Abstract
Signal-transducing adaptor family member-2 (STAP-2) is an adaptor protein that regulates various intracellular signaling pathways and promotes tumorigenesis in melanoma and breast cancer cells. However, the contribution of STAP-2 to the behavior of other types of cancer cells is unclear. Here, we show that STAP-2 promotes tumorigenesis of prostate cancer cells through up-regulation of EGF receptor (EGFR) signaling. Tumor growth of a prostate cancer cell line, DU145, was strongly decreased by STAP-2 knockdown. EGF-induced gene expression and phosphorylation of AKT, ERK, and STAT3 were significantly decreased in STAP-2–knockdown DU145 cells. Mechanistically, we found that STAP-2 interacted with EGFR and enhanced its stability by inhibiting c-CBL–mediated EGFR ubiquitination. Our results indicate that STAP-2 promotes prostate cancer progression via facilitating EGFR activation.
Keywords: adaptor protein, prostate cancer, protein degradation, signal transduction, STAT3, c-CBL
Introduction
Intracellular signaling contributes to various cell fates, such as proliferation, differentiation, and survival/death. Aberrant activation of signaling pathways that promote cell proliferation is a cause of carcinogenesis, and it is, therefore, important to determine the mechanisms underlying this process to facilitate the development of anticancer therapies. Epidermal growth factor receptor (EGFR)3 is the receptor for EGF, TGFα, and HB-EGF, and its activation is required for optimal cell proliferation and development of dermis, lung, and digestive organs (1, 2). Phosphorylated tyrosine and lysine residues of EGFR are associated with JAK and PI3K, inducing ERK- and STAT-mediated gene expression and cell proliferation (2). After ligand binding, EGFR translocates from the plasma membrane to endosomes. Non-ubiquitinated EGFR then returns to the plasma membrane, but ubiquitinated EGFR is degraded in lysosomes (3, 4). EGFR ubiquitination is facilitated by c-CBL, an E3 ubiquitin ligase, which down-regulates EGFR signaling (5, 6). A gain-of-function mutation in EGFR induces aberrant cell proliferation and carcinogenesis, and gefitinib, an EGFR inhibitor, is used to treat lung cancer (2, 7). EGFR signals are mediated by several kinases, and some steps are regulated by specific adaptor proteins. It is possible that proteins relating to EGFR signals may be suitable targets for anticancer drugs.
STAP-2 was originally identified as a protein that interacts with c-Fms, which is a leukemia oncogene that regulates macrophage proliferation and differentiation (8). Our previous studies showed that STAP-2 modulates STAT3- and STAT5-mediated gene expression and promotes Fas-induced caspase-8 activation in T cells (8–10). Furthermore, FcϵRI, and Toll-like receptor signals are also up-regulated in macrophages and dendritic cells by STAP-2 (11, 12). These findings indicate that STAP-2 regulates immune responses and inflammation. In addition, STAP-2 associates with breast tumor kinase and STAT3, leading to proliferation of T47D breast cancer cells (13). STAP-2 also enhances breast tumor kinase-mediated STAT5 activation (14, 15). In B16F10 cells, STAP-2 positively regulates the protein levels of tyrosinase, which determines tumor invasion via controlling chemokine receptor expression (16). STAP-2 can also bind to BCR-ABL, a fusion oncoprotein of chronic myeloid leukemia, leading to significant enhancement of its downstream signals (17). Thus, STAP-2 is also involved in the development and/or progression of some types of malignancies.
In this study, we show that STAP-2 promotes tumorigenesis of prostate cancer cells in the mouse xenograft model. STAP-2 knockdown inhibited prostate cancer cell growth and decreased activation of EGFR signaling. STAP-2 interacted with EGFR and inhibited CBL-mediated EGFR ubiquitination, resulting in up-regulation of EGFR protein levels. Our results indicate that STAP-2 promotes tumorigenesis of prostate cancer cells and indicate that STAP-2 is a valid target for prostate cancer treatments.
Results
STAP-2 is highly expressed in breast cancer and prostate cancer cell lines
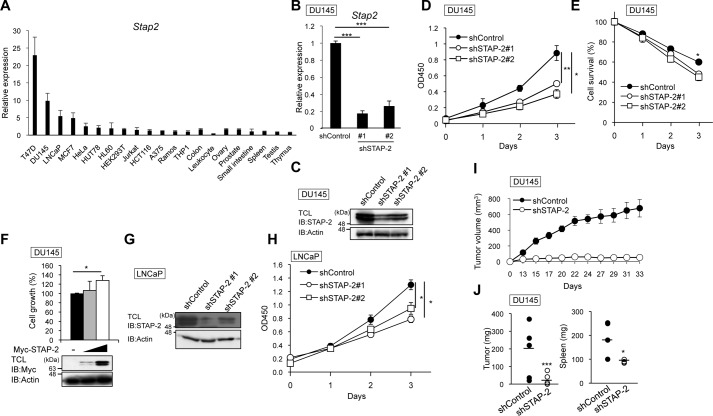
We previously reported that STAP-2 promotes cell proliferation and tumorigenesis of melanoma and breast cancer cells (13, 16). Here, we asked whether STAP-2 contributes to the proliferation of other types of cancer cells. To this end, we investigated STAP-2 expression in various cancer cell lines and human tissues using qPCR (Fig. 1A). Cancer cell lines tended to express higher STAP-2 mRNA than normal cells. Especially, human prostate cancer cell lines (DU145 and LNCaP) expressed STAP-2 at high levels, similarly to breast cancer cell lines (T47D and MCF7). We therefore investigated whether STAP-2 contributes to cell proliferation of prostate cancer cells. STAP-2 shRNA-expressing DU145 cells were prepared, and the knockdown efficiency was confirmed by qPCR and immunoblotting (Fig. 1, B and C). Not only cell proliferation but also survival of STAP-2–knockdown DU145 cells was significantly decreased compared with control shRNA-expressing cells (Fig. 1, D and E), and the impaired growth was recovered by transient overexpression of STAP-2 (Fig. 1F). We then asked whether STAP-2 contributes to cell proliferation of LNCaP cells, an androgen receptor–positive human prostate cell line, by lentiviral knockdown of STAP-2. STAP-2 knockdown decreased cell growth of LNCaP cells, similar to DU145 cells (Fig. 1, G and H). To investigate whether STAP-2 promotes prostate tumor growth in vivo, we injected STAP-2–knockdown DU145 cells into BALB/c nude mice and measured tumor growth. DU145 tumor growth at the sites of injection was significantly reduced by STAP-2 knockdown, and tumor-induced splenomegaly was also decreased in STAP-2–knockdown tumor-bearing mice (Fig. 1, I and J). These findings suggest that STAP-2 is required for tumor formation and proliferation of prostate cancer cells.
