Abstract
Sulfide (H2S, HS− and S2−) oxidation to sulfite and thiosulfate by heterotrophic bacteria, using sulfide:quinone oxidoreductase (SQR) and persulfide dioxygenase (PDO), has recently been reported as a possible detoxification mechanism for sulfide at high levels. Bioinformatic analysis revealed that the sqr and pdo genes were common in sequenced bacterial genomes, implying the sulfide oxidation may have other physiological functions. SQRs have previously been classified into six types. Here we grouped PDOs into three types and showed that some heterotrophic bacteria produced and released H2S from organic sulfur into the headspace during aerobic growth, and others, for example, Pseudomonas aeruginosa PAO1, with sqr and pdo did not release H2S. When the sqr and pdo genes were deleted, the mutants also released H2S. Both sulfide-oxidizing and non-oxidizing heterotrophic bacteria were readily isolated from various environmental samples. The sqr and pdo genes were also common in the published marine metagenomic and metatranscriptomic data, indicating that the genes are present and expressed. Thus, heterotrophic bacteria actively produce and consume sulfide when growing on organic compounds under aerobic conditions. Given their abundance on Earth, their contribution to the sulfur cycle should not be overlooked.
Introduction
Microorganisms are able to metabolize organic sulfur and release H2S under aerobic conditions (Clarke, 1953). Cysteine desulfhydrase and 3-mercaptopyruvate sulfurtransferase are two key enzymes involved in releasing H2S from cysteine catabolism (Morra and Dick, 1991; Oguri et al., 2012). Cystathionine β-synthase and cystathionine γ-lyase, involved in converting methionine to cysteine, catalyze side reactions that release H2S (Shatalin et al., 2011). The self-produced sulfide is known to function as a signaling molecule in mammals (Wagner, 2009) and to act as a general defense against oxidative stress to confer bacteria with resistance to antibiotics (Shatalin et al., 2011; Oguri et al., 2012). However, the prevalence of sulfide production by heterotrophic bacteria during aerobic growth has not been documented.
The produced sulfide may accumulate and inhibit aerobic respiration (Sohn et al., 2000). Mammalian cells prevent the accumulation of sulfide through oxidation by the concerted actions of three enzymes: sulfide:quinone oxidoreductase (SQR), persulfide dioxygenase (PDO), and rhodanese that is a type of sulfurtransferases (Hildebrandt and Grieshaber, 2008). Recently, we have reported that heterotrophic bacteria also possess SQR, PDO and rhodanese, oxidizing sulfide to sulfite and thiosulfate (Liu et al., 2014; Xin et al., 2016). Gram-negative bacteria contain at least two types of PDOs: the type I PDOs include the human PDO (ETHE1), plant PDOs, and bacterial homologs, and the type II PDOs consist of several reported proteins from Proteobacteria (Liu et al., 2014; Xin et al., 2016). Although both types of PDOs use glutathione persulfide (GSSH) as the substrate, they share limited sequence similarity and the substrate binding sites are different between them (Sattler et al., 2015). Further, a PDO (CstB) from the Gram-positive bacterium Staphylococcus aureus uses low-molecular-weight persulfides (RSSH and RSS−), rather than GSSH, as the substrates (Shen et al., 2015). Bacterial SQRs are grouped into six types, I to VI (Marcia et al., 2010). Some PDO-containing heterotrophic bacteria harbor sqr and pdo in a gene cluster (Liu et al., 2014; Shen et al., 2015; Xin et al., 2016). Rhodaneses are universally present in microorganisms, plants, and animals, and a rhodanese domain is often fused to SQR or PDO in heterotrophic bacteria (Guimaraes et al., 2011; Liu et al., 2014; Shen et al., 2015; Xin et al., 2016). In Gram-negative bacteria, SQR oxidizes sulfide to polysulfide, which spontaneously reacts with GSH to produce GSSH, and PDO oxidizes GSSH to sulfite; sulfite spontaneously reacts with polysulfide to produce thiosulfate. Rhodanese speeds up the reaction of polysulfide with GSH to produce GSSH, but the reaction can occur spontaneously (Xin et al., 2016).
