Abstract
Within the plant ATP-binding cassette transporter family, pleiotropic drug resistance (PDR) transporters play essential functions, such as in hormone transport or defense against biotic and abiotic stresses. NtPDR1 from Nicotiana tabacum has been shown to be involved in the constitutive defense against pathogens through the secretion of toxic cyclic diterpenes, such as the antimicrobial substrates cembrene and sclareol from the leaf hairs (trichomes). However, direct evidence of an interaction between NtPDR1 and terpenes is lacking. Here, we stably expressed NtPDR1 in N. tabacum BY-2 suspension cells. NtPDR1 was purified as an active monomer glycosylated at a single site in the third external loop. NtPDR1 reconstitution in proteoliposomes stimulated its basal ATPase activity from 21 to 38 nmol of Pi·mg−1·min−1, and ATPase activity was further stimulated by the NtPDR1 substrates cembrene and sclareol, providing direct evidence of an interaction between NtPDR1 and its two substrates. Interestingly, NtPDR1 was also stimulated by capsidiol, a sesquiterpene produced by N. tabacum upon pathogen attack. We also monitored the transcriptional activity from the NtPDR1 promoter in situ with a reporter gene and found that, although NtPDR1 expression was limited to trichomes under normal conditions, addition of methyl jasmonate, a biotic stress hormone, induced expression in all leaf tissues. This finding indicated that NtPDR1 is involved not only in constitutive but also in induced plant defenses. In conclusion, we provide direct evidence of an interaction between the NtPDR1 transporter and its substrates and that NtPDR1 transports compounds involved in both constitutive (diterpenes) and induced (sesquiterpenes) plant defenses.
Keywords: ABC transporter, glycosylation, liposome, membrane reconstitution, plant defense, protein purification, single particle analysis, BY-2 cells, Nicotiana tabacum, terpenes
Introduction
The ATP-binding cassette (ABC)3 superfamily of proteins is ubiquitous in all kingdoms of life. The family is mainly composed of primary active transporters, which constitute two nucleotide-binding domains (NBDs) and two transmembrane domains (TMDs) (1, 2). While the TMDs bind and translocate substrates across membranes, the NBDs hydrolyze ATP and provide the energy for active transport. The NBDs contain several well-conserved motifs such as Walker A and B; the D, H, and Q loops; and the ABC signature motif (3, 4). The family is particularly large in plants with more than 100 members in both Arabidopsis thaliana and Oryza sativa (5–7). In comparison, the human genome encodes for 49 ABC transporters (8). This large number of ABC proteins in plants can be related to their terrestrial and sessile life style, which makes them particularly exposed to (a)biotic stresses (9–11).
Members of the full-size ABCG subfamily (also called pleiotropic drug resistance (PDR)), which is only found in plants and fungi (12), play various essential roles in plants (for reviews, see Refs. 7, 9, and 12–15)). Known functions of full-size ABCGs include hormone transport (16–20), heavy metal detoxification (21, 22), transport of cuticle precursors (23), plant defense against pathogens (24–29), transport of monoterpene indole alkaloids (30, 31), and transport of β-caryophyllene (32). In contrast, very little information is available concerning the enzymatic properties of plant PDRs or, more generally, plant ABC transporters. To our knowledge, only one plant PDR transporter has been purified and characterized. The full size ABCG NpPDR5 from Nicotiana plumbaginifolia was expressed in Nicotiana tabacum Bright Yellow-2 (BY-2) suspension cells. The purified transporter displayed basal ATPase activity in the absence of substrates, which are still unknown (33).
NtPDR1 from N. tabacum was first identified in N. tabacum BY-2 cells (34) as a gene activated by the INF1 elicitin, a molecule triggering the biotic stress response (35). In BY-2 cells, both the NtPDR1 transcript and protein levels increased upon incubation with methyl jasmonate, a stress-response hormone (25, 35). This induction can be related to plant defense upon infection by pathogens (36). In the plant, NtPDR1 is constitutively expressed in the root epidermis as well as in the long glandular trichomes. These trichomes synthesize and secrete large amounts of antimicrobial cyclic diterpenes that constitute a first and constitutive line of plant defense against pathogens (37). Although these diterpenes freely diffuse through membranes, active transport is necessary to keep the concentration of those toxic compounds as low as possible within the plant cells that synthesize them. Indeed, BY-2 cell lines overexpressing NtPDR1 displayed a stronger tolerance to cyclic diterpenes, and transport assays pointed to the diterpenes sclareol and cembrene as putative substrates (25). NbABCG1, the NtPDR1 ortholog, and NbABCG2, a closely related gene, in the species Nicotiana benthamiana have been shown to be involved in induced defense against the pathogen Phytophthora infestans (28). Plants silenced for both NbABCG1 and NbABCG2 were shown to be more sensitive to the cytotoxic effect of the sesquiterpene capsidiol. Moreover, the secreted/intracellular ratio of capsidiol was reduced, suggesting that NbABCG1/2 might transport capsidiol. Preinvasive defense was also affected in the silenced lines, and one hypothesis is that NbABCG1/2 may also transport diterpenes involved in this type of defense (28). However, direct evidence of the interaction between either NtPDR1 or NbABCG1/2 and sesqui- and/or diterpenes is still lacking to clarify the roles of the transporters.
In this study, we describe the overexpression of NtPDR1 in N. tabacum BY-2 cells, its purification, and its reconstitution into liposomes. We demonstrate that NtPDR1, and not NpPDR5, a transporter involved in plant defense against insects used as a control, had its ATPase activity stimulated by the diterpenes cembrene and sclareol as well as by the sesquiterpene capsidiol. These data provide direct evidence of the interaction between NtPDR1 and these molecules and strongly suggest that NtPDR1 is involved in both constitutive and induced plant defenses.
Results
Expression of NtPDR1 in BY-2 suspension cells
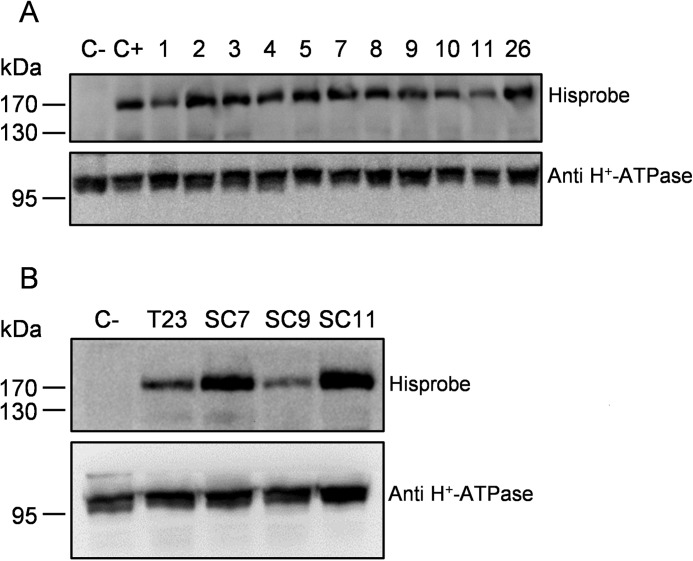
The coding sequence of NtPDR1 (25) was fused to an N-terminal purification tag with the following scheme: His10-StrepII tag-tobacco etch virus protease cleavage site (supplemental Fig. S1). The recombinant gene was inserted in an expression cassette under the control of a strong constitutive promoter (38) and transferred to the N. tabacum BY-2 cell (34) genome using Agrobacterium tumefaciens-mediated transformation. The gene was inserted together with the nptII kanamycin resistance gene for selection and mcherry, a gene encoding a fluorescent protein, which was used for the selection of highly expressing clones. Transformed cell lines were selected on kanamycin, and 60 lines were kept according to their mCherry fluorescence. Then, the lines were screened for NtPDR1 expression through Western blotting using HisProbe (Thermo Scientific), and the constitutively expressed plasma membrane H+-ATPase was used as a loading control. The clones displayed expression of the NtPDR1 at different levels (see Fig. 1A for an example) due to the position effect because the integration in the plant genome occurs randomly. It has been shown that every cell from a primary BY-2 transgenic line does not express the transgene in a homogenous manner. This heterogeneity might result from a double transformation event or appear later as a result of gene silencing. Nevertheless, it is possible to increase the homogeneity via subcloning (39). Because of this additional step performed on the elite line (T23), we obtained two subclone lines (SC7 and SC11) displaying significantly greater NtPDR1 expression (Fig. 1B). Scaling up of NtPDR1 production was performed in a 4-liter stirred-tank bioreactor. A fresh weight of about 1 kg of cells was obtained that typically yielded between 2 and 2.5 g (protein) of microsomal fraction.
Figure 1.
Expression of NtPDR1 in N. tabacum BY-2 cells. Shown are Western blots of 50 μg of microsomal fraction from different clones and a negative control (C−; wild-type BY-2 cells) using HisProbe or H+-ATPase antibodies for the loading control. A, different primary clones. C+ is a line expressing His6-NtPDR1 (25). B, T23 cell line as well as three subcloned lines derived from T23 (SC7, SC9, and SC11) and wild-type BY-2 cells.
