Abstract
A growing body of circumstantial evidence suggests that ice nucleation active (Ice+) bacteria contribute to the initiation of precipitation by heterologous freezing of super-cooled water in clouds. However, little is known about the concentration of Ice+ bacteria in precipitation, their genetic and phenotypic diversity, and their relationship to air mass trajectories and precipitation chemistry. In this study, 23 precipitation events were collected over 15 months in Virginia, USA. Air mass trajectories and water chemistry were determined and 33 134 isolates were screened for ice nucleation activity (INA) at −8 °C. Of 1144 isolates that tested positive during initial screening, 593 had confirmed INA at −8 °C in repeated tests. Concentrations of Ice+ strains in precipitation were found to range from 0 to 13 219 colony forming units per liter, with a mean of 384±147. Most Ice+ bacteria were identified as members of known and unknown Ice+ species in the Pseudomonadaceae, Enterobacteriaceae and Xanthomonadaceae families, which nucleate ice employing the well-characterized membrane-bound INA protein. Two Ice+ strains, however, were identified as Lysinibacillus, a Gram-positive genus not previously known to include Ice+ bacteria. INA of the Lysinibacillus strains is due to a nanometer-sized molecule that is heat resistant, lysozyme and proteinase resistant, and secreted. Ice+ bacteria and the INA mechanisms they employ are thus more diverse than expected. We discuss to what extent the concentration of culturable Ice+ bacteria in precipitation and the identification of a new heat-resistant biological INA mechanism support a role for Ice+ bacteria in the initiation of precipitation.
Introduction
Formation of ice embryos from supercooled water in clouds by heterogeneous ice nucleation initiates precipitation (Murray et al., 2012), particularly in ice clouds and mixed-phase clouds (Hoose and Mohler, 2012) over land masses of mid-latitudes (Mülmenstädt et al., 2015). Circumstantial evidence suggests that ice nuclei (IN) of biological origin may contribute to this process. In fact, some microorganisms can nucleate ice at significantly warmer temperatures compared to non-biological IN. While the nucleation temperature of mineral dusts, carbonaceous combustion products or volcanic ash is mainly limited to temperatures below −12 °C/−15 °C (Murray et al., 2012), some bacteria and fungi nucleate ice at temperatures up to −2 °C (Pouleur et al., 1992; Morris et al., 2004), and pollen at temperatures up to −10 °C (Pummer et al., 2012). Also, as clouds precipitate, the concentration of IN active at −10 °C (and thus most likely biological) decreases faster than the concentration of particles of similar size (Bigg et al., 2015; Stopelli et al., 2015) and the majority of IN in precipitation is heat-sensitive suggesting that they are in fact of biological origin (Diehl et al., 2002; von Blohn et al., 2005; Christner et al., 2008; Pummer et al., 2012). Importantly, ice nucleation active (Ice+) microorganisms have been isolated from clouds (Sands et al., 1982; Attard et al., 2012; Joly et al., 2013), the lower atmosphere (Lindemann et al., 1982; Garcia et al., 2012) and precipitation (Sands et al., 1982; Constantinidou et al., 1990; Morris et al., 2008; Hill et al., 2014; Monteil et al., 2014a; Šantl-Temkiv et al., 2015). Ice+ bacteria are also common in terrestrial environments such as the phyllosphere (Lindow, 1983; Hirano et al., 1985), leaf litter (Maki et al., 1974; Monteil et al., 2012) and soil (Monteil et al., 2014b). Therefore, ice nucleation activity (INA) has been proposed to be an adaptation of microbes to promote their deposition from nutrient-poor clouds back to nutrient-rich vegetation or soil, a theory called bioprecipitation (Sands et al., 1982; Morris et al., 2014).
The first-identified Ice+ bacterium was Pseudomonas syringae (Maki et al., 1974; Arny et al., 1976) and the activity was later identified in additional Pseudomonadaceae, Xanthomonadaceae (Kim et al., 1987) and Enterobacteriaceae (Lindow et al., 1978), which are all members of the Gammaproteobacteria. A small number of studies also reported INA in bacteria outside the Gammaproteobacteria (Ponder et al., 2005; Mortazavi et al., 2008). However, INA of these bacteria was found to be restricted to lower temperatures and was neither further characterized nor confirmed in additional studies.
All Ice+ bacteria whose INA has been molecularly characterized encode orthologues of a single INA protein, which is anchored to the outer membrane (Wolber et al., 1986; Morris et al., 2004) and initiates ice formation by binding water molecules in a conformation resembling an ice lattice (Kajava and Lindow, 1993; Garnham et al., 2011). It was recently proposed that this is accomplished by a hydrophilic-hydrophobic pattern and through the removal of latent heat (Pandey et al., 2016). The molecules that induce ice nucleation in Ice+ fungi and in pollen have not yet been identified, but they have been found to be secreted and to be partially heat resistant (Pouleur et al., 1992; Fröhlich-Nowoisky et al., 2015; O'Sullivan et al., 2015; Pummer et al., 2015).
Studies of temporal and spatial dynamics of Ice+ microorganisms associated with precipitation are needed to determine the environmental conditions promoting dispersal and precipitation of Ice+ bacteria and may also provide further evidence that Ice+ bacteria influence atmospheric processes leading to precipitation. Monteil et al. (2014a) identified chemical features of precipitation and characterized the history of air masses associated with deposition of P. syringae with precipitation, while other studies tested effects of these environmental parameters on the survival and INA of Pseudomonas species in the laboratory (Attard et al., 2012; Amato et al., 2015). However, studies on distribution of Ice+ bacteria in clouds and rain need to be extended to all Ice+ microorganisms to obtain a more complete understanding of the link between Ice+ microorganisms and timing and patterns of precipitation.
Here we extensively sampled culturable bacteria deposited with precipitation to expand our knowledge of the concentration and genetic diversity of Ice+ species in precipitation, the mechanism by which they nucleate ice, and the conditions linked to their deposition. Besides observing a link between the concentration of IN and the history of air masses, measuring the concentration of culturable Ice+ bacteria in precipitation, and elucidating the genetic diversity of the dominant culturable Ice+ populations, we identified two Ice+ Gram-positive strains belonging to the genus Lysinibacillus that produce an INA molecule fundamentally different from the INA protein produced by Gammaproteobacteria. We discuss how our findings improve current estimates of the frequency of INA among bacterial communities in the atmosphere and the putative role of bacterial IN in contributing to the initiation of precipitation.
Materials and methods
Sampling of precipitation and sample processing
Field studies were conducted from February 2013 to April 2014 in Virginia, USA (see Table 1 for details). Locations and an image of one of the precipitation collection sites are shown in Supplementary Figure S1. The number K of total IN at temperatures warmer than −8 °C was determined immediately after sample processing by a droplet-freezing assay (Vali, 1971). Supplementary Materials S1 provide additional details on collection, sample analysis and inference of backward trajectories and climatic conditions of air masses.
Table 1. Water chemistry and concentrations of total culturable bacteria and Ice+ bacteria in precipitation samples in Virginia, USA.