We report here that most bacteria produced sulfide from rich media during aerobic growth and bacteria with SQR and PDO oxidized the self-produced sulfide. Further, the oxidizers also oxidized sulfide produced by bacteria without the enzymes in mixed cultures. Some bacteria contained only SQR or PDO, and they were able to cooperate to oxidize sulfide to thiosulfate. Bioinformatics analysis showed that sqr and pdo genes are common in sequenced bacterial genomes from GenBank, marine metagenomes and metatranscriptomes.
Materials and methods
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table 1. Lysogeny broth (LB) was used for culturing most bacteria. Zymomonas mobilis ATCC 31821 and Gluconobacter oxydans 621H were grown in the Rich Medium (Goodman et al., 1982) and d-sorbitol medium (Yang et al., 2008), respectively. The sulfur contents of yeast extract and tryptone (Oxoid, Thermo, Beijing, China) were measured by using an elemental analyzer (The Vario EL Cube, Elementar Trading, Shanghai, China).
Cloning and gene knockout
The pdo and sqr genes were PCR-amplified from genomic DNA with primers (Supplementary Table 2) and cloned into pBBR1MCS2 or pBBR1MCS5 for expression (Kovach et al., 1995). The vector pK18mobsacBtet was used to generate deletions in Cupriavidus pinatubonensis JMP134 and Pseudomonas aeruginosa PAO1 (Harighi, 2009). Details are given in Supplementary Methods.
Testing bacteria for H2S production
Selected bacteria were used to test for H2S production. Bacteria were transferred into 2 ml of LB or the specified media in a 15-ml glass tube, a paper strip with lead(II)-acetate was affixed at the top of the tube with a rubber stopper. The culture was incubated with shaking. After incubation, the paper strip was photographed to detect any lead(II)-sulfide black precipitates, as a measure of H2S production. LB spiked with known concentrations of NaHS was used to generate dark stains on lead(II)-acetate paper strips for the estimation of H2S production from bacteria cultures. The estimation was done by visually matching the darkness of the paper strips.
The sulfide spiking test with selected strains
Resting cells of C. pinatubonensis JMP134, C. pinatubonensis 2 K (the pdo2 and sqr deletion mutant), P. aeruginosa PAO1, P. aeruginosa 3 K (the pdo, sqr1 and sqr2 deletion mutant), E. coli BL21(DE3), E. coli BL21(DE3)(Papdo-Pasqr2), Bacillus subtilis 168, Agrobacterium tumefaciens C58, Klebsiella pneumonia DSM30104, Serratia fonticola DSM4576 and Corynebacterium vitaeruminis DSM20294 were tested to oxidize spiked sulfide. The selected bacteria were cultured in LB at 30 °C with shaking and harvested at OD600nm of about 1. If the bacteria were induced with isopropyl-β-d-thiogalactopyranoside (IPTG) or sulfide, they were first cultured to OD600nm of about 0.5. E. coli was induced with 0.4 mm IPTG; Serratia fonticola DSM4576, C. vitaeruminis DSM20294, and Sinorhizobium meliloti 1021 were induced with 200 μm NaHS for 2 h before harvesting. The harvested cells were washed with 50 mm Tris buffer (pH 8.0) and suspended in the Tris buffer with 50 μm diethylenetriaminepentaacetic acid (DTPA) at OD600nm of 1. DTPA was added to minimize sulfide oxidation catalyzed by trace transition metals in the buffer (Shen et al., 2012). Then, 100 μm NaHS was added into the cell suspension to initiate sulfide oxidation, and sulfide was determined at various time intervals. Three replicates were done at the same time. Sulfide was analyzed by a colorimetric method (Kamyshny, 2009).