Solubilization and purification
We screened several detergents for solubilization (supplemental Fig. S2), and n-dodecyl β-d-maltoside (DDM) was found to be the most efficient with a solubilization yield above 90% according to gel quantification. We then followed the purification procedure recently used for NpPDR5 (33) with minor modifications as described under “Experimental procedures.” The purification process included two affinity chromatography steps on Ni-NTA resin, the removal of the His10-StrepII tag by the addition of TEV protease, and a final incubation on Ni-NTA (Fig. 2). The resulting flow-through, which contained NtPDR1, was concentrated by ultrafiltration. A single band corresponding to NtPDR1 was obtained (Fig. 2) without visible contamination or degradation fragments upon SDS-PAGE and Coomassie Blue staining. The purification from 1 g of microsomal fraction yielded about 0.4 mg of purified NtPDR1.
Figure 2.
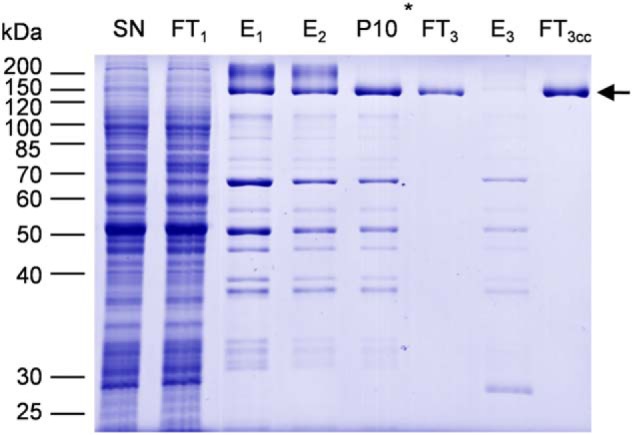
Purification of NtPDR1. NtPDR1 was solubilized from 800 mg of a microsomal fraction and purified as described under “Experimental procedures.” Twenty microliters of solubilized material (SN), flow-through of the first Ni-NTA column (FT1), elutions of the first and second Ni-NTA columns (E1 and E2), elution of the desalting column (P10), the flow-through and elution of the third column (FT3 and E3), and 5 μl of concentrated flow-through 3 were analyzed by SDS-PAGE and Coomassie Blue staining. The star shows at which step the purification tag was cleaved using TEV protease. The arrow indicates the band corresponding to NtPDR1.
Oligomeric state of NtPDR1
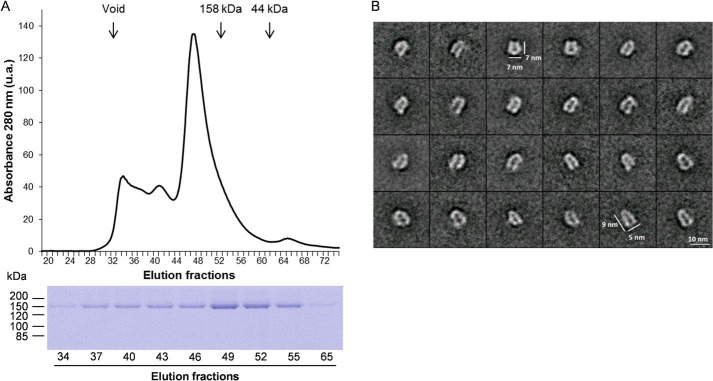
Gel filtration of the purified NtPDR1 was performed to assess the monodispersity and oligomeric state of NtPDR1 (Fig. 3A). Seventy percent of NtPDR1 was eluted in a peak at an apparent size of about 240 kDa, which is consistent with the size of a monomeric NtPDR1 (162 kDa) surrounded by the detergent micelle. Smaller peaks of greater size probably correspond to oligomers or aggregates. We further analyzed the main peak of NtPDR1 by negative-stain electron microscopy (Fig. 3B). The raw images of negatively stained proteins showed that solubilized NtPDR1 was monodisperse. To enhance the signal over noise ratio, 2D classification was carried out using more than 5,300 particles (Fig. 3B). The classes displayed in Fig. 3B most likely represent different views and thereby orientations of the sample adsorbed onto an EM grid. Our unbiased reference-free analysis revealed that NtPDR1 was monomeric. Compared with reported analyses of other ABCs transporters, the particles showed recognizable features of ABCs including U or V shapes with, at one end, greater densities likely corresponding to NBDs.
Figure 3.
Quaternary structure analysis of NtPDR1. A, purified NpPDR1 was analyzed by size-exclusion chromatography on a Superdex 200 column. Protein elution was followed by monitoring the absorbance at 280 nm. The elution peaks corresponding to size markers are indicated by vertical arrows. Below is shown the SDS-PAGE analysis of the indicated fractions. B, single-particle analysis of negatively stained NtPDR1. A gallery of representative 2D class averages is shown. u.a., arbitrary units of absorbance.
Glycosylation of NtPDR1
Five N-glycosylation sites are predicted for NtPDR1 (Asn-5, -641, -738, -1231, and -1386 of the native predicted enzyme (supplemental Fig. S3)). Purified NtPDR1 was proteolyzed either by trypsin or by multienzymatic limited digestions to increase the sequence coverage. The resulting peptides were then analyzed by LC-MS/MS. Peptides covering positions 5, 1231, and 1386 were all detected without any glycosylation. No free or glycosylated peptide covering position 641 was identified. This site is predicted to be localized very close to a membrane-spanning domain (supplemental Fig. S3) and is probably not accessible to the glycosylation machinery or to proteases. In contrast, Asn-738 is localized in an external loop and was found to be glycosylated. The glycosylation profile (Fig. 4) corresponds to a typical plant profile with GnGnXF as the most abundant glycan (supplemental Fig. S4). Deglycosylation was performed using peptide N-glycosidase A to confirm site 738 occupancy. After this step, the deglycosylated peptide (displaying 1 mass unit difference due to the Asn to Asp modification) was observed for site 738, whereas no Asn to Asp modification was observed for other sites, confirming previous results (data not shown). We also tested the glycosylation profile of NpPDR5 purified in a very similar manner as NtPDR1 (33). Two glycosylation sites are predicted, but only free peptides were found covering those positions (data not shown).
Figure 4.
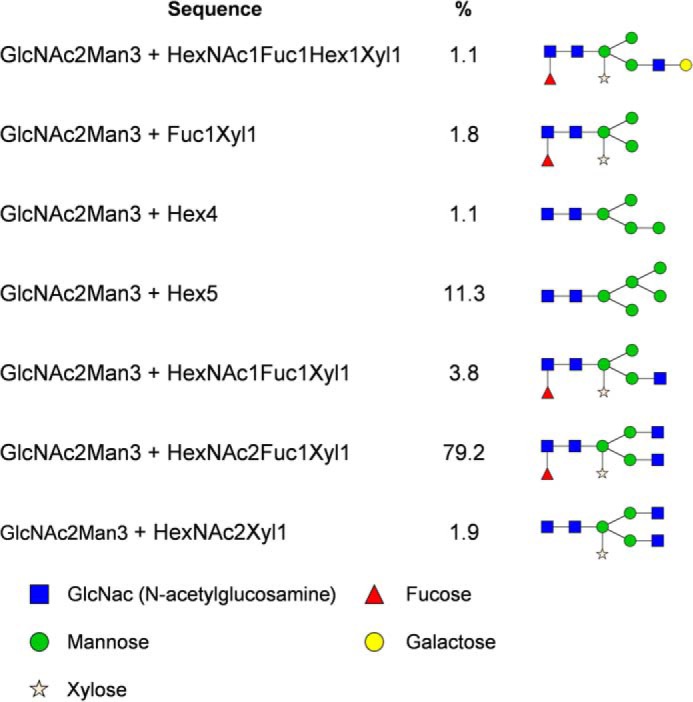
Glycosylation profile of Asn-738. Relative abundances were determined based on the MS1 signal of glycopeptides with WNHIVPGGNETLGSTVVK as a peptide portion. A proposed structure is provided for every glycan composition. Hex, hexose; HexNAc, N-acetylhexosamine; Fuc, fucose; Xyl, xylose.
