Abstract
In their role as primary producers, marine phytoplankton modulate heterotrophic bacterial activities through differences in the types and amounts of organic matter they release. This study investigates the transcriptional response of bacterium Ruegeria pomeroyi, a member of the Roseobacter clade known to affiliate with diverse phytoplankton groups in the ocean, during a shift in phytoplankton taxonomy. The bacterium was initially introduced into a culture of the dinoflagellate Alexandrium tamarense, and then experienced a change in phytoplankton community composition as the diatom Thalassiosira pseudonana gradually outcompeted the dinoflagellate. Samples were taken throughout the 30-day experiment to track shifts in bacterial gene expression informative of metabolic and ecological interactions. Transcriptome data indicate fundamental differences in the exometabolites released by the two phytoplankton. During growth with the dinoflagellate, gene expression patterns indicated that the main sources of carbon and energy for R. pomeroyi were dimethysulfoniopropionate (DMSP), taurine, methylated amines, and polyamines. During growth with the diatom, dihydroxypropanesulfonate (DHPS), xylose, ectoine, and glycolate instead appeared to fuel the bulk of bacterial metabolism. Expression patterns of genes for quorum sensing, gene transfer agent, and motility suggest that bacterial processes related to cell communication and signaling differed depending on which phytoplankton species dominated the co-culture. A remodeling of the R. pomeroyi transcriptome implicating more than a quarter of the genome occurred through the change in phytoplankton regime.
Introduction
Marine bacteria participate in the biogeochemical cycling of all major elements. They are the primary degraders of marine dissolved organic matter (DOM), a pool of carbon equivalent in mass to the atmospheric CO2 reservoir (Hansell, 2013). Phytoplankton constitute the main source of labile carbon for bacteria in the surface ocean via the metabolites they release into the DOM pool, either through exudation or death-related processes such as senescence, predation, or viral lysis (Azam et al., 1983).
Diatoms and dinoflagellates are common phytoplankton groups in the ocean (Hasle, 1976; Bathmann et al., 1997; Taylor et al., 2008), and mixed assemblages or successions of taxa from these groups have been observed in coastal regions and upwelling systems (Seeyave et al., 2009; Ryan et al., 2014; Varaljay et al., 2015). One consequence of fluctuations in primary producer composition is that the supply of labile compounds to heterotrophic bacteria is likely to vary (Becker et al., 2014). Recent work shows that marine bacteria possess strategies to efficiently detect and utilize phytoplankton metabolites by rapidly congregating near particles, including phytoplankton cells (Smriga et al., 2016). Active movement to the enriched phycospheres associated with phytoplankton (Stocker et al., 2008) allows bacteria access to a wide variety of metabolites, a potentially effective strategy for growth in a diverse and patchy chemical environment.
Bacteria may also have a role in shaping the availability of labile organic matter, influencing metabolite release through excretion of diffusible compounds required by phytoplankton; examples of such compounds include vitamins and enzymes (Croft et al., 2005; Grossart and Simon, 2007; Biller et al., 2016). In cases where interactions are species-specific, bacteria are likely to require repertoires of physiological and ecological strategies unique to each primary producer (Sison-Mangus et al., 2014). These interactions may have important consequences for global-scale carbon fluxes (Harvey et al., 2016; Segev et al., 2016).
The Roseobacter clade is a marine alphaproteobacterial group whose members are often found associated with phytoplankton (González et al., 2000). Diatom and dinoflagellate blooms in the ocean are usually associated with high Roseobacter abundance, and laboratory cultures of these phytoplankton groups are frequently populated with Roseobacter strains (Lafay et al., 1995; Schäfer et al., 2002; Jasti et al., 2005; Amin et al., 2012). One hypothesis for why roseobacters co-occur with phytoplankton is that their typically large genomes encode high metabolic versatility and sophisticated regulatory capabilities (Newton et al., 2010) allowing them to take advantage of increased substrate concentrations in phycospheres (Luo and Moran, 2014). The observation that individual Roseobacter strains have different effects on phytoplankton growth and metabolism supports the hypothesis that specific mechanisms exist for recognition and interaction (Seyedsayamdost et al., 2011; Amin et al., 2015), and these could explain the success of roseobacters in phytoplankton blooms.
In this study, we established a three-member model system to examine transcriptional changes in bacteria in response to shifts in the dominant primary producer. The bacterium selected for the model system is Ruegeria pomeroyi DSS-3, a member of the Roseobacter clade for which a genome sequence (Moran et al., 2004) and genetic system (Howard et al., 2006) is available. The two other members are the dinoflagellate Alexandrium tamarense CCMP1771 and the diatom Thalassiosira pseudonana CCMP1335, two phytoplankton strains that are well characterized and readily grow in culture (Armbrust et al., 2004; Moustafa et al., 2010). In the model system, bacterial carbon and nitrogen requirements were satisfied exclusively by metabolites provided first by A. tamarense, and subsequently by T. pseudonana as it outcompeted the dinoflagellate. Here we report changes in the bacterial transcriptome that address three questions: (1) How plastic is gene expression by R. pomeroyi when associating with different phytoplankton taxa? (2) What different metabolites are released from the model dinoflagellate and diatom? (3) Do transcriptional responses suggest a role for interactions or signaling between the bacterium and primary producers?
Materials and methods
Experimental set up
Axenic phytoplankton cultures from the National Center for Marine Algae were maintained without antibiotic addition and examined regularly by microscopy and plating to check for bacterial contamination. Six 20-l LDPE cubitainers (Reliance, Winnipeg, Canada) were filled with 18 l of autoclaved, 0.2 μm-filtered Guillard’s f/2 medium with vitamin B12 added at f/50 concentration. Two liters of an axenic dinoflagellate Alexandrium tamarense CCMP1771 culture grown to exponential phase in the same medium were inoculated into each cubitainer. Cubitainers were maintained at 18 °C with a 16–8 h light-dark cycle under ~160 μmol photons m−2 s−1. After seven days, three cultures were inoculated with Ruegeria pomeroyi DSS-3 while the other remained axenic. Bacteria had been grown overnight in ½YTSS medium, harvested in exponential phase, washed five times with sterile phytoplankton medium, and inoculated at ⩽106 cells ml−1. Five days after bacterial inoculation, 800 μl of an axenic culture of diatom T. pseudonana CCMP1335 prepared as for A. tamarense were inoculated into all six cubitainers, and silicate was added (100 μM final concentration). The order in which the phytoplankton were introduced was determined based on T. pseudonana’s ability to outcompete A. tamarense under the culture conditions used. Cubitainers were maintained for 37 days.
Sampling and monitoring
To monitor growth, triplicate samples of 0.6 ml were mixed with 0.6 ml of glutaraldehyde (2% final concentration) and stored at −80 °C. Once thawed, 500 μl with 5 μm fluorescent beads added (Spherotech, Lake Forest, IL, USA) were stained for 20 min with SYBR Green I (0.75X recommended concentration, Life Technologies, Waltham, MA, USA) and analyzed on a CyAn instrument (Beckman Coulter, Brea, CA, USA) with a 488 nm laser. SYBR Green fluorescence (bacteria) was detected using a FL1–530/30 bandpass filter and chlorophyll a fluorescence (phytoplankton) using a FL4–680/30 bandpass filter. Data were analyzed in FlowJo. Bacterial contamination was checked by streaking onto ½YTSS plates. No-bacteria phytoplankton controls were checked for any growth, while experimental cubitainers were checked for growth of bacteria other than R. pomeroyi. On days 7, 9, 12, 15, 18, 23, 30, and 37, 1-liter samples were collected for RNA sequencing and chemical analysis.