Sulfide oxidation by combined resting cells containing sqr or pdo
The resting were induced and prepared as above. In combined tests, C. vitaeruminis DSM20294 and S. meliloti 1021 were equally mixed, and E. coli BL21 (DE3)(Cpsqr) and E. coli BL21 (DE3)(Cppdo2) were equally mixed at individual OD600nm of 2 and combined OD600nm of 4. NaHS (1 mm) was added into the cell suspension to initiate sulfide oxidation; sulfide, polysulfide, sulfite and thiosulfate were determined at various time points as we previously reported (Xin et al., 2016).
Determine the end products of sulfide oxidation by resting cells
E. coli BL21(DE3) and its recombinant cells were suspended in 100 mm Tris buffer (pH 8) at OD600nm of 2. One mililiter of the cell suspension was transferred into a 15-ml glass tube. Freshly prepared NaHS (500 μm) was added to initiate the reaction. The tube was capped with a rubber stopper and incubated at 25 °C without shaking to minimize autoxidation and volatilization. The sulfide, sulfane sulfur that includes polysulfide and persulfide, sulfite and thiosulfate were analyzed at various time intervals. For details, see Supplementary Methods.
Heterotrophic sulfide oxidizers from various environmental samples
Samples were collected in the summer of 2015. Soil samples were from a wheat field and forest, and freshwater samples were from a lake, around Jinan, Shandong, China; near-shore seawater samples were collected from Qingdao, Shandong, China. To collect soil samples, the top layer of 2 centimeter (cm) was removed and soil from 2–10 cm depth was transferred into a sterile bottle. Water samples were directly collected into sterile bottles. All samples were immediately transported back to lab and processed in the same day.
Two grams of soil were added to a 50-ml centrifugation tube containing 20 ml of 10 mm phosphate buffered saline solution and three glass beads (5 mm in diameter). The samples were vigorously vortexed to disperse bacterial colonies and left on a bench for 2 min. Five ml of the soil leachate, freshwater, or seawater was centrifuged at 8000 × g for 10 min to precipitate bacteria and the pellets were re-suspended in 2 ml of LB and transferred into a 15-ml tube, incubated with shaking at 30 °C for 24 h. H2S was detected with the paper strip containing Lead(II)-acetate in the gas phase. Simultaneously, the soil leachates and water samples were diluted and spread onto LB plates, incubated at 25 °C for 48 h. A circle with twenty colonies on a LB plate was randomly drawn, and the colonies were individually transferred into 2 ml of LB in a 15-ml tube to test for H2S production. The isolated pure bacterial cultures of sulfide-oxidizing and non-oxidizing bacteria were also mixed to various ratios according OD600nm in a fixed volume of 200 microliter (μl) and then transferred into 2 ml of LB medium to test for H2S production with the paper strip method. Seawater samples and bacteria isolated from seawater samples were incubated in LB containing 2% of NaCl and 10 mm MgSO4.
Bioinformatic analysis
The genes coding for PDOs and SQRs were first identified from the sequenced bacterial and archaeal genomes of GenBank by using BLAST, and only the sqr and pdo genes that are located next to each other in a gene cluster were selected to construct phylogenetic trees and establish SQR or PDO groups. These selected SQRs and PDOs were then used as seeds to find SQR and PDO homologs, whose genes are not located next to each other, from the sequenced bacterial and archaeal genomes, and the identified homologs were further confirmed if they were mapped into the established SQR or PDO groups in the phylogenetic trees. Similar strategies were used to identify the genes from marine metagenomes of the Global Ocean Sampling (GOS) Expedition and marine metatranscriptomic data sets (Supplementary Methods).
