Abstract
The mechanism of Nedd4-2 has been quantitatively explored for the first time using biochemically defined kinetic assays examining rates of 125I-polyubiquitin chain assembly as a functional readout. We demonstrate that Nedd4-2 exhibits broad specificity for E2 paralogs of the Ubc4/5 clade to assemble Lys63-linked polyubiquitin chains. Full-length Nedd4-2 catalyzes free 125I-polyubiquitin chain assembly by hyperbolic Michaelis–Menten kinetics with respect to Ubc5B∼ubiquitin thioester concentration (Km = 44 ± 6 nm; kcat = 0.020 ± 0.007 s−1) and substrate inhibition above 0.5 μm (Ki = 2.5 ± 1.3 μm) that tends to zero velocity, requiring ordered binding at two functionally distinct E2∼ubiquitin–binding sites. The Ubc5BC85A product analog non-competitively inhibits Nedd4-2 (Ki = 2.0 ± 0.5 μm), consistent with the presence of the second E2-binding site. In contrast, the isosteric Ubc5BC85S-ubiquitin oxyester substrate analog exhibits competitive inhibition at the high-affinity Site 1 (Ki = 720 ± 340 nm) and non-essential activation at the lower-affinity Site 2 (Kact = 750 ± 260 nm). Additional studies utilizing Ubc5BF62A, defective in binding the canonical E2 site, demonstrate that the cryptic Site 1 is associated with thioester formation, whereas binding at the canonical site (Site 2) is associated with polyubiquitin chain elongation. Finally, previously described Ca2+-dependent C2 domain-mediated autoinhibition of Nedd4-2 is not observed under our reported experimental conditions. These studies collectively demonstrate that Nedd4-2 catalyzes polyubiquitin chain assembly by an ordered two-step mechanism requiring two dynamically linked E2∼ubiquitin–binding sites analogous to that recently reported for E6AP, the founding member of the Hect ligase family.
Keywords: E3 ubiquitin ligase, enzyme kinetics, enzyme mechanism, polyubiquitin chain, protein-protein interaction, ubiquitin, ubiquitylation (ubiquitination), linkage specificity, thiol exchange
Introduction
The homologous to E6AP C terminus (Hect)3 family ubiquitin ligases play critical roles in a variety of fundamental cellular processes, the deregulation or ablation of which is implicated in numerous diseases (1), recently reviewed by Scheffner and Kumar (2). Modification of cellular proteins by ubiquitin occurs through a hierarchical three step pathway as follows: 1) Ubiquitin is activated by an E1 activating enzyme through the formation of a high energy thioester bond at its active-site cysteine in an ATP-coupled step. 2) The activated ubiquitin moiety is transferred to the active-site cysteine of an E2 carrier protein to form an E2∼ubiquitin4 thioester. 3) Transfer of the E2-bound ubiquitin to the target protein substrate is catalyzed by an E3 ubiquitin ligase (1, 3). The Hect family of E3 ligases is unique in requiring the formation of an additional high energy Hect∼ubiquitin thioester intermediate before conjugation of the target protein, distinguishing them from the larger superfamily of RING ligases (1).
In humans, the Hect ligases consist of 28 function-specific paralogs that share a highly conserved 350-residue C-terminal domain responsible for binding their cognate E2∼ubiquitin co-substrate, accepting the ubiquitin moiety from E2∼ubiquitin to form the Hect∼ubiquitin thioester intermediate, and subsequent conjugation of the correctly positioned target protein (1). The crystal structures of Hect domains for several family members have been resolved, demonstrating an L-shaped domain architecture further divided into distinct N-terminal and C-terminal subdomains attached by flexible linker peptides (4–9). The N-terminal subdomain can be further subdivided into large and small subdomains (4–9). Each paralog can be additionally segregated into distinct subfamilies based on shared protein- and membrane-interacting domains located at the N terminus of the protein, distal to the Hect catalytic module (1).
The human Nedd4 family consists of nine members, each containing an N-terminal C2 domain and 2–4 WW domains in addition to the conserved C-terminal Hect domain (1). The C2 domain is a Ca2+-dependent phospholipid-binding domain responsible for localizing the ligase to the inner cell membrane (10–16), whereas the WW domains play a critical role in substrate recruitment through targeting PY motifs on potential protein targets (1, 17–19). The Nedd4-2 ligase is best known for its role in targeting the PY motif-containing amiloride-sensitive epithelial sodium channel (ENaC) in the distal nephron for ubiquitin-mediated endocytic uptake and lysosomal degradation (18, 20–24). Additionally, Nedd4-2 and other Nedd4-family ligases play critical roles in vesicular trafficking and are exploited in viral budding of HIV, Ebola, and Marburg viruses, among others (1, 25, 26). Despite the relative importance of Hect ligases in human health and disease development, their mechanism(s) of action remains speculative.
The standard model of distal sequential addition for Hect ligase conjugation posits binding of the cognate E2∼ubiquitin to a single site at the small N-terminal subdomain of the Hect catalytic module, as suggested by the E6AP Hect domain crystal structure of Huang et al. (4, 27). According to this model, binding of the E2∼ubiquitin is followed by transthiolation to the Hect active-site cysteine to form a Hect∼ubiquitin thioester, which then is transferred to target protein ϵ-amino lysyl residues (4, 27). Repetition of this cycle in turn assembles a polyubiquitin signal by distal elongation from the anchored ubiquitin moiety (4, 5, 27). However, the ∼41-Å distance between the bound donor E2∼ubiquitin thioester and the Hect active-site cysteine nucleophile is inconsistent with the standard model, as has been noted (28). Kamadurai et al. (5) have attempted to resolve this paradox by solving the crystal structure of Nedd4-2 with Ubc5B∼ubiquitin bound at the small N-terminal subdomain predicted by the standard model but required mutagenesis to stabilize the presumptive Michaelis complex; however, the donor and acceptor sulfurs still remain separated by a distance of 8 Å. Marked apparent mobility in the C-terminal subdomain of many Hect ligases has suggested that the C-terminal lobe undergoes a conformational change during the catalytic cycle that allows the donor and acceptor sites to approach within atomic distance for transthiolation (4–9). More recently, our group has exploited biochemically defined kinetic studies to demonstrate that E6AP, the founding member of the Hect ligase family, instead harbors two functionally distinct E2∼ubiquitin–binding sites and that the canonical binding site identified by Huang et al. (4) is not the immediate donor of activated ubiquitin during thioester exchange, potentially resolving the topological paradox of the standard model (28). The latter studies propose a proximal indexation model in which the E2∼ubiquitin binds to a cryptic Site 1 before ubiquitin thioester exchange with the active-site cysteine of the Hect domain, followed by binding of a second E2∼ubiquitin at the canonical site (Site 2) responsible for processive chain elongation by way of proximal addition to a thioester-anchored polyubiquitin chain on the HECT domain (28–30).
Is the emerging two-site mechanism for E6AP unique to this paralog or shared among members of the otherwise markedly conserved Hect superfamily? To address this question, the following studies explore the mechanism of the paralogous Nedd4-2 Hect ligase through the use of similar biochemically defined kinetic assays analyzing the rates of free 125I-polyubiquitin chain assembly as a functional readout of ligase activity. Detailed kinetic analysis was used to define quantitatively the cognate E2 and chain linkage specificity for Nedd4-2. Additionally, we note that Nedd4-2 exhibits hyperbolic Michaelis–Menten kinetics, which show good agreement with kinetics reported previously for E6AP (28, 31). We demonstrate for the first time that Nedd4-2 exhibits two E2∼ubiquitin–binding sites and conjugates ubiquitin through an ordered two-step mechanism. Finally, we show that Nedd4-2–catalyzed free polyubiquitin chain assembly does not exhibit Ca2+-dependent C2 domain–mediated autoinhibition, as suggested previously (15, 16), but shows competitive inhibition by exogenous C2 domain, which tends to ∼60% limiting activity. Competitive inhibition suggests the location of the cryptic E2∼ubiquitin–binding Site 1 associated with Hect domain-linked thioester formation that is consistent with in silico modeling recently reported for E6AP (30).
Results
Nedd4-2 exhibits broad specificity for the Ubc4/5 E2 family in assembling free lysine 63–linked polyubiquitin chains
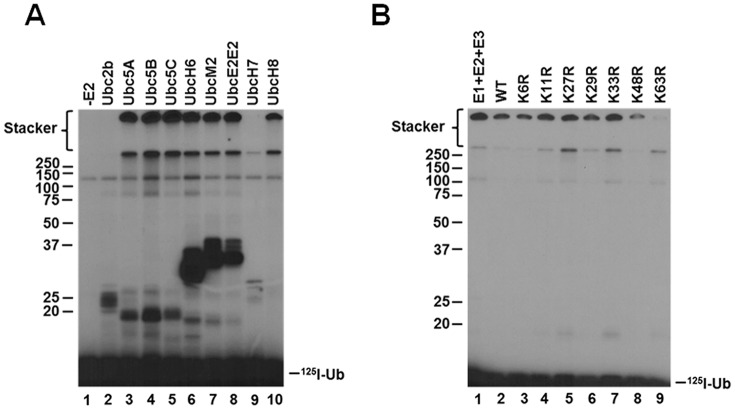
The specificity of ubiquitin ligases for distinct E2 carrier protein families serves a critical role in partitioning activated ubiquitin formed in the E1-catalyzed step to distinct conjugation pathways in the cellular signaling repertoire (31, 32). Typically, ligases are specific for a single or limited number of E2 paralogs (28, 31, 33–35); however, previous studies are ambiguous in defining the E2 specificity of Nedd4-2. Fotia et al. (36) suggested that Nedd4-2 activity is supported by E2 families of the Ubc4/5 clade, including Ubc5A, Ubc5B, UbcH6, UbcH7, and UbcH9, although the latter is recognized as a Sumo-specific E2 for signaling by this unrelated ubiquitin-like protein (37, 38). In contrast, other studies failed to demonstrate support of the Nedd4-2 catalytic cycle by members of the Ubc5 E2 family (39). As we have noted previously (31), identification of cognate E2 carrier protein(s) is complicated by imprecision in the actual concentration of active recombinant protein present in such assays and use of invalid assay conditions, among other factors. Identifying relevant target protein substrates for conjugation in such assays presents an additional barrier; however, we have shown that most ligases are capable of forming polyubiquitin chains in the absence of target protein and that the kinetics of this surrogate reaction recapitulate key properties of target protein conjugation (31). In Fig. 1A, we examined the E2 specificity of Nedd4-2-catalyzed polyubiquitin chain assembly in the absence of target protein substrate using a semiquantitative screen conducted under E3-limiting initial velocity conditions in the presence of a 200 nm5 concentration of the indicated E2 paralogs representing the major carrier protein families, as described previously (28, 31, 35). Active E2 protein concentrations were determined by stoichiometric formation of their respective E2∼125I-ubiquitin thioesters in parallel functional assays (31). Ligase-limiting conditions were confirmed by the independence of rate from Uba1 concentration so that the autoradiographic intensity is proportional to the initial rate for each E2 paralog (28, 31, 35).
Figure 1.
Nedd4-2 exhibits broad specificity for the Ubc4/5 E2 clade in assembling free lysine 63–linked polyubiquitin chains. A, kinetic assays of 125I-polyubiquitin chain assembly were carried out for 20 min at 37 °C containing 2 nm GST-Nedd4-2 in the absence (lane 1) or presence (lanes 2–10) of a 200 nm concentration of the indicated E2 carrier proteins. Reactions were quenched by the addition of SDS sample buffer containing 2% (v/v) β-mercaptoethanol. Reaction products were resolved by SDS-PAGE at 4 °C, and the resulting gel was dried and visualized by autoradiography as described under “Materials and methods.” B, 125I-polyubiquitin conjugation reactions containing 100 nm Uba1, 200 nm Ubc5B, 1 nm GST-Nedd4-2, and 5 μm 125I-ubiquitin (lane 1); 1 μm 125I-ubiquitin and 4 μm unlabeled wild-type ubiquitin (lane 2); or unlabeled single lysine-to-arginine ubiquitin point mutants (lanes 3–9) were carried out for 15 min at 37 °C. Reaction products were resolved by SDS-PAGE and visualized by autoradiography as described in A. The positions of mobility standards are shown to the left; the position for free 125I-ubiquitin is shown to the right.