Figure 1.
STAP-2 enhances proliferation of DU145 cells in vitro and in vivo. A, qPCR analysis for mRNA levels of STAP-2 in various cancer cell lines and human tissues. B and C, expression levels of STAP-2 in STAP-2–knockdown DU145 cells were determined by qPCR (B) and immunoblotting (IB) (C). TCL, total cell lysates. D, viable cells of control or STAP-2 shRNA–expressing DU145 cells were measured by a WST assay. E, control or STAP-2 shRNA–expressing DU145 cells were cultured in DMEM containing 1% FBS for the indicated times, and then the surviving cells were measured by a WST assay. F, STAP-2–knockdown DU145 cells were transfected with pcDNA3.1-Myc-STAP-2, and then their cell proliferation was measured by a WST assay at 72 h post-transfection, as in D. An aliquot of total cell lysates was immunoblotted by anti-Myc and anti-actin antibodies. G, expression levels of STAP-2 in STAP-2–knockdown LNCaP cells were determined by immunoblotting. H, viable cells of control or STAP-2 shRNA–expressing LNCaP cells were measured by a WST assay. I, DU145 cells (5.0 × 106 cells) were subcutaneously injected into BALB/c nude mice (day 0), and tumor volume was measured. Tumors were established in 5 of 5 mice (shControl) and 3 of 5 mice (shSTAP-2). J, weight of tumors and spleens from the tumor-bearing mice at day 33. n = 3; mean values and S.E. (error bars) are depicted. *, p < 0.05; **, p < 0.01; ***, p < 0.005 (paired Student's t test).
STAP-2 up-regulates EGFR signaling
High levels of EGFR expression are associated with high risk and advanced stages of prostate cancer (18). In addition, most metastases of hormone-refractory prostate cancers express EGFR (19). Thus, EGFR is a component of a major transduction pathway for the growth of prostate cancer cells. Our previous studies showed that STAP-2 moves to the plasma membrane after EGF stimulation and that EGF-induced activity of STAT3 is enhanced by STAP-2 (8). Because prostate cancer cell lines express high levels of STAP-2 and respond to EGF stimulation, we hypothesized that STAP-2 may promote prostate cancer growth through up-regulation of EGFR signaling.
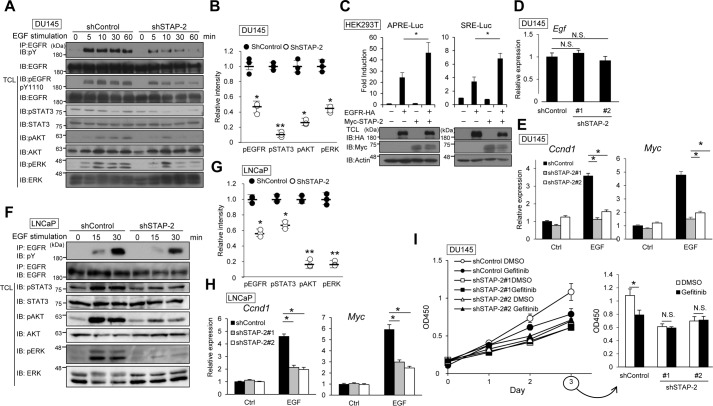
In DU145 cells, STAP-2 knockdown reduces phosphorylation of EGFR and of signaling molecules downstream of EGFR, such as STAT3, AKT, and ERK (Fig. 2, A and B) although the protein amount of EGFR was slightly lower in STAP-2–knockdown cells than in control shRNA-expressing cells after EGF stimulation. We then investigated the contribution of STAP-2 to EGF-induced STAT3 and ERK activation using a luciferase assay with STAT3-responsive (APRE) or ERK-responsive (SRE) reporter vectors. EGFR overexpression induced APRE- and SRE-mediated promoter activities, and the luciferase activities were further enhanced by STAP-2 overexpression in HEK293T cells (Fig. 2C). Although STAP-2 knockdown did not affect EGF mRNA levels, EGF-induced expression of Myc and Ccnd1 was decreased in STAP-2–knockdown DU145 cells (Fig. 2, D and E). Phosphorylation of EGFR, STAT3, AKT, and ERK by EGF was decreased in STAP-2–knockdown LNCaP cells (Fig. 2, F and G). STAP-2–knockdown LNCaP cells showed reduced expression of the EGF-responsible genes Myc and Ccnd1 after EGF stimulation (Fig. 2H). Furthermore, gefitinib, an EGFR inhibitor, significantly inhibited cell proliferation in control shRNA-expressing DU145 cells (Fig. 2I). In STAP-2 shRNA-expressing DU145 and LNCaP cells, proliferation was low and was not affected by gefitinib.
Figure 2.