Chemical analyses
Duplicate 6-ml samples were collected from cubitainers, filtered through 0.45-μm hydrophilic polyethersulfone Acrodisc Supor membranes (Pall Life Sciences, Port Washington, NY, USA), and stored at −20 °C. Samples were analyzed for dissolved inorganic nitrogen (nitrate+nitrite) (APHA Standard Method 4500-NO3 F), ammonium (4500-NH3 G), dissolved inorganic phosphorus (4500-PF), and silicate (spectrophotometric silicomolybdate assays). For DOC, triplicate samples filtered through ashed Whatman GF/F filters were combusted on a Shimadzu TOC-5000A Analyzer. Analyses were performed at the Center for Applied Isotope Studies (University of Georgia, Athens, GA, USA).
Bacterial RNA extraction and sequencing
One liter samples were filtered through 2−μm polycarbonate (PC) membranes to remove eukaryotic cells, and then through 0.2−μm PC membranes to collect bacterial cells. Filters were flash-frozen in liquid nitrogen and stored at −80 °C. For RNA extraction, filters pieces were incubated at 37 °C for 1 h in TE buffer, SDS (0.6% final concentration), and proteinase K (120 ng μl–1 final concentration, Qiagen, Hilden, Germany). Acid phenol:chloroform:isoamylalcohol was added (1:1 volume) and samples were shaken, incubated for 15 min, and centrifuged at 4 °C for 15 min for collection of the supernatant. The process was repeated and the supernatant mixed 1:1 vol:vol with isopropanol, vortexed, incubated for 15 min, centrifuged at 4 °C for 15 min, washed twice with 75% ethanol, and dried and resuspended in RNAse-free water. RNA was precipitated with sodium acetate (0.3 M final concentration) and three volumes of 100% ethanol, and incubated overnight at −20 °C. Pellets were washed twice with 75% ethanol, centrifuged, dried, and resuspended in RNAse-free water.
Potential traces of DNA were removed using the Turbo DNA-free kit (Invitrogen, Waltham, MA, USA). Samples were tested for residual DNA by a 40-cycle PCR targeting the 16 S rRNA gene of R. pomeroyi. Samples were depleted of rRNA using custom probes for small and large subunit rRNA genes from all three microbes (Stewart et al., 2010). Libraries were prepared for two replicate cubitainers at eight time points (16 samples) using the KAPA Stranded mRNA-Seq Kit (Kapa Biosystems, Wilmington, MA, USA) at the Georgia Genomics Facility (University of Georgia) and sequenced on a HiSeq Illumina 2500 at the Hudson Alpha Institute for Biotechnology (AL, USA).
Bioinformatic analysis
Quality control was performed on 249 million 50-bp reads (10±2 million reads per sample; Supplementary Table S1) using the FASTX toolkit, imposing a minimum quality score of 20 over 80% of read length. Reads aligning to an in-house rRNA database were removed (blastn, score cutoff ⩾50). Remaining reads were mapped to the R. pomeroyi genome (Bowtie 2; Langmead and Salzberg, 2012) and counted (HTSeq; Anders et al., 2015), conserving strand information and removing reads that mapped to more than one location (Supplementary Table S1). Counts were converted to transcripts per million (TPM, Supplementary Table S2) and data deposited in the NCBI BioProject database under accession PRJNA381627.
Genes with differential expression between selected time points were determined with DESeq2 (Love et al., 2014), and linear relationships of gene expression with phytoplankton abundance with Pearson correlations. Self-organizing maps (SOM) were built with the Kohonen package in R (Wehrens and Buydens, 2007) using expression of each gene (in TPM) normalized by dividing by the median expression for that gene across the 8 time points followed by a log2 transformation.
DHPS transporter mutant
A deletion mutant of DHPS transporter operon hpsKLM was constructed by homologous recombination (Reisch et al., 2011). Regions up- and downstream of the target region (~1 kb) and tetAR were cloned into pCR2.1 using sequence and ligation-independent cloning (SLIC) (Li and Elledge, 2007) and electroporated into competent R. pomeroyi cells. Recombined clones were selected on tetracycline-amended medium. Triplicate wild type and ΔhpsKLM cultures were incubated at 30 °C in marine basal medium amended with DHPS, glucose, xylose, or acetate as a sole carbon source; or in co-cultures with T. pseudonana. Growth was assessed with OD measurements over 4 days (pure cultures) or on ½YTSS plates, amended with tetracycline in the case of the deletion mutant (co-cultures).
Xylose transporter expression
Triplicate R. pomeroyi cultures were grown on each of four substrates at 30 mM carbon (5 mM glucose, 15 mM acetate, 10 mM pyruvate, 6 mM xylose). Cells were collected in early exponential phase, RNA extracted using the RNeasy mini extraction kit (Qiagen), and DNA removed as described above. xylG (SPO0863) transcripts were assayed by RT-qPCR on an iCycler iQ5 (Bio-Rad, Hercules, CA, USA) using the iTaq SYBR Green one-step kit with a six-fold serially-diluted standard (102 to 107 copies per reaction). Negative controls had no reverse transcriptase addition.
Results and Discussion
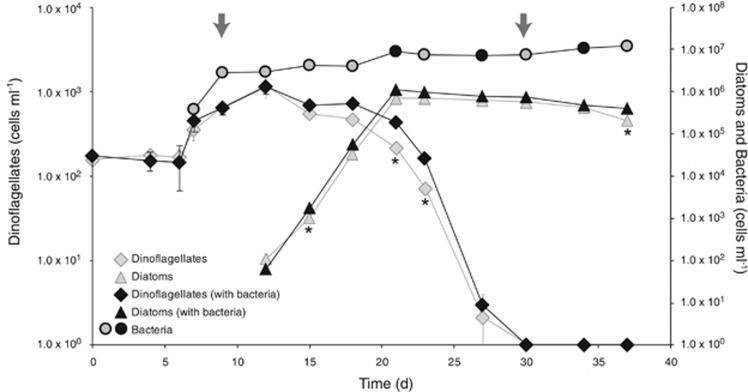
In this three-member co-culture system, the phytoplankton species were the sole source of carbon and usable nitrogen for marine bacterium Ruegeria pomeroyi, since no exogenous carbon source was added and R. pomeroyi cannot assimilate nitrate (Moran et al., 2004). Over the 37 days of the experiment, there was a progression from a dinoflagellate-dominated to a diatom-dominated system. Alexandrium tamarense was the only phytoplankton species present until day 12 when Thalassiosira pseudonana was inoculated into the cultures. At day 15 the diatoms became numerically dominant and remained so until the end of the experiment. During the final stage, dinoflagellate cells decreased dramatically, and they were no longer detectable at day 30 (Figure 1). A biovolume-based estimate accounting for size differences showed nearly equivalent biovolume for each species at their peak abundance, with the switch in biovolume dominance occurring at day 19 (Supplementary Figure S1). Chemical analysis indicated that the co-cultures were never depleted of nitrate or phosphate, although drawdown was evident (Supplementary Table S3). Ammonium remained undetectable, indicating that if any ammonium was produced, it was rapidly utilized. The phytoplankton sustained the growth of R. pomeroyi throughout the experiment, with a ~30-fold increase in bacterial counts between inoculation and day 37 (Figure 1). On the basis of the no-bacteria phytoplankton controls, bacterial presence did not affect maximum phytoplankton abundance but may have extended the duration of the stationary phase for both species (Figure 1, t test, P⩽0.02).
Figure 1.
Cell counts of dinoflagellates (diamonds), diatoms (triangles), and bacteria (circles) as determined by flow cytometry. For phytoplankton, the gray diamonds and triangles correspond to the cell counts measured in no-bacteria controls. For bacteria, gray inner circles indicate RNAseq sample collection. Arrows identify the two time points tested for differential expression comparisons. Asterisks indicate significant differences in phytoplankton cell numbers between co-cultures and no-bacteria controls. Mean value±s.d. for the triplicate cultures are shown.