In the absence of E2, the autoradiogram of Fig. 1A (lane 1) shows only an adduct band at 120 kDa representing the Uba1 monoubiquitin autoconjugate noted previously (28, 31, 35). However, in the presence of selected E2 paralogs, accumulation of high-molecular weight signal at the top of the stacker and resolving gels in the absence of protein target substrate is similar to that observed previously for the human TRIM25 and E6AP ligases and the Shigella flexneri IpaH9.8 bacterial ligase, whereas signal below ∼50 kDa represents E3-independent E2 adducts (28, 32, 35). The data of Fig. 1A indicate that Nedd4-2 polyubiquitin chain formation is robust and exhibits broad specificity for E2 paralogs of the Ubc4/5 clade (lanes 3–8 and 10) but not the more distant Ubc2/Rad6 family (lane 2). Under assay conditions in which autoradiographic intensity is proportional to the initial rate, Nedd4-2-catalyzed polyubiquitin chain assembly is observed with all Ubc4/5 family members except for UbcH7 (lane 9), contrary to Fotia et al. (36). Observation that UbcH8 (lane 10), the ubiquitin-like ISG15-specific E2 carrier protein, functions with Nedd4-2 is unexpected, especially because the closely related ubiquitin-specific UbcH7 paralog does not support the ligase.6
The ubiquitin-specific protease IsoT was employed to distinguish free from anchored forms of the polyubiquitin degradation signal (28, 31, 32, 35). Previous work demonstrates that IsoT disassembles unanchored polyubiquitin chains sequentially from the proximal free C-terminal end, whereas anchored chains are not affected (40). Reactions were carried out as described under “Materials and methods” and quenched by the addition of apyrase. Incubation of the Nedd4-2 reaction products with 34 μm recombinant IsoT resulted in loss of the high-molecular weight signal present in the stacker after a 40-min incubation (not shown), indicating that this autoradiographic signal represents free unanchored chains, as was observed previously for TRIM25, E6AP, and IpaH9.8 (28, 32, 35). In contrast, the autoradiographic signal at the top of the resolving gel was refractory to IsoT incubation, even after 40 min of incubation followed by the addition of a second aliquot of IsoT and incubation for an additional 40 min (not shown). The latter observation indicates that signal at the top of the resolving gel represents anchored 125I-polyubiquitin chains, presumably arising by Nedd4-2 autoubiquitination (28, 31, 35).
An additional layer of signaling diversity is provided by assembly of distinct polyubiquitin chains differing in their linkage specificity between the C-terminal glycine and any of the seven lysines present on the polypeptide or the N-terminal α-amino group (1, 41–44). Specific linkages result in packing of the polyubiquitin chains into distinct structures differentially recognized by dedicated ubiquitin-binding domains, expanding the overall versatility of the polyubiquitin signal (45–48). The Nedd4 ortholog Rsp5 in Saccharomyces cerevisiae assembles Lys63-linked polyubiquitin chains, the canonical signal for endocytosis, vesicular sorting, and degradation through the endosomal/lysosomal pathway, but is not recognized by the 26S proteasome (1, 42, 49–52). To unambiguously test the linkage specificity of Nedd4-2, we employed the isotope dilution/chain termination (ExTerm) assay of Edwards et al. (35) (Fig. 1B). The assay utilizes single lysine-to-arginine ubiquitin point mutants as chain terminators, analogous to 3′-deoxynucleotides of the Sanger sequencing method (53). The 125I-polyubiquitin conjugation reactions were carried out for 15 min in the presence of 5 μm 125I-ubiquitin (lane 1) or 1 μm 125I-ubiquitin and 4 μm unlabeled wild-type ubiquitin (lane 2) or the indicated unlabeled single lysine-to-arginine ubiquitin point mutants (lanes 3–9). Dilution of the radiolabeled polypeptide with unlabeled ubiquitin results in a proportional reduction in signal present in the unanchored chains (lane 2), as would also be expected with single lysine-to-arginine mutants not affecting chain elongation. Marked loss of the autoradiographic signal for the high-molecular weight free 125I-polyubiquitin chains upon the addition of the K63R mutant indicates that Nedd4-2 preferentially assembles Lys63-linked polyubiquitin chains (Fig. 1B, lane 9), consistent with earlier mass spectroscopic analyses by Maspero et al. (54). Slight reduction in the signal in lane 8 suggests that Nedd4-2 is capable of forming alternate linkages through Lys48 at a reduced rate (lane 8). The results of Fig. 1B demonstrate the linkage specificity of Nedd4-2 as being through Lys63 and recapitulate the linkage specificity observed for target protein ligation catalyzed by Nedd4-2 (54).
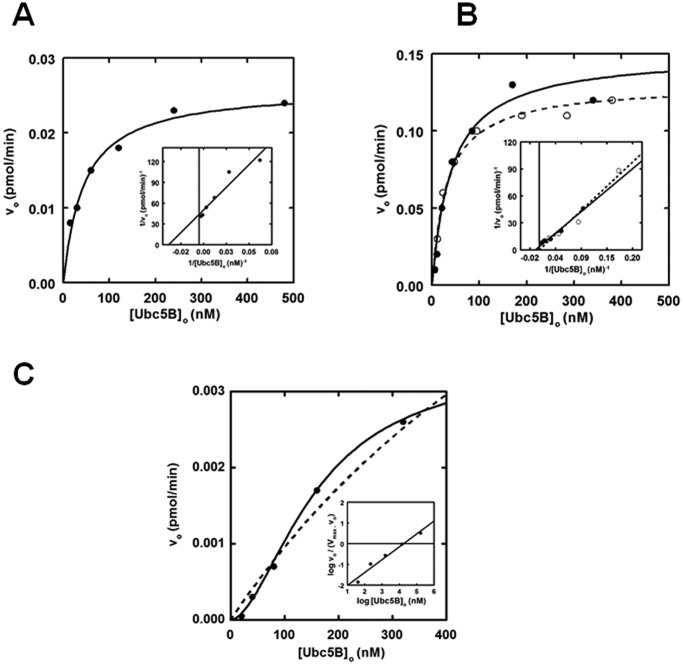
Nedd4-2 exhibits hyperbolic Michaelis–Menten kinetics
Previous studies demonstrate that kinetic analysis of initial rates for 125I-polyubiquitin chain assembly under E3-limiting conditions can serve as a facile reporter for characterizing E3 ligases (28, 31, 35). In the present study, we analyzed the dependence of [Ubc5B]o on the initial rates of GST-Nedd4-2-mediated 125I-polyubiquitin chain assembly following SDS-PAGE resolution of biochemically defined incubations. Chain assembly was quantified by excising the stacker region for each lane, representing free unanchored chains, followed by γ-counting to determine the associated radioactivity (31). From the adjusted specific activity of the 125I-ubiquitin, absolute rates of chain formation were calculated. The data demonstrate that full-length GST-Nedd4-2 exhibits hyperbolic kinetics with respect to [Ubc5B]o (Fig. 2A). Non-linear regression analysis of the data fit to the Michaelis–Menten equation results in a Km = 44 ± 6 nm and kcat = 0.020 ± 0.007 s−1, with kcat defined as Vmax/[E3]o (Table 1) (55). The observed Km and kcat values are comparable with the previously reported values of Km = 58 ± 6 nm and kcat = 0.031 ± 0.009 s−1 for E6AP-catalyzed polyubiquitin chain assembly supported by UbcH7 (28, 56).
Figure 2.
Nedd4-2 exhibits hyperbolic Michaelis–Menten kinetics. 15-min 125I-polyubiquitin conjugation reactions containing 1 nm wild-type or optimized GST-Nedd4-2 or 860 nm GST-Nedd4-2HECT were carried out at 37 °C in the presence of increasing Ubc5B concentrations. A, kinetic analysis of GST-Nedd4-2 under E3-limiting initial velocity conditions. The solid line represents the theoretical non-linear regression fit of the data for Km = 44 ± 6 nm and kcat = 0.020 ± 0.007 s−1. Inset, double reciprocal plot of the data in A. B, kinetic analysis of GST-opt.Nedd4-2 (closed circles) and opt.Nedd4-2 (open circles), as described for A. The solid and dashed lines correspond to the theoretical non-linear regression fit for GST-opt.Nedd4-2 (Km = 41 ± 10 nm; kcat = 0.1 ± 0.01 s−1) and opt.Nedd4-2 (Km = 32 ± 5 nm; kcat = 0.1 ± 0.01 s−1), respectively. The double reciprocal plot of the corresponding data in B is shown in the inset. C, kinetic analysis of GST-Nedd4-2HECT free 125I-polyubiquitin chain assembly. The solid line corresponds to the theoretical non-linear sigmoidal regression fit for [S]½ = 105 ± 20 nm and kcat = 1.9 ± 0.1 × 10−6 s−1. The corresponding Hill plot is shown in the inset. The dashed line corresponds to a hyperbolic non-linear regression fit for Km = 183 ± 123 nm and kcat = 2.5 ± 0.7 × 10−6 s−1. Statistical F-test analysis of the data fit to the Hill equation (solid line) compared with the Michaelis–Menten equation (dashed line) indicates that the data fit to the sigmoidal model is statistically significant (p = 0.034).
Table 1.
Summary of kinetic constants for full-length GST-Nedd4-2
| E2 | Km | kcat | kcat/Km |
|---|---|---|---|
| nm | s−1 | m−1 s−1 | |
| Ubc5B | 44 ± 6 | 2.0 ± 0.7 × 10−2 | 4.5 × 105 |
| UbcH7 | 310 ± 130 | 2.0 ± 0.2 × 10−3 | 6.5 × 103 |
| UbcH8 | 128 ± 43 | 6.0 ± 0.8 × 10−3 | 4.7 × 104 |
Interestingly, we observed subtle cooperative allosteric kinetics (nH = 1.9 ± 0.6) in several enzyme preparations (not shown). Statistical F-test analysis of the kinetic data fit to both the Hill equation and the Michaelis–Menten equations indicated a better fit to the former in several studies but often did not reach statistical significance (p ≤ 0.05). Currently, we do not completely understand why this behavior is observed in certain enzyme preparations and not others. The relatively low Hill coefficients suggest subtle conformational cross-talk within the full-length enzyme. We believe that the high resolution of our radioisotopic kinetic studies allows this behavior to be observed on occasion. Due to the inconsistent reproducibility of these results, we have chosen to analyze the data according to a hyperbolic Michaelis–Menten model for simplicity.