STAP-2 up-regulates EGFR signaling. A, control or STAP-2 shRNA–expressing DU145 cells were stimulated with 30 ng/ml EGF for the indicated periods. The lysate was immunoprecipitated (IP) with anti-EGFR antibody and blotted (IB) with the indicated antibodies. TCL, total cell lysates. B, band intensity of phosphorylated EGFR, STAT3, AKT, and ERK in the DU145 cell lysates was calculated and normalized by the β-actin band using ImageJ software. The data from three independent experiments and S.D. (error bars) are depicted. C, HEK293T cells were transfected with expression vectors of EGFR-HA and Myc-STAP-2 together with APRE or SRE reporter vectors. Luciferase activity was determined at 48 h post-transfection. An aliquot of total cell lysates was immunoblotted by anti-HA, anti-Myc, and anti-actin antibodies. D and E, gene expression was measured by qPCR in control or STAP-2 shRNA–expressing DU145 cells with or without 30 ng/ml EGF stimulation for 1 h. F, control or STAP-2 shRNA–expressing LNCaP cells were stimulated with EGF, and the lysates were blotted as in A. G, band intensity of phosphorylated proteins in the LNCaP cell lysates was calculated as in B. H, control or STAP-2 shRNA–expressing LNCaP cells were stimulated with EGF, and then gene expression was measured by qPCR as in E. I, control or STAP-2 shRNA–expressing DU145 cells were treated with 1 μm gefitinib for 3 days, and then viable cells were measured by a WST assay. n = 3; mean values and S.E. (error bars) are depicted. N.S., not significant; *, p < 0.05; **, p < 0.01 (paired Student's t test).
Our Western blot analysis and luciferase assays strongly indicated that STAP-2 enhances phosphorylation of EGFR and downstream signals after EGF stimulation. The involvement of STAP-2 in EGFR signaling is likely to be required for maximal cell growth of DU145 and LNCaP cells. Of note, STAP-2–knockdown DU145 cells showed similar levels of proliferation under DMSO and gefitinib treatment conditions; likewise, gefitinib inhibited DU145 cell growth only when STAP-2 existed. Therefore, STAP-2 enhances the proliferation of prostate cancer cells through up-regulation of EGFR signaling.
STAP-2 enhances EGFR stability by inhibiting its ubiquitination
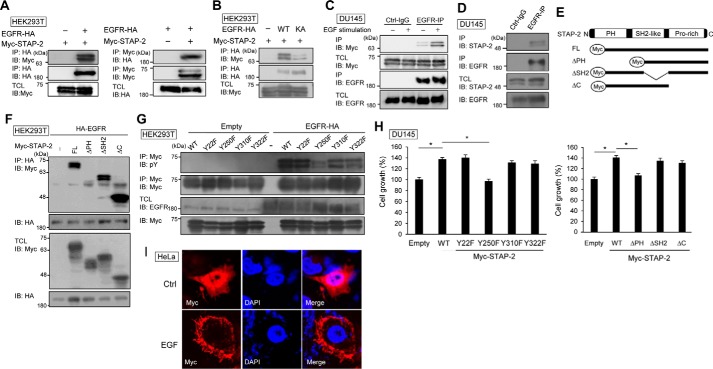
To elucidate the mechanism of STAP-2–mediated up-regulation of EGFR signaling, we investigated the interaction between STAP-2 and EGFR by immunoprecipitation. STAP-2 was co-precipitated with EGFR (Fig. 3A). EGFR K721A is a loss-of-function EGFR mutant because of deficient dimerization and internalization (2). When EGFR K721A was expressed with STAP-2, the binding of the two proteins could not be detected (Fig. 3B). In DU145 cells, endogenous STAP-2–EGFR interaction was enhanced by EGF stimulation (Fig. 3, C and D). STAP-2 contains three functional domains: a PH domain, an Src-homology 2–like domain (SH2-like), and a proline-rich domain (Fig. 3E). We determined which STAP-2 region is required for STAP-2–EGFR interaction by immunoprecipitation. STAP-2–EGFR interaction was decreased by deletion of PH domain of STAP-2 (Fig. 3F), indicating the involvement of a PH domain in their binding. Of importance, EGFR overexpression increased phosphorylation of Tyr-250 in STAP-2 (Fig. 3G). STAP-2 Y250F mutant could not recover cell proliferation of STAP-2–knockdown DU145 cells (Fig. 3H), suggesting that EGFR-mediated STAP-2 phosphorylation facilitates proliferation of DU145 cells. Furthermore, STAP-2 translocated to plasma membrane by EGF stimulation (Fig. 3I). Thus, STAP-2 binds to functionally active EGFR, and EGFR-mediated STAP-2 phosphorylation contributes to full function of STAP-2 on proliferation of DU145 cells.
Figure 3.
STAP-2 interacts with EGFR. A and B, HEK293T cells were transfected with expression vectors of EGFR-HA, EGFR K721A-HA, and Myc-STAP-2, and then the lysates were immunoprecipitated (IP) with anti-HA antibody or anti-Myc antibody at 48 h post-transfection and blotted (IB). C, DU145 cells were transfected with Myc-STAP-2 expression vector, and at 48 h post-transfection, the cells were starved for 2 h following stimulation with 20 ng/ml EGF for 20 min. The lysates were immunoprecipitated with control IgG or anti-EGFR antibody and blotted. D, DU145 cells were stimulated with EGF as in C, and then the lysates were immunoprecipitated and blotted. E, schematic representation of deletion mutants of STAP-2. F and G, HEK293T cells were transfected with expression vectors of EGFR-HA and several mutant Myc-STAP-2s, and then the lysates were immunoprecipitated as in A. H, STAP-2–knockdown DU145 cells were transfected with expression vectors of mutant STAP-2, and their proliferation was measured as in Fig. 1F. I, HeLa cells were transfected with Myc-STAP-2 expression vector and then stimulated with 100 ng/ml EGF for 10 min at 48 h post-transfection. The cells were stained using DAPI (blue) and anti-Myc antibody (red). n = 3; mean values and S.E. (error bars) are depicted. N.S., not significant; *, p < 0.05 (paired Student's t test).