Gene expression clusters
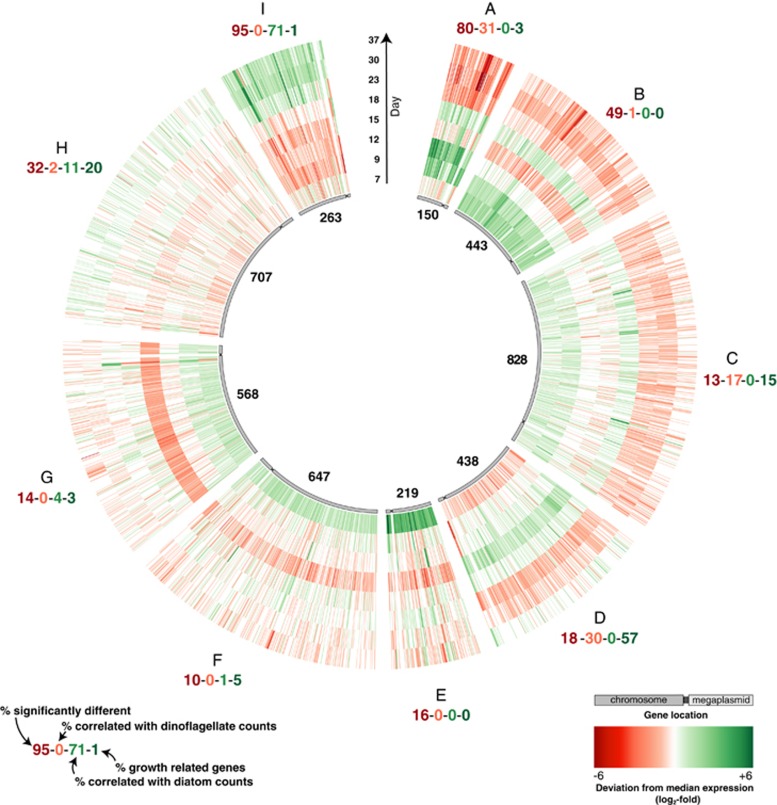
The R. pomeroyi transcriptome changed dramatically above the backdrop of shifting phytoplankton dominance. In a comparison of day 9, when dinoflagellates were the only species present in the co-culture, with day 30, when only diatoms were detectable, 1,179 genes in the R. pomeroyi genome (27%) were differentially expressed (DESeq2, P⩽0.05). All 4,371 R. pomeroyi genes were sorted into clusters based on expression patterns over time using a self-organizing map algorithm (Kohonen, 2001; Wehrens and Buydens, 2007) (Supplementary Figure S2). Of the nine delimited clusters, Clusters A and B had patterns that broadly followed dinoflagellate abundance and Cluster I had patterns that followed diatom abundance (Figure 2, Supplementary Figure S3). Positive linear relationships with dinoflagellate cell numbers were found for 31% of genes in Cluster A and 1% in Cluster B, and with diatom cell numbers for 71% in Cluster I (P⩽0.05, Figure 2). Further, many genes in Clusters A and B had significantly higher expression at dinoflagellate-dominated day 9 time point versus diatom-dominated day 30 time point (80% and 49%), while almost all genes in Cluster I had significantly higher expression at day 30 versus day 9 (95%, Figure 2). Most of the other clusters contained few or no genes having positive linear relationships with phytoplankton abundance and few genes with differential expression between day 9 and 30 (Figure 2).
Figure 2.
Average normalized expression of R. pomeroyi genes from two replicate co-cultures organized into nine expression pattern clusters. The number of genes in each cluster is indicated in the inner circle. Each concentric circle represents one time point.
We also examined 86 bacterial genes whose expression is expected to correlate with cell activity. Assignment of these genes into the putative dinoflagellate or diatom clusters could indicate a growth signal rather than a specific phytoplankton response, since they encode intrinsic processes involved in protein metabolism and secretion, nucleic acid metabolism, and cell cycle and division (Gerdes et al., 2003). Most of the activity-related genes grouped into Cluster D (49 genes), and to a lesser extent in Cluster H (17 genes) and Cluster C (13 genes) (Supplementary Figure S4). Only 1 grouped into the three phytoplankton-related clusters combined, indicating that expression of genes in Clusters A, B, and I does not simply track with bacterial growth rate or activity. Based on these multiple assessments of expression patterns, we subsequently focused analysis on the three gene clusters with clear relationships to dinoflagellate (Clusters A and B) or diatom (Cluster I) dominance in the co-cultures. In this evaluation, we concentrated on biogeochemically-relevant genes whose functions have been experimentally verified in R. pomeroyi in order to minimize speculation built on automated gene function assignment.
Metabolite utilization during growth with dinoflagellates
Several genes involved in degradation of dimethylsulfoniopropionate (DMSP), a phytoplankton osmolyte used by marine bacteria as a source of organic carbon and sulfur, have been experimentally verified in R. pomeroyi (Howard et al., 2006; Reisch et al., 2008, 2011, Todd et al., 2012a, 2012b). The clustering algorithm classified several DMSP-associated genes into dinoflagellate-related Cluster A (Figures 3 and 4), including genes responsible for the committed steps of the DMSP demethylation pathway (dmdA) and cleavage pathway (dddW). In fact, dddW was one of the most differentially expressed genes in the day 9 versus day 30 comparison (72 fold-difference, Table 1). The high expression levels of DMSP degradation genes during R. pomeroyi growth with A. tamarense but not T. pseudonana accords with findings that dinoflagellates typically produce high levels of DMSP, while diatoms do not (Keller, 1989; Keller et al., 1999; Sunda and Hardison, 2008).
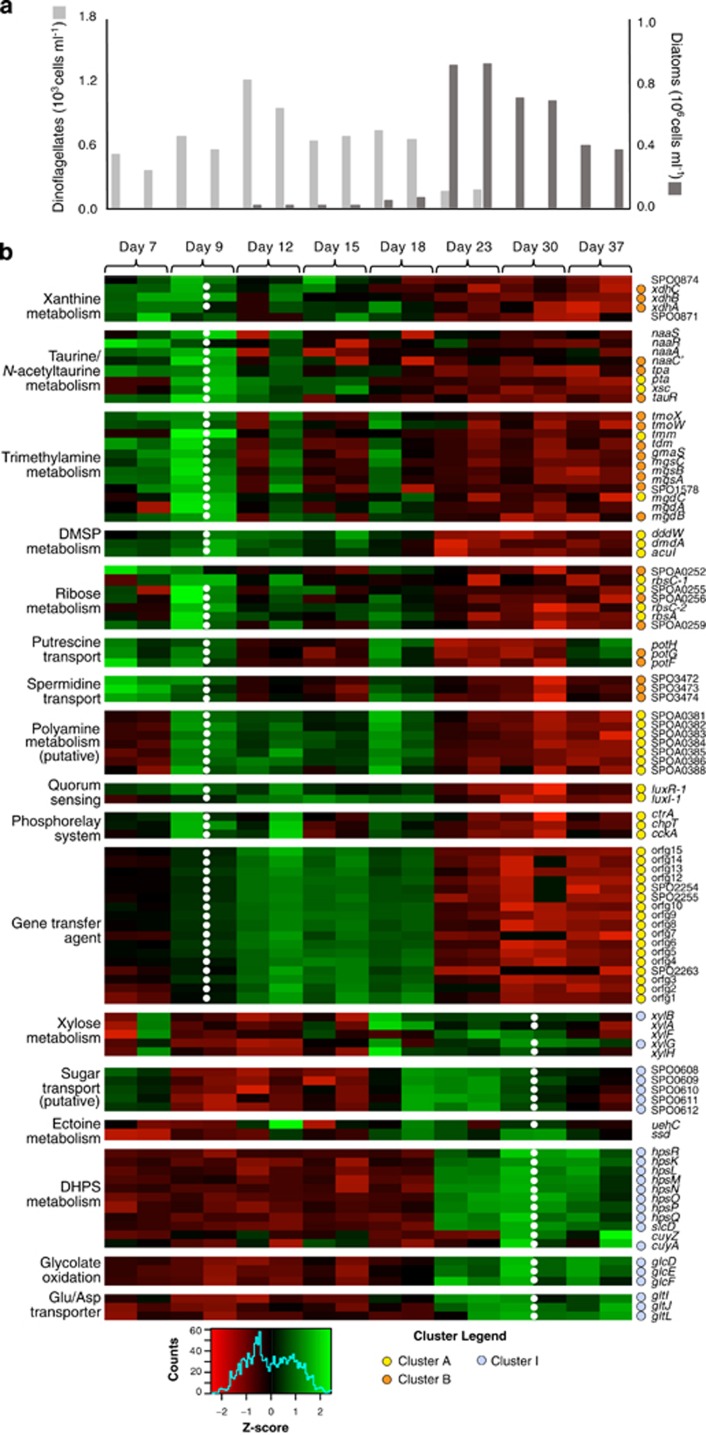
Figure 3.