To determine whether the GST moiety had an effect on Nedd4-2 activity, we subjected the fusion protein to thrombin processing. We observed that full-length thrombin processed Nedd4-2 exhibited an electrophoretic mobility ∼48 kDa smaller than expected. Analysis of the amino acid sequence revealed a cryptic thrombin cleavage site at Arg425. To address this problem, the full-length enzyme was subcloned into a pGEX-6P1 expression vector, allowing GST processing with the Rhinovirus 3C PreScissionTM protease. Additionally, we codon-optimized the Nedd4-2 sequence to yield optimized Nedd4-2 (opt.Nedd4-2), which resulted in higher protein yields and reduced protein fragments during expression and purification. Analysis of GST-opt.Nedd4-2–catalyzed polyubiquitin chain assembly revealed a Km (41 ± 10 nm) comparable with that of wild-type GST-Nedd4-2 (44 ± 6 nm) for binding Ubc5B∼125I-ubiquitin and a 5-fold increase in kcat (0.10 ± 0.01 s−1), probably reflecting an increase in specific activity of the optimized protein (Fig. 2B). Processing the GST moiety did not affect the kinetics of opt.Nedd4-2 (Km = 32 ± 5 nm; kcat = 0.1 ± 0.01 s−1) (Fig. 2B); therefore, the GST fusion protein was retained in subsequent assays. Kinetic analysis of GST-Nedd4-2 with other Ubc4/5 clade family members demonstrates nearly identical Km and kcat values for each E2 carrier protein (not shown), consistent with the similar autoradiographic intensities shown in Fig. 1A. Thus, Nedd4-2 exhibits broad specificity for members of the Ubc4/5 clade, with the exception of UbcH7 and UbcH8. The reduced catalytic competence shown by the ISG15-specific UbcH8 results from a ∼3-fold lower kcat and a slightly increased Km (Table 1). Although UbcH7 appears inactive in supporting GST-Nedd4-2–catalyzed polyubiquitin chain formation in the semiquantitative assay of Fig. 1A, more rigorous kinetic analysis demonstrates that the significantly ablated activity arises from a 10-fold decrease in kcat and a reduced binding affinity for the Ubc5B∼125I-ubiquitin substrate, resulting in an overall 65-fold decrease in catalytic competence, defined as kcat/Km (Table 1).
Prior work exploring the mechanism of the Hect ligases, including Nedd4-2, has predominantly focused on examining the isolated Hect domain, as opposed to full-length enzyme (4, 5, 7–9, 16, 54, 57). Additionally, it has been reported that the Hect E3 ligases exist in an autoinhibited state through intramolecular interactions between the N terminus and the Hect domain by analysis of autoconjugation and/or Hect∼ubiquitin thioester formation (15, 16, 57, 58). To explore the role of the N terminus in Nedd4-2 activity, we truncated GST-Nedd4-2 at Arg597 to generate GST-Nedd4-2HECT (residues 597–955). Interestingly, kinetic analysis of GST-Nedd4-2HECT-catalyzed free 125I-polyubiquitin chain assembly revealed cooperative allosteric kinetics with a Hill coefficient (nH) of 2.6 ± 1, compared with hyperbolic kinetics for full-length enzyme (Fig. 2B). Cooperativity was verified to be statistically significant (p = 0.034) by an F-test analysis comparing the data fit to the Hill equation (χ2 = 5.5 × 10−9) and Michaelis–Menten equation (χ2 = 8.4 × 10−8) (Fig. 2B). Observation of cooperativity requires a priori that the enzyme function as an oligomer, consistent with our previous observations for E6AP (29). The inability to observe consistent statistically significant cooperativity with full-length enzyme compared with GST-Nedd4-2HECT is potentially the result of N-terminal stabilizing interactions dampening the conformational cross-talk within the enzyme. In addition to the assembly of free polyubiquitin chains, analysis of the truncated GST-Nedd4-2HECT mutant demonstrates monoubiquitin autoconjugates not observed for full-length enzyme. To maintain consistency when comparing the kinetics of full-length GST-Nedd4-2 with the truncated GST-Nedd4-2HECT mutant, we restricted our analysis to free polyubiquitin chains. Interestingly, GST-Nedd4-2HECT–catalyzed polyubiquitin chain assembly exhibits a ∼104-fold reduction in kcat (1.9 ± 0.2 × 10−6 s−1), contrary to previous reports of an N-terminal-mediated autoinhibition (15, 16, 57, 58). An [S]½ = 105 ± 20 nm was determined, in relatively good agreement with the Km observed for full-length Nedd4-2 (44 ± 6 nm), suggesting native folding of the truncation, although [S]½ and Km are not directly equatable (56).
Consistent with previous reports examining Hect∼ubiquitin thioester exchange reactions (8, 54), we are unable to demonstrate steady-state formation of full-length GST-Nedd4-2∼125I-ubiquitin thioester. Inability to detect the GST-Nedd4-2∼125I-ubiquitin thioester can result from a slow rate of formation compared with relatively more rapid rates for loss of this intermediate due to incorporation into polyubiquitin chains, transfer to water, and/or transfer to other nucleophiles, such as DTT (Fig. 3, lane 4). In contrast, we observe thioester formation with GST-Nedd4-2HECT, presumably due to the reduced kcat (Fig. 3, lane 5), suggesting that the rate of chain assembly is slower than that of 125I-ubiquitin thioester exchange. Additionally, these findings caution against the use of semiquantitative methods that examine thioester formation as a functional readout of ligase activity, an additional factor probably contributing to previous reports of N terminus–mediated autoinhibition of Hect ligase activity (57, 58). Interestingly, we noted that both full-length GST-Nedd4-2 and GST-Nedd4-2HECT assemble high-molecular weight polyubiquitin chains in 1-min end point reactions under non-reducing conditions, which migrate in the stacker and at the top of the resolving gel, respectively (Fig. 3, lanes 4 and 5). These latter products were eliminated in the presence of β-mercaptoethanol (Fig. 3, lanes 4 and 5 versus lanes 9 and 10), consistent with assembly of thioester-linked polyubiquitin chains on the Hect domain as has been observed for E6AP (30).
Figure 3.
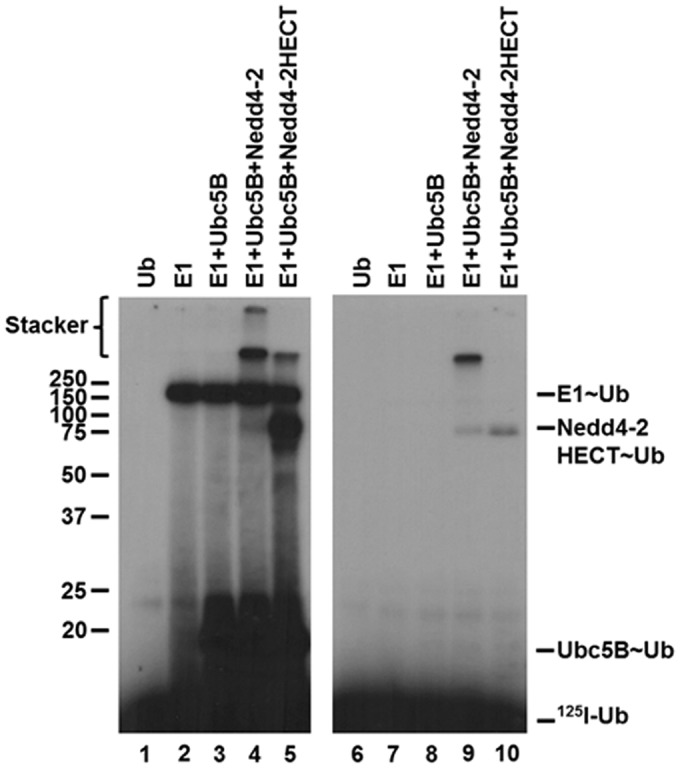
Nedd4-2 forms thioester-linked polyubiquitin chains. Left, autoradiogram of 1-min reactions containing 40 nm each of Uba1, Ubc5B, and GST-Nedd4-2 or GST-Nedd4-2HECT resolved under non-reducing conditions, as indicated under “Materials and methods.” Right, parallel 1-min reactions resolved under reducing conditions. The positions of mobility standards are shown to the left; the position for free 125I-ubiquitin is shown to the right.
The ability to observe steady-state GST-Nedd4-2HECT∼125I-ubiquitin thioester formation (Fig. 3) allowed us directly to analyze the kinetics of Ubc5B-dependent GST-Nedd4-2HECT transthiolation at 37°C, values for which are summarized in Table 2. Kinetic analysis reveals hyperbolic Michaelis–Menten kinetics (not shown) with a Km = 20 ± 5 nm, in good agreement with the Km (44 ± 6 nm) and [S]½ (105 ± 20 nm) observed for chain elongation by full-length and GST-Nedd4-2HECT, respectively. The kcat (3.5 ± 0.2 × 10−4 s−1) observed for GST-Nedd4-2HECT thioester transfer was found to be ∼200-fold faster than that determined for chain assembly (1.9 ± 0.2 × 10−6 s−1), providing additional evidence that chain elongation is rate-limiting in the ubiquitin conjugation reaction.
Table 2.
Summary of kinetic constants for GST-Nedd4-2HECT transthiolation
| E2 | Km | kcat |
|---|---|---|
| nm | s−1 | |
| Ubc5B | 20 ± 5 | 3.5 ± 0.2 × 10−4 |
| Ubc5BF62A | 33 ± 20 | 3.1 ± 0.7 × 10−5 |
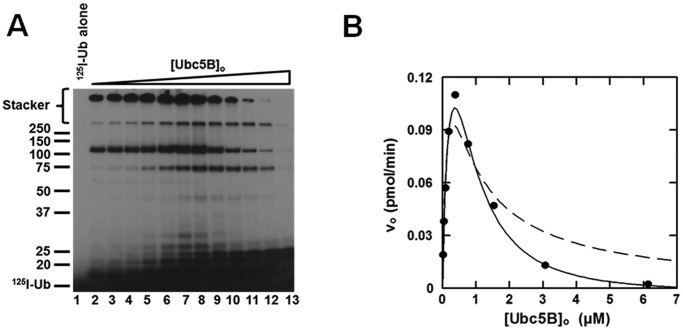
Nedd4-2 displays substrate inhibition
When the concentration dependence of [Ubc5B]o for full-length GST-Nedd4-2–catalyzed polyubiquitin chain assembly is extended above 500 nm, substrate inhibition is observed that tends to zero at infinite substrate concentration (Fig. 4). Similar kinetic behavior has been observed for the E6AP Hect ligase and the IpaH9.8 S. flexneri Hect-like bacterial ligase (28, 35). Substrate inhibition tending to zero velocity requires that the catalytic cycle for Nedd4-2 proceeds through ordered addition at two functionally distinct E2∼ubiquitin–binding sites having different affinities, described kinetically for Nedd4-2 by Equation 1 (28, 35).
| (Eq. 1) |
FIGURE 4.
Nedd4-2 displays substrate inhibition. A, autoradiogram of 15 min 125I-polyubiquitin conjugation reactions containing 1 nm GST-Nedd4-2 and increasing concentrations of Ubc5B (0–14 μm) as described under “Materials and methods.” B, concentration dependence of initial velocity of 125I-polyubiquitin chain assembly on [Ubc5B]o (0–6.1 μm) in a parallel assay. The solid line represents the theoretical non-linear regression fit to Equation 1 for K′1app = 280 ± 50 nm, kcat = 0.20 ± 0.03 s−1, nH = 1.6 ± 0.1, and K′2app = 900 ± 100 nm, where K′2app = [S]1/2nH. The dashed line corresponds to the data fit to Equation 1 excluding the Hill coefficient (nH).
Fundamentally, substrate inhibition is characterized by negative cooperativity in which disordered substrate binding at the apparent low-affinity Site 2 under saturating substrate concentrations competitively inhibits binding at the apparent high-affinity Site 1. Consistent with this mechanism, the data exhibit an excellent fit to the mathematical model for ordered substrate binding at two sites, from which the apparent Km for the leading substrate (K′1app) and trailing inhibitor (K′2app) binding sites were fit by non-linear regression analysis (Fig. 4B). The two sites may be analyzed in isolation by non-linear regression analysis of the ascending (Site 1) and descending (Site 2) segments of the curve independently, demonstrating a K′1 = 80 ± 40 nm and K′2 = 2.5 ± 1.3 μm, respectively. Contrastingly, Equation 1 models the complete system, illustrating the dynamic interplay between Site 1 and Site 2, from which the K′1app and K′2app were fit to values of 280 ± 50 and 900 ± 100 nm, respectively. In contrast to E6AP, the mathematical expression describing substrate inhibition for Nedd4-2 requires inclusion of a Hill coefficient (nH) for the terms representing Site 2, as displayed in the non-linear regression fit of the data in Fig. 4B. The Hill coefficient for Site 2 was fit to a value of 1.6 ± 0.1. Exclusion of the Hill coefficient yields a poor fit to the data for the inhibitory Site 2 (Fig. 4B). These findings demonstrate that the fundamental mechanism of Nedd4-2 is conserved with that of E6AP with respect to a two-site ordered kinetic model, as predicted from the marked conservation of the Hect domain within the superfamily (28, 35).