| Sitea | Date | Time precipitation started | Sample ID | Number of IN L−1 of waterb | Total dissolved solids (ppm) | Salinity (ppm) | pH | Conductivity (μS.cm−1) |
Concentration of total culturable bacteriac
(log(CFU l−1)) |
Frequency of Ina bacteriad
(%) |
Concentration of culturable Ice+
bacteriae
(log(CFU l−1)) |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CA | CA 10% | EMB | MA | R2A | TSA | TSA 10% | CA | CA 10% | EMB | MA | R2A | TSA | TSA 10% | CA | CA 10% | EMB | MA | R2A | TSA | TSA 10% | |||||||||
| Town of Blacksburg | |||||||||||||||||||||||||||||
| 2/23/2013 | 4:45 | VTP01 | na | na | na | na | na | 4.90 | na | 4.48 | 4.79 | 5.03 | 4.81 | na | 2.1 | na | na | — | — | — | na | 3.22 | na | na | — | — | — | na | |
| 2/26/2013 | 8:00 | VTP02 | na | na | na | na | na | 4.11 | na | 4.22 | 3.89 | 4.66 | 4.56 | na | 1.6 | na | 0.5 | 1.0 | — | 0.9 | na | 2.31 | na | 1.94 | 1.91 | — | 2.51 | na | |
| 3/24/2013 | 10:30 | VTP03 | na | na | na | na | na | 4.17 | na | 4.12 | 3.44 | 4.21 | 3.92 | 4.43 | — | na | — | — | — | — | 1.0 | — | na | — | — | — | — | 2.45 | |
| Kentland Farm, Site A | |||||||||||||||||||||||||||||
| 6/13/2013 | 2:30 | VTP07 | 421 | na | na | na | na | 5.63 | 5.79 | 5.40 | 5.70 | 5.79 | 5.68 | 5.71 | — | — | — | — | — | 0.3 | — | — | — | — | — | — | 3.16 | — | |
| 6/18/2013 | 9:30 | VTP08 | 11 | na | na | na | na | 4.16 | 3.97 | 3.82 | 3.90 | 4.22 | 4.03 | 4.12 | 0.3 | 0.3 | 0.7 | 1.7 | — | — | 0.8 | 1.70 | 1.51 | 2.05 | 2.14 | — | — | 2.02 | |
| 7/1/2013 | 21:00 | VTP09 | 441 | na | na | na | na | 5.26 | 5.11 | 4.98 | 5.15 | 5.36 | 5.41 | 5.21 | — | 0.3 | 0.5 | — | — | — | — | — | 2.52 | 2.70 | — | — | — | — | |
| 7/17/2013 | 15:00 | VTP10 | 48 | na | na | na | na | 4.35 | 4.41 | 3.97 | 4.25 | 4.43 | 5.47 | 4.31 | 2.6 | 0.5 | 3.6 | 2.7 | 1.6 | 2.1 | 1.0 | 2.77 | 2.13 | 2.54 | 2.67 | 2.62 | 2.79 | 2.32 | |
| 1/20/2014 | 17:00 | VTP21b | 7440 | 27.50 | 33.1 | 7.74 | 46.3 | 4.26 | 4.35 | 3.93 | 4.16 | 4.25 | 4.25 | 4.40 | — | 1.0 | 0.5 | — | — | — | — | — | 2.37 | 1.64 | — | — | — | — | |
| 3/16/2014 | 13:00 | VTP26b | 103 | 6.20 | 12.6 | 6.34 | 8.7 | 3.63 | 3.59 | 2.90 | 3.27 | 4.02 | 3.51 | 3.63 | — | 1.0 | 5.0 | 1.0 | 0.3 | 1.0 | — | — | 1.60 | 1.60 | 1.29 | 1.56 | 1.53 | — | |
| 4/7/2014 | 7:15 | VTP28b | 11 | 3.70 | 10.9 | 6.97 | 5.3 | 3.23 | 3.37 | 2.94 | 2.90 | 3.56 | 3.18 | 3.31 | 4.2 | 2.6 | 7.5 | 6.3 | 4.9 | 6.9 | 6.3 | 1.85 | 1.78 | 1.82 | 1.70 | 2.25 | 2.01 | 2.11 | |
| Kentland Farm, Site B | |||||||||||||||||||||||||||||
| 4/12/2013 | 2:00 | VTP05 | 441 | na | na | na | na | 4.65 | 4.72 | 4.36 | 4.17 | 4.81 | 4.77 | 4.67 | 5.4 | 2.1 | 5.8 | 6.1 | 2.6 | 3.8 | 2.8 | 3.39 | 3.04 | 4.12 | 2.95 | 3.22 | 3.35 | 3.01 | |
| 11/1/2013 | 1:00 | VTP14b | 1352 | 14.30 | 9.0 | 7.28 | 12.7 | 4.53 | 2.98 | 4.68 | 4.94 | 5.36 | 5.41 | 4.79 | 0.5 | 0.5 | 1.2 | — | — | — | 1.0 | 3.25 | 3.32 | 2.76 | — | — | — | 2.81 | |
| Kentland Farm, Site C | |||||||||||||||||||||||||||||
| 5/6/2013 | 9:30 | VTP06 | na | na | na | 6.58 | 3.5 | 4.22 | 4.16 | 3.90 | 4.90 | 4.26 | 4.07 | 4.29 | 12.8 | 6.3 | 16.7 | 12.0 | 3.6 | 7.6 | 10.4 | 3.18 | 2.95 | 3.12 | 2.98 | 2.82 | 2.95 | 3.31 | |
| Virginia Tech, Parking Lot | |||||||||||||||||||||||||||||
| 8/8/2013 | 16:30 | VTP11 | 557 | 21.80 | 21.6 | 7.94 | 30.9 | 3.67 | 3.92 | 3.56 | 3.68 | 3.95 | 3.81 | 3.90 | 1.4 | 1.4 | 2.8 | — | 2.8 | 2.8 | 2.1 | 1.96 | 2.20 | 2.15 | — | 2.54 | 2.40 | 2.36 | |
| Virginia Tech, Latham Hall | |||||||||||||||||||||||||||||
| 9/21/2013 | 12:00 | VTP12 | 231 | 12.30 | 10.2 | 6.61 | 10.9 | 3.66 | 3.63 | 3.40 | 3.23 | 3.72 | 3.55 | 3.87 | 0.7 | 2.6 | 6.1 | 8.2 | 1.8 | 1.1 | 8.5 | 1.53 | 2.04 | 2.19 | 2.14 | 1.79 | 1.61 | 2.80 | |
| 10/7/2013 | 3:30 | VTP13 | 48 | 15.30 | 12.2 | 7.45 | 17.2 | 3.54 | 3.66 | 3.30 | 3.60 | 3.70 | 3.61 | 3.64 | 1.0 | 1.0 | 5.2 | — | 1.0 | 5.2 | 1.0 | 1.56 | 1.68 | 2.02 | — | 1.72 | 2.32 | 1.66 | |
| 11/1/2013 | 1:00 | VTP14a | 1352 | 44.10 | 54.8 | 7.33 | 76.9 | 3.98 | 2.65 | 4.67 | 5.11 | 5.75 | 5.18 | 5.54 | — | 0.4 | — | 0.5 | — | — | — | — | 2.86 | — | 2.82 | — | — | — | |
| 12/6/2013 | 9:15 | VTP17 | na | 11.10 | 4.2 | 7.88 | 5.8 | 4.17 | na | 4.06 | 4.11 | 4.26 | 4.23 | 4.08 | 2.6 | na | 10.4 | 13.5 | 4.2 | 9.5 | 1.4 | 2.58 | na | 3.07 | 3.24 | 2.88 | 3.20 | 2.23 | |
| 1/2/2014 | 12:30 | VTP19 | na | 13.70 | 9.8 | 7.39 | 13.8 | 4.10 | 4.58 | 4.41 | 3.98 | 4.88 | 4.34 | 4.10 | — | — | 1.0 | — | — | 0.5 | — | — | — | 2.43 | — | — | 2.05 | — | |
| 1/20/2014 | 23:00 | VTP21a | 6080 | 26.40 | 32.3 | 7.91 | 46.6 | 4.34 | 4.52 | 4.18 | 4.28 | 4.30 | 4.44 | 4.27 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | |
| 2/13/2014 | 13:30 | VTP24 | 279 | 11.60 | 4.2 | 6.74 | 5.9 | 2.74 | 3.13 | 2.97 | 3.20 | 3.58 | 2.76 | 3.57 | — | — | — | — | — | — | 0.5 | — | — | — | — | — | — | 1.29 | |
| 3/16/2014 | 12:45 | VTP26a | 170 | 6.60 | 11.0 | 6.80 | 8.6 | 3.58 | 4.70 | 2.64 | 3.42 | 4.58 | 4.33 | 4.89 | — | — | 1.0 | — | 0.5 | — | 1.0 | — | — | 0.66 | — | 2.29 | — | 2.90 | |
| 4/7/2014 | 7:30 | VTP28a | 11 | 9.80 | 3.3 | 6.99 | 4.6 | 3.60 | 4.32 | 3.45 | 3.46 | 4.21 | 3.52 | 3.77 | 1.0 | 1.0 | 1.6 | 4.2 | — | 2.1 | 1.0 | 1.63 | 2.34 | 1.65 | 2.08 | — | 1.84 | 1.78 | |
‘na’ refers to non-analysed data and ‘—’ refers to nul value (under the detection threshold).
GPS coordinates and altitudes for each site are the following: Blacksburg city (37°14'16"N, 80°25'2"W); Kentland Farm—Site A (37°12'9"N, 80°33'51"W); Kentland Farm—Site B (37°11'47"N, 80°34'41"W); Kentland Farm—Site C (37°12'8"N, 80°35'32"W); Virginia Tech—Latham Hall (37°13'29"N, 80°25'22"W); Virginia Tech—Parking Lot (37°13'28"N, 80°25'19"W).
Number of IN active at −8 °C.
CA, CA10%, EMB, MA, R2A, TSA and TSA10% are the abbreviations for Columbia Agar, Columbia Agar 10% concentrated, Ethylene-Methyl-Blue Agar, Marine Agar, Reasoner's 2 Agar, Tryptic Soy Agar, and Tryptic Soy Agar 10% concentrated, respectively.
Corresponds to the frequency of isolates for which ice nucleation activity has been reproduced with purified cultures incubated for 48 h at 20 °C and tested at −8 °C.
Corresponds to the concentration of isolates for which ice nucleation activity has been reproduced with purified cultures incubated for 48 h at 20 °C and tested at −8 °C.
Quantification of culturable bacteria
Each sample was concentrated 100 times with a vacuum filtration system and a nitrocellulose membrane (0.22 μm porosity, GWSP, Millipore, Cork, Ireland) before dilution plating. Seven different types of culture media were used to enumerate total culturable bacteria and to maximize genetic diversity: Columbia Agar and Tryptic Soy Agar (both at 100 and 10% strength), Eosin methylene blue Agar, Marine Agar 2216, Reasoner’s 2A Agar (see Diehl et al. (2002) for details). Plates were incubated at 20 °C for 3–5 days, and then individual colonies were counted, and analysed for INA. Details are provided in Supplementary Methods S1.
