Abstract
Huntington's disease (HD) is caused in large part by a polyglutamine expansion within the huntingtin (Htt) protein. Post-translational modifications (PTMs) control and regulate many protein functions and cellular pathways, and PTMs of mutant Htt are likely important modulators of HD pathogenesis. Alterations of selected numbers of PTMs of Htt fragments have been shown to modulate Htt cellular localization and toxicity. In this study, we systematically introduced site-directed alterations in individual phosphorylation and acetylation sites in full-length Htt constructs. The effects of each of these PTM alteration constructs were tested on cell toxicity using our nuclear condensation assay and on mitochondrial viability by measuring mitochondrial potential and size. Using these functional assays in primary neurons, we identified several PTMs whose alteration can block neuronal toxicity and prevent potential loss and swelling of the mitochondria caused by mutant Htt. These PTMs included previously described sites such as serine 116 and newly found sites such as serine 2652 throughout the protein. We found that these functionally relevant sites are clustered in protease-sensitive domains throughout full-length Htt. These findings advance our understanding of the Htt PTM code and its role in HD pathogenesis. Because PTMs are catalyzed by enzymes, the toxicity-modulating Htt PTMs identified here may be promising therapeutic targets for managing HD.
Keywords: Huntington disease, neurodegeneration, neuron, phosphorylation, toxicity
Introduction
Huntington's disease (HD)3 is an autosomal dominant neurodegenerative disorder caused by a CAG triplet repeat expansion in the huntingtin gene, leading to an abnormal poly(Q) expansion in the huntingtin (Htt) protein (1). At a cellular level, HD is defined by the preferential degeneration and death of striatal medium spiny neurons, as well as cortical pyramidal neurons. Toxicity of Htt and its fragments has been demonstrated in vitro and in vivo, and multiple studies suggest that post-translational modifications (PTMs) of expanded Htt protein are likely to be important modulators of HD pathogenesis.
PTMs of proteins regulate many cellular processes via modulation of protein structure and functions. PTMs may be useful targets for therapeutics, because many PTMs are reversible and controlled by specific enzymes that can be modulated by small molecules.
In neurodegenerative diseases, study of postmortem brains has identified key PTMs believed to be involved in disease pathogenesis. In Alzheimer's disease and related disorders, neurofibrillary tangles contain the hyperphosphorylated microtubule–associated protein Tau (2). Hyperphosphorylated Tau is also ubiquitinated, suggesting a change in its turnover. In Parkinson's disease, postmortem brains showed that the α-synuclein present in Lewy bodies is phosphorylated on serine 129 (3). Recently, TDP-43 phosphorylation has also been shown to be present in postmortem human brain material from frontotemporal dementia and amyotrophic lateral sclerosis (4).
Increasing effort for therapeutic development in HD has focused on mutant Htt itself, with strategies for mutant Htt lowering based on altering production or clearance of the protein and thus decreasing toxicity (5). Identification and characterization of PTMs on Htt represent another strategy, because they can be responsible for the modulation of Htt processing and toxicity and thus may serve as therapeutic targets (6, 7). PTMs on Htt include SUMOylation, acetylation, and palmitoylation, but the most extensively studied involve phosphorylation. Phosphorylation at threonine 3 and serines 13 and 16 can modulate aggregation and toxicity of Htt exon 1, which contains the expanded repeat (8, 9). Phosphorylation at these sites was reported to regulate nuclear localization, clearance, and cleavage of N-terminal fragment of Htt, as well as its interactions with nuclear pore proteins (8, 9). Phosphorylation at serines 13 and 16 also was found to control the nuclear relocalization of N17 Htt induced by stress (10, 11). The simultaneous phosphorylation at serines 13 and 16 together is critical for additional post-translational modifications, including ubiquitination, SUMOylation, and acetylation (12, 13). Overall, the N17 domain of Htt was suggested to be a regulator of Htt phosphorylation and localization by acting as an oxidative stress sensor (14).
Palmitoylation of Htt by HIP14 at cysteine 214 mediates membrane attachment (15), and a reduced palmitoylation of mutant Htt can be responsible for Htt mislocalization and aggregation. The acetylation of lysine 444 has been shown to improve clearance of the mutant protein by macroautophagy by facilitating trafficking of mutant Htt into autophagosomes and reduced Htt toxicity in cell and Caenorhabditis elegans models (16).
In a previous study of Htt PTMs, we identified several phosphorylation sites within the first 586-amino acid (aa) (N586) fragment of Htt (17). Using toxicity assays and other functional measurements, we confirmed sites previously described, and we identified serine 116 as a critical site to control Htt toxicity. Unlike for the other previously identified sites, the phospho-null alteration of Ser116 was protective for neurons against mutant Htt toxicity. Other alterations previously shown to reduce Htt toxicity include serines 421, 536, 1181, and 1201 (17). At serine 421, phosphorylation by Akt and SGK improves axonal transport and protects neurons (18, 19). At serine 536, the cleavage of Htt by calpain reduces mutant Htt toxicity (20). Phosphorylation by cyclin-dependent kinase 5 at serines 1181 and 1201 is induced by DNA damage and is neuroprotective (21).
Many of the PTMs studied in these previous experiments were identified in a context of Htt fragments and in cell lines. There has been relatively little study of the PTMs on full-length Htt. In a recent study (22), we analyzed by mass spectrometry PTMs on full-length Htt immunoprecipitated from human postmortem and mouse HD brains. We identified close to 40 PTMs, including 17 novel sites, throughout full-length Htt.
In the current study, we characterized the functional effects of PTM site modifications on mutant Htt. We introduced in full-length Htt alterations preventing PTMs of the amino acids that we have found modified in human or mouse brain, to determine whether abrogation of single PTM sites could reduce mutant Htt–induced toxicity and mitochondrial dysfunction. We were able to identify several PTM sites that can modulate expanded Htt toxicity and its effects on mitochondria. These sites are located outside of the HEAT repeat domains and appear to cluster in the proteolysis-prone regions. This study is the first step toward deciphering the PTM code of the full-length Htt and validating PTMs on Htt as potential therapeutic targets for HD.
Results
Neuronal toxicity of expanded full-length Htt
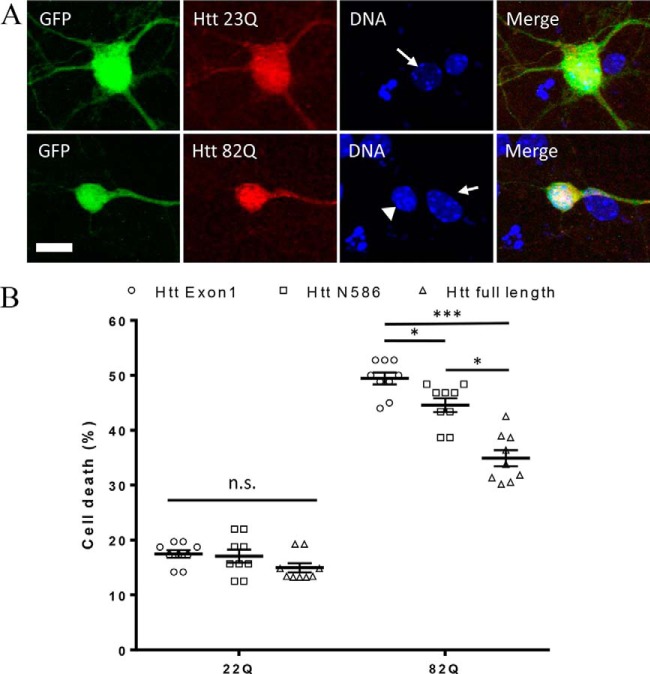
As our primary screen, we have used a previously established cell death assay based on the measurement of nuclear condensation during neuronal cell death using fragments of Htt, Htt Exon1 (23), and Htt aa1–586 (17). In this study, to evaluate the effects of post-translational modifications identified in the full-length Htt, we first have established the toxicity of full-length Htt in our neuronal model. This new model allows comparison of the toxicity levels between the fragments and full-length Htt. First, we transfected neurons with equal amounts of each constructs. After 24 h of transfection, neurons transfected with full-length constructs express detectable levels of protein. Htt is distributed throughout the cell and does not present signs of aggregation (Fig. 1A). When measured by nuclear condensation, full-length normal Htt-23Q induced only a low level of toxicity, comparable in this system with levels observed in cells transfected with Htt fragments, whereas transfection with expanded Htt-82Q elicited a significant amount of cell death (Fig. 1B). However, the total amount of toxicity that is induced by full-length Htt is significantly lower than the toxicity induced by Htt aa 1–586 or by exon 1 containing the same number of glutamines.