(a) Cell numbers of dinoflagellates (light grey) and diatoms (dark grey). (b) Expression patterns of genes involved in key pathways. Cluster assignments are indicated with colored dots. Normalized expression was scaled by row, and both replicates are shown for each time point. White dots indicate significantly higher expression on day 9 or day 30.
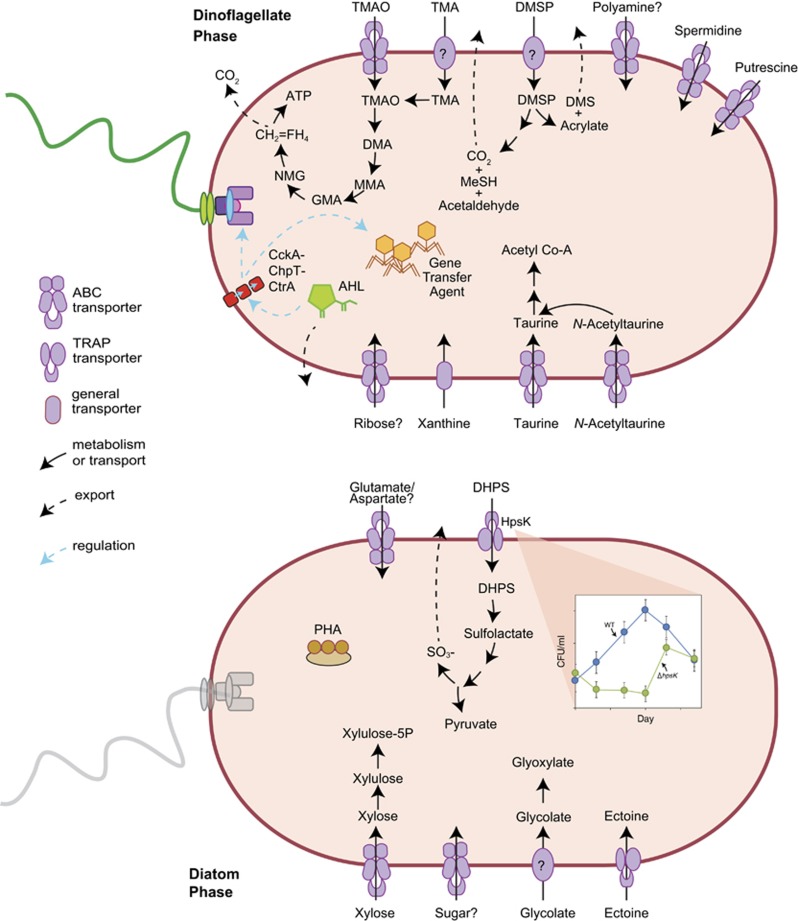
Figure 4.
Selected cell functions differentially regulated by R. pomeroyi during sequential co-culture with dinoflagellates (top) and diatoms (bottom). CH2=FH4, 5,10-methylene tetrahydrofolate; AHL, acylhomoserine lactone; DHPS, dihydroxypropanesulfonate; DMA, dimethylamine; DMS, dimethylsulfide; DMSP, dimethylsulfoniopropionate; GMA, gamma-glutamylmethylamide; MeSH, methanethiol; MMA, monomethylamine; NMG, N-methylglutamate; PHA, polyhydroxyalkanoates; TMA, trimethylamine; TMAO, trimethylamine N-oxide.
Table 1. Ruegeria pomeroyi genes with the highest fold-differences in expression at dinoflagellate-dominated Day 9 (positive fold-change values) and at diatom-dominated Day 30 (negative fold-change values).
| Gene Name | Locus Tag | Annotation | Category | SOM Cluster | Fold Change |
|---|---|---|---|---|---|
| SPO2508 | hypothetical protein | A | 151.6 | ||
| tmm | SPO1551 | TMM monooxygenase | Methylamines | A | 124.6 |
| SPO2259 | head-tail adaptor, putative | GTA | A | 87.8 | |
| SPO0021 | Hpt domain protein | A | 73.2 | ||
| SPO0202 | hypothetical protein | A | 72.7 | ||
| dddW | SPO0453 | DMSP lyase | DMSP | A | 71.8 |
| SPO2260 | hypothetical protein | GTA | A | 59.6 | |
| SPO1554 | ammonium transporter family | A | 59.6 | ||
| SPO2261 | major capsid protein, HK97 family | GTA | A | 59.5 | |
| SPO0931 | response regulator | A | 54.9 | ||
| SPOA0268 | IclR family regulator | A | 51.9 | ||
| SPO0180 | flagellar hook-basal body protein | Motility | A | 50.9 | |
| fliN | SPO0197 | flagellar motor switch protein | Motility | A | 49.3 |
| SPOA0269 | hypothetical protein | A | 48.3 | ||
| SPOA0270 | hypothetical protein | A | 47.2 | ||
| SPOA0271 | methylamine utilization MauG | A | 45.3 | ||
| SPO2263 | hypothetical protein | GTA | A | 44.5 | |
| flgC | SPO0181 | flagellar basal-body rod protein | Motility | A | 39.3 |
| SPOA0381 | polyamine transporter, periplasmic | Polyamines | A | 39.1 | |
| SPOA0382 | polyamine transporter, ATP | Polyamines | A | 38.3 | |
| SPOA0383 | polyamine transporter, permease | Polyamines | A | 37.1 | |
| fliF | SPO0199 | flagellar M-ring protein | Motility | A | 36.6 |
| SPO2256 | hypothetical protein | GTA | A | 34.7 | |
| SPOA0384 | polyamine transporter, permease | Polyamines | A | 33.2 | |
| SPO2264 | portal protein, HK97 family | GTA | A | 31.4 | |
| SPO2258 | hypothetical protein | GTA | A | 30.6 | |
| SPO0930 | hypothetical protein | A | 30.2 | ||
| flgB | SPO0182 | flagellar basal-body rod protein; | Motility | A | 30.1 |
| SPO2313 | hypothetical protein | A | 29.4 | ||
| xsc | SPO3561 | sulfoacetaldehyde acetyltransferase | Taurine | A | 28.4 |
| lpxC | SPO1205 | UDP-3-0-acyl NAG deacetylase | I | −27.5 | |
| SPO1114 | TRAP transporter, DctM | I | −27.9 | ||
| SPO2269 | hypothetical protein | I | −28.2 | ||
| SPOA0369 | copper resistance protein B | I | −28.6 | ||
| SPO1112 | TRAP transporter, periplasmic | I | −29.3 | ||
| slcD | SPO0598 | Sulfolactate dehydrogenase | DHPS | I | −29.9 |
| SPO2180 | hypothetical protein | I | −33.7 | ||
| SPO1372 | hypothetical protein | I | −33.7 | ||
| SPO3301 | LysR family regulator | I | −34.1 | ||
| SPO2657 | D-cysteine desulfhydrase | I | −34.6 | ||
| hpsQ | SPO0597 | UspA stress protein | DHPS | I | −34.7 |
| aspC-1 | SPO0584 | aspartate aminotransferase | I | −38.9 | |
| hpsO | SPO0595 | R or S-DHPS-2-dehydrogenase | DHPS | I | −40.3 |
| SPO2914 | merR family regulator | I | −41.8 | ||
| SPO0636 | EF hand domain protein | I | −42.5 | ||
| hpsL | SPO0592 | DHPS TRAP transporter | DHPS | I | −45.9 |
| cueR | SPO0793 | Cu(I)-responsive regulator | I | −46.7 | |
| glcE | SPO3479 | glycolate oxidase | Glycolate | I | −47.8 |
| SPOA0359 | cytochrome c family protein | I | −52.1 | ||
| SPO0794 | copper-translocating ATPase | I | −55.6 | ||
| hpsN | SPO0594 | DHPS-3-dehydrogenase | DHPS | I | −57.7 |
| SPOA0363 | copper-transporting P-type ATPase ActP | I | −60.6 | ||
| SPOA0368 | hypothetical protein | D | −66.9 | ||
| phaP | SPO1293 | phasin, PhaP | C Storage | I | −73 |
| hpsP | SPO0596 | S or R-DHPS-2-dehydrogenase | DHPS | I | −75.1 |
| copA | SPOA0370 | copper resistance protein A | I | −79.6 | |
| SPOA0371 | hypothetical protein | I | −115.3 | ||
| hpsK | SPO0591 | DHPS TRAP transporter | DHPS | I | −117.2 |
| SPO2237 | hypothetical protein | I | −347.7 | ||
| SPO2236 | hypothetical protein | I | −1063.3 |
Day 9 versus Day 30 comparisons were significantly different (DESeq2, P<0.001) for all genes.
