq data, we identified 586 direct and functional ERα target genes in the female mouse liver (Fig. 4a). Further analysis showed that about 123 ERα direct target genes were differentially regulated between genders after intersecting with 6,612 sexual dimorphic genes (Fig. 4a). The Ingenuity pathway analysis showed that these 123 ERα direct target genes with sexually dimorphic expression were highly correlated to cancer (67%), cell proliferation and growth, hepatic disorders, and tissue development (Fig. 4b). Additionally, we also found that the majority of these 123 genes showed the most significant expression changes at birth during liver development (Fig. 4c). Thus, this is a key list of genes that regulated sexual dimorphism in the liver and are also modulated directly by ERα after birth.
Figure 4.
Direct ERα targets in the mouse liver. (a) 123 ERα targets are identified from liver-specific ERα-deficient livers showing sexually dimorphic expression. (b) The Ingenuity pathway analysis for direct ERα target genes with sexually dimorphic expression in the mouse liver. (c) Expression profiles of direct ERα target genes with sexual dimorphic expression during liver development. E10.5–E16.5, embryonic days 10.5 to 16.5. P0–P60, postnatal days 0 to 60. (d) Validation of selected ERα targets using ChIP-qPCR.
3.5. Direct and functional AR target genes in the male mouse liver
Similar to the ERα analysis, we first identified 3,355 AR-regulated genes by extracting differentially expressed genes in livers between male control and liver-specific AR knockout mice (Fig. 5a). Next, by intersecting them with 4,154 AR-associated target genes, we found about 439 direct and functional AR target genes in the male mouse liver (Fig. 5a). Further intersecting with 6,612 sexually dimorphic genes showed that about 151 direct target genes controlled sexually dimorphic gene expression in the male mouse liver (Fig. 5a). The pathway enrichment analysis showed that these 151 AR direct target genes showing sexually dimorphic expression were highly involved in cancer (72%) and other hepatic disorders (Fig. 5b). Further analysis showed that the majority of these 151 genes had the most significant expression changes at birth during liver development (Fig. 5c). Thus, this is a key list of genes that regulated sexual dimorphism in the liver and are also modulated directly by AR after birth.
Figure 5.
Direct AR targets in the mouse liver. (a) 151 AR targets are identified from liver-specific AR-deficient livers showing sexually dimorphic expression. (b) The Ingenuity pathway analysis for direct AR target genes with sexually dimorphic expression in the mouse liver. (c) Expression profiles of direct AR target genes with sexually dimorphic expression during liver development. E10.5-E16.5, embryonic days 10.5 to 16.5. P0-P60, postnatal days 0 to 60. (d) Validation of selected AR targets using ChIP-qPCR.
3.6. Comparison of ERa and AR direct target genes for sexually dimorphic regulation in the mouse liver
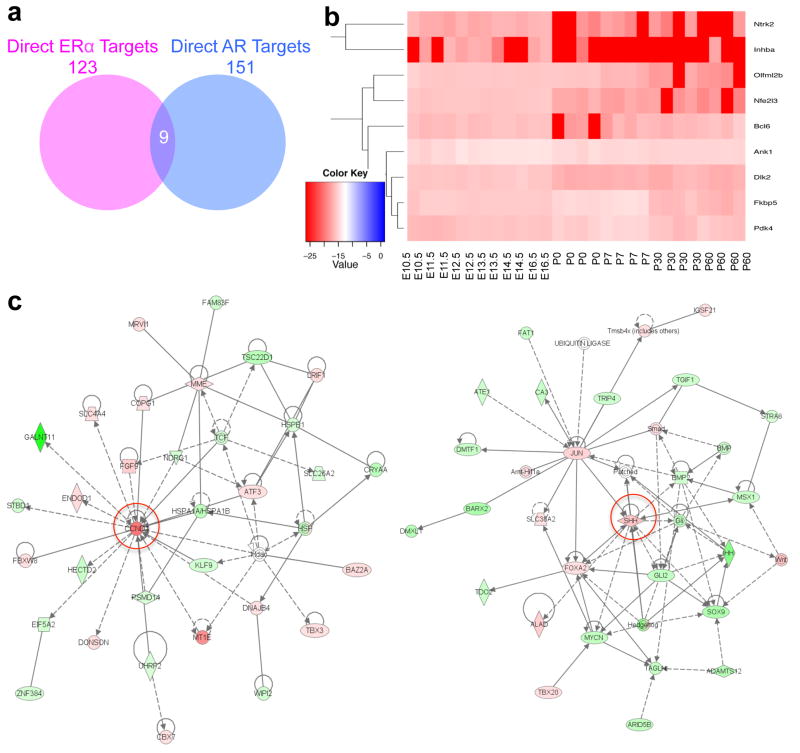