Nedd4-2 exhibits two E2∼ubiquitin thioester-binding sites
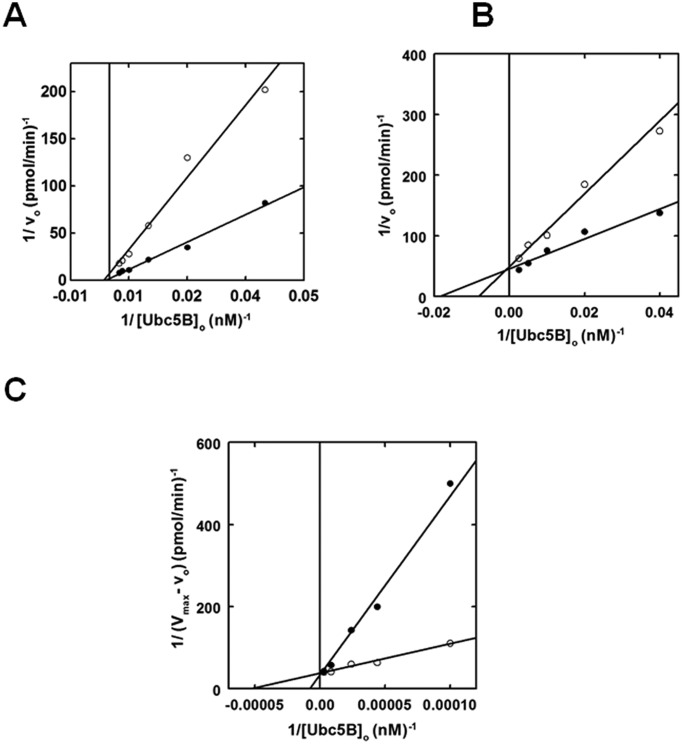
To further explore the catalytic mechanism of Nedd4-2, we expanded our kinetic studies to the effect(s) on polyubiquitin chain assembly of E2 substrate and product analogs (28, 35). According to the standard model in which the catalytic cycle is supported by binding of the E2∼ubiquitin to the single canonical site, the non-reactive Ubc5BC85A product analog should act as a competitive inhibitor of Nedd4-2-catalyzed 125I-polyubiquitin chain assembly. In contrast, at 2 μm, the Ubc5BC85A non-reactive product analog exhibited non-competitive inhibition with respect to wild-type Ubc5B∼125I-ubiquitin binding (Fig. 5A), a priori requiring binding to a second E2-binding site. The calculated Ki of 2.0 ± 0.5 μm for binding of the product analog is in excellent agreement with the K′2 of 2.5 ± 1.2 μm determined by non-linear regression analysis for the isolated Site 2 (Fig. 4B). That the product analog fails to show competitive inhibition suggests that Site 1 has a significantly reduced affinity for uncharged Ubc5B, in good agreement with our previous findings with E6AP and IpaH9.8 (28, 35).
Figure 5.
Nedd4-2 exhibits two E2∼ubiquitin thioester-binding sites. A, kinetic analysis of 1 nm GST-Nedd4-2 identical to that of Fig. 2 examining the rate of 125I-polyubiquitin chain assembly under E3-limiting initial velocity conditions as a function of [Ubc5B]o in the absence (closed circles) or presence (open circles) of 2 μm Ubc5BC85A. B, double reciprocal plot of initial rates of 125I-polyubiquitin chain assembly as a function of [Ubc5B]o as in A, in the absence (closed circles) or presence (open circles) of 550 nm Ubc5BC85S-125I-ubiquitin oxyester. C, double reciprocal plot demonstrating the change in initial velocity (Vmax − vo) as a function of [Ubc5B]o at substrate inhibition concentrations (0.6–4.8 μm) in the absence (closed circles) or presence (open circles) of 550 nm Ubc5BC85S-125I-ubiquitin oxyester.
Because the actual substrate for the Nedd4-2-catalyzed reaction is the charged Ubc5B∼ubiquitin species, we prepared a stable Ubc5BC85S-125I-ubiquitin oxyester substrate analog, as described previously (28, 33, 35). Rates of full-length GST-Nedd4-2-catalyzed polyubiquitin chain assembly were analyzed in the absence or presence of 550 nm Ubc5BC85S-125I-ubiquitin (Fig. 5B), ∼10 times the measured Km for Site 1 determined by non-linear regression analysis (Fig. 2A). The substrate analog exhibited competitive inhibition with respect to the wild-type Ubc5B∼125I-ubiquitin thioester, requiring binding to the same or overlapping binding sites (Fig. 5B). Rapid equilibrium kinetics requires that the intrinsic Ki for binding of the substrate analog to the isolated Site 1 should approximate the Km for binding of the Ubc5B∼ubiquitin thioester substrate. Interestingly, the Ki of 720 ± 340 nm observed was significantly larger than the Km for binding of the Ubc5B∼ubiquitin thioester (44 ± 6 nm), suggesting an apparent reduced affinity for Site 1. Because substrate inhibition (Fig. 4) requires binding of substrate to Site 2, this interaction effectively reduces the affinity for binding at Site 1 in an ordered binding mechanism. Based on this observation, we hypothesized that at 550 nm, the substrate analog additionally populates Site 2, resulting in an apparent Ki for binding in Fig. 5B that is greater than the intrinsic Km for Ubc5B∼125I-ubiquitin. To test this hypothesis, we examined the effect of the substrate analog on the Site 2-dependent inhibition segment of the curve (Fig. 5C). The Ubc5BC85S-125I-ubiquitin oxyester acted as a non-essential activator of the inhibitory Site 2 with a Kact of 750 ± 260 nm (Fig. 5C), satisfying the prediction. Overall, these data are consistent with Nedd4-2 harboring two functionally distinct E2∼ubiquitin–binding sites that catalyze polyubiquitin chain assembly through a dynamic ordered binding mechanism. By analogy to similar mechanisms for E6AP and the bacterial IpaH ligases, we propose that high-affinity Site 1 corresponds to the cryptic E2∼ubiquitin–binding site of E6AP that is associated with Hect∼ubiquitin thioester formation, whereas the lower-affinity Site 2 corresponds to the canonical E2∼ubiquitin–binding site within the small N-terminal subdomain demonstrated by Kamadurai et al. (5) that functions in processive chain elongation (5, 28, 29, 35).
Previous studies have shown that binding of E2 at the canonical site is critically dependent on a hydrophobic interaction with Phe62 in loop 4 of Ubc5B (4, 5, 59–61). According to the standard model, mutation of this residue should preclude binding of Ubc5B to the Nedd4-2 Hect domain, thereby abrogating thioester formation (4, 5, 59–61). To examine the role of the canonical E2-binding site on Nedd4-2-catalyzed polyubiquitin chain assembly, we mutated Phe62 of Ubc5B to alanine to form Ubc5BF62A. As shown in Fig. 6 (lanes 4 and 5), Ubc5BF62A fails to support 125I-polyubiquitin chain assembly compared with wild-type Ubc5B. The observed loss in activity was not due to the inability of Ubc5BF62A to be charged with 125I-ubiquitin by Uba1, as demonstrated by the non-reducing thioester assay in Fig. 6 (lanes 8 and 9). We next asked whether Ubc5BF62A retained the ability to support Nedd4-2–catalyzed 125I-ubiquitin thioester formation using the truncated GST-Nedd4-2HECT, because we are unable to observe thioester with full-length enzyme, as noted earlier. We demonstrated that the Ubc5BF62A mutant supported thioester formation (Fig. 7, lane 6), albeit at a 10-fold lower rate compared with wild-type Ubc5B (Fig. 7, lane 5). Kinetic analysis of initial rates of Ubc5BF62A-dependent transthiolation to GST-Nedd4-2HECT demonstrated hyperbolic kinetics (not shown) and a 10-fold reduction in kcat compared with wild-type Ubc5B, but with no effect on Km at the high-affinity Site 1 (Table 2), as predicted by earlier binding studies at the canonical site (59–61). These findings demonstrate that the canonical binding site (Site 2) requiring Phe62-mediated interaction is associated with polyubiquitin chain assembly but not thioester formation. We cannot currently account for the reduced kcat observed with Ubc5BF62A. Presumably, Phe62 is required for correct positioning of Ubc5B∼ubiquitin at the cryptic Site 1 during thioester exchange with Cys922 of Nedd4-2.
Figure 6.
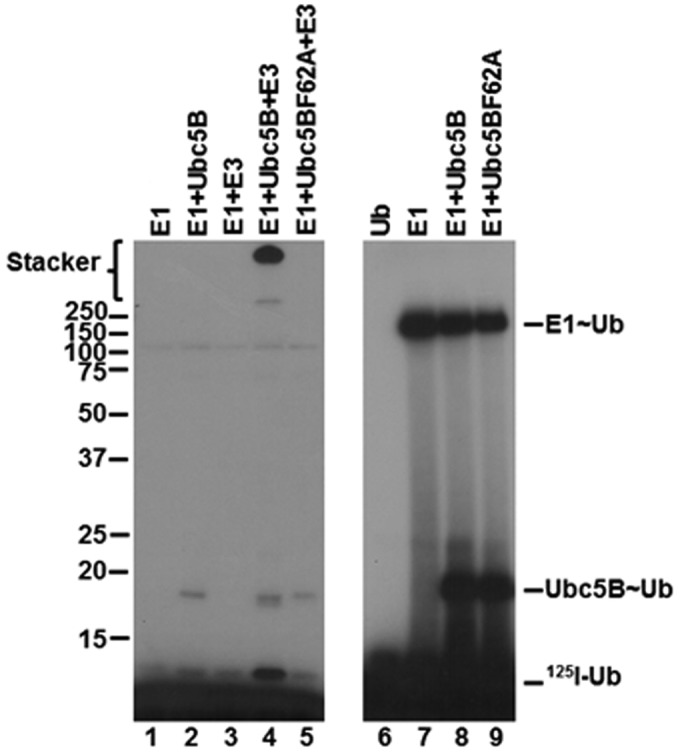
Ubc5BF62A does not support polyubiquitin chain assembly. Left, autoradiogram of 10-min 125I-polyubiquitin conjugation reactions containing 75 nm Uba1, 200 nm Ubc5B or Ubc5BF62A, and 1 nm GST-Nedd4-2 under reducing conditions (lanes 1–5) as described under “Materials and methods.” Right, autoradiogram demonstrating the stoichiometric formation of 125I-ubiquitin thioesters in 1-min reactions under non-reducing conditions containing 1 pmol (40 nm) of Uba1 and 1 pmol of Ubc5B or Ubc5BF62A, as described under “Materials and methods.” The positions of mobility standards are shown to the left; the position for free 125I-ubiquitin is shown to the right.
Figure 7.