Characterization of bacterial INA
Colonies were tested for their ability to freeze sterile water at −8 °C with a droplet-freezing assay (Vali, 1971): well-isolated individual colonies were deposited in 140 μl of sterile water, incubated at 4 °C for at least 1 h, and then three 30 μl droplets of each suspension were deposited on parafilm floating on a −8 °C glycerol bath in a cooling thermostat (Lauda Alpha RA24, LAUDA-Brinkmann, Delran, NJ, USA). After 10 min, the number of frozen drops was recorded for each colony and for positive and negative controls: P. syringae strain CC94 (Morris et al., 2000) and sterile distilled water, respectively. Any isolate with minimal activity (that is, one frozen drop) was considered as a potential ice nucleator. A second step of semi-quantitative INA characterization was performed with standardized suspensions for all strains that had shown at least minimal activity in the first assay: 6 drops of 30 μl suspensions made of 48 h old pure cultures were incubated at 20 °C and adjusted to an optical density (OD) at 600 nm of 0.2 (corresponding approximately to 1.31 × 108±2.12 × 107 CFU ml−1 on average, based on calibration curves performed with different representative species). Four temperatures were tested: −4, −8, −10 and −12 °C. After 10 min of incubation at each temperature, the number of frozen drops was recorded and strains were considered as Ice+ at a given temperature if at least half of the drops froze at that temperature. See Supplementary Methods S1 for details.
Cumulative nucleus spectra of Lysinibacillus sp. strains
The cumulative IN concentrations per cell of Lysinibacillus sp. were determined at every temperature degree from −2 to −12 °C. Cumulative IN concentrations were compared to P. syringae CC94 after both species were grown under the same conditions. The average number of cells per drop was determined by dilution-plating. The cumulative IN concentrations per cell were determined before and after treatments as described in Supplementary Methods S1.
Identification of Ice+ bacteria
All strains for which the initial colony showed at least minimal activity were genotyped. First, a fragment of the cts gene was amplified with primers specific to the Pseudomonas genus (Sarkar and Guttman, 2004). Strains for which the gene was amplified were considered to belong to the genus Pseudomonas. For the strains for which no amplification product was obtained, a 16S rDNA fragment was amplified, sequenced and blasted against the non-redundant nucleotide database in GenBank as described in Supplementary Methods S1. A strain was assigned to a species when sequence similarity with a type strain was greater than 99% over a minimum sequence alignment of 99%. Below these thresholds, strains were identified at either the genus or family level (Supplementary Tables S2 and S3). Phylogenetic trees were built with sequences of strains and of the most similar sequences based on the BLAST results. Our data set was finally enriched with 920 sequences of type strain sequences downloaded from the Ribosomal Database Project (Cole et al., 2014) and GenBank. See Supplementary Methods S1 for details on tree construction. All 16S rDNA sequences were deposited in GenBank: KY073884-KY074547.
Statistical analyses
Statistical analyses were performed in R, version 3.1.2 (R Core Team, 2014), and are described in detail in Supplementary Methods S1. Bacterial concentrations were log transformed to obtain a normal distribution before statistical analyses. For all biological variables, averages were compared pairwise either by the Student’s t-test or by the non-parametric Mann–Whitney U test. Links between concentrations of total bacteria, Ice+ bacteria, and of IN with precipitation sample features were explored testing correlations between variables by building a linear model and analysed using Pearson’s method or Spearman’s method. Differences and correlations were considered significant when P-values were below 0.05.
Results
Concentration of IN, culturable bacteria and Ice+ bacteria varies between precipitation events and correlates with chemical features of precipitation samples
The concentration of culturable bacteria isolated from precipitation samples ranged from 4 × 103 to 1 × 106 CFU l−1 (Table 1). The concentration of culturable bacteria (Table 2) did not change with the type of culture medium except for R2A, on which colony counts were significantly higher compared to EMB (pairwise Welch’s t-test, P<0.05, Table 2). The overall mean and standard error of the lognormal distribution was 4.18±0.13 l−1 when data for all media types were pooled.
Table 2. Comparative analysis of different culture media used for isolating total culturable bacteria and Ice+ bacteria from precipitation collected in Virginia, USA.
| Mediuma |
Concentration of culturable bacteria (log(CFU l−1)) |
Frequency of Ice+ bacteriab
(%) |
Concentration of culturable Ice+ bacteria (log(CFU l−1)) |
|||
|---|---|---|---|---|---|---|
| Mean±s.e.c | Mean±s.e. | Mean±s.e. | ||||
| CA | 4.21±0.16 | ab | 1.58±0.60 | ab | 1.34±0.27 | a |
| CA 10% | 4.37±0.17 | ab | 1.11±0.34 | b | 1.70±0.26 | a |
| EMB | 3.99±0.16 | b | 3.20±0.90 | a | 1.75±0.25 | a |
| MA | 4.03±0.15 | ab | 2.49±0.84 | ab | 1.13±0.27 | a |
| R2A | 4.48±0.13 | a | 0.94±0.32 | b | 1.03±0.26 | a |
| TSA | 4.26±0.15 | ab | 1.91±0.59 | ab | 1.44±0.27 | a |
| TSA 10% | 4.33±0.15 | ab | 1.86±0.64 | ab | 1.57±0.27 | a |
CA, CA10%, EMB, MA, R2A, TSA and TSA10% are the abbreviations for Columbia Agar, Columbia Agar diluted 10 times, Eosin-Methyl-Blue Agar, Marine Agar, Reasoner's 2A Agar, Tryptic Soy Agar and Tryptic Soy Agar diluted 10 times, respectively. Averages and standard errors were estimated from 23 samples for all media, with the exception of CA10% (n=19) and TSA10% (n=21).
Ice nucleation activity of bacteria has been confirmed with purified cultures incubated for 48 h at 20 °C and tested at −8 °C.
For each variable, values followed by the same letter are not significantly different (pairwise Welch's t-test, P<0.05, when data were normally distributed, pairwise Mann–Whitney U test, P<0.05, when data were not normally distributed). Population sizes of total bacteria were normally distributed while frequencies and concentration of culturable Ice+ bacteria were not.
A total of 33 134 colonies were initially tested for INA using the droplet freezing assay. An average of 4±1% of colonies per precipitation sample initiated the freezing of at least one out of three drops at −8 °C. The lowest and highest frequencies of culturable Ice+ bacteria observed in precipitation samples were 1% and 27%, respectively. This corresponds to a range of concentrations of culturable Ice+ bacteria from 40 to 21 840 l−1of precipitation with a mean of 901±240 l−1.
Only 52% of strains that were Ice+ during the first test (593 out of 1144) were confirmed to be Ice+ at −8 °C when purified cultures were re-tested at an OD600 of 0.2. Based on the standardized re-test, culturable Ice+ bacteria active at −8 °C ranged in concentration from 0 CFU l−1 to 13 219 CFU l−1, with a mean of 384±147 CFU l−1. This value was independent of isolation medium (pairwise Mann–Whitney U-tests, Table 2). Interestingly, for an additional 9% and 4%, respectively, of strains that were Ice+ at −8 °C in the initial test, INA was still observed at −10 and −12 °C in the re-test. Moreover, an additional 17% of the strains still had 1 or 2 drops freeze at −12 °C suggesting that they had some minimal INA at lower temperatures.
We also quantified the concentration of total IN in precipitation at −8 °C and found it to range from 11 to above 7.4 × 103 per liter, with a mean of 1117±528 per liter (Table 1), which corresponds to the average value observed in previous studies (Petters and Wright, 2015). The ratio between the concentration of IN and the concentration of confirmed Ice+ bacteria ranged widely from 0.01 to 6080 with a mean of 195. No statistically significant correlation was found between the concentration of IN and that of Ice+ bacteria. However, the concentration of total IN was significantly correlated with the population size of total culturable bacteria (positively) and the frequency of Ice+ strains in that population (negatively) for several media. We finally inferred concentrations of IN of bacterial origin in clouds from the concentrations of cultured Ice+ bacteria in our precipitation samples. With the exception of one sample, the inferred concentration is less than 10 IN m−3 of cloud at −8 °C (Supplementary Table S4), which would not be sufficient to initiate precipitation (Crawford et al., 2012). However, changing some of our assumptions this number would dramatically decrease or increase, revealing how speculative these inferred values are (see a more detailed discussion below).
To learn about the aeroecology of Ice+ bacteria present in precipitation, we determined if the concentrations of total IN and of culturable Ice+ bacteria were associated with selected features of air masses and/or sample characteristics. We found that the concentration of IN, the concentration of culturable bacteria and the frequency of Ice+ bacteria compared to the total number of culturable bacteria were significantly related to (i) several climatic parameters, such as air pressure, relative humidity, radiative fluxes or temperature experienced by air masses along their trajectories, (ii) the distance travelled by air masses and (iii), precipitation water chemistry (see Supplementary Table S5 and Supplementary Results S1). Whenever these relationships were observed, they were consistent for most media types and for the elevations used here to determine air masses trajectories. The most striking result is that we found significant positive correlations between pH, conductivity and concentration in total dissolved solids and concentration of total culturable bacteria and total IN (Pearson’s and Spearman’s correlation tests, coefficients of 0.62 and 0.79, P<0.05).