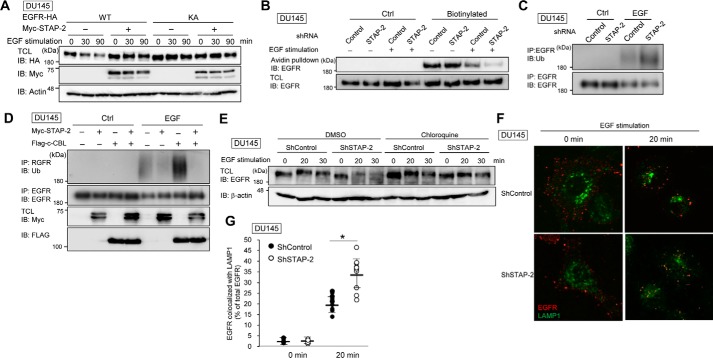
Activated EGFR is ubiquitinated by c-CBL, and ubiquitinated EGFR translocates from the plasma membrane to lysosomes, resulting in its degradation and down-regulation of EGFR signaling (3, 4). Next, we investigated whether this STAP-2–EGFR interaction contributes to EGFR stability because EGFR protein levels were slightly decreased in STAP-2–knockdown cells (Fig. 2, A and G). As shown in Fig. 4A, levels of EGFR rapidly decreased after EGF stimulation. However, EGFR levels were maintained when STAP-2 was overexpressed. In addition, levels of the EGFR K721A mutant did not decrease in DU145 cells independently of STAP-2 overexpression. After EGF stimulation, cell surface–localized EGFR was reduced to a greater extent in STAP-2 shRNA–expressing DU145 cells compared with control shRNA–expressing DU145 cells (Fig. 4B), indicating that STAP-2 stabilizes EGFR by inhibiting EGFR internalization or promoting EGFR recycling from endosomes to the plasma membrane. We previously reported that STAP-2 is a substrate of the E3 ubiquitin ligase, c-CBL, and that STAP-2 is involved in CBL-dependent degradation of focal adhesion kinase (20, 21). Levkowitz et al. also showed that EGFR internalization and degradation in lysosomes are facilitated by c-CBL–mediated EGFR ubiquitination (6). Thus, we hypothesized that STAP-2–mediated EGFR stabilization may arise from down-regulation of EGFR ubiquitination by c-CBL. As shown in Fig. 4C, EGFR was ubiquitinated after EGF stimulation. Importantly, EGF-induced ubiquitination of EGFR was up-regulated in STAP-2–knockdown DU145 cells. EGFR ubiquitination responding to EGF stimulation was enhanced by c-CBL overexpression (Fig. 4D). STAP-2 overexpression completely inhibited c-CBL–dependent EGFR ubiquitination. Under these conditions, c-CBL protein levels were not affected by STAP-2 overexpression (data not shown). Furthermore, EGF-induced EGFR degradation was facilitated by STAP-2 knockdown, and this degradation was blocked by chloroquine treatment, an inhibitor of lysosomal protease (Fig. 4E). EGFR trafficking to lysosomes was also promoted in STAP-2–knockdown DU145 cells (Fig. 4, F and G). These results indicate that STAP-2 interacts with EGFR and enhances its stability by inhibiting c-CBL-mediated EGFR ubiquitination (Fig. 5).
Figure 4.
STAP-2 increases EGFR stability via inhibiting its c-CBL-mediated ubiquitination. A, DU145 cells were transfected with STAP-2 and EGFR expression vectors and then treated with 10 μm cycloheximide in serum-free DMEM for 2 h at 48 h post-transfection. After stimulating the cells with 100 ng/ml EGF for the indicated periods, the cells were lysed and blotted (IB). TCL, total cell lysates. B, control or STAP-2 shRNA–expressing DU145 cells were starved for 2 h and then stimulated with 100 ng/ml EGF for 20 min. After biotinylation of the plasma membrane proteins, the lysates were pulled down with avidin-conjugated Sepharose beads and blotted. C, control or STAP-2 shRNA–expressing DU145 cells were starved for 2 h and then stimulated with 100 ng/ml EGF for 2 min. The lysates were immunoprecipitated (IP) with anti-EGFR antibody and blotted with the indicated antibodies. D, DU145 cells were transfected with expression vectors of FLAG-c-CBL and Myc-STAP-2. Samples were prepared at 48 h post-transfection as in C. E, control or STAP-2 shRNA–expressing DU145 cells were starved for 2 h with or without 100 μm chloroquine and then stimulated with 100 ng/ml EGF for the indicated times. The lysates were blotted as in B. F, control or STAP-2 shRNA–expressing DU145 cells were stimulated with EGF as in B and then stained with anti-EGFR (red) and anti-LAMP1 (green) antibodies. Their localization was observed by confocal microscopy, and the percentage of EGFR co-localized with LAMP1 is depicted in G. n = 10; mean values and S.D. (error bars) are depicted. *, p < 0.05 (paired Student's t test).