The genetic basis of transport and catabolism of taurine and N-acetyltaurine as well as their inducible expression have been verified experimentally in R. pomeroyi (González et al., 2003; Gorzynska et al., 2006; Durham et al., 2015). Gene expression patterns grouped most metabolic genes from these pathways into dinoflagellate-related Clusters A and B, and expression differences were as high as 28-fold between day 9 and day 30 (Table 1, Supplementary Table S4). These included xsc, tpa, and pta, encoding a shared lower pathway for metabolism of both compounds, and naaC′, encoding a transporter component for N-acetyltaurine (Figures 3 and 4). The ability to use taurine appears to be widespread among marine bacteria, while N-acetyltaurine use seems restricted to only a few members of the Roseobacter (Figure 5; Durham et al., 2015) and Rhodospirillales clades based on the diagnostic gene naaS (the amidohydrolase converting N-acetyltaurine to taurine). Transporters for taurine (tauABC) and N-acetyltaurine (naaABB’C) had more consistent levels of expression throughout the co-culture than catabolic genes. While both phytoplankton species may release these metabolites (Durham et al., 2015), gene expression patterns predict they are relatively more important in the A. tamarense exometabolome.
Figure 5.
Genome content of R. pomeroyi and 50 other Roseobacter clade members. For metabolic genes, analysis focused on enzymes responsible for the first degradation step. For transporters, analysis focused on the periplasmic substrate-binding protein. Genomes were analyzed at roseobase.org. The phylogenetic tree is based on Luo and Moran (2014), with white circles marking nodes with bootstrap values ⩾50%. Substrate binding protein genes: gltI, putative glutamate/aspartate; uehC, ectoine; xylF, putative xylose.
A transport and catabolic pathway for the purine xanthine has been proposed for R. pomeroyi that entails conversion to glyoxylate and ammonium (Cunliffe, 2016). Genes encoding the putative xanthine dehydrogenase (xdhA,B,C) were assigned to dinoflagellate-related Cluster B (Figure 3,Supplementary Table S4). Putative xanthine permease (SPO0874) and the catabolic gene 5-hydroxyisourate hydrolase had expression patterns similar to that of xanthine dehydrogenase (Figure 3). Transcripts for xanthine permeases have been observed previously in marine bacterioplankton communities (Poretsky et al., 2010).
While R. pomeroyi cannot grow on trimethylamine (TMA) as a sole carbon source, the substrate induces activities of key enzymes needed for TMA catabolism (TMA monooxygenase, dimethylamine monooxygenase, N-methylglutamate dehydrogenase; Chen et al., 2011) and provides ammonium and energy for the bacterium, with positive effects on biomass production (Lidbury et al., 2015). In the co-culture, multiple genes involved in TMA utilization grouped into dinoflagellate Cluster A (tmm, mgdC) or Cluster B (tmoX,W, tdm, gmaS, mgsA,B,C, mgdB and SPO1578) (Figure 3, Supplementary Table S4). The gene encoding the first step in trimethylamine catabolism (tmm, mediating oxidation of trimethylamine to trimethylamine N-oxide; Figure 4) was one of the most differentially expressed in the day 9 versus day 30 comparison (125-fold higher on day 9; Table 1).
The genome of R. pomeroyi encodes six operons annotated as transporters of polyamines (Moran et al., 2007), organic nitrogen compounds found in phytoplankton metabolomes and marine DOC during phytoplankton blooms (Lee and Jørgensen, 1995; Nishibori et al., 2001). Three of these had expression patterns linked to dinoflagellate abundance. Transporter system SPOA0381-0384 (Cluster A) was the most highly upregulated of the polyamine transporters, with expression differences of up to 39-fold between day 9 and 30 (Figure 3, Table 1). The other two (SPO3467-3469 and SPO3472-3474; Cluster B) both had ~4-fold higher expression on day 9 (Supplementary Table S4). A previous study of R. pomeroyi gene transcription patterns during growth on five polyamines predicted that SPO3467-3469 transports spermidine and SPO3472-3474 transports putrescine, but there was no expression response for SPOA0381-30384 to the polyamines tested (Mou et al., 2010). A. tamarense has been shown to release polyamines (Nishibori and Nishio, 1997), and along with N-acetyltaurine and xanthine, these represent sources of nitrogen to R. pomeroyi in the co-culture. A peak in expression of the putrescine transporter on the last day of the co-cultures indicates decaying diatoms might also release this compound, nuancing the otherwise strong dinoflagellate polyamine signal (Figure 3).
For a number of genes upregulated on day 9 compared to day 30, functional annotations were assigned during genome annotation (Moran et al., 2004, 2007; Rivers et al., 2014) but have not yet been tested experimentally. Six genes encoding a putative ribose transporter and two associated transcriptional regulators were classified into Clusters A and B (Figure 3, Supplementary Table S5). Nineteen other transporter systems were grouped into Cluster B, including four with annotations suggesting opine transport, ten with annotations for amino acids or (di)peptides, and one for an unidentified sulfonate compound (Supplementary Table S5).
Metabolite utilization during growth with diatoms
Expression patterns indicated a different suite of metabolites supporting R. pomeroyi during the diatom-dominated phase of the co-culture. The genetic basis of dihydroxypropanesulfonate (DHPS) uptake (hpsKLM), catabolism (hpsOPN, slcD), and pathway regulation (hpsR) has been established for R. pomeroyi (Denger et al., 2009; Mayer et al., 2010). These genes grouped into diatom-related Cluster I (Figure 3, Table 1). Downstream gene converting cysteate to pyruvate (cuyA) also grouped into diatom-related Cluster I, and the gene exporting the resulting sulfite (cuyZ) (Denger et al., 2006) exhibited similar expression patterns to cuyA (Figure 3). On the basis of the strong positive correlation with diatom abundance (0.72⩽Pearson r⩽0.90, n=8), the extent of upregulation (50- to 117-fold), and a previous finding that T. pseudonana releases DHPS (Durham et al., 2015), this sulfonate appears to one of the most important bacterial substrates provided by the diatoms (Table 1).