Finally, we compared the sexually dimorphic genes of direct ERα target genes (123 genes) with direct AR target genes (151 genes) and found only nine common targets (Fig. 6a). Most of these nine common target genes showed minor changes compared to other direct target genes during liver development (Fig. 4c, 5c, and 6b). Figure 6c showed examples of a direct ERα target gene, cyclin D1 (CCND1), and a direct AR target gene, sonic hedgehog (SSH), regulated a group of sexually dimorphic genes from the total 6,612 sexually dimorphic genes that we identified in the mouse liver (Fig. 1a) using the Ingenuity pathway analysis. These data suggest that these key lists of ERα or AR direct target genes are indeed the central regulators of sexually dimorphic gene expression in female or male livers, respectively and differentially.
Figure 6.
(a) Nine common direct targets between AR and ERα in the mouse liver. (b) Expression profiles of common direct target genes between AR and ERα during liver development. E10.5-E16.5, embryonic days 10.5 to 16.5. P0-P60, postnatal days 0 to 60. (c) Examples of the Ingenuity pathway analysis of ERα- or AR-regulated networks of sexually dimorphic genes using Ingenuity. Left panel, a ERα direct target, CCND1-centered network; right panel, a AR direct target, SSH-centered network. Red, up-regulated genes in females; green, down-regulated genes in females; white, unchanged genes between males and females.
4. Discussion
Males and females always show significant physiological differences that are mainly determined by the signaling of sex hormones and their receptors. These differences include metabolic, emotion, behavior, and so on. However, serological biomarkers representing hepatic functions, such as glucose, bilirubin, bile acids, lipids, and enzymes, are largely the same between males and females. Surprisingly, over 6,000 genes showed differential expression between male and female livers (Fig. 1a), but the liver morphology and functions between sexes are similar. The liver has not been considered the primary organ with sexual dimorphism; however, the liver displays distinct differences in many physiological and pathological processes between the two sexes. For example, the male liver had lower mitochondrial functions and metabolic activities than females (25); sexual dimorphism in liver has been found with significant impacts on the process of drug metabolism (3); the morbidity of several representative hepatic pathologies are different between men and women (4, 5); the incidence rates of fatty liver diseases, chronic hepatitis, and hepatocellular carcinoma are much higher in males than females (2–7), while primary biliary cirrhosis and autoimmune hepatitis are more profound in females (26). The gender-specific liver diseases may result from hepatic gene expression differences between genders. Studies have found sex-based expression differences in several genes, including sulfotransferases, glutathione S-transferases, cytochrome P450, 11β-hydroxysteroid dehydrogenase and UDP-glucuronosyltransferases 24–27 (27–31). In this study, we carried out transcriptomic analyses of the mouse liver from both sexes and displayed comprehensive and integrated information of sexual dimorphism of hepatic gene expression in mice. Our gene expression profiling results for male and female livers clearly showed gene expression differences at the global level. Interestingly, we found many gender-specific genes involved in the cancer pathway, which became more profound in sex hormone receptor target genes. These findings further support the observations of sexual dimorphism in hepatic tumorigenesis.