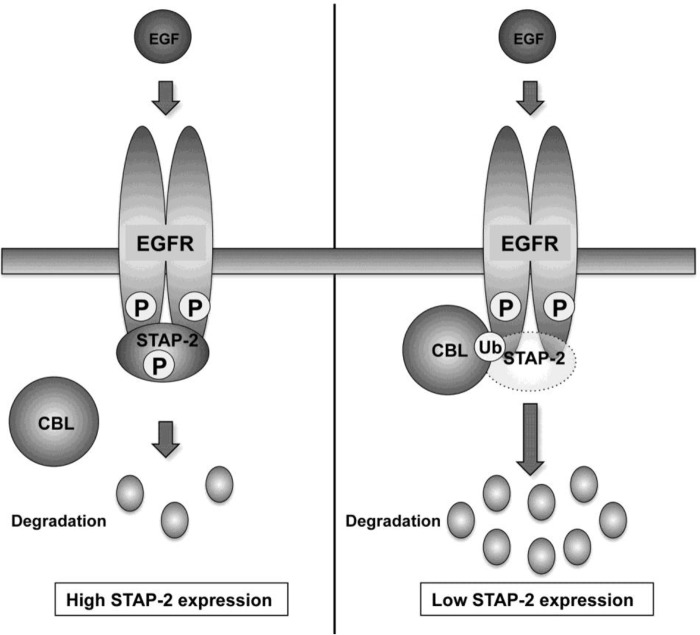
Figure 5.
Proposed model for the STAP-2–mediated up-regulation of EGFR signaling. EGF stimulation induces EGFR phosphorylation, leading to phosphorylation of STAP-2 and activation of its downstream signaling molecules, such as STAT3 and MAPK. Phosphorylated EGFR is ubiquitinated by c-CBL and then sorted to lysosomes, resulting in its degradation and down-regulation of EGFR signaling. In STAP-2 highly expressed cells, STAP-2 increases EGFR stability and activation of its downstream signaling by inhibiting c-CBL–mediated EGFR ubiquitination (left). EGFR ubiquitination and degradation were promoted in STAP-2 low expressing cells (right). Thus, STAP-2 up-regulates EGFR signaling by increasing its stability and enhances prostate cancer cell growth. P and Ub, phosphorylation and ubiquitination, respectively.
Discussion
Autoactivation of EGFR is one of the driving forces of carcinogenesis, and EGFR inhibitors are used clinically for the treatment of some types of malignancy, such as lung cancer (7). However, phase II trials of gefitinib for prostate cancer treatment showed limited efficacy (22, 23). These studies suggest that prostate cancer cells have an unknown up-regulation mechanism of EGFR signaling, which is lacking in lung cancer cells. Thus, cancer treatments with EGFR inhibitors might be more effective by unveiling the detailed mechanism of EGFR. We here showed that STAP-2 enhances EGFR signaling via its stabilization and promotes tumor formation of prostate cancer cells, and our previous work demonstrated that STAP-2 enhances STAT3 transcriptional activity via their direct association (24). These studies indicated that STAP-2 enhances EGFR signaling with two steps, EGFR stabilization and STAT3 up-regulation. STAT3 is activated by not only EGFR but also IL-6R signaling, and blockade of IL-6R inhibits tumor growth, suggesting that STAP-2 knockdown represses prostate tumor growth by synergistic effects of down-regulation of EGFR and IL-6R signaling.
Our previous studies showed that STAP-2 associates with BRK and enhances BRK-mediated STAT3 and STAT5 activation (14, 15). BRK is known to up-regulate EGFR signaling by inhibiting c-CBL–mediated EGFR ubiquitination (25). This BRK inhibition of c-CBL results from competitive binding to EGFR at phosphorylated Tyr-1045. Taken together with the data in this study, STAP-2 function in DU145 cells is indicated to be inhibition of EGFR ubiquitination by c-CBL, resulting in restoration of EGFR on the cell surface after EGF stimulation. EGFR dimerizes after binding of its ligands and then associates with Grb2, resulting in induction of Ras activation at the plasma membrane. Activated Ras induces ERK activation and promotes cancer cell proliferation (26). Thus, the amount of surface EGFR is important for activation of Ras and ERK, and down-regulation of surface EGFR after EGF stimulation is limited by STAP-2 expression (Fig. 4B). This could be a main mechanism for STAP-2 enhancement of EGFR-dependent cell growth in DU145 cells.
EGFR forms homo- or heterodimer with HER2, HER3, or HER4 after EGF binding. These EGFR dimers activate downstream signaling, including AKT and ERK, and are regulated by c-CBL. Ubiquitination of EGFR heterodimer by c-CBL is slower than that of EGFR homodimer; therefore, HER2 and HER3 promote aberrant activation of EGFR signaling and cell proliferation in some HER2- and HER3-overexpressed cancer cells (27–29). Our data showed that STAP-2 knockdown decreased cell proliferation in DU145 cells and LNCaP cells, and STAP-2 overexpression repressed c-CBL–mediated EGFR ubiquitination, leading to restored surface expression of EGFR (Figs. 1 (D and H) and 4B). Moreover, STAP-2 did not associate with EGFR K721A, a dimerization-deficient mutant, indicating that STAP-2 up-regulates EGFR after its dimerization process (Fig. 3C).
STAP-2–knockdown DU145 cells showed similar levels of proliferation in DMSO and gefitinib treatment conditions; likewise, cell growth of gefitinib-treated DU145 cells was not significantly decreased by STAP-2 knockdown (Fig. 2I). Moreover, STAP-2 stabilized wild-type EGFR after EGF stimulation but not the inactive form mutant of EGFR irrespective of EGF stimulation (Fig. 4A). These results suggest that STAP-2 knockdown represses tumor proliferation under EGFR-activating conditions but not its inactivating conditions. Down-regulation of STAP-2 represses EGFR signaling similarly as gefitinib treatment, resulting in tumor growth inhibition, but the mechanisms of their EGFR suppression are different, suggesting that STAP-2 inhibition destabilizes not only wild-type EGFR but also gefitinib-resistant autoactive EGFR. Therefore, inhibitors of STAP-2 function have the possibility of being developed for anticancer drugs for gefitinib-resistant prostate cancers. Although our data are based on overexpression or knockdown of STAP-2, our work implies that further studies on STAP-2, including functional and structural assays, will provide new insights into cancer physiology and support the development of anticancer therapies.