Figure 1.
Neuronal toxicity of expanded full-length Htt. A, example images of primary neurons transfected with full-length Htt containing either 23Q or 82Q and co-transfected with GFP. Transfected Htt was detected by immunocytochemistry using mAb 2166, and DNA is stained with Hoechst. Condensed nuclei (arrowhead) are brighter and smaller than healthy nuclei (arrow). B, quantification of cell death by nuclear condensation assay. Primary cortical neurons were transfected at DIV5 with indicated plasmid and with GFP. The cells were fixed after 48 h, and the nuclei were stained with Hoechst. Automated picture acquisition was performed on Zeiss Axiovert 200 microscope, and automated quantification of the nuclear intensity of transfected cells was performed using Volocity. *, p ≤ 0.05; ***, p ≤ 0.001 (n = 9 independent neuronal preparations).
Expanded full-length Htt PTM alterations
As a first step toward understanding the complexity of PTM of full-length Htt and identifying the potential site for therapeutic intervention on Htt, we studied whether an alteration of a single PTM site may affect toxic and functional properties of mutant Htt. Similar to our previous study with Htt aa 1–586 (17), we developed a series of constructs expressing full-length Htt-82Q with single point mutations at PTM sites. In addition to modification-null alterations done for all the selected sites (Ser to Ala and Lys to Arg), we have also generated modification-mimetic mutations (Ser to Asp, Ser to Glu, and Lys to Gln) for some PTMs of particular interest. To produce these PTM alterations throughout full-length Htt, we used a multistep approach (described under “Experimental procedures”) because of the large size of Htt and the presence of the CAG repeats. All constructs were shown by Western blot analysis in HEK cells (supplemental Fig. S1) and by immunocytochemistry in primary cortical neurons (supplemental Fig. S2) to produce comparable levels of full-length Htt. The list of sites selected for this study was based on the PTM sites identified by mass spectrometry on human and mouse brains (22), as well as some of the previously identified PTMs within the N terminus of Htt and in full-length cell models (summarized in Ref. 7), and is detailed in Table 1. Many of these N-terminal sites have not been previously characterized in the context of full-length Htt.
Table 1.
List of the PTM sites identified in vivo and in vitro
The source in which the PTM has been identified can be human brain (H), HD mouse model (M), or transfected cell lines (HEK). Phos, phosphorylation; Ac, acetylation.
| Huntingtin residue | Modification | Identification source | Reference |
|---|---|---|---|
| Thr3 | Phos | HEK | Aiken et al. (8) |
| Lys6 | Ac | HEK | Steffan et al. (12); Thompson et al. (13) |
| Lys9 | Ac | HEK | Steffan et al. (12); Thompson et al. (13) |
| Ser13/Ser16 | Phos | HEK | Thompson et al. (13) |
| Ser13/Ser16 | Phos | M | Ratovitski et al. (22) |
| Ser116/Ser120 | Phos | HEK | Watkin et al. (17) |
| Ser116/Ser120 | Phos | M | Ratovitski et al. (22) |
| Thr271 | Phos | HEK | Watkin et al. (17) |
| Thr271 | Phos | M, H | Ratovitski et al. (22) |
| Ser421 | Phos | HEK | Humbert et al. (18) |
| Ser421 | Phos | H | Ratovitski et al. (22) |
| Ser432/Ser434 | Phos | HEK | Dong et al. (53); Watkin et al. (17) |
| Ser432/Ser434 | Phos | M | Ratovitski et al. (22) |
| Lys444 | Ac | HEK | Jeong et al. (16) |
| Lys815 | Ac | M, H | Ratovitski et al. (22) |
| Lys818 | Ac | M, HEK | Ratovitski et al. (22) |
| Ser1179 | Phos | M, H | Ratovitski et al. (22) |
| Ser1181 | Phos | HEK | Schilling et al. (20) |
| Ser1181 | Phos | M, H | Ratovitski et al. (22) |
| Lys1190 | Ac | M, H, HEK | Ratovitski et al. (22) |
| Ser1201 | Phos | HEK | Schilling et al. (20) |
| Ser1201 | Phos | M | Ratovitski et al. (22) |
| Lys1204 | Ac | M, H, HEK | Ratovitski et al. (22) |
| Lys1246 | Ac | M, HEK | Ratovitski et al. (22) |
| Ser1864 | Phos | M, H, HEK | Ratovitski et al. (22) |
| Ser1872 | Phos | HEK | Huang et al. (54) |
| Ser1876 | Phos | HEK | Huang et al. (54) |
| Ser1876 | Phos | M, H | Ratovitski et al. (22) |
| Ser2114 | Phos | M, HEK | Ratovitski et al. (22) |
| Ser2116 | Phos | M, HEK | Ratovitski et al. (22) |
| Ser2342 | Phos | H, HEK | Ratovitski et al. (22) |
| Ser2489 | Phos | M | Ratovitski et al. (22) |
| Lys2548 | Ac | M, H, HEK | Ratovitski et al. (22) |
| Lys2615 | Ac | HEK | Ratovitski et al. (22) |
| Ser2652 | Phos | HEK | Ratovitski et al. (22) |
| Ser2653/Ser2657 | Phos | HEK | Schilling et al. (20) |
| Ser2653/Ser2657 | Phos | M, H, HEK | Ratovitski et al. (22) |
| Ser2936 | Phos | M, H | Ratovitski et al. (22) |
| Lys2969 | Ac | H, HEK | Ratovitski et al. (22) |
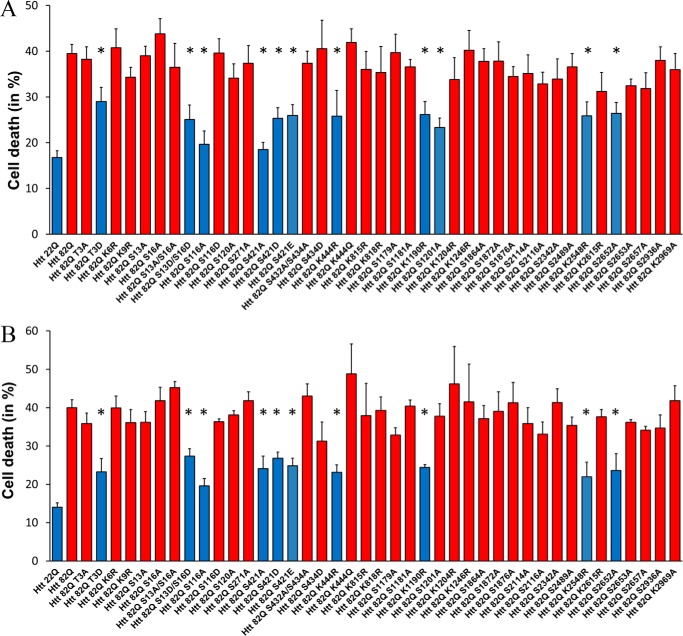
Alterations of PTM modulate Htt toxicity
Toxicity measurements of the point mutation PTMs constructs in primary cortical neurons is shown on Fig. 2A. Similar to previous reports, we found that phospho-mimetic alteration of the Thr3 and Ser13/16 sites ameliorated expanded Htt toxicity (8, 9, 13) and confirmed our previous findings with Htt aa 1–586 (17) that S116A alteration has a neuroprotective effect against the toxicity of expanded Htt to primary neurons. Contrary to previous studies with the 1–480 fragment of Htt (18, 25) but consistent with our previous data obtained using the 1–586 fragment of Htt (17), we found that both phospho-null (S421A) and presumably phospho-mimetic (S421D and S421E) alterations elicited neuroprotection in our system.