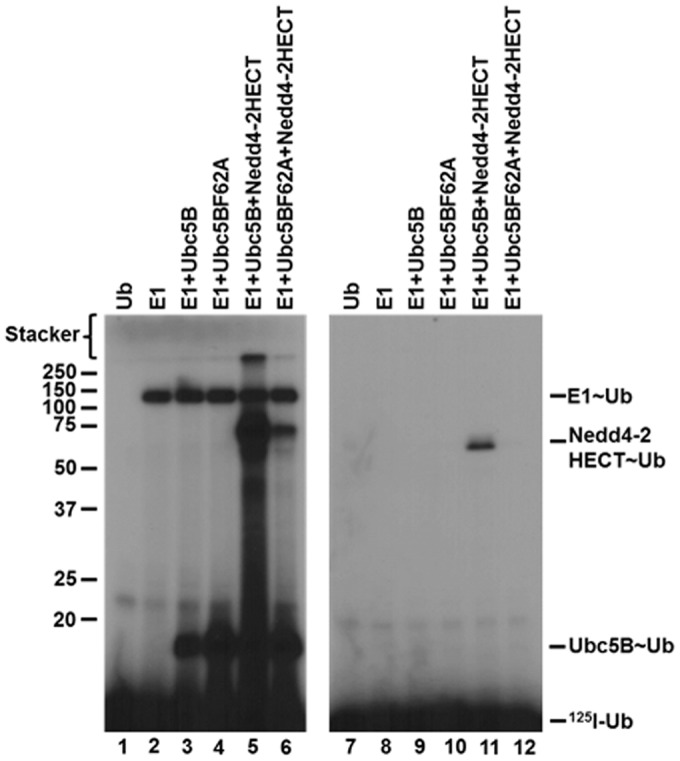
Ubc5BF62A supports GST-Nedd4-2HECT thioester formation. Left, autoradiogram demonstrating 1-min non-reducing reactions containing 1 pmol (40 nm) of E1, Ubc5B, or Ubc5BF62A and GST-Nedd4-2HECT. Right, parallel 1-min reactions conducted under reducing conditions as described under “Materials and methods.” The positions of mobility standards are shown to the left; the position for free 125I-ubiquitin is shown to the right.
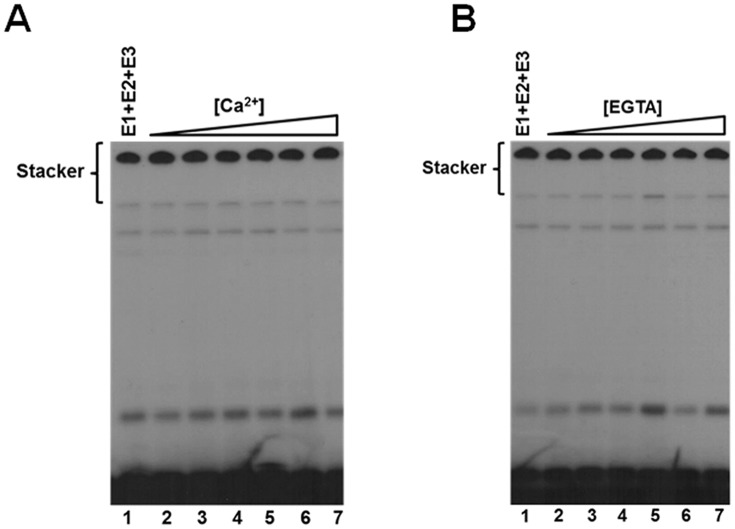
Ca2+ concentration does not affect polyubiquitin chain assembly
Several reports suggest that a hallmark of Nedd4-2 and other Nedd4-family Hect ligases is their existence in an autoinhibited state as a mechanism for self-regulation (15, 16, 57, 58). Specifically, Nedd4-2 is suggested to exhibit Ca2+-dependent C2 domain-mediated autoinhibition through intramolecular interactions between the C2 domain and the HECT domain (15, 16). In our hands, kinetic analysis under E3-limiting initial velocity conditions demonstrated no effect on activity over a physiologically relevant Ca2+ concentration range up to 100 μm (Fig. 8A). Conversely, to ensure that our observed activity of Nedd4-2 was not due to saturating trace Ca2+ contamination in our buffers, we examined the dependence of polyubiquitin chain assembly on EGTA concentration from 1 nm to 100 μm, for which no effect was observed (Fig. 8B). These findings demonstrate that Nedd4-2–catalyzed 125I-polyubiquitin chain assembly does not exhibit Ca2+-dependent activation under the present assay conditions, contrary to previous reports (15, 16).
Figure 8.
Ca2+ does not affect Nedd4-2-catalyzed polyubiquitin chain assembly. A, autoradiogram of 15-min 125I-polyubiquitin chain formation assays containing 100 nm Uba1, 200 nm Ubc5B, 2 nm GST-Nedd4-2, and increasing Ca2+ concentrations (1 nm to 100 μm), as described under “Materials and methods.” B, autoradiogram of 125I-polyubiquitin conjugation assays as described in A (lane 1) in the presence of increasing EGTA concentrations (1 nm to 100 μm).
Truncation of the C2 domain does not affect polyubiquitin chain assembly
The absence of a Ca2+-dependent effect on Nedd4-2 125I-polyubiquitin chain assembly questions a direct role for the C2 domain in regulating the activity of the Nedd4 ligases. Prior structural studies examined direct interaction of the isolated C2 domain with the isolated catalytic domains of Nedd4-2 and other Hect ligases (15, 16, 57, 58). To test such an interaction catalytically, we truncated the N-terminal C2 domain (residues 1–154) from opt.Nedd4-2 to yield opt.ΔC2Nedd4-2, as performed previously (16). Kinetic analysis of opt.ΔC2Nedd4-2–catalyzed polyubiquitin chain assembly demonstrated hyperbolic kinetics (not shown) with a Km = 18 ± 5 nm and kcat = 0.5 ± 0.04 s−1, exhibiting a modest effect on activity compared with full-length ligase (Km = 32 ± 5 nm; kcat = 0.1 ± 0.01 s−1). Because we cannot obtain a precise measure for catalytically active Nedd4-2 protein due to our inability to detect steady-state Hect∼125I-ubiquitin thioester for all but the isolated Hect domain (Fig. 3), the 5-fold difference in kcat for opt.ΔC2Nedd4-2 could be attributed to variability in specific activity between the two preparations. A similar explanation may account for the modest increase in activity previously claimed for comparable C2 domain truncations (15, 57, 58).
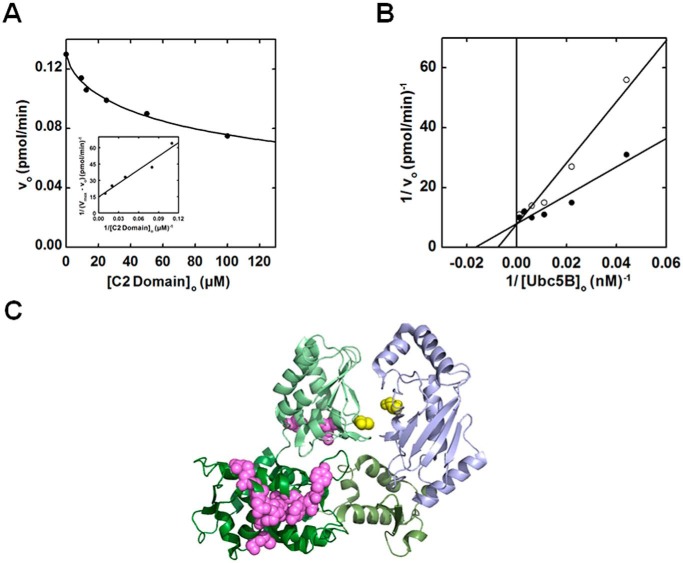
Exogenous C2 domain competitively inhibits Nedd4-2-catalyzed polyubiquitin chain formation
Previous structural studies have reported interactions between the C2 domain and isolated Nedd4-1, Nedd4-2, and Smurf2 Hect domains, suggesting a potential mechanism of physiological inhibition (15, 16, 57). The marginal effect of C2 domain truncation on the rate of polyubiquitin chain assembly by opt. ΔC2Nedd4-2 prompted us to resolve this question for Nedd4-2. We analyzed the dependence of full-length Nedd4-2–catalyzed 125I-polyubiquitin chain assembly on increasing exogenous C2 domain (residues 1–154) concentrations (Fig. 9A). Interestingly, exogenous C2 domain inhibited 125I-polyubiquitin chain assembly with a hyperbolic concentration dependence (Ki = 30 ± 8 μm) tending to ∼60% limiting activity at saturation (Fig. 9A). Parallel experiments between 10 and 100 μm C2 domain confirmed the 60% plateau effect (not shown). We analyzed the dependence of Nedd4-2–catalyzed 125I-polyubiquitin chain assembly on [Ubc5B]o in the absence or presence of near saturating exogenous C2 domain (70 μm) (Fig. 9B). Kinetic analysis revealed that exogenous C2 domain competitively inhibited full-length Nedd4-2–catalyzed 125I-polyubiquitin chain assembly (Fig. 9B). Competitive inhibition requires that the C2 domain bind to the same or overlapping binding site as the Ubc5B∼ubiquitin substrate. The previously reported site for the C2 domain binding identified by NMR is located along the large N-terminal subdomain of the Nedd4-2 Hect domain (Fig. 9C), well removed from the canonical E2∼ubiquitin–binding site within the small N-terminal subdomain reported by Kamadurai et al. (5, 16, 57, 58). Whereas most of the C2-interacting residues identified by Mari et al. (57) map to a single surface on the large N-terminal subdomain, several C2-interacting residues were identified along the “back side” of the Hect C-terminal subdomain, contrary to the expectation of a contiguous binding surface (Fig. 9C). The C-terminal subdomain of Nedd4-2 is rotated relative to that of E6AP (4); however, rotation of the C-terminal subdomain to superimpose on that of E6AP aligns all of the interacting residues to form a contiguous binding region. The observation of competitive inhibition by exogenous C2 domain (Fig. 9B) in context of the previous NMR data is consistent with our finding that Nedd4-2 harbors two E2∼ubiquitin–binding sites (57) while additionally suggesting the potential location of the cryptic E2∼ubiquitin–binding site (Site 1) associated with thioester exchange, as modeled in silico for E6AP (30).
Figure 9.
Exogenous C2 domain competitively inhibits Nedd4-2 polyubiquitination in trans. A, kinetic analysis of rates of 125I-polyubiquitin conjugation under E3-limiting initial velocity conditions in reactions containing 70 nm Uba1, 100 nm Ubc5B, 1 nm opt.Nedd4-2, and increasing exogenous C2 domain concentrations (10–100 μm). B, double reciprocal plot of initial rates of 125I-polyubiquitin chain assembly as a function of [Ubc5B]o (0–500 nm) in the absence (closed circles) or presence (open circles) of 70 μm exogenous C2 domain. C, crystal structure of Nedd4-2HECT (green) in complex with Ubc5B (blue) (Protein Data Bank code 3JW0) reported by Kamadurai et al. (5). The Nedd4-2HECT subdomains are highlighted in different shades of green. The active-site cysteines of Nedd4-2 (Cys922) and Ubc5B (Cys85) are highlighted in yellow spheres. The paralogous C2 domain binding residues to the residues identified for Nedd4-1 and Smurf2 by Mari et al. (57) are highlighted as magenta spheres.
Discussion
The Hect superfamily of ubiquitin ligases share a highly conserved (67% identity) C-terminal catalytic domain responsible for assembly of the polyubiquitin degradation signals appended to target proteins (1). Their marked sequence and structural conservation suggest a common mechanism of conjugation across the entire family. Recently, our group demonstrated that E6AP, the prototypical member of the Hect ligase superfamily, catalyzes polyubiquitin chain assembly on its active-site cysteine through a two-site proximal indexation mechanism that presumably precedes stochastic transfer of the preassembled chain en bloc to a target substrate or competing nucleophile, such as water or DTT (28–30). That an analogous mechanism is observed for the otherwise unrelated IpaH family of bacterial ubiquitin ligases implies convergent evolution and evidence that proximal indexation must be a highly efficient solution to the topological problem of chain assembly on an otherwise anchored target protein (35). These findings suggest that the remaining members of the Hect ligase family probably possess a similar mechanism but perhaps with subtle nuances arising from sequence differences and functional demands. To begin to explore this hypothesis, we have employed for the first time a comprehensive kinetic analysis of the Nedd4-family Hect ligase Nedd4-2.