INA is widespread among the Gammaproteobacteria and has also emerged in the Firmicutes
Because Pseudomonads are already known to be common Ice+ bacteria in precipitation and clouds, we chose to screen our entire collection of 1144 strains that were Ice+ in the initial test by PCR with primers specific to the cts gene of the Pseudomonas genus. We then focused our attention on those Ice+ strains that did not amplify the cts gene, and thus likely belonged to other taxonomic groups with possibly yet unknown Ice+ genera and species, and sequenced their 16S rDNA. However, to gain some insight into the genetic diversity of Ice+ strains belonging to the genus Pseudomonas, 215 out of the 719 Ice+ strains that amplified the cts gene were chosen randomly for 16S rDNA sequencing as well.
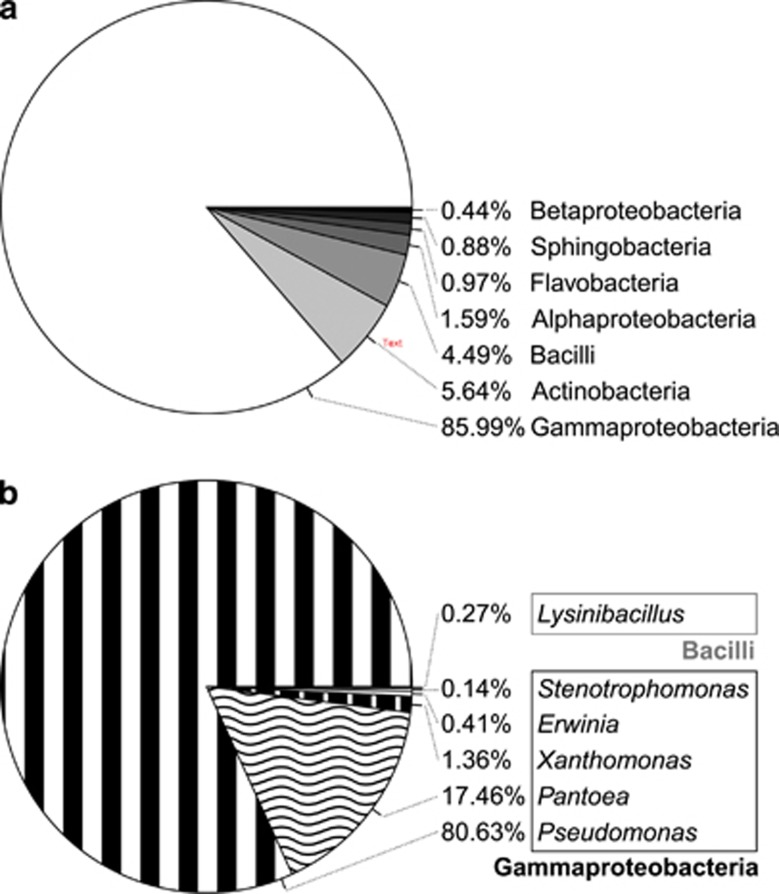
Combining results from cts gene amplification and BLAST searches of 16S rDNA sequences (Supplementary Table S3) showed that culturable Ice+ bacteria that had at least a minimal activity during the initial test belonged to seven different bacterial classes (Figure 1a). Only bacteria belonging to two of these classes were confirmed in the standardized re-test (Figure 1b). A rarefaction curve was built to estimate the richness of these confirmed Ice+ species (Supplementary Figure S2). Since the curve is very close to reaching a horizontal asymptote, we conclude that only a small number of additional culturable Ice+ species remain to be discovered in precipitation using the employed approach, at least at our sampling locations.
Figure 1.
Diversity of culturable Ice+ bacteria in precipitation collected in Virginia, USA. (a) Diversity of culturable Ice+ strains at the level of class for which at least one droplet froze at −8 °C during initial tests of colonies directly cultured from precipitation. (b) Diversity of culturable Ice+ strains at the level of genus for which activity was confirmed via semi-quantitative tests with pure cultures.
Culturable Ice+ strains belonging to the Gammaproteobacteria were present in 96% of the samples and represented up to 86% of the strains that were Ice+ during the first test. Actinobacteria and Bacilli that were Ice+ in the initial test, but could not be confirmed during standardized re-resting, were also regularly found in precipitation (74% and 57% of the samples, respectively) representing a total of 10% of all Ice+ strains. However, strains confirmed as Ice+ during standardized re-testing were almost exclusively Gammaproteobacteria (99.73%, Figure 1b) and were isolated from all but one of the precipitation samples. Within this class, members of the Pseudomonadaceae were the most frequent Ice+ bacteria (80.7%), followed by Enterobacteriaceae (17.1%) and Xanthomonadaceae (1.3%). While these three families include previously identified Ice+ species (Morris et al., 2008; Mortazavi et al., 2008; Joly et al., 2013; Šantl-Temkiv et al., 2015), we also confirmed two Ice+ strains in the Firmicute Lysinibacillus, a Gram-positive genus not previously known to include Ice+ bacteria (Figure 1b,Supplementary Table S3).
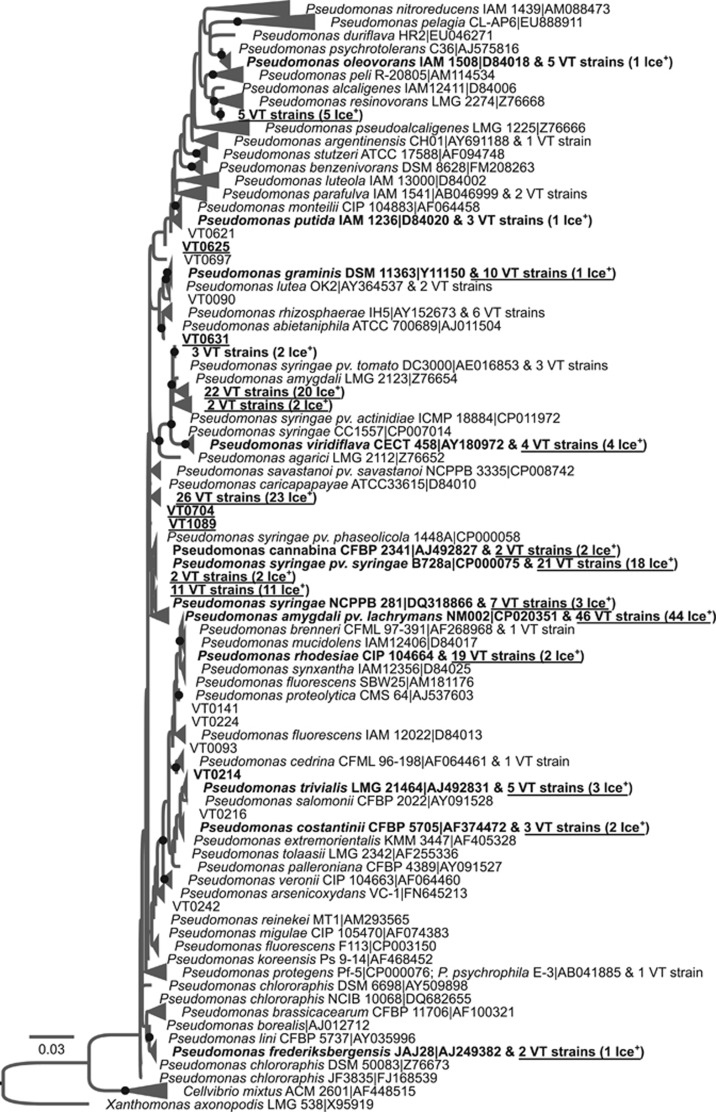
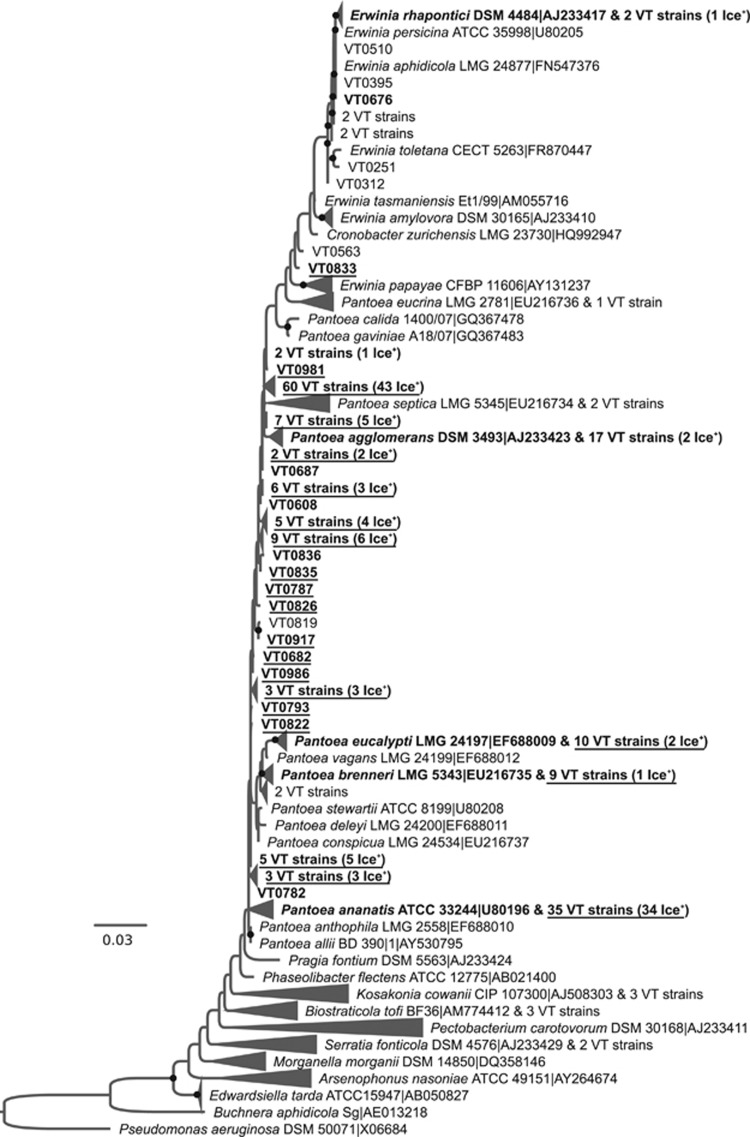
To better determine the evolutionary relationships between the diversity of Ice+ strains and characterized type strains, phylogenetic trees using 16S RNA gene sequences were built (Figures 2,3,4 and Supplementary Figures S3–S6). Pseudomonadaceae strains for which minimal INA during the first test was observed are located in several clades containing known environmental species. Most of the strains belong to the P. syringae complex and, to a smaller extent, to the Pseudomonas fluorescens complex (Garrido-Sanz et al., 2016) (Figure 2). While the diversity of Ice+ strains is well known in the P. syringae complex (Berge et al., 2014) and to some extent in the P. fluorescens complex (Hill et al., 2014), here we found confirmed Ice+ strains closely related to additional species in the P. fluorescens complex (Pseudomonas