Results
The identification and distribution of pdo and sqr genes in sequenced bacterial and archaeal genomes
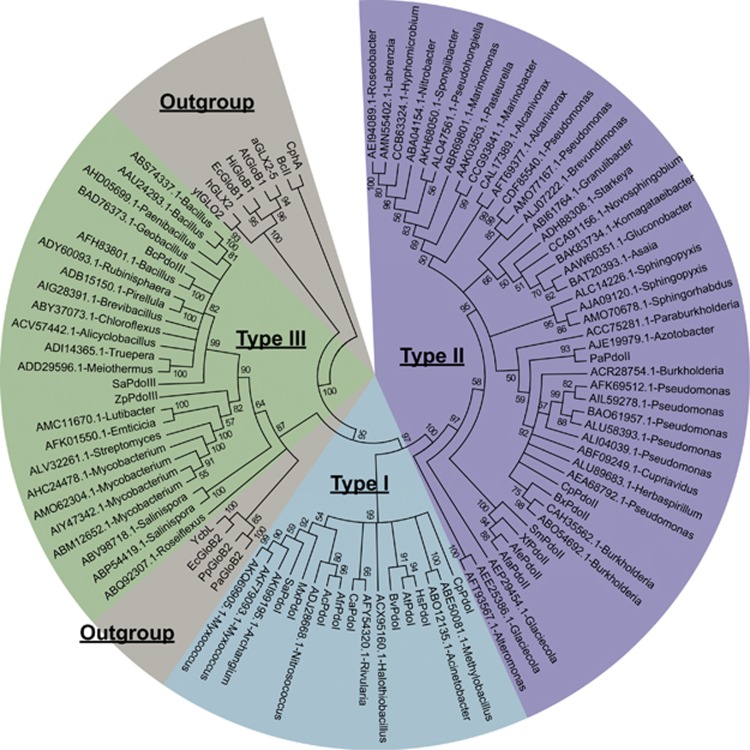
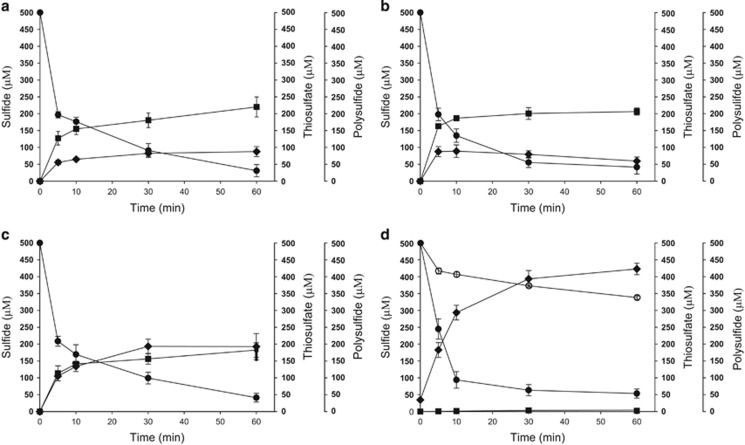
The reports that sqr and pdo are often present in a gene cluster (Liu et al., 2014; Shen et al., 2015; Xin et al., 2016) prompted us to search for adjacent pdo and sqr genes in the 4929 genomes from GenBank (updated until 15 April 2016). 454 pdo and 455 sqr genes were adjacently located in 441 genomes (Supplementary Tables 3–4). After the reconstruction of phylogenetic tree with known SQRs and PDOs, our data supported the topology of six types of SQRs (Marcia et al., 2010; Supplementary Figure 1) and yielded three types of PDOs (Figure 1). The types I and II PDOs are well characterized and present only in Gram-negative bacteria (Liu et al., 2014; Sattler et al., 2015; Xin et al., 2016). The type III PDOs are present in both Gram-negative an positive bacteria, of which only the PDO from Staphylococcus aureus ATCC 6538 P has been characterized to use small organic persulfides, rather than GSSH, as substrates (Shen et al., 2015). As the type III PDOs were diverse and shared about 30% sequence identity between the PDOs from Gram-negative bacteria and Gram-positive bacteria, we tested their activity. When the type III PDOs of Bacillus cereus ATCC10876, S. aureus ATCC 6538 P, and the Gram-negative bacterium Zunongwangia profunda SM-A87 were cloned together with Pasqr2, the recombinant E. coli oxidized sulfide to thiosulfate (Figures 2a–c). E. coli with only SQR oxidized sulfide to sulfane sulfur, for example, polysulfide and persulfide (Figure 2d). Sulfide oxidation by E. coli with the vector or the cloned PDOs was slow with no apparent production of sulfane sulfur or thiosulfate (Figure 2d). The results demonstrate that the type III PDOs are functional in the E. coli host.