To test this hypothesis, we constructed an R. pomeroyi mutant in which the DHPS transporter (hpsK,L,M) was deleted. The mutant was unable to grow at the expense of DHPS, but grew at comparable rates to the wild type on other substrates (Supplementary Figure S5). Following inoculation into a co-culture with T. pseudonana, cell numbers were compared for mutant and wild-type R. pomeroyi over a period of 18 days. Lower cell numbers were observed for the ΔhpsKLM strain compared to the wild type by as much as six-fold (a difference of 1.2 × 106 cells ml−1 after 10 days) suggesting that DHPS supports the majority of bacterial biomass during co-growth (Figure 4, Supplementary Figure S5). Mutant cells recovered to similar levels as the culture aged, possibly due to a shift in the composition of organic matter released from the diatoms.
The three genes encoding subunits of glycolate oxidase (glcDEF) grouped in Cluster I, with 16- to 48-fold higher expression at day 30 compared to day 9 (Table 1, Supplementary Table S4) and strong positive correlations with diatom abundance (Pearson r⩾0.7). Glycolate is a product of phytoplankton photorespiration, with excretion rate dependent on nutrient and light conditions and varying by species (Hellebust, 1965). The T. pseudonana strain used here was previously shown to release glycolate, particularly when nitrate was the dominant form of available nitrogen (Parker and Armbrust, 2005), as was the case in the co-cultures (Supplementary Table S3). Once released, the metabolite is thought to be used by marine bacteria primarily for energy production (Wright and Shah, 1977; Lau and Armbrust, 2006).
A component of a putative xylose transporter system was assigned to expression Cluster I (Figure 3, Supplementary Table S4), but the function of this transporter had not yet been verified. We therefore grew R. pomeroyi on xylose and three other substrates (glucose, acetate, pyruvate) as sole carbon and energy sources and compared transcription levels under each condition. As was found in a proteomic analysis of an orthologous transporter in Roseobacter member Phaeobacter inhibens (Wiegmann et al., 2014), both xylose and glucose induced expression of the transporter but xylose had a higher signal (five-fold higher in the case of R. pomeroyi; Supplementary Figure S6). The placement of xylB (xylulokinase) in Cluster I, along with similar expression patterns for xylose catabolism genes such as xylA (xylose isomerase, Figure 3) provided additional transcriptional evidence that xylose is a component of the diatom’s exometabolome.
Ectoine is a nitrogen-rich organic osmolyte whose transport and catabolic genes have been experimentally verified in R. pomeroyi (Schulz et al., 2017). The substrate binding protein for the ectoine TRAP transporter had 5-fold higher expression on day 30 compared to day 9 and, along with ectoine catabolic genes, its expression pattern was consistent with a diatom-derived source of carbon and nitrogen (Figure 3).
Genes with unverified substrate annotations that were upregulated during the diatom phase involved those encoding a putative amino acid transporter potentially involved in the uptake of glutamate or aspartate (putative gltIJL, Cluster I, Figure 3,Supplementary Table S5), and two ABC transporters annotated as sugar uptake systems (Supplementary Table S5). The remaining diatom-related transporters with unverified function included two annotated for amino acid uptake (polar and branched chain) and two for carboxylic acid uptake. Of the 94 R. pomeroyi transporter systems predicted to function in organic compound uptake, 26 were assigned to dinoflagellate-related clusters and 10 to diatom-related clusters (Supplementary Table S5).
Evidence for signaling-driven processes
The expression patterns of genes predicted to be involved in cell–cell communication and signaling suggests that R. pomeroyi may be regulating transcription in response to A. tamarense. The bacterial genome encodes two experimentally verified quorum-sensing systems consisting of luxI/luxR pairs that are controlled by acylhomoserine lactone (AHL)-type signal molecules (Moran et al., 2004). The luxR-1/luxI-1 system was recruited into Cluster A with 26-fold (luxR-1) and 14-fold (luxI-1) higher expression on day 9 compared to day 30; luxI-1 expression was also positively correlated with dinoflagellate abundance (Pearson r=0.75, Supplementary Table S4). Quorum sensing was initially described as a regulation system driven by bacterial density, but there is growing evidence for triggers other than cell numbers, some of which include eukaryote metabolites (Miller and Bassler, 2001; Williams, 2007; Natrah et al., 2011). In our co-culture experiment, quorum sensing expression was decoupled from bacterial abundance. AHL production in R. pomeroyi has been shown to be induced by DMSP (Johnson et al., 2016), and it is hypothesized that DMSP signals the presence of phytoplankton and stimulates rearrangement of the bacterium's metabolism (Bürgmann et al., 2007; Johnson et al., 2016).
In Rhodobacter capsulatus and Roseobacter strains Ruegeria TM1040 and KLH11, quorum sensing was shown to induce a phosphorelay system mediated by the cckA-chpT-ctrA genes, which in turn induces expression of motility (Miller and Belas, 2006; Leung et al., 2012; Mercer et al., 2012; Zan et al., 2013). In Ruegeria sp. TM1040 this regulation cascade is involved in the bacterium’s symbiont-like relationship with dinoflagellate Pfiesteria piscicida (Miller and Belas, 2006), while in Ruegeria sp. KLH11 it is implicated in a symbiosis with a marine sponge (Zan et al., 2013). In R. capsulatus, the phosphorelay system is responsible for the expression of the transduction-like gene transfer agent (GTA), a 15-gene cassette that transfers random 4–5 kb genome fragments between cells (Lang and Beatty, 2002). In R. pomeroyi, the function of GTA in gene transfer has also been demonstrated experimentally (Biers et al., 2008). In the co-culture, quorum sensing genes, the cckA-chpT-ctrA phosphorelay genes, flagellar assembly genes, and GTA genes were all assigned to dinoflagellate-related Cluster A and had higher expression at day 9 versus day 30 by 2- to 88-fold (Figure 3, Table 1,Supplementary Table S4). The classification of these genes into the same expression cluster suggests that a similar regulation cascade could exist in R. pomeroyi (Figure 4), and interactions between R. pomeroyi and A. tamarense might be influenced at the level of the luxR-1/luxI-1 quorum sensing system. None of the genes linked in this regulatory cascade had expression patterns that tracked with T. pseudonana abundance.
Conclusions
The abundance and taxonomy of phytoplankton, the chief producers of labile substrates for marine bacteria, vary on scales of days to weeks in the surface ocean (Teeling et al., 2012; Needham and Fuhrman, 2016) and provide heterotrophic bacteria with a dynamic repertoire of organic substrates. Roseobacter member R. pomeroyi changed expression of 27% of its genes when co-grown with a dinoflagellate versus diatom, with many expression shifts linked to non-overlapping suites of metabolites fulfilling bacterial C and N requirements. Further development of chemical methods for identifying low-concentration plankton-derived metabolites in seawater (Moran et al., 2016), in concert with the mining of biological signals through genomics, transcriptomics, and proteomics, will continue to improve our understanding of the flux and fate of marine carbon.