trivalis, Pseudomonas costantinii and Pseudomonas orientalis) and in the Pseudomonas mandelii group (VT0005, closely related to Pseudomonas frederiksbergensis and Pseudomonas lini). Additional Ice+ strains clustered with the known Ice+ species Pseudomonas abietaniphila, Pseudomonas graminis and Pseudomonas putida (Lee et al., 1995), and even in species in the Pseudomonas aeruginosa group never described as Ice+ before (closely related to Pseudomonas alcaligenes, Pseudomonas resinovorans and Pseudomonas oleovorans). In the Enterobacteriaceae family, confirmed Ice+ were distributed in the Pantoea and Erwinia genera and clustered with several species that had already been described as Ice+ (Lindow et al., 1978) (Figure 3). Most of the collection was closely related to Pa. ananatis and Pa. agglomerans, but some strains also clustered with Pa. brenneri, Pa. eucalypti, E. rhapontici or E. aphidicola. Because 16S rDNA sequence polymorphism was too low to determine a robust phylogeny, species-level identification of some of these strains may slightly change using additional genes or whole genomes. The phylogenetic tree of the Xanthomonadaceae family showed that most Ice+ strains clustered with Xanthomonas campestris (Supplementary Figure S5). However, three strains were closely related to X. translucens and one strain clustered with the genus Stenotrophomonas, a genus not previously known to be INA at −8 °C. Some strains that only showed minimal INA at the first characterization were identified as members of the genera Luteibacter and Luteimonas, and as relatives of Pseudoxanthomonas spadix.
Figure 2.
Evolutionary relationships between 229 Ice+ strains isolated from precipitation in Virginia, USA (labeled 'VT#') and 151 type strains of the Pseudomonadaceae family based on the alignment of partial 930bp-long 16S rDNA sequences. A maximum likelihood tree was built using the GTR model and 100 bootstrap replicates. Gaps were considered as missing data and were partially deleted with a site coverage cut-off of 95%. The strain Xanthomonas axonopodis LMG 538-T was used as a root. Only bootstrap values over 50% were included and are symbolized by black dots. Strains that were Ice+ in standardized re-tests at −8 °C are labelled in bold and are underlined, those active at −10 °C or −12 °C in standardized re-tests are labelled in bold only, and those that were only active in the initial test but not in the re-tests are neither underlined nor in bold. When more than one VT# strain had the same 16S rDNA sequence, only the number of strains is indicated and Ice+ strains that were confirmed in repeated tests are labelled ‘Ice+’. RDP names are composed of the species name, the strain code and the accession number in the public database NCBI. The name of the type strain closest to Ice+ strains was chosen to represent collapsed branches. A version of the tree in which all VT strains are listed is given in Supplementary Figure S3.
Figure 3.
Evolutionary relationships between Ice+ strains isolated from precipitation in Virginia, USA, and other members of the Enterobacteriaceae family based on the alignment of partial 16S rDNA sequences. The phylogenetic tree was built from 212 strains isolated in this study and 241 type strains. The strain Pseudomonas aeruginosa DSM 50071 was used as a root. See Figure 2 for details about tree construction and labels. A version of the tree in which all VT# strains are listed is given in Supplementary Figure S4.
Figure 4.
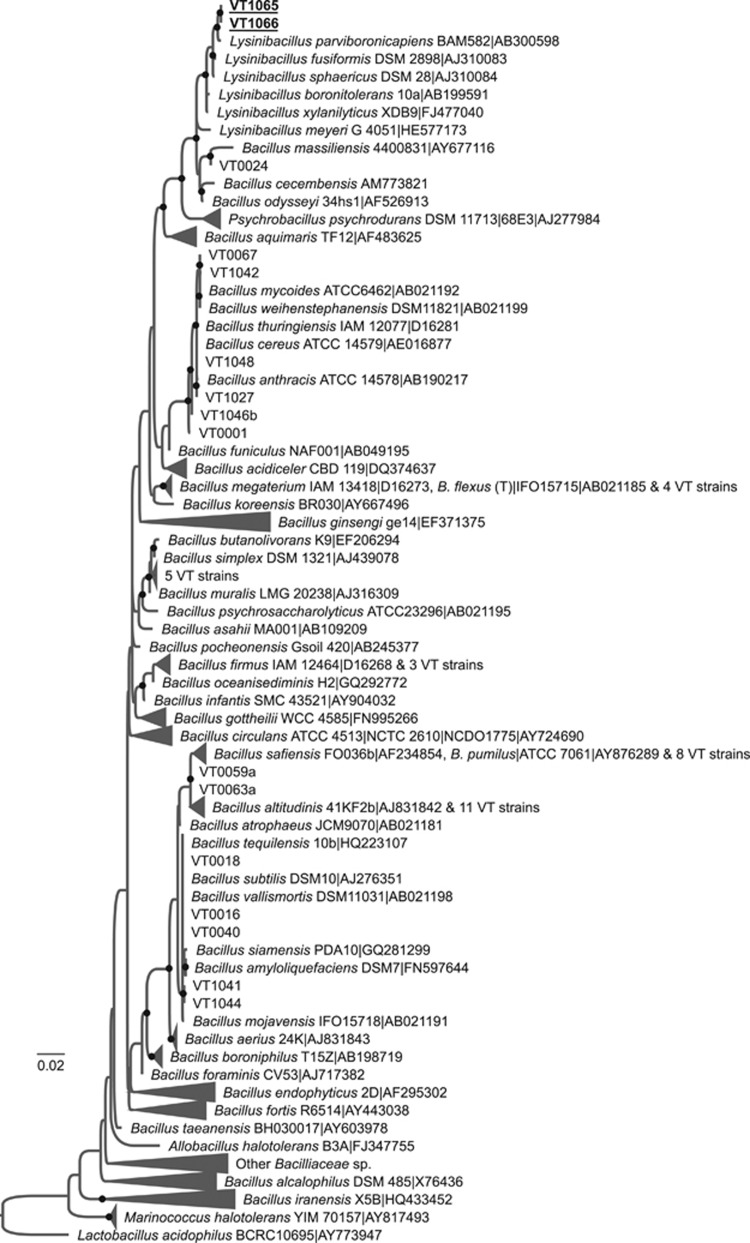
Evolutionary relationships between Ice+ strains isolated from precipitation and other members of the Bacilliaceae family based on the alignment of partial 16S rDNA sequences. The phylogenetic tree was built from 47 strains isolated in this study and 353 type strains. The strain Lactobacillus acidophilus BCRC10695 was used as a root. See Figure 2 for details about tree construction and labels. A version of the tree in which all VT# strains are listed is given in Supplementary Figure S6.