Figure 1.
The phylogenetic tree of representative PDOs whose genes are physically linked to sqr in bacterial genomes. 69 representative PDOs were used for phylogenetic tree construction with reference sequences. PDOs belong to the metallo-beta-lactamase superfamily, and several related proteins, such as glyoxalase II (GloB) proteins, were also included as references. The representative proteins were labeled with their GenBank accession numbers and bacterial genera. These sequences were aligned by using ClustalW, and the tree was built by using MEGA6. Reference proteins with accession number were given below. Type I PDOs: SaPdoI (YP_003957083.1), CpPdoI (YP_297536.1), MxPdoI (YP_633997.1), HsPdoI (NP_055112.2), CaPdoI (YP_007162862.1), BvPdoI (ZP_00420127.1), AtPdoI (NP_974018.3), AcPdoI (AEK59246.1). Type II PDOs: XfPdoII (NP_298058.1), SmPdoII (NP_435818.1), CpPdoII (YP_297791.1), PaPdoII (NP_251605.1), BxPdoII (YP_554628.1), AfaPdoII (AAK89929.1), AfePdoII (ZP_11421028.1), AfrPdoII (YP_002424776.1). Type III PDOs: ZpPdoIII (ADF52140.1), SaPdoIII (WP_000465474.1), BcPdoIII (EEK49737.1). Glyoxalase II and related proteins: aGLX2-5 (NP_850166.1), AtGloB1 (NP_356997.2), BcII (AAA22276.1), CphA (CAA40386.1), EcGloB1 (NP_414748.1), EcGloB2 (NP_415447.1), hGLX2 (CAA62483.1), HiGloB1 (ADO96205.1), PaGloB2 (NP_249523.1), PpGloB2 (ABQ76961.1), YcbL (CAD05397.1) and ytGLO2 (CAA71335.1).
Figure 2.
Sulfide oxidation by recombinant E. coli BL21(DE3) cells. Cells were suspended in 100 mm Tris buffer (pH 8) with 50 μm DTPA at OD600nm of 2. Sulfide (500 μm) was added to initiate the reaction. (a) E. coli BL21(DE3) (Ec) with cloned Zppdo-Pasqr2, Ec(Zppdo-Pasqr2); (b) Ec(Bcpdo-Pasqr2); (c) Ec(Sapdo-Pasqr2); (d) Ec(Pasqr2) and Ec(pMCS5). Sulfide, ● thiosulfate, ▪ and sulfane sulfur,  In Figure 2d, control Ec(pMCS5): sulfide, ○ thiosulfate, □ and sulfane sulfur,
In Figure 2d, control Ec(pMCS5): sulfide, ○ thiosulfate, □ and sulfane sulfur,  Sulfide oxidation, polysulfide production and thiosulfate production by E. coli with cloned Zppdo, Bcpdo or Sapdo were essentially the same as Ec(pMCS5) (Data not shown).
Sulfide oxidation, polysulfide production and thiosulfate production by E. coli with cloned Zppdo, Bcpdo or Sapdo were essentially the same as Ec(pMCS5) (Data not shown).