Purified NtPDR1 is active
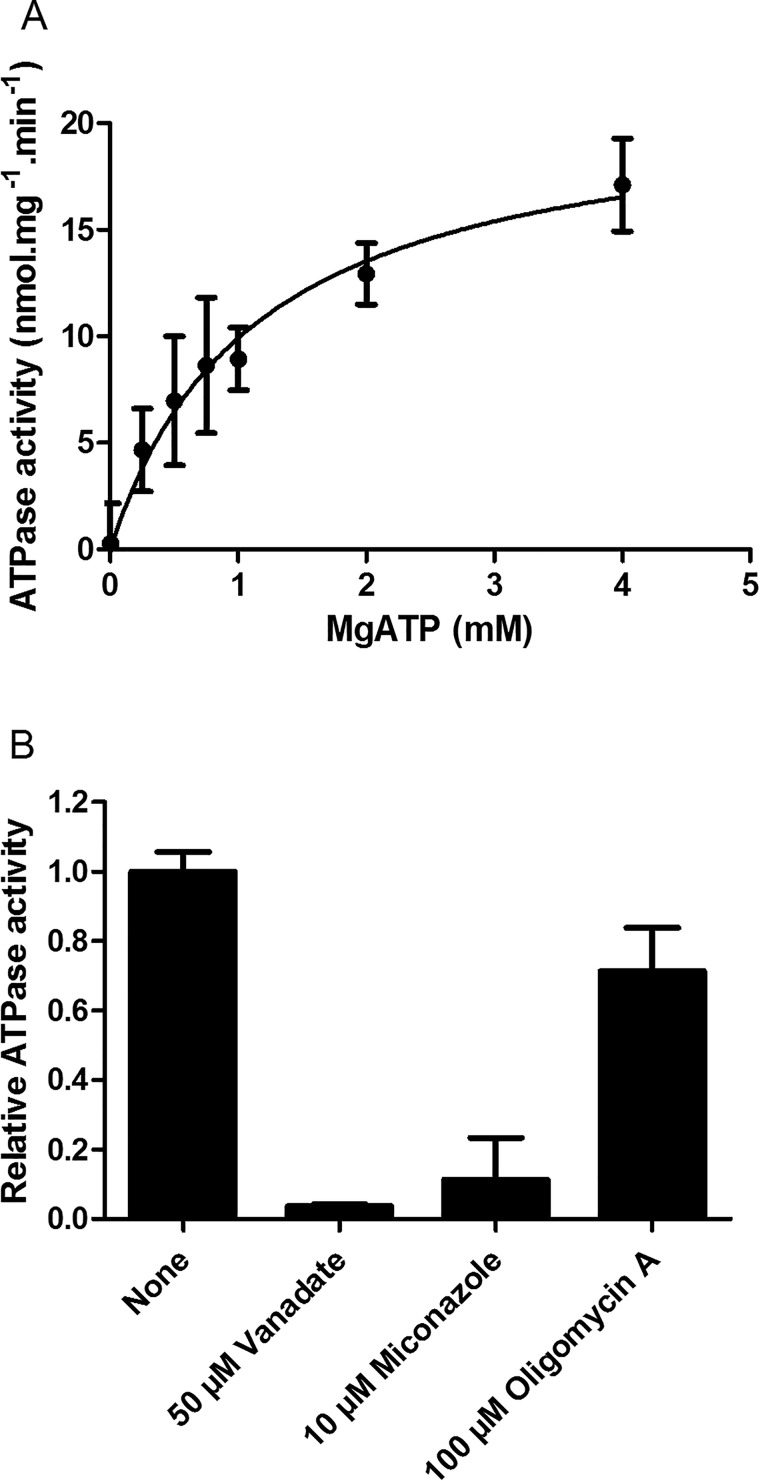
ABC proteins are known to exhibit basal ATPase activity. NtPDR1 was purified in an active form because it was able to hydrolyze ATP (Fig. 5A). In the presence of DDM and asolectin, a mixture of phospholipids, NtPDR1 displays a Km of 1.14 ± 0.30 mm Mg-ATP and a Vmax of 21.24 ± 2.39 nmol of Pi·min−1·mg−1. In addition, NtPDR1 basal ATPase activity was inhibited by the well-known ABC inhibitors vanadate, miconazole, and, to a lesser extent, oligomycin A (Fig. 5B).
Figure 5.
Basal ATPase activity of purified NtPDR1. A, the ATPase activity of purified NtPDR1 was measured in the presence of the indicated Mg-ATP concentration. Free Mg2+ was kept at 1 mm. Km and Vmax are 1.14 ± 0.30 mm Mg-ATP and 21.24 ± 2.39 nmol·min−1·mg−1, respectively. B, the specific activity was assessed in the presence and absence of 50 μm vanadate, 10 μm miconazole, or 100 μm oligomycin A. Error bars represent S.D.
Reconstitution into liposomes
To assess the activity of NtPDR1 in a lipid environment, purified NtPDR1 was reconstituted in asolectin liposomes. The reconstitution efficiency was assessed by flotation centrifugation on a sucrose gradient. More than 90% of the NtPDR1 signal was found in the top fractions of the gradient (supplemental Fig. S5A), demonstrating that the transporter was reconstituted in liposomes. A protease assay using trypsin demonstrated that the proteoliposomes were well-sealed (supplemental Fig. S5B). Indeed, the antibody against NtPDR1 was obtained using a peptide corresponding to a region present in the second NBD. Upon reconstitution, part of the transporter is expected to be oriented in the membrane with its NBDs inside the liposome. In that case, only partial digestion by trypsin should occur because few cleavage sites are available outside of the vesicles. Indeed, different bands corresponding to partial digestion of NtPDR1 were visible when the proteoliposomes were incubated with trypsin. All these bands were not present when the liposomes were destabilized by Triton X-100. Altogether, these results show that NtPDR1 was well-reconstituted in sealed vesicles. The same approach was used to reconstitute NpPDR5, and similar results were obtained (supplemental Fig. S6). These NpPDR5 proteoliposomes served as a control in ATPase activity tests.
Reconstitution of NtPDR1 increases its basal ATPase activity
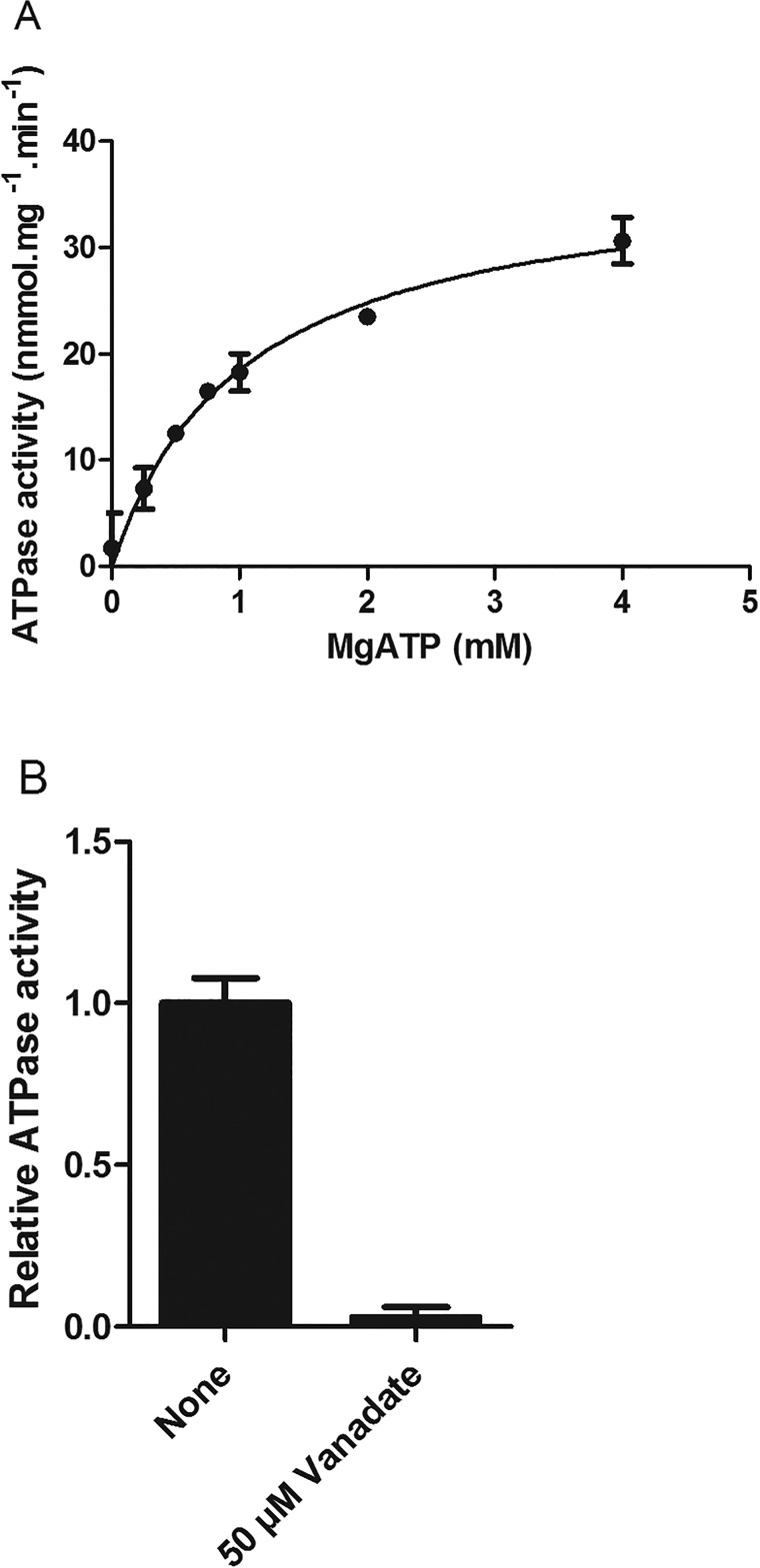
Basal ATPase activity of reconstituted NtPDR1 was first assessed in the absence of putative substrates (Fig. 6A). NtPDR1 displayed a Km of 1.00 ± 0.14 mm ATP, which is in the range of the typical ATP concentration of plant cells (40). The apparent Vmax was 37.6 ± 2.0 nmol of Pi·mg−1·min−1, a 77% increase compared with the value observed for the solubilized enzyme. It has to be noted that this Vmax is underestimated because only the proteins with their NBDs at the external side of the membranes have access to ATP. The reconstituted NtPDR1 ATPase activity was almost completely abolished by vanadate (Fig. 6B).
Figure 6.
Basal ATPase activity of reconstituted NtPDR1. A, the ATPase activity of reconstituted NtPDR1 (4 μg) was measured in the presence of the indicated Mg-ATP concentration. Free Mg2+ was kept at 1 mm. Km and Vmax are 1.04 ± 0.14 mm Mg-ATP and 37.64 ± 2.01 nmol·min−1·mg−1, respectively. B, the specific activity was assessed in the presence and absence of 50 μm vanadate. Error bars represent S.D.