Most of the Bacillaceae strains that did not have reproducible INA were related to the Bacillus pumilus group, the Bacillus megaterium group and the Bacillus subtilis group (Figure 4). The two strains with reproducible INA, VT1065 and VT1066, had identical 16S rDNA sequences and showed over 99% DNA identity to the type strains of Lysinibacillus parviboronicapiens, Lysinibacillus fusiformis, and Lysinibacillus sphaericus.
INA of Lysinibacillus is heat, lysozyme, and proteinase-resistant and secreted
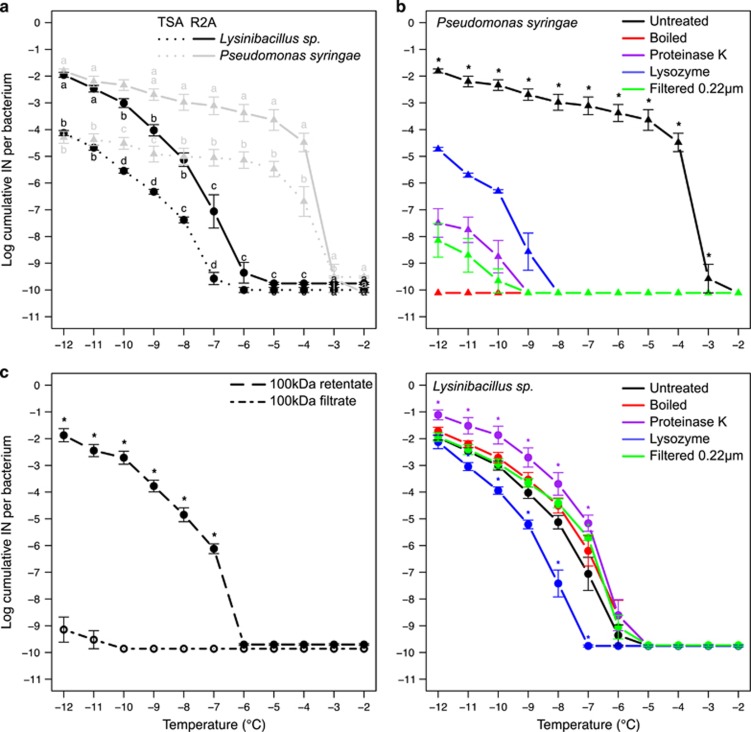
Since INA in Gram-positive bacteria has not been characterized, we sought to determine the nature of the Lysinibacillus INA (LINA) molecule in comparison with the well-known INA protein (INP) of P. syringae. First, we showed that when grown on the same culture medium (R2A), and when tested under the same conditions, the Lysinibacillus strains produced at least seven orders of magnitude fewer IN active between −6 and −3 °C compared to P. syringae. However, this difference decreased rapidly when lowering temperatures and became equal below −11 °C (Figure 5a). To test whether nutrient availability may impact the production of the INP and of the LINA molecule, we compared these ice nucleation spectra with those obtained from TSA cultures. Growing bacteria on TSA, a medium richer in nutrients, lowered the production of INA molecules by two orders of magnitude for both bacterial classes (Figure 5b), which indicates that growth in oligotrophic conditions promotes the production of both types of INA molecules.
Figure 5.
Ice nucleation spectra of Lysinibacillus sp. in different growth conditions and after different treatments. For each temperature, average values were calculated from at least four independent cultures and bars correspond to standard errors. Lysinibacillus sp. curves were obtained using both strains, VT1065 and VT1066. The minimum detection threshold obtained for P. syringae and Lysinibacillus sp. were 10−10.11 and 10−9.86 IN per bacterium respectively. (a) Ice nucleation spectra of Lysinibacillus sp. • compared to Pseudomonas syringae strain CC94 ▴, using cultures grown for 24 h on either R2A or TSA. Same letters associated to all values for each temperature mean that values were not significantly different according to multiple Student t-tests comparisons. (b) Ice nucleation spectra of cell suspensions obtained from cultures grown on R2A that had been incubated with Proteinase K, lysozyme, filtered through a 0.22 μm membrane or heated at 99 °C for 1 h. For P. syringae CC94, the number of cumulative IN per bacterium was significantly different for all treatments compared to the untreated cell suspension between −12 and −3 °C (denoted by an asterisk). For Lysinibacillus sp., stars were used to label values of treated cell suspensions that were significantly different from the untreated suspension. (c) Ice nucleation activity of the Lysinibacillus sp. 0.22 μm membrane filtrate after being either resuspended from a 100 kDa membrane (the retentate) or filtered through the 100 kDa membrane (the filtrate).
INA of Ice+ Gammaproteobacteria is due to the INA protein, which is anchored to the membrane and sensitive to heat (>95 °C) and lysozyme treatment (Morris et al., 2004; Christner et al., 2008). As expected, by (i) heating cells, (ii) cleaving peptidic chains with Proteinase K, (iii) breaking peptidoglycans with lysozymes, and (iv) filtering the cell suspensions through a 0.22 μm membrane, we observed a significant decrease, or even total suppression, of INA of P. syringae (Figure 5b). Applying the same treatments to the Lysinibacillus strains, very different results were obtained; ice nucleation spectra of boiled or filtered cell suspensions were not significantly different from those of untreated cell suspensions. Therefore, the LINA molecule is secreted and its activity is heat-resistant. This resistance to heat strongly suggests that LINA is not a protein. This was confirmed with cell suspensions incubated with Proteinase K, after which INA was even higher by one order of magnitude between −7 and −12 °C (Figure 5b). Lysozyme treatment slightly reduced INA of Lysinibacillus at all temperatures. Therefore, part of LINA may either depend on glycosydic bonds, or lysozyme treatment of Lysinibacillus releases molecules that interfere with INA. Finally, to get an indication of the size of the LINA molecule, the 0.22 μm filtrate was filtered through a 100 kDa pore-sized membrane. Retentate from the 100 kDa pore-sized membrane (which retains molecules larger than 30–90 nm) showed an INA spectrum similar to that of the original cell suspension, but the filtrate did not (Figure 5c). Therefore, the LINA molecule appears to be larger than 30–90 nm or the molecule strongly interacts with the membrane interfering with its passage.
Discussion
Although Ice+ bacteria have been well known to be the most active IN at warm temperatures since the 1970s (Maki et al., 1974), information about their diversity and the conditions associated with their deposition with precipitation is limited. To fill this knowledge gap, Ice+ bacteria were isolated from 23 precipitation samples in Virginia on seven different media types. This study revealed that the most common culturable Ice+ bacteria in precipitation are Gammaproteobacteria. For this class, INA had been observed previously and the underlying mechanism is well characterized (Morris et al., 2004). However, additional Ice+ species and one additional Ice+ genus, Stenotrophomonas, were found for the first time to nucleate ice at −8 °C. Although precipitation samples were only collected in one geographic area, an astonishing species richness (>30 OTUs) of known and unknown Ice+ species was measured. Importantly, two Ice+ Lysinibacillus sp. strains provide the first evidence to date of a Gram-positive bacterium exhibiting reproducible INA with magnitudes comparable to P. syringae at −8 °C. Ponder et al. (2005) and Mortazavi et al. (2008) had previously reported INA for some Gram-positive bacteria but at lower temperatures and with lower magnitude. Moreover, two Exiguobacterium strains and a Psychrobater strain identified by Ponder et al. (2005) were inactive under our assay conditions even at −12 °C (data not shown).
Although the LINA molecule still needs to be identified and purified to determine its structure, we showed that the active molecule is not a protein and is secreted, contrary to the INA protein produced by Gammaproteobacteria that is part of the outer membrane (with the exception of Erwinia herbicola which can shed some IN (Phelps et al., 1986)). From an evolutionary perspective, this means that INA emerged at least twice over the course of Eubacteria evolution through two independent mechanisms. This is a remarkable example of convergent evolution, in which INA has evolved in phylogenetically unrelated organisms. Determining the species niche and ecology of Ice+ Lysinibacillus will be required to better understand the role of INA in the life history of this organism. Since the Ice+ Lysinibacillus strains are most closely related to soil-borne bacteria and entomopathogens (Ahmed et al., 2007; Miwa et al., 2009) and the type strain of L. parviboronicapiens, a soil-borne bacterium as well (Miwa et al., 2009), is also Ice+ (data not shown), INA in these strains may not have evolved as a primary adaptation to atmospheric dissemination as proposed in the bioprecipitation theory for INA of P. syringae (Morris et al., 2014).
The discovery of INA in Lysinibacillus underscores the need to research other INA microorganisms and mechanisms. Although the rarefaction curve suggests that only a small number of INA species remain to be discovered (at least at our sample location in Virginia), we may still be underestimating the true biodiversity of Ice+ bacteria. In fact, many microorganisms are not culturable or only express a small set of phenotypes in culture (Barer and Harwood, 1999; Kaeberlein et al., 2002). INA is possibly one of these phenotypes, as suggested by the effect of incubation temperature on INA observed by Ponder et al. (2005) in psychroactive permafrost strains and based on studies reviewed by Murray et al. (2012). Consequently, discrepancies we observed between results from INA tests of colonies directly plated from precipitation samples and those from purified strains may be explained in part by growth conditions not conducive for the expression of INA, even if it is likely that some colonies for which only one droplet froze at −8 °C during the initial tests were simply false positives. Moreover, since nutrient limitation induced higher INA in Lysinibacillus sp. (Figure 5c) and the same was observed previously for P. syringae (Nemecek-Marshall et al., 1993), INA molecules might generally be produced in higher amounts in oligotrophic conditions, like those found in the atmosphere. Therefore, detection of new Ice+ species/mechanisms can probably still be improved by optimizing culture media and growth and incubation conditions.