Unambiguously defining the E2 substrate specificity of ubiquitin ligases poses a significant technical challenge (31). When assayed under semiquantitative biochemically defined conditions, Nedd4-2 shows broad specificity for E2 paralogs of the Ubc4/5 clade, explicating the seemingly contradictory observations reported previously (36, 39) (Fig. 1A). Kinetic characterization of the functional paralogs shows good agreement among their Km and kcat values and with that observed for E6AP, suggesting a common binding mechanism and transition state for E2-Hect thiol exchange (28, 31). Typically, ubiquitin ligases exhibit narrow E2 specificity, especially among RING ligases for which direct interaction of the E3-bound E2∼ubiquitin thioester with the target protein is required for conjugation. However, transfer from a Hect∼ubiquitin intermediate obviates such target-E2 interactions, precluding an overt requirement for E2 specificity. Therefore, restricted E2 specificity must serve regulatory functions. Broad E2 specificity potentially promotes a steady cellular supply of E2∼ubiquitin substrate for Nedd4-2, consistent with its critical role in regulating a wide array of cellular processes (1). This model suggests the possibility that two E2 paralogs of similar affinity could support polyubiquitin chain assembly by acting at Site 1 and/or Site 2 in concert. Additional experiments are required to explore this hypothesis but remain outside the scope of this paper. Evidence that Nedd4-2 exhibits a functional bias toward the more recently evolved ISG15-specific UbcH8 rather than its closest ubiquitin-specific UbcH7 paralog (Fig. 1A and Table 1) suggests that Nedd4-2 may additionally function with ISG15, potentially accounting for previous reports of ISGylation-dependent Nedd4-family inactivation (62, 63).
Kinetic analysis of Nedd4-2–catalyzed 125I-polyubiquitin chain assembly reveals hyperbolic Michaelis–Menten kinetics (Fig. 2A), demonstrating a Km and kcat in good agreement with our previous observations for E6AP (28). Additionally, Nedd4-2 demonstrates substrate inhibition at E2∼ubiquitin concentrations greater that 500 nm that tends to zero at infinite substrate concentrations (Fig. 4). These results require Nedd4-2 to harbor two functionally distinct E2∼ubiquitin–binding sites of different affinities (28). The standard model with a single canonical binding site for Ubc5B∼ubiquitin requires that the Ubc5BC85A product analog act as a competitive inhibitor; in contrast, we demonstrate that Ubc5BC85A non-competitively inhibits Nedd4-2 with a Ki = 2.0 ± 0.5 μm (Fig. 5A), similar to the observed K′2 = 2.5 ± 1.3 μm for the isolated inhibitory Site 2 observed by substrate inhibition (Fig. 4B). The Ubc5BC85S-125I-ubiquitin substrate analog competitively inhibits Nedd4-2 (Ki = 720 ± 340 nm), requiring its binding to Site 1 responsible for E2-Hect thioester exchange (Fig. 5B). Accounting for the otherwise unexpected increase in apparent Ki (720 ± 340 nm) for binding of the substrate analog to Site 1 compared with the Km for Ubc5B∼125I-ubiquitin (44 ± 6 nm), we demonstrate that Ubc5BC85S-125I-ubiquitin at 550 nm serves as a non-essential activator of the inhibitory Site 2 (Fig. 5C), effectively reducing the affinity for substrate binding at Site 1. The observed non-essential activation at Site 2 by Ubc5BC85S-125I-ubiquitin and non-competitive inhibition by the Ubc5BC85A product analog suggest that the ubiquitin moiety probably contributes to the competitive inhibition at Site 1 when E2∼ubiquitin is bound at Site 2. In the absence of the ubiquitin moiety, inhibition is probably driven by allosteric conformational changes. In this scenario, two potential regulatory strategies are employed: ordered binding driven by dynamic changes in affinity between Sites 1 and 2 required for assembly of the polyubiquitin chain and product inhibition at Site 2 in a negative feedback loop.
Earlier structural studies demonstrate that Phe62 of Ubc5B is critical for binding at the canonical E2-binding site in the small N-terminal subdomain (4, 5). Work by Nuber and Scheffner (59) shows that introduction of a Phe residue in the paralogous position of specific non-cognate E2 chimeras is sufficient to support thioester formation and conjugation. Additionally, fluorescence polarization studies conducted by Eletr and Kuhlman (61) demonstrate that mutation of the paralogous phenylalanine in UbcH7 (Phe63) reduces binding of free UbcH7 to the E6AP Hect domain at the canonical low-affinity binding site (28). Whereas these studies highlight the importance of Phe62 in binding of E2∼ubiquitin at the canonical site to support Hect ligase activity, they fail to dissect its role mechanistically, resolving thioester exchange from conjugation (5, 59). Through the use of quantitative kinetic methods, we demonstrate for the first time the actual role of the canonical E2-binding site in the two-step kinetic mechanism. Our results show that mutation of Phe62 of Ubc5B inhibits polyubiquitin chain assembly (Fig. 6, lanes 4 and 5) but does not prevent thioester exchange (Fig. 7, lane 6). Kinetic analysis of Ubc5BF62A-catalyzed transthiolation reveals that mutation of Phe62 has no effect on binding at the high-affinity Site 1 (Table 2), consistent with conclusions from the substrate and product analog kinetics (Fig. 5, A and B). These findings require thioester formation to occur through binding of the E2∼ubiquitin to a cryptic Site 1 not represented in the structural data, whereas polyubiquitin chain elongation is supported by binding at the canonical E2-binding site (Site 2). The use of non-cognate E2 mutants and uncharged E2s that selectively bind Site 2 (Fig. 5A) probably obscured these observations in previous studies (59, 61). Interestingly, we also observe a 10-fold reduction in kcat (Table 2) for Ubc5BF62A-catalyzed thioester exchange, suggesting a potential role of Phe62 in stabilizing transition state geometry at the cryptic Site 1 during transthiolation.
The N-terminal C2 domain has been reported to inhibit several Nedd4-family ligases through intramolecular interactions with the catalytic Hect domain (15, 16, 57, 58). This phenomenon was first reported by Wiesner et al. (58) for Smurf2, Nedd4-1, and WWP2. Later, Nedd4-2 was reported to exhibit a similar C2 domain–mediated autoinhibition (15). In contrast to Smurf2, Nedd4-2 autoinhibition was reported to exhibit Ca2+ dependence (15, 16). Interestingly, a number of closely related C2 domain-containing Hect ligase family members, including Rsp5, Itch/AIP4, and Smurf1, were not found to exhibit this autoinhibitory behavior (58, 64). These observations illustrate the complexity of Hect ligase regulation, along with the limited understanding of the actual mechanism of action. Our results demonstrate that the rate of free 125I-polyubiquitin chain assembly under rigorous E3-limiting initial velocity conditions does not exhibit a Ca2+ dependence (Fig. 8), contrary to previous reports (15, 16). We observed that truncation of the C2 domain (residues 1–154) of opt.Nedd4-2 to form opt.ΔC2 Nedd4-2 leads to a modest 5-fold increase in the rate of 125I-polyubiquitin chain assembly, in good agreement with previous observations by Wang et al. (15) but well within the expected variability in Nedd4-2 specific activity among different enzyme preparations, especially in the absence of a stoichiometric Hect∼125I-ubiquitin thioester assay for full-length protein.
We demonstrate that exogenous C2 domain competitively inhibits (Ki = 30 ± 8 μm) Nedd4-2–catalyzed 125I-polyubiquitin chain assembly to ∼60% limiting activity (Fig. 9, A and B). Our observed Ki for C2 domain binding to full-length Nedd4-2 (Fig. 9A) is lower than the previously reported Kd = 200 μm for C2 domain binding to the isolated Nedd4 Hect domain (57). This significant discrepancy in Ki values could reflect different affinities for binding of the C2 domain to full-length enzyme versus the isolated Hect domain. This observation suggests N-terminal contributions to the C2 domain binding surface or binding to an alternative site. Additionally, differences in the quality of the C2 domain preparations (i.e. specific activity) may account for the observed disparity in apparent binding affinity. Interpretation is further complicated when considering the marginal increase in kcat observed by truncation of the C2 domain compared with the effect on Km on the addition of exogenous C2 domain (Fig. 9B), suggesting two independent mechanisms affecting activity. Finally, the possibility that the effect of exogenous C2 domain represents an artifact cannot be excluded based on interactions with the isolated Hect domain at non-physiological protein concentrations (15, 16, 57, 58).
Observation of competitive inhibition of full-length Nedd4-2 by the C2 domain is inconsistent with the canonical E2∼ubiquitin–binding site serving as the donor of activated ubiquitin in Nedd4-2∼ubiquitin thioester formation (57, 58). The residues involved in C2 domain binding extend along the large N-terminal subdomain into the C-terminal lobe containing the active-site cysteine (57), Fig. 9C. Curiously, Mari et al. (57) noted that the candidate C2 domain binding residues located in the C-lobe of Nedd4-1 were not in proximity to the binding residues identified along the N-terminal subdomain, as we have illustrated for Nedd4-2 (Fig. 9C). When the C2 domain-binding residues located in the C-terminal lobe were modeled to form a continuous surface with the N-terminal subdomain residues, the catalytic cysteine adopts a conformation resembling E6AP, in which the catalytic cysteine is oriented away from the canonical E2∼ubiquitin–binding site (4, 57). This finding was interpreted as a potential regulatory strategy in which the C2 domain functions by restricting the orientation of the C-terminal lobe (57). Interestingly, the latter E6AP-like orientation is also observed for Smurf2 (57). This model predicts the C2 domain to exhibit non-competitive inhibition by conformationally altering the orientation of the active enzyme. In contrast, observation of competitive inhibition refutes the model in favor of the C2 domain binding to the same or an overlapping site with the E2∼ubiquitin substrate. The latter conclusion is consistent with our observation that Nedd4-2 contains two distinct E2∼ubiquitin–binding sites and that the cryptic Site 1 with which the C2 domain competes is paralogous with that identified by in silico modeling of the E6AP Site 1 (30).
The present studies represent the first rigorous kinetic analysis of the mechanism of the Nedd4-2 Hect ubiquitin ligase. Our findings demonstrate an intricate mechanism for free polyubiquitin chain assembly orchestrated through dynamic interactions between two ordered E2∼ubiquitin–binding sites of different affinities and suggest conservation of this catalytic cycle among members of the Hect ligase superfamily.
Materials and methods
Bovine ubiquitin and creatine phosphokinase were purchased from Sigma; apyrase was purchased from New England Biolabs. The ubiquitin was purified to apparent homogeneity and radioiodinated by Chloramine-T using carrier-free Na125I obtained from PerkinElmer Life Sciences (65). Human erythrocyte Uba1 was purified from outdated human blood and quantified by the stoichiometric formation of 125I-ubiquitin thioester (66, 67). Human recombinant Ubc2b (UBE2B), Ubc5A (UBE2D1), Ubc5B (UBE2D2), Ubc5C (UBE2D3), UbcH6 (UBE2E1), UbE2E2 (UBE2E2), UbcM2 (UBE2E3), UbcH7 (UBE2L3), and UbcH8 (UBE2L6) were those used previously (68). Active E2 proteins were determined by their stoichiometric formation of 125I-ubiquitin thioester (31). All E2 proteins were stored at −80 °C in small aliquots to minimize loss of activity with repeated freeze-thaw cycles (31, 33).