Figure 2.
Neuronal toxicity of expanded full-length Htt is modulated by PTMs. Quantification of cell death by nuclear condensation assay is shown. Primary cortical (A) and striatal (B) neurons were transfected at DIV5 with indicated plasmid and with GFP. The cells were fixed after 48 h, and the nuclei were stained with Hoechst. Automated picture acquisition was performed on Zeiss Axiovert 200 microscope, and automated quantification of the nuclear intensity of transfected cells was performed using Volocity. The results are expressed as means ± S.E. of the percentage of dead cells. *, p < 0.05 versus Htt-82Q (n = 4 independent neuronal preparations).
The acetylation mimetic Htt K444Q alteration showed no protection, whereas the acetylation-null alteration K444R ameliorated expanded Htt toxicity measured by nuclear condensation. Previously, acetylation at this site has been shown to be protective (16). Other modification-null alterations that reduced toxic properties of expanded Htt in cortical neurons are K1190R, S1201A, K2548R, and S2652A. Notably, an absence of Htt phosphorylation at position Ser1201 (within 1301 amino acids fragment of Htt with normal poly(Q) range) was previously found to make Htt toxic at levels similar to those of expanded protein, whereas it had no effect on poly(Q)-Htt–induced cell death in striatal neurons (21).
Because striatum appears to be most affected in HD (26–28), we tested whether primary striatal neurons are as sensitive to acute toxicity of full-length Htt as cortical neurons. Overexpression of full-length Htt-82Q with and without PTM alterations in striatal neurons induced levels of toxicity comparable with the levels observed in cortical neurons transfected with same constructs (Fig. 2B). These results show that PTM alterations that ameliorated toxicity of expanded Htt in cortical neurons also protected striatal neurons. The only exception is the S1201A substitution, which, unlike in cortical neurons, did not result in a protective effect in striatal neurons, consistent with a previous report (21).
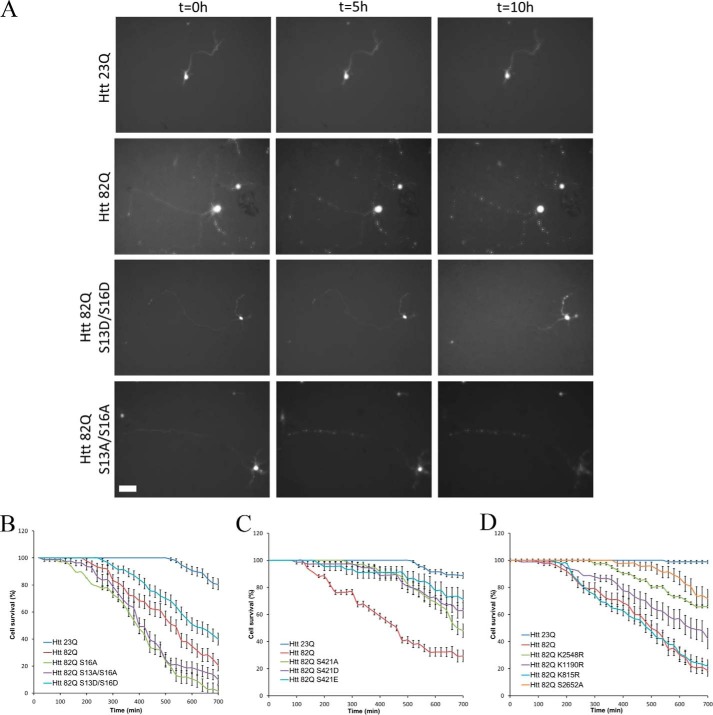
In addition to the nuclear condensation assay, we also conducted a direct observation of cell death using previously established (17) time-lapse video imaging of primary neurons (Fig. 3). We followed cells over time by video microscopy, beginning at hour 24 and ending at hour 34 post-transfection. Morphology of every neuron was quantified blindly using ImageJ software. The cells were considered dead when the majority of the neurites detached from the soma or were fragmented or when the soma became totally round. Morphological changes of neurons transfected with expanded full-length Htt started to appear after 3 h of imaging (27 h post-transfection). Cellular extensions retracted, fragmented, and detached from the soma. The cell soma progressively got rounder and in the most extreme cases disappeared. The morphology of the neurons showed a strong and steady decline over the course of the experiment (Fig. 3A). By the end of the imaging, most cells transfected with Htt-82Q were considered dead according to morphological criteria. The viability of control neurons transfected with normal full-length Htt showed a relatively small change in morphology by the end of the experiment. We previously showed that this late-stage decline is not specifically due to Htt because cells transfected with GFP alone undergo the same process (17). Examples of morphological changes of single transfected with the Ser13/Ser16 constructs and the controls observed over time are shown on Fig. 3A.
Figure 3.
Time-lapse imaging quantification of neuronal survival. Primary cortical neurons were co-transfected at DIV5 with indicated full-length constructs and GFP. After 24 h, GFP positive neurons were image every 20 min for 10 h. A, examples of neuronal morphology over time for selected Htt constructs. Scale bar, 100 μm. B–D, morphology of every neuron was quantified as described under “Experimental procedures” for the Ser13/Ser16 (B), Ser421 (C), and downstream new sites (D) series of constructs. The results are expressed as means ± S.E. (n = 200 cells analyzed over five independent neuronal preparations).
Consistent with the results of nuclear condensation assay, we observed that S13D/S16D phospho-mimetic alteration slows down the progression of the neuronal death induced by expanded Htt (Fig. 3B), whereas a phospho-null S13A/S16A mutation renders expanded Htt even more toxic to the neurons than unmodified full-length Htt-82Q protein. Thus, unlike the nuclear condensation assay, the time-lapse imaging allows detection of a potential increase in toxic properties of expanded Htt with certain PTM alterations. We also tested the effect of the alterations of Ser421 in the same system (Fig. 3C). Because both phospho-null and phospho-mimetic mutations at Ser421 proved to be protective in our nuclear condensation assay, we investigated the possibility of a different effect on neuronal morphology. Both S421A and S421D alterations appeared to ameliorate expanded Htt toxicity when assayed by time-lapse imaging, in agreement with the nuclear condensation results. The other PTM alterations, K1190R, K2548R and S2652A, that showed a neuroprotective effect in nuclear condensation assay significantly decreased toxicity of expanded Htt, as measured by direct imaging of neuronal death (Fig. 3D). In contrast, the K815R substitution, a randomly selected negative control, which did not affect the nuclear condensation induced by expanded Htt, had also no effect on neuronal death, as measured by time-lapse imaging (Fig. 3D).