The size and the chemical properties of the LINA molecule also raises questions about the way abundance of bacterial IN has been estimated in the atmosphere. In fact, because it has been assumed that bacterial IN are exclusively of proteinaceous origin (Morris et al., 2004; O'Sullivan et al., 2015), bacterial IN are often quantified comparing IN concentrations before and after boiling samples that have been filtered and concentrated with a 0.22 μm membrane (Christner et al., 2008). Yet, the size of the LINA molecule is only nanometers in scale and its activity resists boiling and is independent of lipidic membranes, similar to the ice nucleation particle produced by Fusarium (O'Sullivan et al., 2015), Mortierella (Fröhlich-Nowoisky et al., 2015) and pollen (Pummer et al., 2015). This also means that single cells of these organisms could produce many IN over their residence time in the atmosphere. This differs from the Gammaproteobacteria, for which INA is dependent on cell integrity and for which the number of IN is limited to the number of bacterial cells in the atmosphere. Moreover, because of their size, bacterial LINA-like molecules could represent some of the abundant nanometer-sized Ice+ particles active at warm temperatures identified by Vali (1966) and O'Sullivan et al. (2015) and more recently by Du et al. (2017) or Šantl-Temkiv et al. (2015).
In regard to conditions linked to higher concentrations of total IN active at −8 °C in precipitation (and thus assumed to be biological), the most striking results were: (i) alkaline and mineralized precipitation water and (ii) precipitation initiated within air masses in which relative humidity was lower and that travelled over larger distances. These results are consistent with other experimental and modelling studies suggesting that the frequency of deposition of biological IN like P. syringae increases with alkalinity and conductivity of precipitation that is derived from rapidly travelling air masses (Monteil et al., 2014a). It is therefore possible that the link between the concentration of IN and the distance travelled by air masses is due to a greater uptake of IN from Earth surfaces by upward air fluxes.
Importantly, do our results support a role for bacterial IN in the initiation of precipitation? Crawford et al. (2012) estimated that a small amount of primary ice, as low as 10 IN per m3, would be enough to initiate precipitation at temperatures as high as −8 °C. We thus estimated the number of IN produced by Ice+ bacteria in clouds based on the mean concentration of Ice+ bacteria in our precipitation samples (Supplementary Table S4). Interestingly, we inferred fewer than 10 IN of bacterial origin per m3 of cloud in all samples but one when we assume that (i) Ice+ bacteria are neither lost nor scavenged while precipitation falls, (ii) expression of genes that determine INA in the clouds is similar to that under our growth conditions, (iii) all bacteria in precipitation are culturable, and (iv) one Ice+ colony plated from precipitation corresponds to one IN in the clouds. However, changing these assumptions our estimate increases or decreases by orders of magnitudes. In particular, the number of IN may be an underestimation if many Ice+ bacteria in precipitation were not culturable or may not have expressed INA genes under the conditions we employed. At the other extreme, inferring the number of IN produced by a single bacterial cell in clouds from our droplet freezing assay, the concentration of IN in clouds would only be 1.3 × 10−4 per m3, by far insufficient to induce precipitation. Even assuming that the concentration of total IN in our precipitation samples (based on the droplet freezing assay) were all of bacterial origin, there would not be a sufficient number of bacterial IN in clouds to initiate precipitation at −8 °C (they may be sufficient at lower temperatures but we did not determine concentrations of IN below −8 °C). Moreover, the droplet freezing assay may underestimate the number of IN (Emersic et al., 2015; Whale et al., 2015). Therefore, developing laboratory assays that accurately quantify IN under conditions to which IN are exposed in clouds needs to be prioritized to further investigate the role of biological IN in atmospheric processes.
In summary, we expanded our knowledge of the genetic diversity of Ice+ bacteria and the factors that correlate with their deposition. Most importantly, we uncovered a new INA mechanism in Gram-positive bacteria. This novel mechanism, which is based on a heat-stable and proteinase-resistant secreted molecule, challenges our current view of bacterial INA as being exclusively proteinaceous and thus heat-sensitive. This discovery of the first new bacterial INA mechanism since P. syringae over 40 years ago will require re-interpretation of some previous data on biological and non-biological INA detected in clouds, aerosols and precipitation. Our results also ask for optimization of protocols for detecting and isolating Ice+ species and for quantifying IN under cloud-like conditions. Finally, identification and characterization of the LINA molecule and surveys of its concentration (and the concentration of Ice+ Lysinibacillus) will be important to determine if Ice+ Lysinibacillus bacteria influence atmospheric processes leading to precipitation.
Acknowledgments
The authors gratefully acknowledge the NOAA Air Resources Laboratory (ARL) for the provision of the HYSPLIT transport and dispersion model and/or READY website (http://www.ready.noaa.gov) used in this publication. This research was supported by the National Science Foundation under grant DEB-1241068. Funding to Boris A. Vinatzer and David G. Schmale III was also provided in part by the Virginia Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, US Department of Agriculture.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Ahmed I, Yokota A, Yamazoe A, Fujiwara T. (2007). Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int J Syst Evol Micr 57: 1117–1125. [DOI] [PubMed] [Google Scholar]
- Amato P, Joly M, Schaupp C, Attard E, Möhler O, Morris CE et al. (2015). Survival and ice nucleation activity of bacteria as aerosols in a cloud simulation chamber. Atmos Chem Phys Discuss 15: 6455–6465. [Google Scholar]
- Arny DC, Lindow SE, Upper CD. (1976). Frost sensitivity of Zea mays increased by application of Pseudomonas syringae. Nature 262: 282–284. [Google Scholar]
- Attard E, Yang H, Delort AM, Amato P, Poeschl U, Glaux C et al. (2012). Effects of atmospheric conditions on ice nucleation activity of Pseudomonas. Atmos Chem Phys 12: 10667–10677. [Google Scholar]
- Barer MR, Harwood CR. (1999). Bacterial viability and culturability. Adv Microb Physiol 41: 93–137. [DOI] [PubMed] [Google Scholar]
- Berge O, Monteil CL, Bartoli C, Chandeysson C, Guilbaud C, Sands DC et al. (2014). A user's guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. Plos One 9: e105547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigg EK, Soubeyrand S, Morris CE. (2015). Persistent after-effects of heavy rain on concentrations of ice nuclei and rainfall suggest a biological cause. Atmos Chem Phys 15: 2313–2326. [Google Scholar]
- Christner BC, Morris CE, Foreman CM, Cai RM, Sands DC. (2008). Ubiquity of biological ice nucleators in snowfall. Science 319: 1214–1214. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarell DM, Sun Y et al. (2014). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 1: D633–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinidou HA, Hirano SS, Baker LS, Upper CD. (1990). Atmospheric dispersal of ice nucleation active bacteria—the role of rain. Phytopathology 80: 934–937. [Google Scholar]
- Crawford I, Bower KN, Choularton TW, Dearden C, Crosier J, Westbrook C et al. (2012). Ice formation and development in aged, wintertime cumulus over the UK: observations and modelling. Atmos Chem Phys 12: 4963–4985. [Google Scholar]
- Diehl K, Matthias-Maser S, Jaenicke R, Mitra SK. (2002). The ice nucleating ability of pollen: Part II. Laboratory studies in immersion and contact freezing modes. Atmos Res 61: 125–133. [Google Scholar]
- Du R, Du P, Lu Z, Ren W, Liang Z, Qin S et al. (2017). Evidence for a missing source of efficient ice nuclei. Sci Rep 7: 39673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emersic C, Connolly PJ, Boult S, Campana M, Li Z. (2015). Investigating the discrepancy between wet-suspension- and dry-dispersion-derived ice nucleation efficiency of mineral particles. Atmos Chem Phys 15: 11311–11326. [Google Scholar]
- Fröhlich-Nowoisky J, Hill TCJ, Pummer BG, Yordanova P, Franc GD, Pöschl U. (2015). Ice nucleation activity in the widespread soil fungus Mortierella alpina. Biogeosciences 12: 1057–1071. [Google Scholar]
- Garcia E, Hill TC, Prenni AJ, DeMott PJ, Franc GD, Kreidenweis SM. (2012). Biogenic ice nuclei in boundary layer air over two US high plains agricultural regions. J Geophys Res 117. [Google Scholar]