Sex hormone signaling is mediated by estrogen receptors and androgen receptors and involves the binding of sex hormones and receptors, the translocation of hormone-receptor complex into the nuclei, the binding to DNA response elements in the promoter or enhancer region of the target genes to activate or repress gene transcription. Our functional analysis of sexually dimorphic ERα and AR targeting in the mouse liver provides a comprehensive understanding of dimorphic sex hormone signaling in the regulation of hepatic gene expression in mice. In combination with the loss-of-function analysis using liver-specific ERα or AR knockout mice, our ERα and AR ChIP-Seq data give a unique view to depict the global functions of sex hormone receptors in hepatic sexual dimorphism. Thus, sex hormones play vital roles in regulating hepatic gene expression differences between sexes.
123 out of 5,193 ERα target genes and 151 out of 4,154 AR target genes were found to be direct and functional target genes of ERα and AR in the mouse liver, respectively. These core target genes drive differential expression of over 6,600 genes directly and indirectly through the regulatory cascades in the mouse liver between sexes as evidenced from our Ingenuity pathway analysis (Fig. 6c). Additionally, only nine common genes found in 123 ERα and 151 AR direct target genes suggests that sexually dimorphic gene expression between female and male livers is regulated by independent ERα- and AR-mediated sex hormone signaling mechanisms in females and males, respectively. Although only very few ERα or AR target genes were involved in sexually dimorphic gene expression in the normal mouse liver, we could not exclude the possibility that additional ERα or AR binding events could turn into direct and functional targets during multiple pathological processes as evidenced from the pathway analysis. Indeed, increased functional ERα and AR targeting has been observed during hepatic tumorigenesis (5). Thus, our study provide lists of potential and functional direct target genes that are regulated by ERα or AR in normal mouse liver, which will be beneficial for future studies of sexual dimorphism of hepatic disorders, such as hepatocellular carcinoma, steatosis, and hepatitis. These direct and functional target genes of hormone receptors could be the future research objectives for gender-specific medicine on liver diseases.
Regulation of sexually dimorphic gene expression in tissue development is a new topic in sex hormone signaling research. We discuss the role of the sexually dimorphic gene expression from the embryonic stage to the postnatal stage during liver development. Gene expression profiles in the embryonic and postnatal livers from both genders would be ideal for addressing our question. Nevertheless, we found that the birth could be the key regulatory factor to trigger sexual dimorphic gene expression in the mouse liver.
Overall, this global and integrative study demonstrates that sex hormone receptor signaling plays the essential role in hepatic sexually dimorphic gene regulation during liver development.
Highlights.
Numerous sexual differences in hepatic gene expression in the normal mouse liver;
Hepatic estrogen receptor alpha and androgen receptor differentially drive sexual dimorphic gene expression;
The most significant sexually dimorphic gene expression occurs at birth during liver development.
Acknowledgments
We thank Dr. Klaus H. Kaestner (University of Pennsylvania) kindly provided us the AlfpCre mice. We thank staffs from the Sequencing Core and Bioinformatics Core facilities at Mayo Clinic for their great technical support.
Funding: This project was supported by the NIH Pathways to Independence Award (K99/R00) to Z.L. (grant # R00CA168983).
Footnotes
Author Contributions: Z.L. designed and wrote the manuscript. D.Z., X.W., P. A. and J.G. performed the experiments and/or wrote a part of manuscript.