Mitochondrial abnormalities caused by expanded Htt are modulated by PTMs
Metabolism and mitochondrial dysfunction have been implicated in HD pathogenesis (29–34). Mutant Htt was shown to directly associate with mitochondria, impair mitochondrial dynamics, and decrease mitochondrial function in affected in HD brain regions (35, 36).
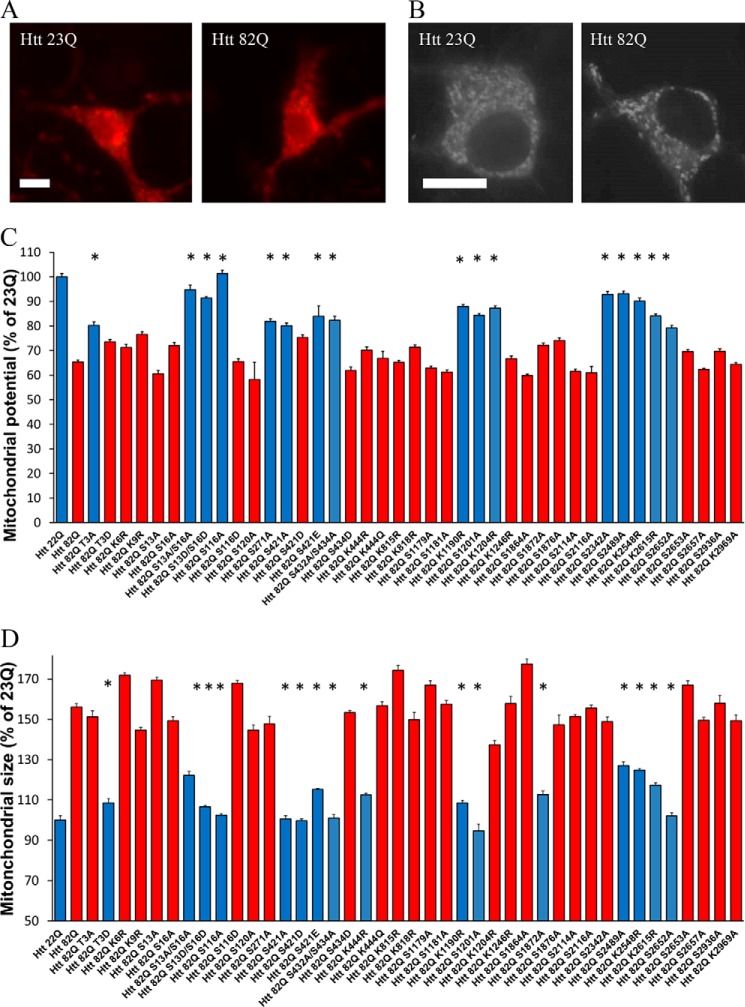
Mitochondrial potential integrity is important for cell survival, and depolarization is a sign of cell toxicity (37). We have established an assay to measure the mitochondrial potential in primary neurons using tetramethyl rhodamine methyl ester (TMRM), a live mitochondrial dye sensitive to the potential (examples of staining appear in Fig. 4A). Using high magnification live microscopy and the Volocity software, we measured the average intensity of TMRM staining in primary neurons transfected with full-length Htt constructs with and without PTM alterations.
Figure 4.
Mitochondrial abnormalities caused by expanded Htt are modulated by PTMs. A, example images of primary neurons transfected with full-length Htt containing either 23Q or 82Q and stained with the potential sensitive mitochondrial dye TMRM. Scale bar, 10 μm. B, example images of primary neurons transfected with full-length Htt containing either 23Q or 82Q and stained with MitoTracker. C, cells were then imaged at 24 h post-transfection, and individual cell mitochondrial potential was measured using Volocity. *, p ≤ 0.05 versus Htt-82Q (n = 3 independent neuronal preparations). D, mitochondrial swelling assay. Primary cortical neurons were transfected at DIV5 with indicated full-length constructs and GFP. After 24 h, the cells were loaded with MitoTracker Green for 45 min at 37 °C. The cells were then fixed and imaged. Individual mitochondria size was measured using Volocity. *, p ≤ 0.05 versus Htt-82Q (n = 3 independent neuronal preparations).
After quantification using Volocity, as shown on Fig. 4C, there is a significant decrease of the mitochondrial potential of the neurons transfected with expanded full-length Htt-82Q compared with cells transfected with normal repeat Htt-23Q. We found that several alterations of previously studied sites within the N-terminal domain (T3A, S13A/S16A, S13D/S16D, S116A, S271A, S421A, and S432/S434A) were able to partially rescue the loss of mitochondrial potential, observed in primary cortical neurons transfected with expanded Htt. A few modifications within the middle part of protein (K1190R, S1201A, and K1204R) and within its C-terminal domain (S2342A, S2489A, K2548R, and K2615R) may also modulate a drop in mitochondrial potential induced by expanded Htt (Fig. 4C). When the survival of the cell and the mitochondrial potential values are normalized to the controls (scale from 0 for Htt-82Q to 100 for Htt-23Q), the resulting correlation is moderate (r = 0.5714; supplemental Fig. 3A).
Mitochondrial swelling is also an early sign of cell toxicity (37). We used MitoTracker Red dye and confocal microscopy to measure mitochondrial size in primary neurons transfected with full-length Htt constructs with and without PTM alterations. We found significant mitochondrial swelling in neurons transfected with expanded Htt-82Q, compared with cells transfected with normal repeat Htt (examples of staining appear in Fig. 4B), whereas little to no fragmentation of the mitochondria was observed. Some of the PTM alterations were able to protect the mitochondria from the toxic effect of expanded Htt (Fig. 4D). Unlike changes in potential, there was a better correlation between the alterations that were protective against the mitochondrial swelling and those that ultimately protected from cell death. When the survival of the cell and the mitochondrial size values are normalized to the controls (scale from 0 for Htt-82Q to 100 for Htt-23Q), the resulting correlation is moderate (r = 0.7677; supplemental Fig. 3B).
Overall, with changes in both morphology and potential, our results suggest an early involvement of the mitochondria in the toxic mechanisms of expanded Htt. We observed a partial overlap between PTM alterations in expanded Htt that may modulate mitochondrial function with the sites that appear to affect neuronal toxicity of expanded Htt, as measured by nuclear condensation assay or by direct imaging of neuronal death. Notably, most of PTM alterations that rescued toxicity also improved the mitochondrial phenotype.
Discussion
In our previous studies, we demonstrated the toxicity of mutant exon 1 and Htt aa 1–586 fragments of Htt in primary cortical neurons measured by nuclear condensation assay (23, 17). Here, we extended the study of Htt toxicity and the effects of PTM site modifications to full-length Htt. In accordance with mouse models of HD that show a faster and stronger phenotype in models expressing shorter fragments of mutant Htt (38), we find greater toxicity of mutant Htt N-terminal fragments compared with full-length mutant Htt. The Htt protein is very large and likely serves a scaffold, with many protein interaction partners. It has a complex structure, with intramolecular interactions and multiple sites of PTMs, with potential for cross-talk that may affect the structure Htt and its interactions with other proteins, as well as influencing Htt subcellular localization. Thus, it is essential to characterize and study PTMs within the context of the full-length protein, as we have done in this study.