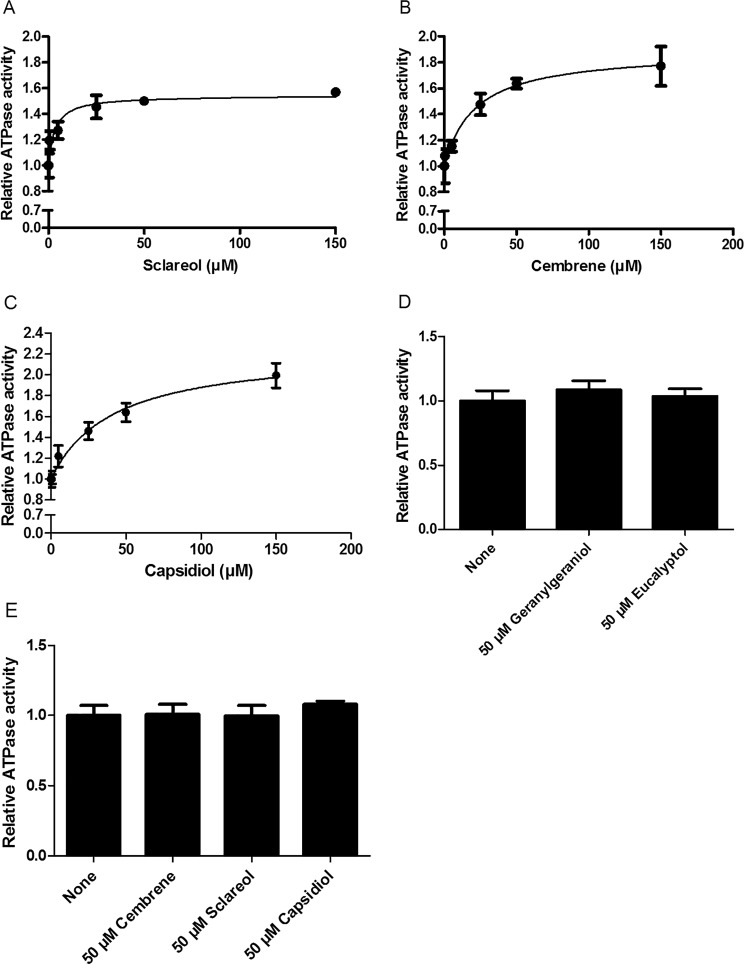
NtPDR1 is specifically activated by defense terpenoids
In vivo toxicity and transport assays (25) supported the hypothesis that NtPDR1 is involved in the transport of cyclic diterpenes such as sclareol and cembrene but could not discriminate between direct and indirect effects. It is well-known that the ATPase activity of ABC transporters can be stimulated by their substrates. However, this is not the case for all ABC transporters. For example, yeast PDR proteins are not stimulated by their substrates (41). Here, we investigated the potential stimulation of NtPDR1 upon the addition of different terpenoids. NtPDR1 ATPase activity was stimulated by the cyclic diterpenes sclareol (55%) and cembrene (89%) with Km values of 3.4 ± 1.4 and 21.3 ± 6 μm, respectively (Fig. 7, A and B). Because its N. benthamiana homologs NbPDR1 and NbPDR2 have been proposed to transport capsidiol, the effect of this cyclic sesquiterpene on ATPase activity was also tested on NtPDR1. Capsidiol stimulated NtPDR1 (120%) with a Km of 41.7 ± 10.2 μm (Fig. 7C). In contrast, neither the monoterpene eucalyptol nor the linear diterpene geranylgeraniol stimulated NtPDR1 activity (Fig. 7D). In addition, none of the NtPDR1-stimulating compounds increased the NpPDR5 basal ATPase activity (Fig. 7E), indicating that the stimulation of NtPDR1 activity was specific.
Figure 7.
ATPase activity of reconstituted NtPDR1 (A–D) or NpPDR5 (E) activity in the presence of different terpenoids. A, sclareol. Km = 3.36 ± 1.36 μm, and the maximal stimulation is 54 ± 4%. B, cembrene. Km = 21.27 ± 6.02 μm, and the maximal stimulation is 89 ± 7%. C, capsidiol. Km = 41.47 ± 10.19 μm, and the maximal stimulation is 125 ± 12%. D, geranylgeraniol and eucalyptol at 50 μm. E, relative activity of reconstituted NpPDR5 in the presence or absence of 50 μm cembrene, sclareol, or capsidiol. Error bars represent S.D.
NtPDR1 expression is induced in leaf tissues by methyl jasmonate
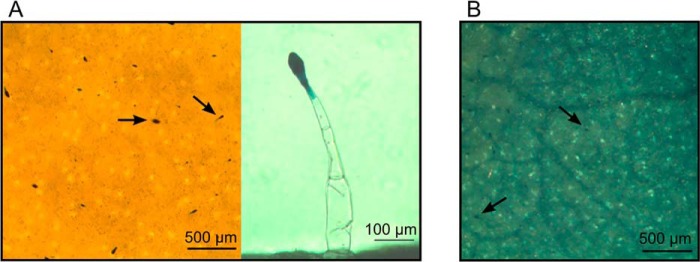
In leaf tissues, NtPDR1 is constitutively expressed in the head cells of long glandular trichomes where the defense diterpenes are synthesized, secreted, and take part in the constitutive plant defense (25). The stimulation of NtPDR1 by capsidiol raises the hypothesis that this transporter is also involved in induced defense in leaf tissues because capsidiol was shown to be produced upon pathogen attack (42). We thus tested whether the β-glucuronidase (GUS) reporter gene controlled by the NtPDR1 transcription promoter was activated in the presence of methyl jasmonate, a typical hormone involved in plant defense. Although GUS activity was confined to trichomes in control conditions, leaf incubation with methyl jasmonate induced GUS expression in the whole leaf tissue (Fig. 8).
Figure 8.
Stimulation of NtPDR1:GUS expression by methyl jasmonate. Leaf discs of N. tabacum plants expressing the GUS reporter gene under the control of the NtPDR1 promoter (25) were incubated with water or with 250 μm methyl jasmonate, and the in situ GUS activity was monitored 24 h later. A, control leaf disk incubated with water. The left panel shows a top view of a leaf disk. GUS expression is limited to trichomes. The right panel shows a trichome in detail. B, leaf disk incubated with 250 μm methyl jasmonate. Expression was observed in the whole leaf tissue. Arrows indicate examples of stained trichomes head cells.
Discussion
Despite their essential roles, plant ABC transporters have been poorly characterized at the enzymatic level. One explanation for this lack of data resides in the difficulty of achieving heterologous expression in organisms such as bacteria or yeast (9). N. tabacum BY-2 cells have been used to functionally characterize different ABC transporters such as Arabidopsis ABCB4 (43, 44) and ABCG40 (19), MtABCG10 from Medicago truncatula (45), and PhABCG1 from Petunia hybrida (46). For the latter as well as NpPDR5 (33), purification of the protein was also carried out. BY-2 cells offer among others the advantages of proper membrane composition and glycosylation. The lipid composition of the membranes has an impact on protein folding, structure, and function (47, 48). In plants, the plasma membrane is particularly rich in sterols and polar sphingolipids (49, 50), which might be crucial for membrane protein stability. It would be interesting to test these lipids during reconstitution in liposomes.
Size-exclusion chromatography and single-particle analysis by electron microscopy of NtPDR1 showed that solubilized NtPDR1 is a monomer. The length of ∼9 nm is similar to lengths of other ABCG transporters including the plant NpPDR5 (33) as well as the mammalian ABCG2 (Protein Data Bank code 5NJ3) and ABCG5/G8 (Protein Data Bank code 5DO7). This is shorter than the 12–13 nm reported for several ABCs of other subfamilies such as ABCA1 (Protein Data Bank code 5XJY), ABCB1 (Protein Data Bank code 5KOY), and ABCC1 (Protein Data Bank code 5UJ9). This is related to shorter intracytoplasmic helices in ABCG transporters, suggesting a similar topology for NtPDR1.
Glycosylation is a common co- and post-translational modification of proteins moving through the secretory pathway and plays a crucial role in protein folding, trafficking, stability, and activity (51). Because glycosylation is not strictly conserved between plants and other eukaryotes (52), glycosylation defects might explain the difficulty in overexpressing some plant ABC transporters in these organisms. Interestingly, glycosylation has also been shown to affect the transport activity of the human ABCC4 (MRP4) transporter (53). The authors demonstrated that the absence of glycosylation was not deleterious for ABCC4 localization but affected its transport activity in a substrate-specific manner. Because our analysis demonstrated that NtPDR1 is indeed glycosylated, we believe it was safer to use a homologous expression system to ensure correct glycosylation. The single glycosylation site is localized within a ∼60-residue external loop in the first TMD. Curiously, the corresponding predicted site in the second TMD was not found glycosylated. Glycosylation asymmetry was also found in the external loops of the human heterodimer ABCG5/ABCG8 for which two and one glycosylation sites were identified, respectively (54). In contrast, no glycosylation was found for NpPDR5.