Competing Interest: The authors disclose no potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18(2):175–89. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Ng MK, Jessup W, Celermajer DS. Sex-related differences in the regulation of macrophage cholesterol metabolism. Curr Opin Lipidol. 2001;12(5):505–10. doi: 10.1097/00041433-200110000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Wolbold R, Klein K, Burk O, Nussler AK, Neuhaus P, Eichelbaum M, Schwab M, Zanger UM. Sex is a major determinant of CYP3A4 expression in human liver. Hepatology. 2003;38(4):978–88. doi: 10.1053/jhep.2003.50393. [DOI] [PubMed] [Google Scholar]
- 4.Lee YH, Kim SH, Kim SN, Kwon HJ, Kim JD, Oh JY, Jung YS. Sex-specific metabolic interactions between liver and adipose tissue in MCD diet-induced non-alcoholic fatty liver disease. Oncotarget. 2016;7(30):46959–71. doi: 10.18632/oncotarget.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148(1–2):72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bigsby RM, Caperell-Grant A. The role for estrogen receptor-alpha and prolactin receptor in sex-dependent DEN-induced liver tumorigenesis. Carcinogenesis. 2011;32(8):1162–6. doi: 10.1093/carcin/bgr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang SH, Yeh SH, Lin WH, Wang HY, Chen DS, Chen PJ. Identification of androgen response elements in the enhancer I of hepatitis B virus: a mechanism for sex disparity in chronic hepatitis B. Hepatology. 2009;50(5):1392–402. doi: 10.1002/hep.23163. [DOI] [PubMed] [Google Scholar]
- 8.Zheng W, Xu H, Lam SH, Luo H, Karuturi RK, Gong Z. Transcriptomic analyses of sexual dimorphism of the zebrafish liver and the effect of sex hormones. PloS one. 2013;8(1):e53562. doi: 10.1371/journal.pone.0053562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiao Q, Le Manach S, Sotton B, Huet H, Duvernois-Berthet E, Paris A, Duval C, Ponger L, Marie A, Blond A, Matheron L, Vinh J, Bolbach G, Djediat C, Bernard C, Edery M, Marie B. Deep sexual dimorphism in adult medaka fish liver highlighted by multi-omic approach. Scientific reports. 2016;6:32459. doi: 10.1038/srep32459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conforto TL, Waxman DJ. Sex-specific mouse liver gene expression: genome-wide analysis of developmental changes from pre-pubertal period to young adulthood. Biology of sex differences. 2012;3:9. doi: 10.1186/2042-6410-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Nas A, Guhathakurta D, Wang SS, Yehya N, Horvath S, Zhang B, Ingram-Drake L, Chaudhuri G, Schadt EE, Drake TA, Arnold AP, Lusis AJ. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology. 2009;150(3):1235–49. doi: 10.1210/en.2008-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Li Z. Interplay of estrogen receptors and FOXA factors in the liver cancer. Molecular and cellular endocrinology. 2015 doi: 10.1016/j.mce.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen EV, Block GE, Smith S, Kyser K, DeSombre ER. Estrogen receptors and breast cancer response to adrenalectomy. Natl Cancer Inst Monogr. 1971;34:55–70. [PubMed] [Google Scholar]
- 14.Whitacre CC. Sex differences in autoimmune disease. Nat Immunol. 2001;2(9):777–80. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 15.Benz V, Bloch M, Wardat S, Bohm C, Maurer L, Mahmoodzadeh S, Wiedmer P, Spranger J, Foryst-Ludwig A, Kintscher U. Sexual dimorphic regulation of body weight dynamics and adipose tissue lipolysis. PloS one. 2012;7(5):e37794. doi: 10.1371/journal.pone.0037794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simanainen U, Ryan T, Li D, Suarez FG, Gao YR, Watson G, Wang Y, Handelsman DJ. Androgen receptor actions modify skin structure and chemical carcinogen-induced skin cancer susceptibility in mice. Horm Cancer. 2015;6(1):45–53. doi: 10.1007/s12672-014-0210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyake H, Hara I, Gleave ME, Eto H. Protection of androgen-dependent human prostate cancer cells from oxidative stress-induced DNA damage by overexpression of clusterin and its modulation by androgen. Prostate. 2004;61(4):318–23. doi: 10.1002/pros.20087. [DOI] [PubMed] [Google Scholar]