Our toxicity experiments using full-length Htt expression in primary neurons support previous reports that some phospho-mimetic alterations of expanded Htt (T3D, S13D/S16D, and S421D) are neuroprotective. However, we observed several differences compared with published studies that used fragments of Htt. In contrast with earlier reports (18, 19), but in line with our previous data (17), we found neuroprotection with both phospho-null (S421A) and phospho-mimetic (S421D and S421E) alterations of S421, suggesting the possibility that Asp acts as a phospho-null rather than phospho-mimetics at this site and that phosphorylation may enhance toxicity.
Previous reports (18, 19) have suggested that phosphorylation is protective. However, the experimental systems used in the previous studies were quite different from ours and often did not directly assay neuronal toxicity of mutant Htt. Recently, the importance of the potential phosphorylation at serine 421 for modulation of pathogenesis has been confirmed in vivo by the analysis of mutant HTT-S421D BACHD mice. However, a difference in expression levels in the mutant HTT-S421A did not allow a definitive comparison among the three constructs (39). Overall, there is generally consensus on which are the relevant PTM sites, with discrepancies among models regarding whether phosphorylation is protective or pathogenic, in part because of the limitations of using amino acid substitutions to mimic or remove a phosphorylation. More experiments, both in vitro and in vivo, will be required in the future.
In our full-length mutant Htt neuronal model, we also confirmed the activity of several phospho-null modifications. S116A, S1201A, and S2652A were protective alterations. The effect of Ser116 confirmed our previous results with Htt aa 1–586 (17). These sites for which the phospho-null mutation is protective are of particular interest, because they are potential candidates for small molecule therapeutic development targeting the potential kinases active at the site. Using a kinase prediction software, NetPhos 3.1 (40), we identified candidates kinases, which include PKC, p38 MAPK, and CKII.
Interestingly, we found that S1201A yielded different results in striatal neurons than in cortical neurons. This is consistent with the idea of cell type–specific vulnerability and protective factors. Therefore exploring mechanisms of Ser1201 phosphorylation protection could uncover potential neuronal cell type–specific protective pathways. Previously, the modification of Ser1201 was reported to have no effect on cell death in striatal neurons, while turning normal Htt into pro-apoptotic protein (21).
It has been suggested by several groups (35, 36, 41), including our own (33), that Htt may be localized to mitochondria (possibly on the outer membrane, but may also enter and interact with mitochondrial proteins). The effect of Htt phosphorylation on the mitochondrial localization and function of Htt is largely unknown and could be the subject of future studies. We did not observe a complete correlation between toxicity and mitochondrial measures, suggesting that mitochondrial pathways represent just one portion of pathogenesis or neuroprotection.
In this full-length model, we also identified several acetylation sites that can modulate Htt toxicity and effects on mitochondria. Acetylation at Lys444 has been reported to be protective (16); however, putative acetylation-mimetic Htt K444Q alteration showed no effect in our system, whereas acetylation-null alteration K444R ameliorated expanded Htt toxicity. These discrepancies of our data with some previous findings may be accounted for by using the full-length Htt system, preserving potential short-range and distant PTM interactions within the whole protein. Interestingly, each of the acetylation sites we observed having an effect on toxicity was located near one of the identified phosphorylation sites, suggesting the possibility of cross-talk between acetylation and phosphorylation of Htt, which merits further investigation.
Biophysical and cross-linking mass spectroscopy experiments indicated that Htt adopts a compact conformation with intramolecular contacts (42), both short and long range. It is likely that PTMs can modulate these interactions. The physical interaction between matching N-terminal and C-terminal fragments of Htt may be important for Htt toxicity. When the N-terminal and C-terminal fragments do not interact, there may be enhanced toxicity of the fragment containing the polyglutamine, but also it is conceivable that the C-terminal fragment may become toxic (43).
Predictions of structure of Htt based on its sequence show that there are unstructured proteolysis-prone PEST domains between highly structured α-helix supercoiled HEAT repeat domains (44). Hayden and co-workers (6) suggested that most of the PTM identified on Htt are found in these PEST domains and not in the HEAT repeat domains, and our data confirm and extend this concept.
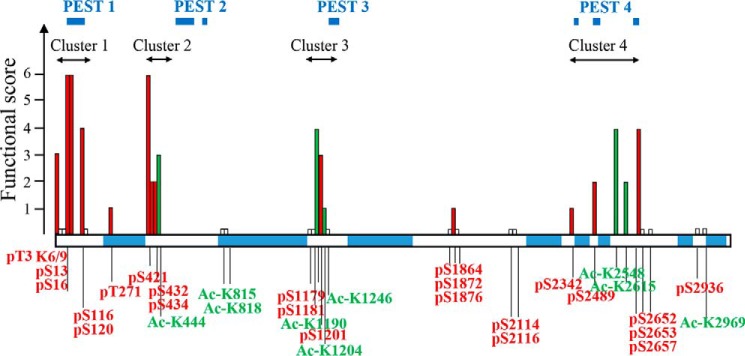
Locations of the Htt PTMs with functional effects are shown in a schematic diagram of Htt in Fig. 5. Because we have used several approaches to functionally assess PTMs identified on the full-length Htt, we combined the results to summarize the properties of each site. At each PTM, a score of 1 was given for each assay where significant changes were observed (and score 0 if there were no significant changes).
Figure 5.
Functional assessment of PTMs on Htt. PTMs clustering within proteolytic domains modulate mutant Htt toxicity. The functional score has been calculated for each PTM, based on the outcome of all the assays (toxicity in cortex, toxicity in striatum, mitochondrial potential, and mitochondrial size). Each PTM was given a score of 1 per assay if significant changes were observed and 0 if there was no difference. For multiple alterations of the same amino acid, a combined score was recorded. Ser13 and Ser16 are scored based on the single and combined alterations results. Based on this scoring, we found four clusters of PTMs that largely modulate expanded Htt functional or toxic properties. These clusters are located within protease-sensitive (PEST) regions (predicted by epestfind-EMBOSS Explorer) as indicated. Phosphorylation sites are shown in red, acetylation sites are in green, and sites that did not score in any of the assays are indicated by short white bars.
Based on this scoring, we found four clusters of PTMs that modulate expanded Htt functional or toxic properties: the previously well characterized N-terminal domain (cluster 1, residues 1–116), the region around Ser421 (cluster 2), the mid region (cluster 3, residues Ser1201–Lys1246), and C-terminal cluster 4 (residues Ser2342–Ser2657). Notably, it appears that PTM clusters modulating Htt properties are grouped within predicted protease-sensitive regions (http://emboss.bioinformatics.nl/cgi-bin/emboss/epestfind; Fig. 5).4 Interestingly, three of these clusters of PTMs lie closely to the boundaries between previously suggested “distinct domains defined by protease-sensitive sites,” as described by Seong and co-workers (42, 45): our cluster 2 falls between N-terminal domains I and II; cluster 3 is close to the location of the protease-sensitive major hinge region (residues 1184–1254) between N-terminal domain II and C-terminal domain I; and cluster 4 maps to the C-terminal domain II boundary.
Proteolytic cleavage into a toxic N-terminal fragment containing the mutant polyglutamine stretch has been proposed to contribute to HD pathogenesis (6, 7, 28, 43, 46). Cleavage sites appear to be preferentially in the PEST regions (44).