Experimental procedures
Reagents and cells
Cycloheximide was purchased from WAKO. MG132 was purchased from Calbiochem. Gefinitib was purchased from Cayman Chemical. Recombinant human EGF was purchased from PeproTech. DU145 and HEK293T cells were cultured in DMEM (Sigma) supplemented with 10% FBS (Sigma) and 0.05 mm 2-mercaptoethanol (Nacalai Tesque) at 37 °C in a humidified 5% CO2, 95% air atmosphere.
Plasmid construction
Construction of expression vectors of STAP-2, c-CBL, and ubiquitin was described previously (20). EGFR expression vectors were kind gifts from Dr. J. N. Ihle (St. Jude Children's Research Hospital, Memphis, TN) and Dr. H Sakurai (Toyama University, Toyama Japan) (30).
Knockdown
For knockdown of STAP-2 expression in DU145 cells, shRNA-expressing vectors were constructed by inserting the oligonucleotide DNA into the AgeI/EcoRI sites of pLKO.1. Oligonucleotide DNA sequences were as follows: shControl, 5′-tctcgcttgggcgagagtaagctcgagcttactctcgcccaagcgaga-3′; shSTAP-2#1, 5′-ctacaatagcaatcgggacttctcgagaagtcccgattgctattgtag-3′; shSTAP-2#2, 5′-catcctgaagccaaagaagttctcgagaacttctttggcttcaggatg-3′. shRNA-expressing lentivirus was prepared by transfecting these vectors and pVSV-G into HEK293T cells using Lipofectamine 2000 (Thermo Scientific Inc.) according to the manufacturer's instructions. DU145 cells were infected with the virus and then selected with 2.5 μg/ml puromycin for 48 h.
qPCR
Total RNA was isolated from DU145 cells using TRIzol (Thermo Scientific Inc.) according to the manufacturer's instructions. cDNA was synthesized from the isolated RNA using ReverTra Ace (TOYOBO) with random hexamer primers according to the manufacturer's instructions. Semiquantitative analysis of mRNA levels was performed with a MX3000P (Agilent Technologies). Primer sets for qPCR analysis were as follows: GAPDH fwd, 5′-gaaatcccatcaccatcttccagg-3′; GAPDH rev, 5′-cagtagaggcagggatgatgttc-3′; STAP-2 fwd, 5′-ggagaatggcgagaatgtgt-3′; STAP-2 rev, 5′-gcaacttctttggcttcagg-3′; Myc fwd, 5′-tcggattctctgctctcctc-3′; Myc rev, 5′-tcggattctctgctctcctc-3′; Ccnd1 fwd, 5′-gatgccaacctcctcaacga-3′; Ccnd1 rev, 5′-cacttctgttcctcgcagacc-3′.
Immunoprecipitation
DU145 cells were grown in 60-mm dishes semiconfluently and then transfected with the indicated expression vectors using Lipofectamine 2000 according to the manufacturer's instructions. The cells were cultured in serum-free DMEM for 2 h and then stimulated with 30 or 100 ng/ml human recombinant EGF (Peprotech) for the indicated periods at 48 h post-transfection and then lysed in lysing buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, protease inhibitor mixture (Sigma)). For the ubiquitination assay, the cells were lysed in radioimmune precipitation buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS, and protease inhibitor mixture). The lysate was immunoprecipitated for 2 h with anti-EGFR antibody (Ab-1, Millipore), anti-HA antibody (HA-7, Sigma), or anti-Myc antibody (9E10, Sigma) together with protein A-Sepharose beads (GE Healthcare). The protein was separated with SDS-PAGE and then blotted with the following antibodies; anti-FLAG antibody (M2, Sigma), anti-EGFR antibody (Santa Cruz Biotechnology, Inc.), anti-ubiquitin antibody (P4D1, Santa Cruz Biotechnology), anti-phosphotyrosine antibody (4G10, Millipore), anti-phospho-EGFR antibody (pY1173, Santa Cruz Biotechnology), anti-STAT3 antibody (Cell Signaling Technology), anti-phospho-STAT3 antibody (Cell Signaling Technology), anti-AKT antibody (B-1, Santa Cruz Biotechnology), anti-phospho-AKT antibody (Cell Signaling Technology), anti-ERK antibody (Cell Signaling Technology), anti-phospho-ERK antibody (Cell Signaling Technology), and anti-β-actin antibody (Sigma).
Luciferase assay
HEK293T cells were grown in a 24-well plate semiconfluently and then transfected with pCI-EGFR-HA, pcDNA3.1-Myc-STAP-2, pTK-RLuc, pGL3-APRE, and pGL3-SRE using Lipofectamine 2000 according to the manufacturer's instructions. The cells were lysed at 48 h post-transfection, and then luciferase activity was determined using the Dual-Luciferase reporter assay system (Promega) with a luminometer (ATTO). pGL3-APRE and pGL3-SRE were kind gifts from Dr. H. Maita (Hokkaido University, Japan).
Cell proliferation assay
Cell proliferation of D145 cells was measured using Cell Counting Kit-8 (WAKO) as described previously (16). Briefly, DU145 cells (5000 cells/well) were seeded on a 96-well plate and cultured for the indicated periods. Then 10 μl of WST solution was added to the cells. After culturing the cells for 3 h, absorbance at 450 nm was measured with a plate reader (Bio-Rad).
In vivo tumor progression
DU145 cells (5.0 × 106 cells) were subcutaneously injected into the right flank of BALB/c nude mice (female, 4 weeks old). Tumor diameter was measured with a caliper, and the tumor volume was calculated by the formula, volume = 0.52 × (width)2 × length.