The PDOs and SQRs encoded by adjacent genes were further used to search for potential PDOs and SQRs from the 4929 bacterial and 242 archaeal genomes. The potential PDOs and SQRs were then checked via phylogenetic analysis, and 1908 PDOs and 1310 SQRs were stably conserved on the phylogenetic trees (Supplementary Figures 2 and 3). The type II PDOs were the most abundant but limited to Proteobacteria; the type I PDOs had a wider distribution in Gram-negative bacteria; the type III PDOs had the widest distribution present in both Gram-positive and -negative bacteria (Table 1). SQRs were widely distributed among bacterial phyla, and the type II proteins were the most abundant (Table 1). Of the sequenced bacterial genomes, 646 genomes had only pdo, 208 genomes had only sqr, and 806 possessed both. The genomes carrying both sqr and pdo mainly distributed in the phyla of Proteobacteria, Cyanobacteria and Firmicutes (Supplementary Figure 4; Table 1). 733 of the 806 genomes with both sqr and pdo belonged to the genera of known aerobic heterotrophs. 122 of 208 genomes with only sqr belonged to the genera of known aerobic heterotrophs.
Table 1. The classification and distribution of SQRs and PDOs from sequenced bacterial genomes at the phylum level.
| Phylum |
SQR types |
PDO types |
Total genomes at phylum levels | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type I | Type II | Type III | Type IV | Type V | Type VI | Type I | Type II | Type III | ||
| Acidobacteria | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 8 |
| Actinobacteria | 1 | 61 | 11 | 13 | 0 | 0 | 0 | 0 | 210 | 531 |
| Aquificae | 10 | 0 | 5 | 3 | 0 | 10 | 0 | 0 | 0 | 16 |
| Armatimonadetes | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Bacteroidetes | 1 | 22 | 2 | 5 | 0 | 1 | 0 | 0 | 194 | 185 |
| Candidate division SR1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
| Chlorobi | 0 | 0 | 9 | 13 | 2 | 4 | 0 | 0 | 0 | 11 |
| Chloroflexi | 0 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 8 | 25 |
| Cyanobacteria | 15 | 24 | 0 | 0 | 0 | 0 | 58 | 0 | 0 | 91 |
| Deinococcus-Thermus | 1 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 19 | 24 |
| Firmicutes | 2 | 158 | 10 | 2 | 6 | 0 | 1 | 0 | 246 | 1086 |
| Gemmatimonadetes | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| Ignavibacteriae | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 2 |
| Nitrospirae | 7 | 0 | 4 | 0 | 0 | 3 | 0 | 0 | 2 | 8 |
| Planctomycetes | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 6 |
| Proteobacteria | 130 | 532 | 143 | 15 | 8 | 53 | 272 | 852 | 34 | 2504 |
| Spirochaetes | 2 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 65 |
| Thermobaculum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Verrucomicrobia | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 9 |
From the 242 archaeal genomes, 86 SQRs and 10 PDOs were identified. 10 Crenarchaeota genomes contained pdo and sqr, and 51 genomes of Crenarchaeota and Euryarchaeota had only sqr. Of the SQRs, 59 belonged to the type III, and the rest belonged to the type V. The 10 PDOs were all of the type III.
Heterotrophic bacteria with sqr and pdo oxidized self-produced and exogenous H2S
A total of 24 heterotrophic bacteria with sequenced genomes were tested for releasing H2S during growth in LB or the recommended rich medium for the bacterium. The sulfur contents of the yeast extract and tryptone were determined as 0.43 and 0.85%, and the calculated sulfur in LB was 3.3 mm. Nine bacteria with sqr and pdo did not release H2S (Table 2), but the strains without sqr or pdo did. Of the 15 H2S producers, S. fonticola DSM4576 and C. vitaeruminis DSM20294 possess only sqr; A. tumefaciens C58, S. meliloti 1021, and Streptomyces griseus NBRC13350 contain only pdo (Guimaraes et al., 2011; Liu et al., 2014); others do not have the genes.