A number of fragments of Htt can be observed in different systems, and several proteases have been proposed to cleave Htt. Caspases, including Caspases 2, 3, and 6, generate fragments around 500 amino acids in length. Caspase 6, which cleaves at 586, has been suggested by in vivo experiments to be especially relevant (47, 48). Calpain, cathepsins, and the matrix metalloproteinase MMP10 have also been proposed (49). The potential cleavage fragments near the N terminus termed cp-A and cp-B or cp-1 and cp-2 (50, 51), possibly generated by Bleomycin hydrolase BLMH or cathepsin Z (52), are especially interesting, because they are near the Ser116 site, shown in this study with full-length Htt and in our previous study with Htt aa 1–586 fragments (17) to modulate toxicity.
To conclude, we have employed here a functional analysis of the individual PTM sites on full-length mutant Htt. This approach enabled us to identify individual modifications, as well as clusters of PTMs that modulate mutant Htt cellular toxicity, and thus provide insight into HD pathogenesis. Identification of the kinases and other enzymes that catalyze these PTMs (especially when inhibiting the formation of the PTM would reduce toxicity) may provide excellent therapeutic targets to abrogate HD pathogenesis.
Experimental procedures
Plasmid generation and mutagenesis
Mammalian expression constructs encoding full-length Htt with the N-terminal PTM alterations (of Thr3, Lys6, Lys9, Ser13, Ser16, Ser116, and Ser120) were generated in two steps. As a first step, these alterations were introduced within an existing Htt-N586–82Q(1–586) plasmid (obtained from D. Borchelt and described in Ref. 24), using site-directed mutagenesis and the QuikChange II XL kit (Stratagene) according to the manufacturer's protocol, except that Stbl2TM competent cells (Thermo Fisher Scientific), specifically designed for cloning of unstable inserts, were used throughout this study. The presence of PTM mutations and the entire N586–82Q sequences were confirmed by sequencing. As a second step, the fragments comprising the first 171 aa of Htt with corresponding mutations were generated from the above N586 constructs using digestion at 5′ flanking NotI and Htt-internal XhoI restriction sites. These N-terminal fragments were subcloned into a vector comprising the rest of the downstream sequence of full-length Htt. This vector was constructed by subcloning XhoI/SalI downstream Htt fragment (of the original synthetic plasmid containing the entire full-length Htt-23Q) in the XhoI site of pcDNA vector. The synthetic FL-Htt-23Q plasmid was generated (DNA 2.0) previously and was checked for expression of full-length Htt protein in our laboratory. PTM alterations of Thr271, Ser421, Ser434, and Lys444 were also introduced within an existing Htt-N586–82Q plasmid, as described above. As a second step, the fragments containing N586–82Q with corresponding mutations were generated by PCR of the above Htt-586–82Q constructs using the primers incorporating KpnI restriction sites on both 5′ and 3′ ends. Purified PCR products were digested with the above enzyme and subcloned into a vector comprising the rest of the downstream sequence of full-length Htt (described above), prepared upon digestion with KpnI (utilizing 5′ flanking and Htt-internal KpnI sites). The rest of the PTM alterations (C-terminal of aa 586) were generated using a newly synthesized big C-terminal Htt fragment (downstream of aa 586) with incorporated convenient unique restriction sites (DNA2.0), allowing isolation of five domains separated by these restriction sites. These domains were subcloned into pJ cloning vectors (DNA2.0) and used to introduce alterations by site-directed mutagenesis and the QuikChange II XL kit (Stratagene). These mutagenized domains were subcloned into the big C-terminal Htt fragment using unique restriction sites (swapping with corresponding domains without mutations). At the final step we reintroduced the N-terminal 586-aa fragment with poly(Q) into mutagenized C-terminal fragment (for each PTM alteration).
Primary neuron preparation
Primary neurons from CD1 mice were prepared from embryonic days 15–17 embryos. Cells from both cortex and striatum were independently prepared from the same embryos and plated at 106 cells/mm2 in Neurobasal medium with 2% B27, 2 mm GlutaMAX, and 1% penicillin/streptomycin in a 24-well plate. The cells were maintained at 37 °C with 5% CO2 throughout the experiments. All cell culture supplies were obtained from Corning, and all the media were from Thermo Fisher Scientific.
Transfection of primary neurons
Neurons were co-transfected using Lipofectamine 2000 (Thermo Fisher Scientific) at day in vitro (DIV) 5 or 6. Per well of 24-well plates, 1 μg of Htt plasmid, and 0.1 μg of GFP was used with a ratio DNA/Lipofectamine of 1/1.75. DNA and Lipofectamine were diluted separately in 50 μl of OptiMEM/well for each and incubated for 5 min before being combined. After being combined, the transfecting solution is incubated for 90 min at room temperature. During the incubation of the transfecting solution, the plates are emptied, and the medium is replaced with 400 μl of fresh OptiMEM. After the incubation, 100 μl of transfection solution is deposited in each well. The plates are returned to the incubator for 4.5 h when the wells are empty, and the medium was replaced with fresh Neurobasal medium.
Nuclear condensation cell death assay
The toxicity experiments were performed in primary cortical and primary striatal neurons according to our established protocol (16). Nuclei were stained using Hoechst 3342 (Sigma; 0.2 μg/ml in PBS for 5 min). Automated picture acquisition was performed using a Zeiss Axiovert 200 inverted microscope with a 10× objective. Automatic quantification of the nuclear intensity of transfected cells was performed using Volocity. The cells were considered dead when their nuclear intensity was higher than the average intensity plus two standard deviations. Each condition was performed in quadruplicate within each experiment, and each experiment was repeated in at least four independent neuronal preparations for each construct studied.
Time-lapse imaging of cell death
Primary cortical neurons were transfected with full-length Htt constructs and GFP as described above. 24 h after transfection, neurons expressing GFP were randomly chosen and followed for 10 h with a picture taken every 20 min. Morphology of every neuron was quantified blindly using ImageJ software. Neurons were quantified as 100 when alive and 0 when dead for each frame. The cells were considered dead when most of the neurites detached from the soma or were fragmented or when the soma became totally round. The results are expressed as means ± S.E. (n = 200 cells analyzed in three independent experiments).
Mitochondrial function assays
For the potential measurement, primary cortical neurons were co-transfected at DIV5 with the full-length Htt constructs and GFP, as described above. 24 h after transfection, the cells were loaded with 20 nm TMRM stain for 45 min at 37 °C. After three washes with PBS, the medium was changed to Neurobasal + B27 without phenol red for imaging. The cells were imaged on an Axiovert 200 microscope. Average measure of the TMRM intensity of transfected cells was quantified using Volocity. For mitochondrial size measurement of the mitochondria, primary cortical neurons were co-transfected at DIV5 with the full-length Htt constructs and GFP, as described above. 24 h after transfection, the cells were loaded with 500 nm MitoTracker Red (Molecular Probes) for 45 min at 37 °C. After three washes with PBS, the cells were fixed with 4% paraformaldehyde, 1× PBS for 30 min. The cells were imaged on a laser-scanning confocal Zeiss LSM 510-meta microscope. Average sizes of individual mitochondria of transfected cells were measured using Volocity. The results were normalized to the average size of mitochondria of cells transfected with Htt-82Q.