The cyclic diterpenes cembrene and sclareol have been identified as putative substrates of NtPDR1. However, direct evidence of an interaction between these compounds and the protein was lacking. The same is true for any other plant ABC transporter. We thus cannot exclude that transport that was observed at the cell level (25) was an indirect consequence of NtPDR1 expression. To solve this question, we first tried to monitor the transport of a radiolabeled analog of sclareol (55) that was shown to be transported in N. tabacum BY-2 cells overexpressing NtPDR1 (25). To do so, we followed the accumulation of the sclareol analog inside proteoliposomes, but we could not obtain reliable transport data. This illustrates the difficulty in showing direct transport of hydrophobic substrates by in vitro assays. For instance, close to half of the human ABC transporters are thought to facilitate lipids or lipid-related compounds. In many cases, the substrates have been indirectly identified by monitoring accumulation of compounds in humans with specific diseases or in knock-out mice (56). The lack of evident transport is probably due to the high hydrophobicity of the substrates, which readily diffuse and accumulate inside the membrane. We indeed observed a fast and huge background of the diterpenes associated with the liposomes in the absence of ATP. Even in the case of active transport toward the liposome lumen, the compounds will likely re-enter the membrane by passive diffusion. In planta, lipid transfer proteins have been shown to be involved in the transport of hydrophobic diterpenes and sesquiterpenes from the membrane vicinity to lipid droplets to avoid re-entering in the plasma membrane (57, 58).
For some ABC transporters, an alternative to transport assays is the stimulation of the ATPase activity by the substrate (e.g. Refs. 59 and 60). Indeed, the addition of the diterpenes cembrene and sclareol as well as the sesquiterpene capsidiol did stimulate the ATPase activity with Km values in the micromolar range, indicating direct interaction between NtPDR1 and these substrates. None of these compounds were found to have an impact on NpPDR5 basal activity, indicating that the interaction with NtPDR1 was specific. Neither the monoterpene eucalyptol nor the linear diterpene geranylgeraniol stimulated the NtPDR1 ATPase activity. This is consistent with in vivo data (25).
The observation that the diterpenes sclareol and cembrene as well as the sesquiterpene capsidiol stimulate NtPDR1 ATPase activity is very interesting because these two types of compounds are involved in different steps of plant defense. Diterpenes are constitutively synthesized in, and secreted from, the head cells of glandular trichomes (61) where NtPDR1 is constitutively expressed (25). Capsidiol and other sesquiterpenes are synthesized as a response to pathogen attack in tissues next to the site of pathogen invasion (62, 63). This is in agreement with the induced expression of the GUS reporter under the control of the NtPDR1 promoter in whole leaf tissues upon addition of the defense hormone methyl jasmonate, whereas expression was confined to the trichome secretory cells in the absence of this hormone. NtPDR1 is thus involved in transporting compounds involved in both constitutive (diterpenes) and induced (sesquiterpenes) defenses.
An important question that remains to be addressed is the subcellular localization of NtPDR1. This transporter has been localized to the plasma membrane in BY-2 cells, and this is in agreement with its transport activity (25) as well with the fact that diterpenes are secreted from the trichome head cells (64). However, one hypothesis might be that NtPDR1 is localized to internal vesicles in which substrates are transported before being released out of the cell upon fusion of the vesicles with the plasma membrane. The advantage would be to considerably increase the secretion capacity, which would be in agreement with the massive secretion of diterpenes from tobacco trichomes. Indeed, diterpenes together with sucrose esters also secreted from the trichomes might represent more than 10% of the dry weight of the whole tobacco leaf (64). Future work should therefore focus on the subcellular localization of NtPDR1 in trichomes but also in other leaf tissues upon pathogen attack.
In conclusion, this work provides direct evidence of the interaction between a plant ABC transporter and its substrates. We demonstrated in vitro that the activity of purified NtPDR1 is specifically stimulated by both cyclic di- and sesquiterpenes involved in constitutive and induced plant defenses. We also provide another example of an ABC protein for which N. tabacum BY-2 cells are a convenient host for protein expression, purification, reconstitution, and biochemical characterization. Our pipeline could be used for other plant ABC proteins for which very little is known at the biochemical level (65).
Experimental procedures
Plant materials
N. tabacum L. cv. BY-2 suspension cells (34) were cultured in MS medium (0.44% (w/v) Murashige and Skoog salts (MP Biomedicals), 0.02% (w/v) KH2PO4, 5·10−3% (w/v) myoinositol, 5·10−4% (w/v) thiamine, and 2·10−5% (w/v) 2,4-dichlorophenoxyacetic acid, pH 5.8 (KOH)) supplemented with 100 μg/ml kanamycin. The cultures were grown in the dark at 25 °C and shaken at 90 rpm. The lines were subcultured (diluted 10 times in fresh medium) every week.
Genetic engineering and BY-2 cell transformation
The sequence coding for the His10-StrepII-TEV cleavage site tag was synthesized in pUC57 (Genscript) and fused to the N terminus of the NtPDR1 coding sequence. The resulting amino acid sequence is depicted in supplemental Fig. S1. This sequence was inserted in a modified pAUX3131 vector (66) under the control of the strong constitutive promoter En2PMA4 (38) and the nos terminator. The cassette was inserted in the binary vector for A. tumefaciens-mediated plant transformation using the vector pPZP-RCS2 (66) provided with nptII (kanamycin resistance) and mcherry (gene encoding a fluorescent protein). The final construct (pPZP-RCS2-nptII-mcherry-ntpdr1) was introduced in A. tumefaciens LBA4404virG (67) via electroporation, and N. tabacum BY-2 cells were transformed as described (38).
Screening of the lines and cloning
Transformed plant cells were selected on solid MS medium supplemented with 100 μg/ml kanamycin. The resulting calli were preliminarily screened according to their mCherry fluorescence under a binocular microscope (Nikon Intensilight C-HGFI). Sixty calli were selected and transferred to liquid medium. The greatest expressing line (T23) was selected by Western blot analysis of a microsomal fraction (as described in Ref. 25) using HisProbe according to the manufacturer's protocol. For subcloning, a 6-day-old culture of the elite line (T23) was diluted 5,000 times with wild-type BY-2 cells and plated on kanamycin-containing MS medium, and the selection process was repeated as mentioned above on 15 secondary calli.
Immunoblotting
Samples were analyzed by SDS-PAGE (10% polyacrylamide) and transferred onto poly(vinylidene difluoride) membranes. Antibodies were diluted as follows: anti-NtPDR1 (25), 1:1,000; anti-NpABCG5/NpPDR5 antibodies (27), 1:500; HisProbe-horseradish peroxidase (HRP) conjugate (Thermo Fisher Scientific), 1:1,000; anti-H+-ATPase antibodies (68), 1:100,000; HRP-labeled goat anti-rabbit IgG antibodies (Biognost), 1:10,000.
NtPDR1 production in a 4-liter bioreactor
Three 100-ml 6-day-old cultures of BY-2 cells were transferred into a 4-liter stirred-tank bioreactor (Sartorius Biostat A). The culture was grown at 25 °C in the dark under agitation with a propeller (60 rpm) and air bubbling to provide oxygen.
Membrane preparation
The isolation of the microsomal fraction was performed as described (33). Briefly, the cells from a 6-day-old bioreactor were harvested by filtration through Miracloth (Calbiochem) and washed with ice-cold homogenization buffer (50 mm Tris-HCl (pH 8.0), 2 mm EDTA, 0.25 m sorbitol, 20 mm DTT, and protease inhibitor mixture (1 mm PMSF and 1·10−4% (w/v) each of leupeptin, pepstatin, aprotinin, antipain, and chymostatin)). The cells were resuspended in 1 volume of homogenization buffer and ground twice with glass beads (0.8–1-mm diameter; VWR) in a DYNO-MILL Type KDL. The homogenate was centrifuged at 3,700 × g (Beckman-Coulter Avanti J-E centrifuge and JLA-9.1000 rotor) for 5 min, and the supernatant was centrifuged at 6,000 × g for 5 min (same rotor) to remove the cell debris and the heavy subcellular fraction. The supernatant was centrifuged at 100,000 × g for 40 min to pellet the membranes (microsomal fraction). The pellet was suspended in suspension buffer (330 mm sucrose, 3 mm KH2PO4 (pH 7.8), and 3 mm KCl). Protein concentration was assessed using the Bradford method (69) with bovine serum albumin as the standard.
Solubilization assays
The solubilization assay was adapted from Toussaint et al. (33). Microsomal proteins were diluted at 2.5 mg/ml in the solubilization buffer (20 mm Tris-HCl (pH 7.5), 50 mm NaCl, 20 mm imidazole, 10% (w/v) glycerol, and protease inhibitor mixture) supplemented with 1% detergent (DDM, decyl maltose neopentyl glycol (MNG), octyl β-glucoside, Triton X-100, tetradecyl octaethylene glycol ether (C14E8), CHAPS, lauryldimethyl amine oxide, or SDS). All samples were incubated for 30 min under agitation on a wheel at 4 °C. After centrifugation for 15 min at 100,000 × g and 4 °C (Beckman Optima MAX ultracentrifuge and TLA-55 rotor), the supernatant was kept, and the pellet was resuspended in the same volume of solubilization buffer by sonication (ultrasonic processor Sonics Vibra-Cell 75022; amplitude, 30). The supernatants and pellets were analyzed by SDS-PAGE and Western blotting.