Biotinylation of plasma membrane proteins
Control or STAP-2 shRNA–expressing DU145 cells were grown in 100-mm dishes semiconfluently and cultured in serum-free DMEM for 2 h. After EGF stimulation, the cells were washed with cold PBS and then incubated with 3 ml of HEPES, pH 8.0, 150 mm NaCl, 0.5 mg/ml Biotin-Sulfo-NHS on ice for 1 h. The cells were incubated with PBS containing 50 mm glycine for 5 min on ice and then lysed with 800 μl of lysing buffer for 10 min. Cellular debris in the lysates was disrupted by sonication and clarified by centrifugation. The supernatants were mixed with 25 μl of avidin-Sepharose (Sigma) and incubated for 2 h. The beads were washed three times with lysing buffer, and then biotinylated proteins were eluted by boiling with SDS sample buffer.
Immunofluorescence
HeLa cells were grown semiconfluently on 12-mm glass coverslips (Matsunami Glass) and transfected with pcDNA3.1-Myc-STAP-2 using Lipofectamine 2000. The cells were starved for 2 h at 48 h post-transfection and then stimulated with 100 ng/ml EGF for 10 min. Control or STAP-2 shRNA–expressing DU145 cells were stimulated with 100 ng/ml EGF for 20 min. The cells were fixed and stained using anti-EGFR (Ab-1, Millipore) and anti-LAMP1 antibody (Cell Signaling Technology) as described previously (31).
Author contributions
Y. K., M. I., K. S., S. T., and S. I. performed experiments; Y. S., R. M., and K. J. analyzed data; A. Y. distributed materials; Y. K. and K. O. wrote the paper; and T. M. designed experiments, supervised the project, and wrote the paper.
This study was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. The authors declare that they have no conflicts of interest with the contents of this article.
- EGFR
- EGF receptor
- qPCR
- quantitative PCR
- PH
- Pleckstrin homology
- WST
- water-soluble tetrazolium salt.
References
- 1. Threadgill D. W., Dlugosz A. A., Hansen L. A., Tennenbaum T., Lichti U., Yee D., LaMantia C., Mourton T., Herrup K., and Harris R. C. (1995) Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269, 230–234 [DOI] [PubMed] [Google Scholar]
- 2. Jorissen R. N., Walker F., Pouliot N., Garrett T. P., Ward C. W., and Burgess A. W. (2003) Epidermal growth factor receptor: mechanisms of activation and signalling. Exp. Cell Res. 284, 31–53 [DOI] [PubMed] [Google Scholar]
- 3. Sorkin A., and von Zastrow M. (2009) Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 10, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marmor M. D., and Yarden Y. (2004) Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene 23, 2057–2070 [DOI] [PubMed] [Google Scholar]
- 5. Grøvdal L. M., Stang E., Sorkin A., and Madshus I. H. (2004) Direct interaction of Cbl with pTyr 1045 of the EGF receptor (EGFR) is required to sort the EGFR to lysosomes for degradation. Exp. Cell Res. 300, 388–395 [DOI] [PubMed] [Google Scholar]
- 6. Levkowitz G., Waterman H., Zamir E., Kam Z., Oved S., Langdon W. Y., Beguinot L., Geiger B., and Yarden Y. (1998) c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12, 3663–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maemondo M., Inoue A., Kobayashi K., Sugawara S., Oizumi S., Isobe H., Gemma A., Harada M., Yoshizawa H., Kinoshita I., Fujita Y., Okinaga S., Hirano H., Yoshimori K., Harada T., et al. (2010) Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 362, 2380–2388 [DOI] [PubMed] [Google Scholar]
- 8. Minoguchi M., Minoguchi S., Aki D., Joo A., Yamamoto T., Yumioka T., Matsuda T., and Yoshimura A. (2003) STAP-2/BKS, an adaptor/docking protein, modulates STAT3 activation in acute-phase response through its YXXQ motif. J. Biol. Chem. 278, 11182–11189 [DOI] [PubMed] [Google Scholar]
- 9. Sekine Y., Yamamoto C., Kakisaka M., Muromoto R., Kon S., Ashitomi D., Fujita N., Yoshimura A., Oritani K., and Matsuda T. (2012) Signal-transducing adaptor protein-2 modulates Fas-mediated T cell apoptosis by interacting with caspase-8. J. Immunol. 188, 6194–6204 [DOI] [PubMed] [Google Scholar]
- 10. Sekine Y., Yamamoto T., Yumioka T., Sugiyama K., Tsuji S., Oritani K., Shimoda K., Minoguchi M., Yoshimura A., and Matsuda T. (2005) Physical and functional interactions between STAP-2/BKS and STAT5. J. Biol. Chem. 280, 8188–8196 [DOI] [PubMed] [Google Scholar]
- 11. Sekine Y., Nishida K., Yamasaki S., Muromoto R., Kon S., Kashiwakura J., Saitoh K., Togi S., Yoshimura A., Oritani K., and Matsuda T. (2014) Signal-transducing adaptor protein-2 controls the IgE-mediated, mast cell-mediated anaphylactic responses. J. Immunol. 192, 3488–3495 [DOI] [PubMed] [Google Scholar]
- 12. Sekine Y., Yumioka T., Yamamoto T., Muromoto R., Imoto S., Sugiyma K., Oritani K., Shimoda K., Minoguchi M., Akira S., Yoshimura A., and Matsuda T. (2006) Modulation of TLR4 signaling by a novel adaptor protein signal-transducing adaptor protein-2 in macrophages. J. Immunol. 176, 380–389 [DOI] [PubMed] [Google Scholar]