Author contributions
N. A. designed, performed, and analyzed the experiments and wrote the paper. T. R., E. R., E. C., and J. C. S. designed and engineered all the plasmids, performed the expression experiments, and prepared the figures. T. R. compiled the results and designed Fig. 5. M. R. and X. W. provided technical assistance and contributed to the preparation of the neurons. D. J. L. coordinated the study. C. A. R. conceived and coordinated the study. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by the CHDI Foundation, Inc. and by National Institutes of Health Grant R01 NS086452. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- HD
- Huntington's disease
- PTM
- post-translational modification
- aa
- amino acid(s)
- TMRM
- tetramethyl rhodamine methyl ester
- DIV
- day in vitro.
References
- 1. The Huntington's Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 2. Iqbal K., Liu F., Gong C. X., and Grundke-Iqbal I. (2010) Tau in Alzheimer disease and related tauopathies. Curr. Alzheimer Res. 7, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xu Y., Deng Y., and Qing H. (2015) The phosphorylation of α-synuclein: development and implication for the mechanism and therapy of the Parkinson's disease. J. Neurochem. 135, 4–18 [DOI] [PubMed] [Google Scholar]
- 4. Hasegawa M., Nonaka T., Tsuji H., Tamaoka A., Yamashita M., Kametani F., Yoshida M., Arai T., and Akiyama H. (2011) Molecular dissection of tdp-43 proteinopathies. J. Mol. Neurosci. 45, 480–485 [DOI] [PubMed] [Google Scholar]
- 5. Aronin N., and DiFiglia M. (2014) Huntingtin-lowering strategies in Huntington's disease: antisense oligonucleotides, small RNAs, and gene editing. Mov. Disord 29, 1455–1461 [DOI] [PubMed] [Google Scholar]
- 6. Ehrnhoefer D. E., Sutton L., and Hayden M. R. (2011) Small changes, big impact: posttranslational modifications and function of huntingtin in Huntington disease. Neuroscientist 17, 475–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saudou F., and Humbert S. (2016) The biology of huntingtin. Neuron 89, 910–926 [DOI] [PubMed] [Google Scholar]
- 8. Aiken C. T., Steffan J. S., Guerrero C. M., Khashwji H., Lukacsovich T., Simmons D., Purcell J. M., Menhaji K., Zhu Y. Z., Green K., Laferla F., Huang L., Thompson L. M., and Marsh J. L. (2009) Phosphorylation of threonine 3: implications for huntingtin aggregation and neurotoxicity. J. Biol. Chem. 284, 29427–29436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gu X., Greiner E. R., Mishra R., Kodali R., Osmand A., Finkbeiner S., Steffan J. S., Thompson L. M., Wetzel R., and Yang X. W. (2009) Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in hd mice. Neuron 64, 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Atwal R. S., Desmond C. R., Caron N., Maiuri T., Xia J., Sipione S., and Truant R. (2011) Kinase inhibitors modulate huntingtin cell localization and toxicity. Nat. Chem. Biol. 7, 453–460 [DOI] [PubMed] [Google Scholar]
- 11. Maiuri T., Woloshansky T., Xia J., and Truant R. (2013) The huntingtin N17 domain is a multifunctional crm1 and ran-dependent nuclear and cilial export signal. Hum. Mol. Genet. 22, 1383–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steffan J. S., Agrawal N., Pallos J., Rockabrand E., Trotman L. C., Slepko N., Illes K., Lukacsovich T., Zhu Y. Z., Cattaneo E., Pandolfi P. P., Thompson L. M., and Marsh J. L. (2004) Sumo modification of huntingtin and Huntington's disease pathology. Sci. 304, 100–104 [DOI] [PubMed] [Google Scholar]
- 13. Thompson L. M., Aiken C. T., Kaltenbach L. S., Agrawal N., Illes K., Khoshnan A., Martinez-Vincente M., Arrasate M., O'Rourke J. G., Khashwji H., Lukacsovich T., Zhu Y. Z., Lau A. L., Massey A., Hayden M. R., et al. (2009) Ikk phosphorylates huntingtin and targets it for degradation by the proteasome and lysosome. J. Cell Biol. 187, 1083–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DiGiovanni L. F., Mocle A. J., Xia J., and Truant R. (2016) Huntingtin N17 domain is a reactive oxygen species sensor regulating huntingtin phosphorylation and localization. Hum. Mol. Genet. 25, 3937–3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yanai A., Huang K., Kang R., Singaraja R. R., Arstikaitis P., Gan L., Orban P. C., Mullard A., Cowan C. M., Raymond L. A., Drisdel R. C., Green W. N., Ravikumar B., Rubinsztein D. C., El-Husseini A., et al. (2006) Palmitoylation of huntingtin by hip14 is essential for its trafficking and function. Nat. Neurosci. 9, 824–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeong H., Then F., Melia T. J. Jr., Mazzulli J. R., Cui L., Savas J. N., Voisine C., Paganetti P., Tanese N., Hart A. C., Yamamoto A., and Krainc D. (2009) Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell 137, 60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watkin E. E., Arbez N., Waldron-Roby E., O'Meally R., Ratovitski T., Cole R. N., and Ross C. A. (2014) Phosphorylation of mutant huntingtin at serine 116 modulates neuronal toxicity. PLoS One 9, e88284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Humbert S., Bryson E. A., Cordelières F. P., Connors N. C., Datta S. R., Finkbeiner S., Greenberg M. E., and Saudou F. (2002) The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves huntingtin phosphorylation by Akt. Dev. Cell 2, 831–837 [DOI] [PubMed] [Google Scholar]
- 19. Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X. J., Saudou F., and Humbert S. (2008) Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 27, 2124–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schilling B., Gafni J., Torcassi C., Cong X., Row R. H., LaFevre-Bernt M. A., Cusack M. P., Ratovitski T., Hirschhorn R., Ross C. A., Gibson B. W., and Ellerby L. M. (2006) Huntingtin phosphorylation sites mapped by mass spectrometry, modulation of cleavage and toxicity. J. Biol. Chem. 281, 23686–23697 [DOI] [PubMed] [Google Scholar]
- 21. Anne S. L., Saudou F., and Humbert S. (2007) Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J. Neurosci. 27, 7318–7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ratovitski T., O'Meally R. N., Jiang M., Chaerkady R., Chighladze E., Stewart J. C., Wang X., Arbez N., Roby E., Alexandris A., Duan W., Vijayvargia R., Seong I. S., Lavery D. J., Cole R. N., et al. (2017) Post-translational modifications (PTMs), identified on endogenous huntingtin, cluster within proteolytic domains between HEAT repeats. J. Proteome Res. 16, 2692–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nucifora L. G., Burke K. A., Feng X., Arbez N., Zhu S., Miller J., Yang G., Ratovitski T., Delannoy M., Muchowski P. J., Finkbeiner S., Legleiter J., Ross C. A., and Poirier M. A. (2012) Identification of novel potentially toxic oligomers formed in vitro from mammalian-derived expanded huntingtin exon-1 protein. J. Biol. Chem. 287, 16017–16028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tebbenkamp A. T., Crosby K. W., Siemienski Z. B., Brown H. H., Golde T. E., and Borchelt D. R. (2012) Analysis of proteolytic processes and enzymatic activities in the generation of huntingtin N-terminal fragments in an HEK293 cell model. PLoS One 7, e50750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Colin E., Régulier E., Perrin V., Dürr A., Brice A., Aebischer P., Déglon N., Humbert S., and Saudou F. (2005) Akt is altered in an animal model of Huntington's disease and in patients. Eur. J. Neurosci 21, 1478–1488 [DOI] [PubMed] [Google Scholar]
- 26. Halliday G. M., McRitchie D. A., Macdonald V., Double K. L., Trent R. J., and McCusker E. (1998) Regional specificity of brain atrophy in Huntington's disease. Exp. Neurol 154, 663–672 [DOI] [PubMed] [Google Scholar]