Purification of NtPDR1 and NpPDR5
The purification was performed as described (33). Briefly, the proteins were solubilized in DDM and purified twice by nickel-affinity chromatography. Then, the His-StrepII tag was removed, the proteins were incubated a third time with Ni-NTA, and purified NtPDR1 or NpPDR5 were recovered in the flow-through and concentrated by centrifugal ultrafiltration. One difference between NtPDR1 and NpPDR5 purification was that the imidazole concentration used for the incubation on the third Ni-NTA column was 10 mm (NtPDR1) or 15 mm (NpPDR5) due to the greater tendency of NtPDR5 to bind to Ni-NTA with its endogenous histidine residues.
Size-exclusion chromatography
Purified proteins were loaded on a Superdex 200 10/300 GL size-exclusion chromatography column (GE Healthcare) linked to the ÅKTA Explorer system. The column was pre-equilibrated with 20 mm Tris-HCl (pH 7.5), 50 mm NaCl, and 10% (w/v) glycerol supplemented with 0.03% MNG. The elution was carried out at 0.45 ml/min, and the absorbance at 280 nm was recorded.
Reconstitution of NtPDR1 and NpPDR5
The reconstitution protocol was adapted from Geertsma et al. (70). Fifty mg of asolectin from soybean (Sigma, reference number 11145) were dissolved in chloroform and dried under N2. The lipidic film was dissolved in diethyl ether and dried again under N2 to remove traces of chloroform. The lipid film was suspended in 1 ml of reconstitution buffer (20 mm Tris-HCl (pH 7.5) and 50 mm NaCl) and sonicated on ice for 15 min (ultrasonic processor Sonics Vibra-Cell 75022; amplitude, 30; pulses of 6 s). The suspension was submitted to five cycles of flash freezing in liquid nitrogen and slow thawing at room temperature, and the liposomes were flash frozen and stored at −80 °C. When needed, 1 ml of liposomes at 50 mg/ml was slowly thawed at room temperature and extruded 11 times through a 400-nm polycarbonate filter (Whatman) using a Mini Extruder (Avanti Polar Lipids). The suspension was diluted to 4 mg/ml asolectin with reconstitution buffer (total volume, 12.5 ml) and destabilized by 10 successive additions of 3.75 mg of Triton X-100 (final concentration, 3 mg/ml) with 1 min of gentle agitation in between. Purified NtPDR1 or NpPDR5 was added to 3 ml of destabilized liposomes at a protein/lipid ratio of 1:100 (w/w) and agitated for 15 min at 4 °C. To remove detergents, the suspension was agitated for 16 h at 4 °C with successive additions of 40 mg/ml SM-2 Bio-Beads (Bio-Rad) after 0, 30, 90, and 840 min. The suspension was filtered on a Poly-Prep column (Bio-Rad), and the beads were washed with 2 ml of reconstitution buffer. The proteoliposomes were collected by centrifugation for 20 min at 267,000 × g (Beckman Optima MAX ultracentrifuge and MLA-80 rotor). The proteoliposomes were suspended in reconstitution buffer to 10 mg/ml lipids and used for further analysis.
Protease assay
Thirteen microliters of reconstitution buffer supplemented or not with 1 μg of trypsin (Sigma), 1.6% Triton X-100 (Sigma), and 20 μg of trypsin inhibitor (Sigma) were incubated for 10 min on ice. Fifteen microliters of proteoliposomes (1.5 μg of proteins) were added, and the solution was incubated for 20 min at 37 °C. The reaction was stopped by the addition of 20 μg of trypsin inhibitor and incubation for 10 min on ice, and then the samples were analyzed by immunoblotting.
Flotation assay
The sucrose flotation assay was adapted from Pernstich et al. (71). Proteoliposomes (5 μg of reconstituted proteins) in 150 μl of reconstitution buffer supplemented with 50% (w/v) sucrose were layered at the bottom of a 2-ml centrifugation tube (Beckman Optima MAX ultracentrifuge and TLS-55 rotor) and overlaid with 1.75 ml of reconstitution buffer supplemented with 40% (w/v) sucrose and 200 μl of reconstitution buffer. The tube was centrifuged for 12 h at 200,000 × g. Aliquots of 300 μl were recovered, and 30 μl of each aliquot were analyzed by immunoblotting. Proteoliposomes (0.5 μg of reconstituted proteins) were used as the input control.
ATPase activity
For solubilized NtPDR1, the ATPase assay was performed as described (33) in 50 mm Tris-HCl (pH 7.5), 50 mm NaCl, 60 μg/ml pyruvate kinase (Roche Applied Science), 32 μg/ml lactate dehydrogenase (Roche Applied Science), 0.1% (w/v) asolectin and DDM, 20 mm KNO3, 0.2 mm (NH4)2MoO4, 10 mm NaN3, 4 mm phosphoenol pyruvate, 0.4 mm NADH, 4 mm ATP, and 5 mm MgCl2 except when stated otherwise. The ATP hydrolytic activity of reconstituted NtPDR1 or NpPDR5 was assessed according to Galián et al. (60). The reaction mixture (100 μl) contained 20 mm Tris-HCl (pH 7.5), 50 mm NaCl, 60 μg/ml pyruvate kinase, 32 μg/ml lactate dehydrogenase, 4 mm phosphoenol pyruvate, 0.4 mm NADH, 20 mm KNO3, 0.2 mm (NH4)2MoO4, 10 mm NaN3, 4 mm ATP, and 5 mm MgCl2. The reaction was started by the addition of 40 μl of proteoliposomes (4 μg of proteins), and the absorbance at 340 nm was measured at 37 °C using a microplate reader (SPECTROstar Nano, BMG Labtech). All activity data represent the mean ± S.D. of at least three replicates
Electron microscopy and single-particle analysis
The purified proteins were diluted into a detergent-containing buffer (20 mm Tris (pH 7.5), 50 mm NaCl, and 0.03% MNG) at a concentration of 0.02 mg·ml−1. The sample was adsorbed on a carbon-coated glow-discharged EM grid (300 mesh; Electron Microscopy Science) for 30 s before being stained with a 2% uranyl formate solution. The data were collected using a LaB6 G2 Tecnai (FEI, Eindhoven, The Netherlands) electron microscope operated at an acceleration voltage of 200 kV. The images were acquired using a 4,000 × 4,000 CMOS F416 camera (Tietz Video and Image Processing Systems, Germany) in an automated manner using the EMTools software suite (Tietz Video and Image Processing Systems) with defocus varying from −1 to −3 μm. The images were collected at a magnification of 50,000, a dose of 10 electrons/Å2, and a pixel size of 2.13 Å.
From 28 images, 5,301 boxes of 135 × 135 pixels were manually windowed out using the boxer EMAN software suite (72). The picked particles were normalized against the background and appended into a single SPIDER file (73). A first round of reference-free alignment and classification was performed using SPIDER to obtain 40 unbiased classes. Those classes were then used as references for a multireference alignment followed by a classification that was iterated four times until no change was observed. Eventually 129 classes were obtained.
Glycosylation profiling
Purified NtPDR1 was reduced, alkylated, and purified as described (74). The protein was then resuspended in proteolysis buffer supplemented with 0.1% (w/w) Rapigest (Waters). Two proteolysis methods were performed: trypsin digestion as described previously (74) and multienzymatic (using a trypsin, chymotrypsin, and Glu-C mixture) limited proteolytic digestions (for 2 h at 37 °C). An aliquot of the resulting peptides was then subjected to deglycosylation at 37 °C for 24 h using peptide N-glycosidase A (0.2 milliunit/100 μg; Roche Applied Science) in a citrate/phosphate buffer adjusted to pH 5 and supplemented with 0.005% (w/w) Rapigest.
Peptides were then analyzed by LC-MS/MS. Briefly, peptides were separated by 1D reverse-phase chromatography (M-Class, HSS T3 C18 column, Waters) using a 70-min gradient (5–40% acetonitrile/water and 0.1% formic acid) at 600 nl/min. Eluted peptides were then analyzed by nanoelectrospray ionization-MS on a quadrupole Orbitrap hybrid mass spectrometer (Q Exactive Plus, Thermo Fischer Scientific) operated in positive mode. Survey scans were recorded at 70,000 resolving power (full width at half-maximum) from 400 to 1,750 m/z. MS/MS spectra were recorded using a “data-dependent analysis” scheme in which the 12 most abundant ions observed in MS mode were selected for fragmentation experimentation (normalized collision energy set at 28).
MS/MS spectra were searched for typical oxonium ions (m/z 204.09 and 366.14) indicating the presence of glycopeptides. For all candidates, the presence of the Y1 ion (fragment containing peptide + N-acetylhexosamine) was checked to confirm the glycosylation site. The composition of the assigned spectra was determined using Glycomod software (available at http://web.expasy.org/glycomod/)4 and manual interpretation of MS/MS spectra. Relative abundances were then determined by integrating MS1 signals using Skyline 3.0.1. Alternatively, MS/MS spectra were subjected to a database search using the Mascot 2.2 search engine and the TrEMBL database restricted to the Solanales taxon (downloaded on November 23, 2015).
GUS reporter activity
Leaf discs of N. tabacum plants expressing the GUS reporter gene under the control of the NtPDR1 promoter (25) were submerged in water or 250 μm methyl jasmonate for 24 h, and GUS reporter activity was measured as described (75).
Author contributions
M. B. conceived and coordinated the study. B. P. and F. T. performed most of the experiments. A. B. and D. L. performed the electron microscopy analysis. N. S. and E. D. P. performed the glycosylation analysis. All authors reviewed the results and wrote the paper.
Supplementary Material
Acknowledgment
We thank Joseph Nader for excellent technical help.