Other members of the Roseobacter clade may have similar metabolic links to phytoplankton exudates. For example, genes for use of putative dinoflagellate metabolites DMSP, taurine, xanthine, and spermidine were found in over half of 50 Roseobacter isolates surveyed, as were the genes for use of putative diatom metabolites DHPS, xylose, and glycolate (Figure 5). These metabolites may support a non-trivial component of Roseobacter heterotrophy in the surface ocean. Other genetic capabilities were infrequent within the clade, including N-acetyltaurine and trimethylamine utilization, and these may be examples of currencies of more specific relationships between individual roseobacters and phytoplankton species. Further, metabolites not used as substrates by R. pomeroyi would have escaped detection in our experimental system. Thus the surprising diversity and divergence of labile metabolites produced from two phytoplankton strains, each representing just one member of extensive marine groups, and detected by just one bacterial strain highlights the need for better understanding of the compounds that sustain bacterial heterotrophy in the ocean.
The capacity for extensive bacterial transcriptome remodeling observed in response to a shifting mixture of metabolites may be a requirement for successful interactions with diverse phytoplankton. The evidence for a possible recognition response by R. pomeroyi during co-growth with the dinoflagellate further suggests that we understand only the tip of an intricate iceberg of autotroph-heterotroph interactions that influence carbon flux through surface ocean plankton.
Acknowledgments
We appreciate advice and assistance from B Nowinski, S Sharma, C Smith, B Durham, and A Vorobev. This work was funded by NSF grant OCE-1342694 and Gordon and Betty Moore Foundation grant #5503.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522: 98–101. [DOI] [PubMed] [Google Scholar]
- Amin SA, Parker MS, Armbrust EV. (2012). Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 76: 667–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306: 79–86. [DOI] [PubMed] [Google Scholar]
- Azam F, Fenchel T, Field J, Gray J, Meyer-Reil L, Thingstad F. (1983). The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 10: 257–263. [Google Scholar]
- Bathmann UV, Scharek R, Klaas C, Dubischar CD, Smetacek V. (1997). Spring development of phytoplankton biomass and composition in major water masses of the Atlantic sector of the Southern Ocean. Deep Sea Res Part II Top Stud Oceanogr 44: 51–67. [Google Scholar]
- Becker JW, Berube PM, Follett CL, Waterbury JB, Chisholm SW, DeLong EF et al. (2014). Closely related phytoplankton species produce similar suites of dissolved organic matter. Front Microbiol 5: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biers EJ, Wang K, Pennington C, Belas R, Chen F, Moran MA. (2008). Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl Environ Microbiol 74: 2933–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Coe A, Chisholm SW. (2016). Torn apart and reunited: impact of a heterotroph on the transcriptome of Prochlorococcus. ISME J 10: 2831–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürgmann H, Howard EC, Ye W, Sun F, Sun S, Napierala S et al. (2007). Transcriptional response of Silicibacter pomeroyi DSS-3 to dimethylsulfoniopropionate (DMSP). Environ Microbiol 9: 2742–2755. [DOI] [PubMed] [Google Scholar]
- Chen Y, Patel NA, Crombie A, Scrivens JH, Murrell JC. (2011). Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc Natl Acad Sci 108: 17791–17796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438: 90–93. [DOI] [PubMed] [Google Scholar]
- Cunliffe M. (2016). Purine catabolic pathway revealed by transcriptomics in the model marine bacterium Ruegeria pomeroyi DSS-3. FEMS Microbiol Ecol 92: 150. [DOI] [PubMed] [Google Scholar]
- Denger K, Mayer J, Buhmann M, Weinitschke S, Smits THM, Cook AM. (2009). Bifurcated degradative pathway of 3-sulfolactate in Roseovarius nubinhibens ISM via sulfoacetaldehyde acetyltransferase and (S)-cysteate sulfolyase. J Bacteriol 191: 5648–5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denger K, Smits THM, Cook AM. (2006). L-Cysteate sulpho-lyase, a widespread pyridoxal 5′-phosphate-coupled desulphonative enzyme purified from Silicibacter pomeroyi DSS-3 T. Biochem J 394: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ et al. (2015). Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci 112: 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes SY, Scholle MD, Campbell JW, Balázsi G, Ravasz E, Daugherty MD et al. (2003). Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol 185: 5673–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JM, Covert JS, Whitman WB, Henriksen JR, Mayer F, Scharf B et al. (2003). Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int J Syst Evol Microbiol 53: 1261–1269. [DOI] [PubMed] [Google Scholar]
- González JM, Simó R, Massana R, Covert JS, Casamayor EO, Pedrós-Alió C et al. (2000). Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol 66: 4237–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzynska AK, Denger K, Cook AM, Smits THM. (2006). Inducible transcription of genes involved in taurine uptake and dissimilation by Silicibacter pomeroyi DSS-3 T. Arch Microbiol 185: 402. [DOI] [PubMed] [Google Scholar]
- Grossart H, Simon M. (2007). Interactions of planktonic algae and bacteria: effects on algal growth and organic matter dynamics. Aquat Microb Ecol 47: 163–176. [Google Scholar]
- Hansell DA. (2013). Recalcitrant dissolved organic carbon fractions. Annu Rev Mar Sci 5: 421–445. [DOI] [PubMed] [Google Scholar]
- Harvey EL, Deering RW, Rowley DC, El Gamal A, Schorn M, Moore BS et al. (2016). A bacterial quorum-sensing precursor induces mortality in the marine coccolithophore, Emiliania huxleyi. Front Microbiol 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasle G. (1976). The biogeography of some marine planktonic diatoms. Deep Sea Res Oceanogr Abstr 23: 319–338. [Google Scholar]
- Hellebust JA. (1965). Excretion of some organic compounds by marine phytoplankton. Limnol Ocean 10: 192–206. [Google Scholar]
- Howard EC, Henriksen JR, Buchan A, Reisch CR, Bürgmann H, Welsh R et al. (2006). Bacterial taxa that limit sulfur flux from the ocean. Science 314: 649–652. [DOI] [PubMed] [Google Scholar]
- Jasti S, Sieracki ME, Poulton NJ, Giewat MW, Rooney-Varga JN. (2005). Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl Environ Microbiol 71: 3483–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WM, Kido Soule MC, Kujawinski EB. (2016). Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J 10: 2304–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MD. (1989). Dimethyl sulfide production and marine phytoplankton: the importance of species composition and cell size. Biol Oceanogr 6: 375–382. [Google Scholar]
- Keller MD, Kiene RP, Matrai PA, Bellows WK. (1999). Production of glycine betaine and dimethylsulfoniopropionate in marine phytoplankton. I. Batch cultures. Mar Biol 135: 237–248. [Google Scholar]
- Kohonen T. (2001) Self-organizing Maps. Springer: Germany. [Google Scholar]
- Lafay B, Ruimy R, Rausch De Traubenberg C, Breittmayer V, Gauthier MJ, Christen R. (1995). Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellate Prorocentrum lima. Int J Syst Evol Microbiol 45: 290–296. [DOI] [PubMed] [Google Scholar]
- Lang AS, Beatty JT. (2002). A bacterial signal transduction system controls genetic exchange and motility. J Bacteriol 184: 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau WWY, Armbrust EV. (2006). Detection of glycolate oxidase gene glcD diversity among cultured and environmental marine bacteria. Environ Microbiol 8: 1688–1702. [DOI] [PubMed] [Google Scholar]
- Lee C, Jørgensen NOG. (1995). Seasonal cycling of putrescine and amino acids in relation to biological production in a stratified coastal salt pond. Biogeochemistry 29: 131–157. [Google Scholar]
- Leung MM, Brimacombe CA, Spiegelman GB, Beatty JT. (2012). The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol Microbiol 83: 759–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. (2007). Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4: 251–256. [DOI] [PubMed] [Google Scholar]