Table 2. H2S production by bacteria in rich media.
| Strains | sqr | pdo | Adjacently linkeda | H2S production |
|---|---|---|---|---|
| Agrobacterium tumefaciens C58 | 0 | 1 | NA | + |
| Bacillus subtilis 168 | 0 | 0 | NA | + |
| Corynebacterium glutamicum RES167 | 0 | 1 | NA | + |
| Corynebacterium vitaeruminis DSM20294 | 2 | 0 | NA | + |
| Enterobacter cloacae ATCC 13047 | 0 | 0 | NA | + |
| Enterococcus faecalis ATCC29200 | 0 | 0 | NA | + |
| Escherichia coli BL21(DE3) | 0 | 0 | NA | + |
| Klebsiella pneumonia DSM30104 | 0 | 0 | NA | + |
| Serratia fonticola DSM4576 | 1 | 0 | NA | + |
| Sinorhizobium meliloti 1021 | 0 | 2 | NA | + |
| Sphingobium chlorophenolicum L-1 | 0 | 0 | NA | + |
| Staphylococcus sciuri Z8 | 0 | 0 | NA | + |
| Streptomyces coelicolor M145 | 0 | 0 | NA | + |
| Streptomyces griseus NBRC13350 | 0 | 1 | NA | + |
| Zymomonas mobilis ATCC 31821 | 0 | 0 | NA | + |
| Burkholderia cepacia ATCC 25416 | 1 | 2 | + | − |
| Cupriavidus pinatubonensis JMP134 | 1 | 2 | + | − |
| Gluconobacter oxydans 621H | 2 | 1 | + | − |
| Pseudomonas aeruginosa PAO1 | 2 | 1 | − | − |
| Pseudomonas putida ML2 | 1 | 1 | + | − |
| Serratia marcescens ATCC13880 | 2 | 1 | − | − |
| Bacillus cereus ATCC10876 | 1 | 1 | + | − |
| Staphylococcus aureus ATCC6538P | 1 | 1 | + | − |
| Zunongwangia profunda SM-A87 | 1 | 4 | + | − |
Abbreviation: NA, not applicable.
The accession numbers of PDOs and SQRs are given in Supplementary Table 5.
The sqr and pdo genes are adjacently located on the chromosome.
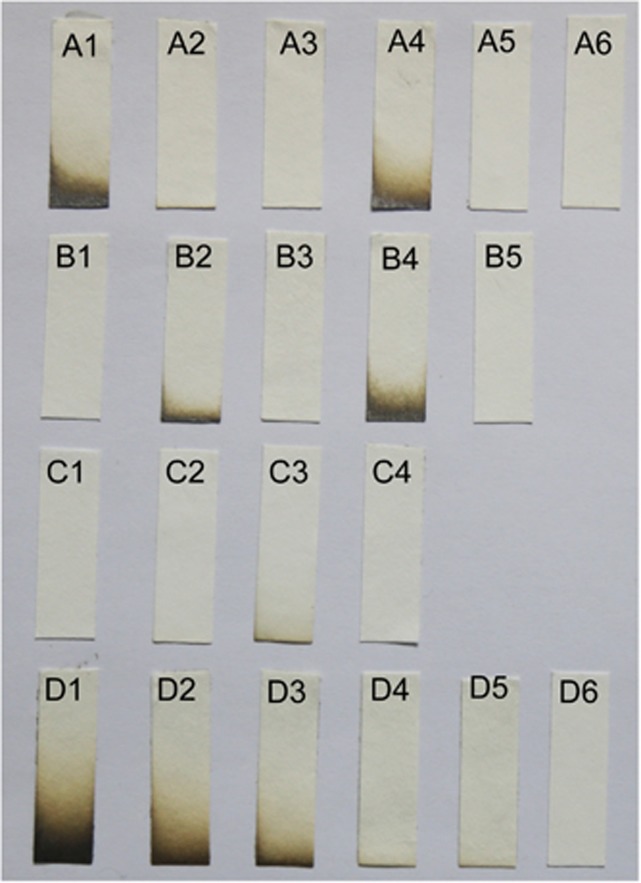
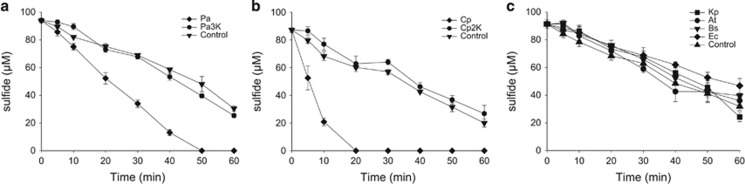
The role of sqr and pdo was further tested with recombinants and mutants. During aerobic growth in LB, E. coli BL21(DE3) produced and released 50–100 μm H2S into the headspace; however, E. coli BL21(DE3) with cloned pdo-sqr did not release H2S (Figure 3a). P. aeruginosa PAO1 and C. pinatubonensis JMP134 (Supplementary Figure 5) did not release H2S (Figures 3b and c), but the P. aeruginosa triple mutant Δsqr1Δsqr2Δpdo (Pa3K) released 25–50 μm H2S, and the C. pinatubonensis double mutant