This work was supported by grants from the French National Research Agency through the “Investments for the Future” (France-BioImaging, Grant ANR-10-INSB-04), the Plateforme d'Imagerie Cellulaire et Tissulaire - Infrastructures Biologie Santé et Agronomie (PICT-IBiSA) (Institut Curie, Paris, France), the Belgian Fund for Scientific Research, and the Interuniversity Poles of Attraction Program (Belgian State, Scientific, Technical and Cultural Services). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S6.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- ABC
- ATP-binding cassette
- NBD
- nucleotide-binding domain
- TMD
- transmembrane domain
- DDM
- n-dodecyl β-d-maltoside
- PDR
- pleiotropic drug resistance
- MNG
- decyl maltose neopentyl glycol
- TEV
- tobacco etch virus
- Nt
- N. tabacum
- Np
- N. plumbaginifolia
- BY-2
- Bright Yellow-2
- Nb
- N. benthamiana
- Ni-NTA
- nickel-nitrilotriacetic acid
- GUS
- β-glucuronidase.
References
- 1. Higgins C. F. (2001) ABC transporters: physiology, structure and mechanism-an overview. Res. Microbiol. 152, 205–210 [DOI] [PubMed] [Google Scholar]
- 2. Bouige P., Laurent D., Piloyan L., and Dassa E. (2002) Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr. Protein Pept. Sci. 3, 541–559 [DOI] [PubMed] [Google Scholar]
- 3. Holland I. B. (2011) ABC transporters, mechanisms and biology: an overview. Essays Biochem. 50, 1–17 [DOI] [PubMed] [Google Scholar]
- 4. Higgins C. F., and Linton K. J. (2004) The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 11, 918–926 [DOI] [PubMed] [Google Scholar]
- 5. Sánchez-Fernández R., Davies T. G., Coleman J. O., and Rea P. A. (2001) The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 276, 30231–30244 [DOI] [PubMed] [Google Scholar]
- 6. International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436, 793–800 [DOI] [PubMed] [Google Scholar]
- 7. Verrier P. J., Bird D., Burla B., Dassa E., Forestier C., Geisler M., Klein M., Kolukisaoglu U., Lee Y., Martinoia E., Murphy A., Rea P. A., Samuels L., Schulz B., Spalding E. J., et al. (2008) Plant ABC proteins-a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159 [DOI] [PubMed] [Google Scholar]
- 8. Vasiliou V., Vasiliou K., and Nebert D. W. (2009) Human ATP-binding cassette (ABC) transporter family. Hum. Genomics 3, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hwang J. U., Song W. Y., Hong D., Ko D., Yamaoka Y., Jang S., Yim S., Lee E., Khare D., Kim K., Palmgren M., Yoon H. S., Martinoia E., and Lee Y. (2016) Plant ABC transporters enable many unique aspects of a terrestrial plant's lifestyle. Mol. Plant 9, 338–355 [DOI] [PubMed] [Google Scholar]
- 10. Rea P. A. (2007) Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol. 58, 347–375 [DOI] [PubMed] [Google Scholar]
- 11. Shoji T. (2014) ATP-binding cassette and multidrug and toxic compound extrusion transporters in plants. A common theme among diverse detoxification mechanisms. Int. Rev. Cell Mol. Biol. 309, 303–346 [DOI] [PubMed] [Google Scholar]
- 12. Crouzet J., Trombik T., Fraysse A. S., Boutry M. (2006) Organization and function of the plant pleiotropic drug resistance ABC transporter family. FEBS Lett. 580, 1123–1130 [DOI] [PubMed] [Google Scholar]
- 13. Kang J., Park J., Choi H., Burla B., Kretzschmar T., Lee Y., and Martinoia E. (2011) Plant ABC transporters. Arabidopsis Book 9, e0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yazaki K., Shitan N., Sugiyama A., and Takanashi K. (2009) Cell and molecular biology of ATP-binding cassette proteins in plants. Int. Rev. Cell Mol. Biol. 276, 263–299 [DOI] [PubMed] [Google Scholar]
- 15. Kretzschmar T., Burla B., Lee Y., Martinoia E., and Nagy R. (2011) Functions of ABC transporters in plants. Essays Biochem. 50, 145–160 [DOI] [PubMed] [Google Scholar]
- 16. Ruzicka K., Strader L. C., Bailly A., Yang H., Blakeslee J., Langowski L., Nejedlá E., Fujita H., Itoh H., Syono K., Hejátko J., Gray W. M., Martinoia E., Geisler M., Bartel B., et al. (2010) Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc. Natl. Acad. Sci. U.S.A. 107, 10749–10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ito H., and Gray W. M. (2006) A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 142, 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strader L. C., and Bartel B. (2009) The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21, 1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kang J., Hwang J.-U., Lee M., Kim Y.-Y., Assmann S. M., Martinoia E., and Lee Y. (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kretzschmar T., Kohlen W., Sasse J., Borghi L., Schlegel M., Bachelier J. B., Reinhardt D., Bours R., Bouwmeester H. J., and Martinoia E. (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483, 341–344 [DOI] [PubMed] [Google Scholar]
- 21. Kim D.-Y., Bovet L., Maeshima M., Martinoia E., and Lee Y. (2007) The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 50, 207–218 [DOI] [PubMed] [Google Scholar]
- 22. Lee M., Lee K., Lee J., Noh E. W., and Lee Y. (2005) AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol. 138, 827–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bessire M., Borel S., Fabre G., Carraça L., Efremova N., Yephremov A., Cao Y., Jetter R., Jacquat A.-C., Métraux J.-P., and Nawrath C. (2011) A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 23, 1958–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bultreys A., Trombik T., Drozak A., and Boutry M. (2009) Nicotiana plumbaginifolia plants silenced for the ATP-binding cassette transporter gene NpPDR1 show increased susceptibility to a group of fungal and oomycete pathogens. Mol. Plant Pathol. 10, 651–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crouzet J., Roland J., Peeters E., Trombik T., Ducos E., Nader J., and Boutry M. (2013) NtPDR1, a plasma membrane ABC transporter from Nicotiana tabacum, is involved in diterpene transport. Plant Mol. Biol. 82, 181–192 [DOI] [PubMed] [Google Scholar]
- 26. Banasiak J., Biała W., Staszków A., Swarcewicz B., Kȩpczyńska, Figlerowicz E. M., and Jasiński M. (2013) A Medicago truncatula ABC transporter belonging to subfamily G modulates the level of isoflavonoids. J. Exp. Bot. 64, 1005–1015 [DOI] [PubMed] [Google Scholar]
- 27. Bienert M. D., Siegmund S. E., Drozak A., Trombik T., Bultreys A., Baldwin I. T., and Boutry M. (2012) A pleiotropic drug resistance transporter in Nicotiana tabacum is involved in defense against the herbivore Manduca sexta. Plant J. 72, 745–757 [DOI] [PubMed] [Google Scholar]
- 28. Shibata Y., Ojika M., Sugiyama A., Yazaki K., Jones D. A., Kawakita K., and Takemoto D. (2016) The full-size ABCG transporters Nb-ABCG1 and Nb-ABCG2 function in pre- and post-invasion defense against Phytophthora infestans in Nicotiana benthamiana. Plant Cell 28, 1163–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sasse J., Schlegel M., Borghi L., Ullrich F., Lee M., Liu G. W., Giner J. L., Kayser O., Bigler L., Martinoia E., and Kretzschmar T. (2016) Petunia hybrida PDR2 is involved in herbivore defense by controlling steroidal contents in trichomes. Plant Cell Environ. 39, 2725–2739 [DOI] [PubMed] [Google Scholar]
- 30. Demessie Z., Woolfson K. N., Yu F., Qu Y., and De Luca V. (2017) The ATP binding cassette transporter, VmTPT2/VmABCG1, is involved in export of the monoterpenoid indole alkaloid, vincamine in Vinca minor leaves. Phytochemistry 140, 118–124 [DOI] [PubMed] [Google Scholar]
- 31. Yu F., and De Luca V. (2013) ATP-binding cassette transporter controls leaf surface secretion of anticancer drug components in Catharanthus roseus. Proc. Natl. Acad. Sci. U.S.A. 110, 15830–15835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fu X., Shi P., He Q., Shen Q., Tang Y., Pan Q., Ma Y., Yan T., Chen M., Hao X., Liu P., Li L., Wang Y., Sun X., and Tang K. (2017) AaPDR3, a PDR transporter 3, is involved in sesquiterpene β-caryophyllene transport in Artemisia annua. Front. Plant Sci. 8, 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Toussaint F., Pierman B., Bertin A., Lévy D., and Boutry M. (2017) Purification and biochemical characterization of NpABCG5/NpPDR5, a plant pleiotropic drug resistance transporter expressed in Nicotiana tabacum BY-2 suspension cells. Biochem. J. 474, 1689–1703 [DOI] [PubMed] [Google Scholar]
- 34. Nagata T., Nemoto Y., and Hasezawas S. (1992) Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int. Rev. Cytol. 132, 1–30 [Google Scholar]
- 35. Sasabe M., Toyoda K., Shiraishi T., Inagaki Y., and Ichinose Y. (2002) cDNA cloning and characterization of tobacco ABC transporter: NtPDR1 is a novel elicitor-responsive gene. FEBS Lett. 518, 164–168 [DOI] [PubMed] [Google Scholar]
- 36. Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227 [DOI] [PubMed] [Google Scholar]
- 37. Champagne A., and Boutry M. (2016) Proteomics of terpenoid biosynthesis and secretion in trichomes of higher plant species. Biochim. Biophys. Acta 1864, 1039–1049 [DOI] [PubMed] [Google Scholar]