Generation and purification of Ubc5B point mutants
Recombinant Ubc5BC85A, Ubc5BC85S, and Ubc5BF62A point mutants were generated from pGEX4T1-HsUbc5B as described previously, and their coding regions were directly sequenced to obviate cloning artifacts and to confirm the correct point mutation (33). The recombinant mutant proteins were expressed in Escherichia coli BL21 (DE3) cells harboring the desired pGEX-E2 plasmid, purified by glutathione-Sepharose affinity chromatography, treated with 50 units/ml thrombin (GE Life Sciences) for 6 h at room temperature to remove the GST tag, and then purified to apparent homogeneity as described previously (68). Active Ubc5BC85S and Ubc5BF62A protein concentrations were quantified by the Uba1-dependent stoichiometric formation of 125I-ubiquitin oxyester/thioester, respectively, and compared with total E2 protein as determined spectrophotometrically using their calculated 280-nm extinction coefficients (31, 33). Because active Ubc5BC85A could not be similarly determined by stoichiometric 125I-ubiquitin adduct formation, native protein was estimated from total protein determined spectrophotometrically at 280 nm and the fraction of active protein determined in parallel for wild-type Ubc5B (33).
Generation and purification of wild-type and mutant Nedd4-2
Human Nedd4-2 (isoform 3- NM015277.5; generous gift of Dr. Daniela Rotin, The Hospital for Sick Children and Departments of Biochemistry and Molecular Genetics, University of Toronto) was subcloned into a pDEST15 expression vector to yield pDEST15-Nedd4-2 and then expressed in E. coli BL21 AI cells (Invitrogen). opt.Nedd4-2 was obtained from GenScript, subcloned into a pGEX6P1 GST expression vector, and sequenced to validate the absence of cloning artifacts. Recombinant opt.ΔC2Nedd4-2 (residues 155–955) was generated by PCR amplification from pGEX4T1-opt.Nedd4-2 and subcloned into a pGEX6P1 expression vector. Optimized Nedd4-2 and opt.ΔC2Nedd4-2 were expressed in E. coli BL21 (DE3) cells (Invitrogen). Recombinant Nedd4-2HECT (residues 597–955) was subcloned from full-length pGEX4T1-Nedd4-2 and was expressed from a pGEX4T1 vector in E. coli BL21 (DE3) cells (Invitrogen). The exogenous C2 domain (residues 1–154) truncation mutant was generated from pGEX4T1-Nedd4-2 by QuikChange (Agilent) site-directed mutagenesis, sequenced to verify the correct mutation and absence of cloning artifacts, and expressed from a pGEX4T1 vector in E. coli BL21 (DE3) cells (Invitrogen). Cultures were grown at 37 °C to an A600 of 0.4. The temperature was reduced to 15 °C, and cultures were grown until an A600 of 0.6, after which protein expression was induced by adjusting cultures to 0.2% (w/v) l-arabinose (BL21 AI cells) or 0.4 mm isopropyl 1-thio-β-d-galactopyranoside (BL21 DE3 cells). After 20 h at 15 °C (A600 ∼2.5), cells were harvested by centrifugation at 6,000 × g for 30 min at 4 °C and then resuspended in ice-cold 50 mm Tris-HCl (pH 7.5) containing 150 mm NaCl and 1 mm DTT. Cells were lysed by Emulsiflex (Avestin) and then centrifuged at 100,000 × g for 30 min at 4 °C. Recombinant GST-Nedd4-2 was purified by glutathione-Sepharose affinity chromatography from the resulting supernatant (33). Recombinant GST-Nedd4-2 was used directly or processed with 50 units/ml PreScission ProteaseTM (GE Healthcare) for 6 h at 4 °C and further purified on a second glutathione-Sepharose column to remove free GST and GST-protease. Exogenous GST-C2 domain was processed with 50 units/ml thrombin (GE Life Sciences) for 6 h at room temperature followed by passing over a second glutathione-Sepharose column to remove free GST. Total protein content of the apparently homogeneous preparations was determined spectrophotometrically from the theoretical 280-nm extinction coefficient, with typical yields of ∼5 mg/liter of medium. Protein was flash-frozen and stored at −80 °C in small aliquots to avoid activity loss due to repeated freeze-thaw cycles.
Nedd4-2-catalyzed 125I-ubiquitin conjugation assay
The ligase activities of recombinant Nedd4-2 proteins were quantified in biochemically defined kinetic assays of 125I-polyubiquitin chain formation under E3-limiting initial velocity conditions. Reactions were carried out at 37 °C in incubations of 25-μl final volume containing 50 mm Tris-HCl (pH 7.5), 1 mm ATP, 10 mm MgCl2, 1 mm DTT, 10 mm creatine phosphate, 1 IU of creatine phosphokinase, 4 μm 125I-ubiquitin, 100 nm human Uba1, the indicated concentrations of E2 protein, and 1 nm Nedd4-2 protein or the indicated concentration of the respective mutant (31, 33, 69). Reactions were initiated by the addition of 125I-ubiquitin. After 10–20 min, reactions were quenched by the addition of 25 μl of 2× SDS sample buffer containing 4.0% (v/v) β-mercaptoethanol followed by incubation at 100 °C for 2 min. Samples were resolved by 12% (w/v) SDS-PAGE under reducing conditions at 4 °C and visualized by autoradiography of the dried gels (31, 33, 70). Unanchored polyubiquitin chain formation was measured by excising polyubiquitin conjugates in the stacker and quantifying 125I-ubiquitin by γ-counting. Rates were determined using the corrected specific radioactivity of the 125I-ubiquitin (31, 33). Kinetic data were calculated by non-linear regression analysis using GraFit version 5.0 (Erithacus Software Ltd.). Because we cannot demonstrate steady-state Nedd4-2∼125I-ubiquitin thioester formation for full-length protein, total protein was used to estimate concentration for kinetic analysis of all Nedd4-2 constructs, providing consistency when comparing kinetic data.
Ubc5B-catalyzed GST-Nedd4-2HECT 125I-ubiquitin transthiolation assay
The reduced kcat for GST-Nedd4-2HECT allowed for direct observation of steady-state GST-Nedd4-2HECT∼125I-ubiquitin thioester formation. Ubc5B-catalyzed GST-Nedd4-2HECT thioester exchange was analyzed by kinetic assays of 125I-ubiquitin thioester formation under biochemically defined E3-limiting initial velocity conditions. 25-μl reactions containing 50 mm Tris-HCl (pH 7.5), 1 mm ATP, 10 mm MgCl2, 1 mm DTT, 10 mm creatine phosphate, 1 IU of creatine phosphokinase, 4 μm 125I-ubiquitin, 40 nm human Uba1, increasing concentrations of Ubc5B or Ubc5BF62A (0–400 nm), and 10 μm GST-Nedd4-2HECT (total protein) were carried out at 37 °C for 10 s. Reactions were quenched by the addition of 25 μl of 2× SDS sample buffer and resolved by SDS-PAGE under non-reducing conditions at 4 °C. Gels were dried and visualized by autoradiography. Rates of GST-Nedd4-2HECT∼ubiquitin thioester formation were measured by excising the respective band in the resolving gel, quantifying 125I-ubiquitin by γ-counting, and correcting for the specific activity of the 125I-ubiquitin, as indicated above. Kinetic data were determined by non-linear regression analysis using GraFit version 5.0 (Erithacus Software). Total GST-Nedd4-2HECT protein was used to maintain consistency with full-length protein assays.
Structural modeling
Structures were visualized using the PyMOL Molecular Graphics System version 1.8 (Schrödinger, LLC).
Author contributions
D. R. T., A. C. A.-W., and A. L. H. conceived and coordinated the studies, and the first two performed the experiments. J. M. K. performed the study outlined in Fig. 1A, along with cloning and expression of proteins and preliminary experiments leading to the study.
Acknowledgments
We thank Dr. Daniela Rotin for generously providing the Nedd4-2 (isoform 3) clone.
This work was supported by National Institutes of Health Grant GM034009 (to A. L. H.) and an appropriation from the Louisiana State University School of Medicine (New Orleans, LA). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The symbol ∼ denotes a high-energy thioester bond.
We chose a uniform E2 concentration of 200 nm for the screen assays because this approximates typical Km values noted previously for various ligases, obviating the potential to artifactually force an otherwise unfavorable reaction at abnormally high E2 concentration (31).
Although UbcH8 is an ISG15-specific E2 carrier protein, it can be forced to support the Uba1 reaction and be charged with 125I-ubiquitin at sufficient Uba1 concentration (28).
- Hect
- homologous to E6AP C terminus
- opt.Nedd4-2
- optimized Nedd4-2
- Ub
- ubiquitin
- E1
- generic name for activating enzymes of class 1 ubiquitin-like proteins
- E2 or Ubc
- generic name for ubiquitin carrier protein/ubiquitin-conjugating enzyme
- E6AP
- E6-associated protein (gene name UBE3A)
- Nedd4-1
- neural precursor cell expressed developmentally down-regulated 4–1 (gene name NEDD4)
- Nedd4-2
- neural precursor cell expressed developmentally down-regulated 4-2 (gene name NEDD4L)
- Rsp5
- Nedd4-family E3 ubiquitin-protein ligase in S. cerevisiae (gene name RSP5)
- Smurf2
- SMAD-specific E3 ubiquitin protein ligase 2 (gene name SMURF2)
- Uba1
- ubiquitin-activating enzyme (gene name UBE1)
- Ubc2b
- B isoform of the human/rabbit ortholog of S. cerevisiae Rad6/Ubc2 (gene name UBE2B)
- Ubc5A
- A isoform of the human ortholog of the S. cerevisiae Ubc5 (gene name UBE2D1)
- Ubc5B
- B isoform of the human ortholog of the S. cerevisiae Ubc5 (gene name UBE2D2)
- Ubc5C
- C isoform of the human ortholog of the S. cerevisiae Ubc5 (gene name UBE2D3)
- UbcE2E2
- human E2 carrier protein (gene name UBE2E2)
- UbcH6
- human E2 carrier protein (gene name UBE2E1)
- UbcH7
- human E2 carrier protein (gene name UBE2L3)
- UbcH8
- human E2 carrier protein (gene name UBE2L6)
- UbcM2
- human E2 carrier protein (gene name UBE2E3)
- WWP-2
- WW domain containing E3 ubiquitin protein lipase 2 (gene name WWP2).