- 27. Vonsattel J. P. (2008) Huntington disease models and human neuropathology: similarities and differences. Acta Neuropathol 115, 55–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ross C. A., and Tabrizi S. J. (2011) Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 10, 83–98 [DOI] [PubMed] [Google Scholar]
- 29. Jin J., Gu H., Anders N. M., Ren T., Jiang M., Tao M., Peng Q., Rudek M. A., and Duan W. (2016) Metformin protects cells from mutant huntingtin toxicity through activation of ampk and modulation of mitochondrial dynamics. Neuromolecular Med. 18, 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duan W., Guo Z., Jiang H., Ware M., Li X. J., and Mattson M. P. (2003) Dietary restriction normalizes glucose metabolism and BDNF levels, slows disease progression, and increases survival in huntingtin mutant mice. Proc. Natl. Acad. Sci. U.S.A. 100, 2911–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duan W., Jiang M., and Jin J. (2014) Metabolism in hd: still a relevant mechanism? Mov. Disord. 29, 1366–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin M. T., and Beal M. F. (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795 [DOI] [PubMed] [Google Scholar]
- 33. Ratovitski T., Chighladze E., Arbez N., Boronina T., Herbrich S., Cole R. N., and Ross C. A. (2012) Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle 11, 2006–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ratovitski T., Chaerkady R., Kammers K., Stewart J. C., Zavala A., Pletnikova O., Troncoso J. C., Rudnicki D. D., Margolis R. L., Cole R. N., and Ross C. A. (2016) Quantitative proteomic analysis reveals similarities between Huntington's disease (HD) and Huntington's disease-like 2 (HDL2) human brains. J. Proteome Res. 15, 3266–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Orr A. L., Li S., Wang C. E., Li H., Wang J., Rong J., Xu X., Mastroberardino P. G., Greenamyre J. T., and Li X. J. (2008) N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci. 28, 2783–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shirendeb U., Reddy A. P., Manczak M., Calkins M. J., Mao P., Tagle D. A., and Reddy P. H. (2011) Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington's disease: implications for selective neuronal damage. Hum. Mol. Genet. 20, 1438–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kasahara A., and Scorrano L. (2014) Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 24, 761–770 [DOI] [PubMed] [Google Scholar]
- 38. Ehrnhoefer D. E., Butland S. L., Pouladi M. A., and Hayden M. R. (2009) Mouse models of Huntington disease: variations on a theme. Dis. Model. Mech. 2, 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kratter I. H., Zahed H., Lau A., Tsvetkov A. S., Daub A. C., Weiberth K. F., Gu X., Saudou F., Humbert S., Yang X. W., Osmand A., Steffan J. S., Masliah E., and Finkbeiner S. (2016) Serine 421 regulates mutant huntingtin toxicity and clearance in mice. J. Clin. Invest. 126, 3585–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., and Brunak S. (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649 [DOI] [PubMed] [Google Scholar]
- 41. Dickey A. S., Pineda V. V., Tsunemi T., Liu P. P., Miranda H. C., Gilmore-Hall S. K., Lomas N., Sampat K. R., Buttgereit A., Torres M. J., Flores A. L., Arreola M., Arbez N., Akimov S. S., Gaasterland T., et al. (2016) Ppar-delta is repressed in Huntington's disease, is required for normal neuronal function and can be targeted therapeutically. Nat. Med. 22, 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vijayvargia R., Epand R., Leitner A., Jung T. Y., Shin B., Jung R., Lloret A., Singh Atwal R., Lee H., Lee J. M., Aebersold R., Hebert H., Song J. J., and Seong I. S. (2016) Huntingtin's spherical solenoid structure enables polyglutamine tract-dependent modulation of its structure and function. Elife 5, e11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. El-Daher M. T., Hangen E., Bruyère J., Poizat G., Al-Ramahi I., Pardo R., Bourg N., Souquere S., Mayet C., Pierron G., Lévêque-Fort S., Botas J., Humbert S., and Saudou F. (2015) Huntingtin proteolysis releases non-polyQ fragments that cause toxicity through dynamin 1 dysregulation. EMBO J. 34, 2255–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Warby S. C., Doty C. N., Graham R. K., Carroll J. B., Yang Y. Z., Singaraja R. R., Overall C. M., and Hayden M. R. (2008) Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum. Mol. Genet. 17, 2390–2404 [DOI] [PubMed] [Google Scholar]
- 45. Seong I. S., Woda J. M., Song J. J., Lloret A., Abeyrathne P. D., Woo C. J., Gregory G., Lee J. M., Wheeler V. C., Walz T., Kingston R. E., Gusella J. F., Conlon R. A., and MacDonald M. E. (2010) Huntingtin facilitates polycomb repressive complex 2. Hum. Mol. Genet. 19, 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Landles C., Sathasivam K., Weiss A., Woodman B., Moffitt H., Finkbeiner S., Sun B., Gafni J., Ellerby L. M., Trottier Y., Richards W. G., Osmand A., Paganetti P., and Bates G. P. (2010) Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J. Biol. Chem. 285, 8808–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Graham R. K., Deng Y., Slow E. J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., Warby S. C., Doty C. N., Roy S., Wellington C. L., Leavitt B. R., et al. (2006) Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125, 1179–1191 [DOI] [PubMed] [Google Scholar]
- 48. Aharony I., Ehrnhoefer D. E., Shruster A., Qiu X., Franciosi S., Hayden M. R., and Offen D. (2015) A huntingtin-based peptide inhibitor of caspase-6 provides protection from mutant huntingtin-induced motor and behavioral deficits. Hum. Mol. Genet. 24, 2604–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miller J. P., Holcomb J., Al-Ramahi I., de Haro M., Gafni J., Zhang N., Kim E., Sanhueza M., Torcassi C., Kwak S., Botas J., Hughes R. E., and Ellerby L. M. (2010) Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington's disease. Neuron 67, 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lunkes A., Lindenberg K. S., Ben-Haïem L., Weber C., Devys D., Landwehrmeyer G. B., Mandel J. L., and Trottier Y. (2002) Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol. Cell 10, 259–269 [DOI] [PubMed] [Google Scholar]
- 51. Ratovitski T., Gucek M., Jiang H., Chighladze E., Waldron E., D'Ambola J., Hou Z., Liang Y., Poirier M. A., Hirschhorn R. R., Graham R., Hayden M. R., Cole R. N., and Ross C. A. (2009) Mutant huntingtin N-terminal fragments of specific size mediate aggregation and toxicity in neuronal cells. J. Biol. Chem. 284, 10855–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ratovitski T., Chighladze E., Waldron E., Hirschhorn R. R., and Ross C. A. (2011) Cysteine proteases bleomycin hydrolase and cathepsin Z mediate N-terminal proteolysis and toxicity of mutant huntingtin. J. Biol. Chem. 286, 12578–12589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dong G., Callegari E., Gloeckner C. J., Ueffing M., and Wang H. (2012) Mass spectrometric identification of novel posttranslational modification sites in Huntingtin. Proteomics 12, 2060–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang B., Lucas T., Kueppers C., Dong X., Krause M., Bepperling A., Buchner J., Voshol H., Weiss A., Gerrits B., and Kochanek S. (2015) Scalable production in human cells and biochemical characterization of full-length normal and mutant huntingtin. PLoS One 10, e0121055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.