- 38. De Muynck B., Navarre C., Nizet Y., Stadlmann J., and Boutry M. (2009) Different subcellular localization and glycosylation for a functional antibody expressed in Nicotiana tabacum plants and suspension cells. Transgenic Res. 18, 467–482 [DOI] [PubMed] [Google Scholar]
- 39. Nocarova E., and Fischer L. (2009) Cloning of transgenic tobacco BY-2 cells; an efficient method to analyse and reduce high natural heterogeneity of transgene expression. BMC Plant Biol. 9, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jeter C. R., Tang W., Henaff E., Butterfield T., and Roux S. J. (2004) Evidence of a novel cell signaling role for extracellular adenosine triphosphates and diphosphates in Arabidopsis. Plant Cell 16, 2652–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ernst R., Kueppers P., Klein C. M., Schwarzmueller T., Kuchler K., and Schmitt L. (2008) A mutation of the H-loop selectively affects rhodamine transport by the yeast multidrug ABC transporter Pdr5. Proc. Natl. Acad. Sci. U.S.A. 105, 5069–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burden R. S., Bailey J. A., and Vincent G. G. (1975) Glutinosone, a new antifungal sesquiterpene from Nicotiana glutinosa infected with tobacco mosaic virus. Phytochemistry 14, 221–223 [Google Scholar]
- 43. Cho M., Lee S. H., and Cho H. T. (2007) P-glycoprotein4 displays auxin efflux transporter like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19, 3930–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kubeš M., Yang H., Richter G. L., Cheng Y., Młodzińska E., Wang X., Blakeslee J. J., Carraro N., Petrášek J., Zažímalová E., Hoyerová K., Peer W. A., and Murphy A. S. (2012) The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J. 69, 640–654 [DOI] [PubMed] [Google Scholar]
- 45. Biała W., Banasiak J., Jarzyniak K., Pawela A., and Jasiński M. (2017) Medicago truncatula ABCG10 is a transporter of 4-coumarate and liquiritigenin in the medicarpin biosynthetic pathway. J. Exp. Bot. 68, 3231–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adebesin F., Widhalm J. R., Boachon B., Lefèvre F., Pierman B., Lynch J. H., Alam I., Junqueira B., Benke R., Ray S., Porter J. A., Yanagisawa M., Wetzstein H. Y., Morgan J. A., Boutry M., et al. (2017) Emission of volatile organic compounds from petunia flowers is facilitated by an ABC transporter. Science 356, 1386–1388 [DOI] [PubMed] [Google Scholar]
- 47. Laganowsky A., Reading E., Allison T. M., Ulmschneider M. B., Degiacomi M. T., Baldwin A. J., and Robinson C. V. (2014) Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klappe K., Hummel I., Hoekstra D., and Kok J. W. (2009) Lipid dependence of ABC transporter localization and function. Chem. Phys. Lipids 161, 57–64 [DOI] [PubMed] [Google Scholar]
- 49. Furt F., Simon-Plas F., and Mongrand S. (2010) Lipids of the plant plasma membrane. Plant Cell Monogr. 19, 3–30 [Google Scholar]
- 50. Cacas J.-L., Buré C., Grosjean K., Gerbeau-Pissot P., Lherminier J., Rombouts Y., Maes E., Bossard C., Gronnier J., Furt F., Fouillen L., Germain V., Bayer E., Cluzet S., Robert F., et al. (2016) Re-visiting plant plasma membrane lipids in tobacco: a focus on sphingolipids. Plant Physiol. 170, 367–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moremen K. W., Tiemeyer M., and Nairn A. V. (2012) Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Strasser R. (2016) Plant protein glycosylation. Glycobiology 26, 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miah M. F., Conseil G., and Cole S. P. (2016) N-Linked glycans do not affect plasma membrane localization of multidrug resistance protein 4 (MRP4) but selectively alter its prostaglandin E 2 transport activity. Biochem. Biophys. Res. Commun. 469, 954–959 [DOI] [PubMed] [Google Scholar]
- 54. Lee J.-Y., Kinch L. N., Borek D. M., Wang J., Wang J., Urbatsch I. L., Xie X.-S., Grishin N. V., Cohen J. C., Otwinowski Z., Hobbs H. H., and Rosenbaum D. M. (2016) Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature 533, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jasiński M., Stukkens Y., Degand H., Purnelle B., Marchand-Brynaert J., and Boutry M. (2001) A plant plasma membrane ATP binding cassette–type transporter is involved in antifungal terpenoid secretion. Plant Cell 13, 1095–1107 [PMC free article] [PubMed] [Google Scholar]
- 56. Tarling E. J., de Aguiar Vallim T. Q., and Edwards P. A. (2013) Role of ABC transporters in lipid transport and human disease. Trends Endocrinol. Metab. 24, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang B., Kashkooli A. B., Sallets A., Ting H. M., de Ruijter N. C. A., Olofsson L., Brodelius P., Pottier M., Boutry M., Bouwmeester H., and van der Krol A. R. (2016) Transient production of artemisinin in Nicotiana benthamiana is boosted by a specific lipid transfer protein from A. annua. Metab. Eng. 38, 159–169 [DOI] [PubMed] [Google Scholar]
- 58. Choi Y. E., Lim S., Kim H. J., Han J. Y., Lee M. H., Yang Y., Kim J. A., and Kim Y. S. (2012) Tobacco NtLTP1, a glandular-specific lipid transfer protein, is required for lipid secretion from glandular trichomes. Plant J. 70, 480–491 [DOI] [PubMed] [Google Scholar]
- 59. Herget M., Kreissig N., Kolbe C., Schölz C., Tampé R., and Abele R. (2009) Purification and reconstitution of the antigen transport complex TAP. A prerequisite determination of peptide stoichiometry and ATP hydrolysis. J. Biol. Chem. 284, 33740–33749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Galián C., Manon F., Dezi M., Torres C., Ebel C., Lévy D., and Jault J.-M. (2011) Optimized purification of a heterodimeric ABC transporter in a highly stable form amenable to 2-D crystallization. PLoS One 6, e19677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wagner G. J., Wang E., and Shepherd R. W. (2004) New approaches for studying and exploiting an old protuberance, the plant trichome. Ann. Bot. 93, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hammerschmidt R. (1999) Phytoalexins: what have we learned after 60 years? Annu. Rev. Phytopathol. 37, 285–306 [DOI] [PubMed] [Google Scholar]
- 63. Maldonado-Bonilla L. D., Betancourt-Jiménez M., and Lozoya-Gloria E. (2008) Local and systemic gene expression of sesquiterpene phytoalexin biosynthetic enzymes in plant leaves. Eur. J. Plant Pathol. 121, 439–449 [Google Scholar]
- 64. Wagner G. J. (1991) Secreting glandular trichomes: more than just hairs. Plant Physiol. 96, 675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lefèvre F., Baijot A., and Boutry M. (2015) Plant ABC transporters: time for biochemistry? Biochem. Soc. Trans. 43, 931–936 [DOI] [PubMed] [Google Scholar]
- 66. Goderis I. J., De Bolle M. F., François I. E., Wouters P. F., Broekaert W. F., and Cammue B. P. (2002) A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol. Biol. 50, 17–27 [DOI] [PubMed] [Google Scholar]
- 67. van der Fits L., Deakin E. A., Hoge J. H., and Memelink J. (2000) The ternary transformation system: constitutive virG on a compatible plasmid dramatically increases Agrobacterium-mediated plant transformation. Plant Mol. Biol. 43, 495–502 [DOI] [PubMed] [Google Scholar]
- 68. Morsomme P., Dambly S., Maudoux O., and Boutry M. (1998) Single point mutations distributed in 10 soluble and membrane regions of the Nicotiana plumbaginifolia plasma membrane PMA2 H+-ATPase activate the enzyme and modify the structure of the C-terminal region. J. Biol. Chem. 273, 34837–34842 [DOI] [PubMed] [Google Scholar]
- 69. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 70. Geertsma E. R., Nik Mahmood N. A., Schuurman-Wolters G. K., and Poolman B. (2008) Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc. 3, 256–266 [DOI] [PubMed] [Google Scholar]
- 71. Pernstich C., Senior L., MacInnes K. A., Forsaith M., and Curnow P. (2014) Expression, purification and reconstitution of the 4-hydroxybenzoate transporter PcaK from Acinetobacter sp. ADP1. Protein Expr. Purif. 101, 68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ludtke S. J., Baldwin P. R., and Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 73. Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., and Leith A. (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 74. Navarre C., Smargiasso N., Duvivier L., Nader J., Far J., De Pauw E., and Boutry M. (2017) N-Glycosylation of an IgG antibody secreted by Nicotiana tabacum BY-2 cells can be modulated through co-expression of human β-1,4-galactosyltransferase. Transgenic Res. 26, 375–384 [DOI] [PubMed] [Google Scholar]
- 75. Moriau L., Michelet B., Bogaerts P., Lambert L., Michel A., Oufattole M., and Boutry M. (1999) Expression analysis of two gene subfamilies encoding the plasma membrane H+-ATPase in Nicotiana plumbaginifolia reveals the major transport functions of this enzyme. Plant J. 19, 31–41 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.