References
- 1. Rotin D., and Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 2. Scheffner M., and Kumar S. (2014) Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochim. Biophys. Acta 1843, 61–74 [DOI] [PubMed] [Google Scholar]
- 3. Pickart C. M. (2001) Mechanism underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 4. Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., and Pavletich N. P. (1999) Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286, 1321–1326 [DOI] [PubMed] [Google Scholar]
- 5. Kamadurai H. B., Souphron J., Scott D. C., Duda D. M., Miller D. J., Stringer D., Piper R. C., and Schulman B. A. (2009) Insights into ubiquitin transfer cascades from a structure of a UbcH5B∼ubiquitin-HECT(NEDD4L) complex. Mol. Cell 36, 1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., and Wrana J. L. (2005) Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol. Cell 19, 297–308 [DOI] [PubMed] [Google Scholar]
- 7. Verdecia M. A., Joazeiro C. A., Wells N. J., Ferrer J. L., Bowman M. E., Hunter T., and Noel J. P. (2003) Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol. Cell 11, 249–259 [DOI] [PubMed] [Google Scholar]
- 8. Maspero E., Mari S., Valentini E., Musacchio A., Fish A., Pasqualato S., and Polo S. (2011) Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 12, 342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gong W., Zhang X., Zhang W., Li J., and Li Z. (2015) Structure of the HECT domain of human WWP2. Acta Crystallogr. F Struct. Biol. Commun. 71, 1251–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rizo J., and Südhof T. C. (1998) C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 273, 15879–15882 [DOI] [PubMed] [Google Scholar]
- 11. Cho W., and Stahelin R. V. (2006) Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta 1761, 838–849 [DOI] [PubMed] [Google Scholar]
- 12. Plant P. J., Yeger H., Staub O., Howard P., and Rotin D. (1997) The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+-dependent plasma membrane localization. J. Biol. Chem. 272, 32329–32336 [DOI] [PubMed] [Google Scholar]
- 13. Garrone N. F., Blazer-Yost B. L., Weiss R. B., Lalouel J. M., and Rohrwasser A. (2009) A human polymorphism affects NEDD4L subcellular targeting by leading to two isoforms that contain or lack a C2 domain. BMC Cell Biol. 10, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunn R., Klos D. A., Adler A. S., and Hicke L. (2004) The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J. Cell Biol. 165, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J., Peng Q., Lin Q., Childress C., Carey D., and Yang W. (2010) Calcium activates Nedd4 E3 ubiquitin ligases by releasing the C2 domain-mediated auto-inhibition. J. Biol. Chem. 285, 12279–12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Escobedo A., Gomes T., Aragón E., Martín-Malpartida P., Ruiz L., and Macias M. J. (2014) Structural basis of the activation and degradation mechanisms of the E3 ubiquitin ligase Nedd4L. Structure 22, 1446–1457 [DOI] [PubMed] [Google Scholar]
- 17. Sudol M., Chen H. I., Bougeret C., Einbond A., and Bork P. (1995) Characterization of a novel protein-binding module—the WW domain. FEBS Lett. 369, 67–71 [DOI] [PubMed] [Google Scholar]
- 18. Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., and Rotin D. (1996) WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15, 2371–2380 [PMC free article] [PubMed] [Google Scholar]
- 19. Kanelis V., Rotin D., and Forman-Kay J. D. (2001) Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat. Struct. Biol. 8, 407–412 [DOI] [PubMed] [Google Scholar]
- 20. Goel P., Manning J. A., and Kumar S. (2015) NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene 557, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou R., Patel S. V., and Snyder P. M. (2007) Nedd4-2 catalyzes ubiquitination and degradation of cell surface ENaC. J. Biol. Chem. 282, 20207–20212 [DOI] [PubMed] [Google Scholar]
- 22. Harvey K. F., Dinudom A., Cook D. I., and Kumar S. (2001) The Nedd4-like protein KIAA0439 is a potential regulator of the epithelial sodium channel. J. Biol. Chem. 276, 8597–8601 [DOI] [PubMed] [Google Scholar]
- 23. Snyder P. M., Steines J. C., and Olson D. R. (2004) Relative contribution of Nedd4 and Nedd4-2 to ENaC regulation in epithelia determined by RNA interference. J. Biol. Chem. 279, 5042–5046 [DOI] [PubMed] [Google Scholar]
- 24. Kabra R., Knight K. K., Zhou R., and Snyder P. M. (2008) Nedd4-2 induces endocytosis and degradation of proteolytically cleaved epithelial Na+ channels. J. Biol. Chem. 283, 6033–6039 [DOI] [PubMed] [Google Scholar]
- 25. Morita E., and Sundquist W. I. (2004) Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20, 395–425 [DOI] [PubMed] [Google Scholar]
- 26. Chung H. Y., Morita E., von Schwedler U., Müller B., Kräusslich H. G., and Sundquist W. I. (2008) NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J. Virol. 82, 4884–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hochstrasser M. (2006) Lingering mysteries of ubiquitin-chain assembly. Cell 124, 27–34 [DOI] [PubMed] [Google Scholar]
- 28. Ronchi V. P., Klein J. M., and Haas A. L. (2013) E6AP/UBE3A ubiquitin ligase harbors two E2∼ubiquitin binding sites. J. Biol. Chem. 288, 10349–10360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ronchi V. P., Klein J. M., Edwards D. J., and Haas A. L. (2014) The active form of E6-associated protein (E6AP)/UBE3A ubiquitin ligase is an oligomer. J. Biol. Chem. 289, 1033–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ronchi V. P., Summa C. M., Kim E. D., Klein J. M., and Haas A. L. (September 18, 2017) In silico modeling of the cryptic E2∼ubiquitin–binding site of E6-associated protein (E6AP)/UBE3A reveals the mechanism of polyubiquitin chain assembly. J. Biol. Chem. 10.1074/jbc.M117.813477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ronchi V. P., and Haas A. L. (2012) Measuring rates of ubiquitin chain formation as a functional readout of ligase activity. Methods Mol. Biol. 832, 197–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Streich F. C. Jr., and Haas A. L. (2010) Activation of ubiquitin and ubiquitin-like proteins. Subcell. Biochem. 54, 1–16 [DOI] [PubMed] [Google Scholar]
- 33. Siepmann T. J., Bohnsack R. N., Tokgöz Z., Baboshina O. V., and Haas A. L. (2003) Protein interactions within the N-end rule ubiquitin ligation pathway. J. Biol. Chem. 278, 9448–9457 [DOI] [PubMed] [Google Scholar]
- 34. Streich F. C. Jr., Ronchi V. P., Connick J. P., and Haas A. L. (2013) TRIM ligases catalyze polyubiquitin chain formation through a cooperative allosteric mechanism. J. Biol. Chem. 288, 8209–8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edwards D. J., Streich F. C. Jr., Ronchi V. P., Todaro D. R., and Haas A. L. (2014) Convergent evolution in the assembly of polyubiquitin degradation signals by the Shigella flexneri IpaH9.8 ligase. J. Biol. Chem. 289, 34114–34128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fotia A. B., Cook D. I., and Kumar S. (2006) The ubiquitin-protein ligases Nedd4 and Nedd4-2 show similar ubiquitin-conjugating enzyme specificities. Int. J. Biochem. Cell Biol. 38, 472–479 [DOI] [PubMed] [Google Scholar]
- 37. Johnson E. S., and Blobel G. (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272, 26799–26802 [DOI] [PubMed] [Google Scholar]
- 38. Schwarz S. E., Matuschewski K., Liakopoulos D., Scheffner M., and Jentsch S. (1998) The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. U.S.A. 95, 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Debonneville C., and Staub O. (2004) Participation of the ubiquitin-conjugating enzyme UBE2E3 in Nedd4-2-dependent regulation of the epithelial Na+ channel. Mol. Cell Biol. 24, 2397–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilkinson K. D., Tashayev V. L., O'Connor L. B., Larsen C. N., Kasperek E., and Pickart C. M. (1995) Metabolism of the polyubiquitin degradation signal- Structure, mechanism, and role of isopeptidase T. Biochemistry 34, 14535–14546 [DOI] [PubMed] [Google Scholar]
- 41. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 42. Erpapazoglou Z., Walker O., and Haguenauer-Tsapis R. (2014) Versatile roles of K63-linked ubiquitin chains in trafficking. Cells 3, 1027–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim H. C., and Huibregtse J. M. (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell Biol. 29, 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ikeda F., and Dikic I. (2008) Atypical ubiquitin chains: new molecular signals. EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dikic I., Wakatsuki S., and Walters K. J. (2009) Ubiquitin-binding domains: from structures to functions. Nat. Rev. Mol. Cell Biol. 10, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hurley J. H., Lee S., and Prag G. (2006) Ubiquitin-binding domains. Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Winget J. M., and Mayor T. (2010) The diversity of ubiquitin recognition: hot spots and varied specificity. Mol. Cell 38, 627–635 [DOI] [PubMed] [Google Scholar]
- 48. Perica T., and Chothia C. (2010) Ubiquitin-molecular mechanisms for recognition of different structures. Curr. Opin. Struct. Biol. 20, 367–376 [DOI] [PubMed] [Google Scholar]
- 49. Duncan L. M., Piper S., Dodd R. B., Saville M. K., Sanderson C. M., Luzio J. P., and Lehner P. J. (2006) Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 25, 1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rotin D., Staub O., and Haguenauer-Tsapis R. (2000) Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176, 1–17 [DOI] [PubMed] [Google Scholar]
- 51. Hicke L., and Riezman H. (1996) Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84, 277–287 [DOI] [PubMed] [Google Scholar]
- 52. Hicke L., and Dunn R. (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 53. Sanger F., Niklen S., and Coulson A. R. (1977) DNA sequencing with chain-termination inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maspero E., Valentini E., Mari S., Cecatiello V., Soffientini P., Pasqualato S., and Polo S. (2013) Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat. Struct. Mol. Biol. 20, 696–701 [DOI] [PubMed] [Google Scholar]
- 55. Maddika S., Kavela S., Rani N., Palicharla V. R., Pokorny J. L., Sarkaria J. N., and Chen J. (2011) WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 13, 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Segal I. A. (1975) Enzyme Kinetics, Wiley-Interscience, New York [Google Scholar]
- 57. Mari S., Ruetalo N., Maspero E., Stoffregen M. C., Pasqualato S., Polo S., and Wiesner S. (2014) Structural and functional framework for the autoinhibition of Nedd4-family ubiquitin ligases. Structure 22, 1639–1649 [DOI] [PubMed] [Google Scholar]
- 58. Wiesner S., Ogunjimi A. A., Wang H. R., Rotin D., Sicheri F., Wrana J. L., and Forman-Kay J. D. (2007) Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130, 651–662 [DOI] [PubMed] [Google Scholar]
- 59. Nuber U., and Scheffner M. (1999) Identification of determinants in E2 ubiquitin-conjugating enzymes required for hect E3 ubiquitin-protein ligase interaction. J. Biol. Chem. 274, 7576–7582 [DOI] [PubMed] [Google Scholar]
- 60. Eletr Z. M., Huang D. T., Duda D. M., Schulman B. A., and Kuhlman B. (2005) E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat. Struct. Mol. Biol. 12, 933–934 [DOI] [PubMed] [Google Scholar]
- 61. Eletr Z. M., and Kuhlman B. (2007) Sequence determinants of E2-E6AP binding affinity and specificity. J. Mol. Biol. 369, 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Malakhova O. A., and Zhang D. E. (2008) ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J. Biol. Chem. 283, 8783–8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Okumura A., Pitha P. M., and Harty R. N. (2008) ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc. Natl. Acad. Sci. U.S.A. 105, 3974–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lu K., Li P., Zhang M., Xing G., Li X., Zhou W., Bartlam M., Zhang L., Rao Z., and He F. (2011) Pivotal role of the C2 domain of the Smurf1 ubiquitin ligase in substrate selection. J. Biol. Chem. 286, 16861–16870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Baboshina O. V., and Haas A. L. (1996) Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2epf and Rad6 are recognized by the 26S proteasome subunit 5. J. Biol. Chem. 271, 2823–2831 [DOI] [PubMed] [Google Scholar]
- 66. Haas A. L., and Bright P. M. (1988) The resolution and characterization of putative ubiquitin carrier protein isozymes from rabbit reticulocytes. J. Biol. Chem. 263, 13258–13267 [PubMed] [Google Scholar]
- 67. Haas A. L. (2005) Purification of E1 and E1-like enzymes. Methods Mol. Biol. 301, 23–35 [DOI] [PubMed] [Google Scholar]
- 68. Tokgöz Z., Siepmann T. J., Streich F. C. Jr., Kumar B., Klein J. M., and Haas A. L. (2012) E1-E2 interactions in the ubiquitin and Nedd8 ligation pathways. J. Biol. Chem. 287, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kumar B., Lecompte K. G., Klein J. M., and Haas A. L. (2010) Ser120 of Ubc2/Rad6 regulates ubiquitin-dependent N-end rule targeting by E3α/Ubr1. J. Biol. Chem. 285, 41300–41309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Baboshina O. V., Crinelli R., Siepmann T. J., and Haas A. L. (2001) N-end rule specificity within the ubiquitin/proteasome pathway is not an affinity effect. J. Biol. Chem. 276, 39428–39437 [DOI] [PubMed] [Google Scholar]