- Lidbury ID, Murrell JC, Chen Y. (2015). Trimethylamine and trimethylamine N-oxide are supplementary energy sources for a marine heterotrophic bacterium: implications for marine carbon and nitrogen cycling. ISME J 9: 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Moran MA. (2014). Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev 78: 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J, Huhn T, Habeck M, Denger K, Hollemeyer K, Cook AM. (2010). 2,3-Dihydroxypropane-1-sulfonate degraded by Cupriavidus pinatubonensis JMP134: purification of dihydroxypropanesulfonate 3-dehydrogenase. Microbiology 156: 1556–1564. [DOI] [PubMed] [Google Scholar]
- Mercer RG, Quinlan M, Rose AR, Noll S, Beatty JT, Lang AS. (2012). Regulatory systems controlling motility and gene transfer agent production and release in Rhodobacter capsulatus. FEMS Microbiol Lett 331: 53–62. [DOI] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. (2001). Quorum sensing in bacteria. Annu Rev Microbiol 55: 165–199. [DOI] [PubMed] [Google Scholar]
- Miller TR, Belas R. (2006). Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ Microbiol 8: 1648–1659. [DOI] [PubMed] [Google Scholar]
- Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S et al. (2007). Ecological genomics of marine roseobacters. Appl Environ Microbiol 73: 4559–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, Buchan A, González JM, Heidelberg JF, Whitman WB, Kiene RP et al. (2004). Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432: 910–913. [DOI] [PubMed] [Google Scholar]
- Moran MA, Kujawinski EB, Stubbins A, Fatland R, Aluwihare LI, Buchan A et al. (2016). Deciphering ocean carbon in a changing world. Proc Natl Acad Sci 113: 3143–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou X, Sun S, Rayapati P, Moran MA. (2010). Genes for transport and metabolism of spermidine in Ruegeria pomeroyi DSS-3 and other marine bacteria. Aquat Microb Ecol 58: 311–321. [Google Scholar]
- Moustafa A, Evans AN, Kulis DM, Hackett JD, Erdner DL, Anderson DM et al. (2010). Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS ONE 5: e9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natrah FMI, Kenmegne MM, Wiyoto W, Sorgeloos P, Bossier P, Defoirdt T. (2011). Effects of micro-algae commonly used in aquaculture on acyl-homoserine lactone quorum sensing. Aquaculture 317: 53–57. [Google Scholar]
- Needham DM, Fuhrman JA. (2016). Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol 1: 16005. [DOI] [PubMed] [Google Scholar]
- Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE et al. (2010). Genome characteristics of a generalist marine bacterial lineage. ISME J 4: 784–798. [DOI] [PubMed] [Google Scholar]
- Nishibori N, Nishio S. (1997). Occurrence of polyamines in the bloom forming toxic dinoflagellate Alexandrium tamarense. Fish Sci 63: 319–320. [Google Scholar]
- Nishibori N, Yuasa A, Sakai M, Fujihara S, Nishio S. (2001). Free polyamine concentrations in coastal seawater during phytoplankton bloom. Fish Sci 67: 79–83. [Google Scholar]
- Parker MS, Armbrust EV. (2005). Synergistic effects of light, temperature, and nitrogen source on transcription of genes for carbon and nitrogen metabolism in the centric diatom Thalassiosira pseudonana (Bacillariophyceae). J Phycol 41: 1142–1153. [Google Scholar]
- Poretsky RS, Sun S, Mou X, Moran MA. (2010). Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ Microbiol 12: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisch CR, Moran MA, Whitman WB. (2008). Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J Bacteriol 190: 8018–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB. (2011). Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473: 208–211. [DOI] [PubMed] [Google Scholar]
- Rivers AR, Smith CB, Moran MA. (2014). An updated genome annotation for the model marine bacterium Ruegeria pomeroyi DSS-3. Stand Genomic Sci 9: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JP, McManus MA, Kudela RM, Lara Artigas M, Bellingham JG, Chavez FP et al. (2014). Boundary influences on HAB phytoplankton ecology in a stratification-enhanced upwelling shadow. Deep Sea Res Part II Top Stud Oceanogr 101: 63–79. [Google Scholar]
- Schäfer H, Abbas B, Witte H, Muyzer G. (2002). Genetic diversity of ‘satellite’ bacteria present in cultures of marine diatoms. FEMS Microbiol Ecol 42: 25–35. [DOI] [PubMed] [Google Scholar]
- Schulz A, Stöveken N, Binzen IM, Hoffmann T, Heider J, Bremer E. (2017). Feeding on compatible solutes: a substrate-induced pathway for uptake and catabolism of ectoines and its genetic control by EnuR. Environ Microbiol 19: 926–946. [DOI] [PubMed] [Google Scholar]
- Seeyave S, Probyn TA, Pitcher GC, Lucas MI, Purdie DA. (2009). Nitrogen nutrition in assemblages dominated by Pseudo-nitzschia spp., Alexandrium catenella and Dinophysis acuminata off the west coast of South Africa. Mar Ecol Prog Ser 379: 91–107. [Google Scholar]
- Segev E, Wyche TP, Kim KH, Petersen J, Ellebrandt C, Vlamakis H et al. (2016). Dynamic metabolic exchange governs a marine algal-bacterial interaction. eLife 5: e17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. (2011). The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem 3: 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sison-Mangus MP, Jiang S, Tran KN, Kudela RM. (2014). Host-specific adaptation governs the interaction of the marine diatom, Pseudo-nitzschia and their microbiota. ISME J 8: 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smriga S, Fernandez VI, Mitchell JG, Stocker R. (2016). Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. Proc Natl Acad Sci 113: 1576–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart FJ, Ottesen EA, DeLong EF. (2010). Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J 4: 896–907. [DOI] [PubMed] [Google Scholar]
- Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF. (2008). Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci 105: 4209–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunda WG, Hardison DR. (2008). Contrasting seasonal patterns in dimethylsulfide, dimethylsulfoniopropionate, and chlorophyll a in a shallow North Carolina estuary and the Sargasso Sea. Aquat Microb Ecol 53: 281–294. [Google Scholar]
- Taylor FJR, Hoppenrath M, Saldarriaga JF. (2008). Dinoflagellate diversity and distribution. Biodivers Conserv 17: 407–418. [Google Scholar]
- Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611. [DOI] [PubMed] [Google Scholar]
- Todd JD, Curson ARJ, Sullivan MJ, Kirkwood M, Johnston AWB. (2012. a). The Ruegeria pomeroyi acuI gene has a role in DMSP catabolism and resembles yhdH of E. coli and other bacteria in conferring resistance to acrylate. PLoS ONE 7: e35947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JD, Kirkwood M, Newton-Payne S, Johnston AWB. (2012. b). DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J 6: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varaljay VA, Robidart J, Preston CM, Gifford SM, Durham BP, Burns AS et al. (2015). Single-taxon field measurements of bacterial gene regulation controlling DMSP fate. ISME J 9: 1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrens R, Buydens LM. (2007). Self-and super-organizing maps in R: the Kohonen package. J Stat Softw 21: 1–19. [Google Scholar]
- Wiegmann K, Hensler M, Wöhlbrand L, Ulbrich M, Schomburg D, Rabus R. (2014). Carbohydrate catabolism in Phaeobacter inhibens DSM 17395, a member of the marine Roseobacter clade. Appl Environ Microbiol 80: 4725–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. (2007). Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153: 3923–3938. [DOI] [PubMed] [Google Scholar]
- Wright RT, Shah NM. (1977). The trophic role of glycolic acid in coastal seawater. II. Seasonal changes in concentration and heterotrophic use in Ipswich Bay, Massachusetts, USA. Mar Biol 43: 257–263. [Google Scholar]
- Zan J, Heindl JE, Liu Y, Fuqua C, Hill RT. (2013). The CckA-ChpT-CtrA phosphorelay system is regulated by quorum sensing and controls flagellar motility in the marine sponge symbiont Ruegeria sp. KLH11. PLOS One 8: e66346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.