- 13. Ikeda O., Sekine Y., Mizushima A., Nakasuji M., Miyasaka Y., Yamamoto C., Muromoto R., Nanbo A., Oritani K., Yoshimura A., and Matsuda T. (2010) Interactions of STAP-2 with Brk and STAT3 participate in cell growth of human breast cancer cells. J. Biol. Chem. 285, 38093–38103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ikeda O., Miyasaka Y., Sekine Y., Mizushima A., Muromoto R., Nanbo A., Yoshimura A., and Matsuda T. (2009) STAP-2 is phosphorylated at tyrosine-250 by Brk and modulates Brk-mediated STAT3 activation. Biochem. Biophys. Res. Commun. 384, 71–75 [DOI] [PubMed] [Google Scholar]
- 15. Ikeda O., Mizushima A., Sekine Y., Yamamoto C., Muromoto R., Nanbo A., Oritani K., Yoshimura A., and Matsuda T. (2011) Involvement of STAP-2 in Brk-mediated phosphorylation and activation of STAT5 in breast cancer cells. Cancer Sci. 102, 756–761 [DOI] [PubMed] [Google Scholar]
- 16. Sekine Y., Togi S., Muromoto R., Kon S., Kitai Y., Yoshimura A., Oritani K., and Matsuda T. (2015) STAP-2 protein expression in B16F10 melanoma cells positively regulates protein levels of tyrosinase, which determines organs to infiltrate in the body. J. Biol. Chem. 290, 17462–17473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sekine Y., Ikeda O., Mizushima A., Ueno Y., Muromoto R., Yoshimura A., Kanakura Y., Oritani K., and Matsuda T. (2012) STAP-2 interacts with and modulates BCR-ABL-mediated tumorigenesis. Oncogene 31, 4384–4396 [DOI] [PubMed] [Google Scholar]
- 18. van Bokhoven A., Varella-Garcia M., Korch C., Johannes W. U., Smith E. E., Miller H. L., Nordeen S. K., Miller G. J., and Lucia M. S. (2003) Molecular characterization of human prostate carcinoma cell lines. Prostate 57, 205–225 [DOI] [PubMed] [Google Scholar]
- 19. Gan Y., Shi C., Inge L., Hibner M., Balducci J., and Huang Y. (2010) Differential roles of ERK and Akt pathways in regulation of EGFR-mediated signaling and motility in prostate cancer cells. Oncogene 29, 4947–4958 [DOI] [PubMed] [Google Scholar]
- 20. Sekine Y., Yamamoto C., Ikeda O., Muromoto R., Nanbo A., Oritani K., Yoshimura A., and Matsuda T. (2009) The protein content of an adaptor protein, STAP-2 is controlled by E3 ubiquitin ligase Cbl. Biochem. Biophys. Res. Commun. 384, 187–192 [DOI] [PubMed] [Google Scholar]
- 21. Sekine Y., Tsuji S., Ikeda O., Sugiyma K., Oritani K., Shimoda K., Muromoto R., Ohbayashi N., Yoshimura A., and Matsuda T. (2007) Signal-transducing adaptor protein-2 regulates integrin-mediated T cell adhesion through protein degradation of focal adhesion kinase. J. Immunol. 179, 2397–2407 [DOI] [PubMed] [Google Scholar]
- 22. Canil C. M., Moore M. J., Winquist E., Baetz T., Pollak M., Chi K. N., Berry S., Ernst D. S., Douglas L., Brundage M., Fisher B., McKenna A., and Seymour L. (2005) Randomized phase II study of two doses of gefitinib in hormone-refractory prostate cancer: a trial of the National Cancer Institute of Canada-Clinical Trials Group. J. Clin. Oncol. 23, 455–460 [DOI] [PubMed] [Google Scholar]
- 23. Small E. J., Fontana J., Tannir N., DiPaola R. S., Wilding G., Rubin M., Iacona R. B., and Kabbinavar F. F. (2007) A phase II trial of gefitinib in patients with non-metastatic hormone-refractory prostate cancer. BJU Int. 100, 765–769 [DOI] [PubMed] [Google Scholar]
- 24. Matsuda T., Muromoto R., Sekine Y., Togi S., Kitai Y., Kon S., and Oritani K. (2015) Signal transducer and activator of transcription 3 regulation by novel binding partners. World J. Biol. Chem. 6, 324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li X., Lu Y., Liang K., Hsu J. M., Albarracin C., Mills G. B., Hung M. C., and Fan Z. (2012) Brk/PTK6 sustains activated EGFR signaling through inhibiting EGFR degradation and transactivating EGFR. Oncogene 31, 4372–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanafusa H., Torii S., Yasunaga T., and Nishida E. (2002) Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat. Cell Biol. 4, 850–858 [DOI] [PubMed] [Google Scholar]
- 27. Muthuswamy S. K., Gilman M., and Brugge J. S. (1999) Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol. Cell. Biol. 19, 6845–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mellinghoff I. K., Tran C., and Sawyers C. L. (2002) Growth inhibitory effects of the dual ErbB1/ErbB2 tyrosine kinase inhibitor PKI-166 on human prostate cancer xenografts. Cancer Res. 62, 5254–5259 [PubMed] [Google Scholar]
- 29. Carrión-Salip D., Panosa C., Menendez J. A., Puig T., Oliveras G., Pandiella A., De Llorens R., and Massaguer A. (2012) Androgen-independent prostate cancer cells circumvent EGFR inhibition by overexpression of alternative HER receptors and ligands. Int. J. Oncol. 41, 1128–1138 [DOI] [PubMed] [Google Scholar]
- 30. Nishimura M., Shin M. S., Singhirunnusorn P., Suzuki S., Kawanishi M., Koizumi K., Saiki I., and Sakurai H. (2009) TAK1-mediated serine/threonine phosphorylation of epidermal growth factor receptor via p38/extracellular signal-regulated kinase: NF-κB-independent survival pathways in tumor necrosis factor α signaling. Mol. Cell. Biol. 29, 5529–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kitai Y., Takeuchi O., Kawasaki T., Ori D., Sueyoshi T., Murase M., Akira S., and Kawai T. (2015) Negative regulation of melanoma differentiation-associated gene 5 (MDA5)-dependent antiviral innate immune responses by Arf-like protein 5B. J. Biol. Chem. 290, 1269–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]