- 18.Nohara K, Liu S, Meyers MS, Waget A, Ferron M, Karsenty G, Burcelin R, Mauvais-Jarvis F. Developmental androgen excess disrupts reproduction and energy homeostasis in adult male mice. The Journal of endocrinology. 2013;219(3):259–68. doi: 10.1530/JOE-13-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong KW, Zhao JC, Kim J, Li S, Yang YA, Song B, Rittie L, Hu M, Yang XJ, Perbal B, Yu J. Polycomb-mediated disruption of an androgen receptor feedback loop drives castration-resistant prostate cancer. Cancer research. 2016 doi: 10.1158/0008-5472.CAN-16-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiewer MJ, Knudsen KE. Linking DNA Damage and Hormone Signaling Pathways in Cancer. Trends Endocrinol Metab. 2016;27(4):216–25. doi: 10.1016/j.tem.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma WL, Hsu CL, Wu MH, Wu CT, Wu CC, Lai JJ, Jou YS, Chen CW, Yeh S, Chang C. Androgen receptor is a new potential therapeutic target for the treatment of hepatocellular carcinoma. Gastroenterology. 2008;135(3):947–55. 55 e1–5. doi: 10.1053/j.gastro.2008.05.046. Epub 2008/07/22 S0016-5085(08)00867-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matic M, Bryzgalova G, Gao H, Antonson P, Humire P, Omoto Y, Portwood N, Pramfalk C, Efendic S, Berggren PO, Gustafsson JA, Dahlman-Wright K. Estrogen signalling and the metabolic syndrome: targeting the hepatic estrogen receptor alpha action. PloS one. 2013;8(2):e57458. doi: 10.1371/journal.pone.0057458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otu HH, Naxerova K, Ho K, Can H, Nesbitt N, Libermann TA, Karp SJ. Restoration of liver mass after injury requires proliferative and not embryonic transcriptional patterns. J Biol Chem. 2007;282(15):11197–204. doi: 10.1074/jbc.M608441200. [DOI] [PubMed] [Google Scholar]
- 24.Lee JS, Ward WO, Knapp G, Ren H, Vallanat B, Abbott B, Ho K, Karp SJ, Corton JC. Transcriptional ontogeny of the developing liver. BMC genomics. 2012;13:33. doi: 10.1186/1471-2164-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Justo R, Boada J, Frontera M, Oliver J, Bermudez J, Gianotti M. Gender dimorphism in rat liver mitochondrial oxidative metabolism and biogenesis. Am J Physiol Cell Physiol. 2005;289(2):C372–8. doi: 10.1152/ajpcell.00035.2005. [DOI] [PubMed] [Google Scholar]
- 26.Gossard AA, Lindor KD. Development of autoimmune hepatitis in primary biliary cirrhosis. Liver international : official journal of the International Association for the Study of the Liver. 2007;27(8):1086–90. doi: 10.1111/j.1478-3231.2007.01538.x. [DOI] [PubMed] [Google Scholar]
- 27.Liu L, Klaassen CD. Ontogeny and hormonal basis of female-dominant rat hepatic sulfotransferases. J Pharmacol Exp Ther. 1996;279(1):386–91. [PubMed] [Google Scholar]
- 28.Liu L, Klaassen CD. Ontogeny and hormonal basis of male-dominant rat hepatic sulfotransferases. Mol Pharmacol. 1996;50(3):565–72. [PubMed] [Google Scholar]
- 29.Srivastava PK, Waxman DJ. Sex-dependent expression and growth hormone regulation of class alpha and class mu glutathione S-transferase mRNAs in adult rat liver. Biochem J. 1993;294( Pt 1):159–65. doi: 10.1042/bj2940159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Low SC, Chapman KE, Edwards CR, Wells T, Robinson IC, Seckl JR. Sexual dimorphism of hepatic 11 beta-hydroxysteroid dehydrogenase in the rat: the role of growth hormone patterns. The Journal of endocrinology. 1994;143(3):541–8. doi: 10.1677/joe.0.1430541. [DOI] [PubMed] [Google Scholar]
- 31.Buckley DB, Klaassen CD. Mechanism of gender-divergent UDP-glucuronosyltransferase mRNA expression in mouse liver and kidney. Drug metabolism and disposition: the biological fate of chemicals. 2009;37(4):834–40. doi: 10.1124/dmd.108.024224. [DOI] [PMC free article] [PubMed] [Google Scholar]