- Garnham CP, Campbell RL, Walker VK, Davies PL. (2011). Novel dimeric beta-helical model of an ice nucleation protein with bridged active sites. Bmc Structural Biol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Sanz D, Meier-Kolthoff J, Göker M, Martín M, Rivilla R, Redondo-Nieto M. (2016). Genomic and genetic diversity within the Pseudomonas fluorescens Complex. PLoS ONE 11: e0150183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TC, Moffett B, DeMott PJ, Georgakopoulos DG, Stump WL, Franc GD. (2014). Measurement of ice nucleation-active bacteria on plants and in precipitation by quantitative PCR. Appl Env Microbiol 80: 1256–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano SS, Baker LS, Upper CD. (1985). Ice nucleation temperature of individual leaves in relation to population sizes of ice nucleation active bacteria and frost injury. Plant Physiol 77: 259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoose C, Mohler O. (2012). Heterogeneous ice nucleation on atmospheric aerosols: a review of results from laboratory experiments. Atmos Chem Phys 12: 9817–9854. [Google Scholar]
- Joly M, Attard E, Sancelme M, Deguillaume L, Guilbaud C, Morris CE et al. (2013). Ice nucleation activity of bacteria isolated from cloud water. Atmos Environ 70: 392–400. [Google Scholar]
- Kaeberlein T, Lewis K, Epstein SS. (2002). Isolating ‘uncultivable’ microorganims in pure culture in a simulated natural environment. Science 296: 1127–1129. [DOI] [PubMed] [Google Scholar]
- Kajava A, Lindow S. (1993). A model of the three-dimensional structure of ice nucleation proteins. J Mol Biol 232: 709–717. [DOI] [PubMed] [Google Scholar]
- Kim HK, Orser C, Lindow SE, Sands DC. (1987). Xanthomonas campestris pv. translucens strains active in ice nucleation. Plant Dis 71: 994–997. [Google Scholar]
- Lee MR, Lee RE, Strong-Gunderson JM, Minges SR. (1995). Isolation of ice-nucleating active bacteria from the freeze-tolerant frog Rana sylvatica. Cryobiology 32: 358–365. [DOI] [PubMed] [Google Scholar]
- Lindemann J, Constantinidou HA, Barchet WR, Upper CD. (1982). Plants as sources of airborne bacteria, including ice nucleation active bacteria. Appl Env Microbiol 44: 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow S, Arny DC, Upper CD. (1978). Erwinia herbicola: a bacterial ice nucleus active in increasing frost injury to corn. Phytopathology 68: 523–527. [Google Scholar]
- Lindow SE. (1983). The role of bacterial ice nucleation in frost injury to plants. Annu Rev Phytopathol 21: 363–384. [Google Scholar]
- Maki LR, Galyan EL, Chang-Chien MM, Caldwell DR. (1974). Ice nucleation induced by Pseudomonas syringae. Appl Microbiol 28: 456–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H, Ahmed I, Yokota A, Fujiwara T. (2009). Lysinibacillus parviboronicapiens sp. nov., a low-boron-containing bacterium isolated from soil. Int J Syst Evol Micr 59: 1427–1432. [DOI] [PubMed] [Google Scholar]
- Monteil CL, Guilbaud C, Glaux C, Lafolie F, Soubeyrand S, Morris CE. (2012). Emigration of the plant pathogen Pseudomonas syringae from leaf litter contributes to its population dynamics in alpine snowpack. Environ Microbiol 14: 2099–2112. [DOI] [PubMed] [Google Scholar]
- Monteil CL, Bardin M, Morris CE. (2014. a). Features of air masses associated with the deposition of Pseudomonas syringae and Botrytis cinerea by rain and snowfall. ISME J 8: 2290–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil CL, Lafolie F, Laurent J, Clement JC, Simler R, Travi Y et al. (2014. b). Soil water flow is a source of the plant pathogen Pseudomonas syringae in subalpine headwaters. Environ Microbiol 16: 2038–2052. [DOI] [PubMed] [Google Scholar]
- Morris CE, Glaux C, Latour X, Gardan L, Samson R, Pitrat M. (2000). The relationship of host range, physiology, and genotype to virulence on cantaloupe in Pseudomonas syringae from cantaloupe blight epidemics in France. Phytopathology 90: 636–646. [DOI] [PubMed] [Google Scholar]
- Morris CE, Georgakopoulos DG, Sands DC. (2004). Ice nucleation active bacteria and their potential role in precipitation. J Phys IV 121: 87–103. [Google Scholar]
- Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffiere A et al. (2008). The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J 2: 321–334. [DOI] [PubMed] [Google Scholar]
- Morris CE, Conen F, Huffman JA, Phillips V, Pöschl U, Sands DC. (2014). Bioprecipitation: feedbacks linking Earth history, ecosystem dynamics and land use through biological ice nucleators in the atmosphere. Global Change Biol 20: 341–351. [DOI] [PubMed] [Google Scholar]
- Mortazavi R, Hayes CT, Ariya PA. (2008). Ice nucleation activity of bacteria isolated from snow compared with organic and inorganic substrates. Environ Chem 5: 373–381. [Google Scholar]
- Mülmenstädt J, Sourdeval O, Delanoë J, Quaas J. (2015). Frequency of occurrence of rain from liquid-, mixed-, and ice-phase clouds derived from A-Train satellite retrievals. Geophys Res Lett 42: 6502–6509. [Google Scholar]
- Murray BJ, O'Sullivan D, Atkinson JD, Webb ME. (2012). Ice nucleation by particles immersed in supercooled cloud droplets. Chem Soc Rev 41: 6519–6554. [DOI] [PubMed] [Google Scholar]
- Nemecek-Marshall M, Laduca R, Fall R. (1993). High level expression of ice nuclei in a Pseudomonas syringae strain is induced by nutrient limitation and low temperature. J Bacteriol 175: 4062–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan D, Murray BJ, Ross JF, Whale TF, Price HC, Atkinson JD et al. (2015). The relevance of nanoscale biological fragments for ice nucleation in clouds. Sci Rep 5: 8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Usui K, Livingstone RA, Fischer SA, Pfaendtner J, Backus EHG et al. (2016). Ice-nucleating bacteria control the order and dynamics of interfacial water. Sci Adv 2: e1501630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petters MD, Wright TP. (2015). Revisiting ice nucleation from precipitation samples. Geophys Res Lett 42: 8758–8766. [Google Scholar]
- Phelps P, Giddings TH, Prochoda M, Fall R. (1986). Release of cell-free ice nuclei by Erwinia herbicola. J Bacteriol 167: 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder MA, Gilmour SJ, Bergholz PW, Mindock CA, Hollingsworth R, Thomashow MF et al. (2005). Characterization of potential stress responses in ancient Siberian permafrost psychroactive bacteria. FEMS Microbiol Ecol 53: 103–115. [DOI] [PubMed] [Google Scholar]
- Pouleur S, Richard C, Martin JG, Antoun H. (1992). Ice nucleation activity in Fusarium acuminatum and Fusarium avenaceum. Appl Env Microbiol 58: 2960–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pummer BG, Bauer H, Bernardi J, Bleicher S, Grothe H. (2012). Suspendable macromolecules are responsible for ice nucleation activity of birch and conifer pollen. Atmos Chem Phys 12: 2541–2550. [Google Scholar]
- Pummer BG, Budke C, Augustin-Bauditz S, Niedermeier D, Felgitsch L, Kampf CJ et al. (2015). Ice nucleation by water-soluble macromolecules. Atmos Chem Phys 15: 4077–4091. [Google Scholar]
- R Core Team. (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Sands DC, Langhans VE, Scharen AL, de Smet G. (1982). The association between bacteria and rain and possible resultant meteorological implications. J Hungarian Meteorol Ser 86: 148–152. [Google Scholar]
- Šantl-Temkiv T, Sahyoun M, Finster K, Hartmann S, Augustin-Bauditz S, Stratmann F et al. (2015). Characterization of airborne ice-nucleation-active bacteria and bacterial fragments. Atmos Environ 109: 105–117. [Google Scholar]
- Sarkar SF, Guttman DS. (2004). Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl Env Microbiol 70: 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopelli E, Conen F, Morris CE, Herrmann E, Bukowiecki N, Alewell C. (2015). Ice nucleation active particles are efficiently removed by precipitating clouds. Sci Rep 5: 16433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vali G. (1966). Sizes of atmospheric ice nuclei. Nature Biotechnol 212: 384–385. [Google Scholar]
- Vali G. (1971). Quantitative evaluation of experimental results on the heterogeneous freezing nucleation of supercooled liquids. J Atmos Sci 28: 402–409. [Google Scholar]
- von Blohn N, Mitra SK, Diehl K, Borrmann S. (2005). The ice nucleating ability of pollen Part III: New laboratory studies in immersion and contact freezing modes including more pollen types. Atmos Res 78: 182–189. [Google Scholar]
- Whale TF, Murray BJ, O'Sullivan D, Wilson TW, Umo NS, Baustian KJ et al. (2015). A technique for quantifying heterogeneous ice nucleation in microlitre supercooled water droplets. Atmos Meas Tech 8: 2437–2447. [Google Scholar]
- Wolber PK, Deininger CA, Southworth MW, Vandekerckhove J, Vanmontagu M, Warren GJ. (1986). Identification and purification of a bacterial ice nucleation protein. Proc Natl Acad Sci USA 83: 7256–7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.