CpΔpdo2Δsqr (Cp2K) released about 10 μm H2S. However, the complementation strains Pa3K (Papdo-Psqr2) or Cp2K (Cppdo2-Cpsqr) did not release H2S (Figures 3b and c). Further, the resting cells of P. aeruginosa PAO1 and C. pinatubonensis JMP134 oxidized spiked sulfide, but Pa3K and Cp2K did not (Figures 4a and b). None of the tested H2S-releasing bacteria was able to oxidize spiked sulfide (Figure 4c), and the slow decrease of sulfide is likely due to volatilization (Xin et al., 2016) and non-specific enzymatic reactions (Luther et al., 2011a). C. vitaeruminis DSM20294 with only sqr oxidized sulfide after induction with sulfide (Supplementary Figure 6A), but S. fonticola DSM4576 carrying a sqr gene did not metabolize spiked sulfide even after induction with sulfide (Supplementary Figure 6B).
Figure 3.
Testing H2S Production by bacteria in LB. Bacteria were inoculated in 2 ml of LB in 15-ml tubes with the lead-acetate filter paper fixed at the top of the headspace. (Row A) A1, E. coli BL21(DE3) (Ec); A2, Ec(Pasqr1); A3, Ec(Pasqr2); A4, Ec(Papdo); A5, Ec(Papdo-Pasqr2); A6, Ec(Cppdo2-Cpsqr). (Row B) B1, P. aeruginosa PAO1 (Pa); B2, PaΔsqr1Δsqr2; B3, PaΔpdo; B4, PaΔsqr1Δsqr2Δpdo (Pa3K), B5: Pa3K(Papdo-Pasqr2). (Row C) C1, C. pinatubonensis JMP134 (Cp); C2, CpΔsqr; C3, CpΔpdo2Δsqr (Cp2K); C4, Cp2K(Cppdo2-Cpsqr). (Row D) NaHS standards in LB: D1, 100 μm; D2, 50 μm; D3, 25 μm; D4, 10 μm; D5, 5 μm; D6, 0 μm. E. coli and P. aeruginosa were incubated at 37 °C, and C. pinatubonensis was incubated at 30 °C for 18 h.
Figure 4.
Resting bacterial cells oxidize spiked sulfide. Cells were suspended in 50 mm Tris buffer, pH 8.0, with 50 μm DTPA at OD600nm of 1, and NaHS was added to initiate the reaction. Controls were done in the same buffer without bacterial cells. All data are average of three samples with standard deviation (error bar). (a) C. pinatubonensis JMP134 (Cp) and C. pinatubonensis 2 K (Cp2K); (b) P. aeruginosa PAO1 (Pa) and P. aeruginosa 3 K (Pa3K); (c) B. subtilis 168 (Bs), A. tumefaciens C58 (At), K. pneumonia DSM30104 (Kp) and E. coli BL21(DE3) (Ec).
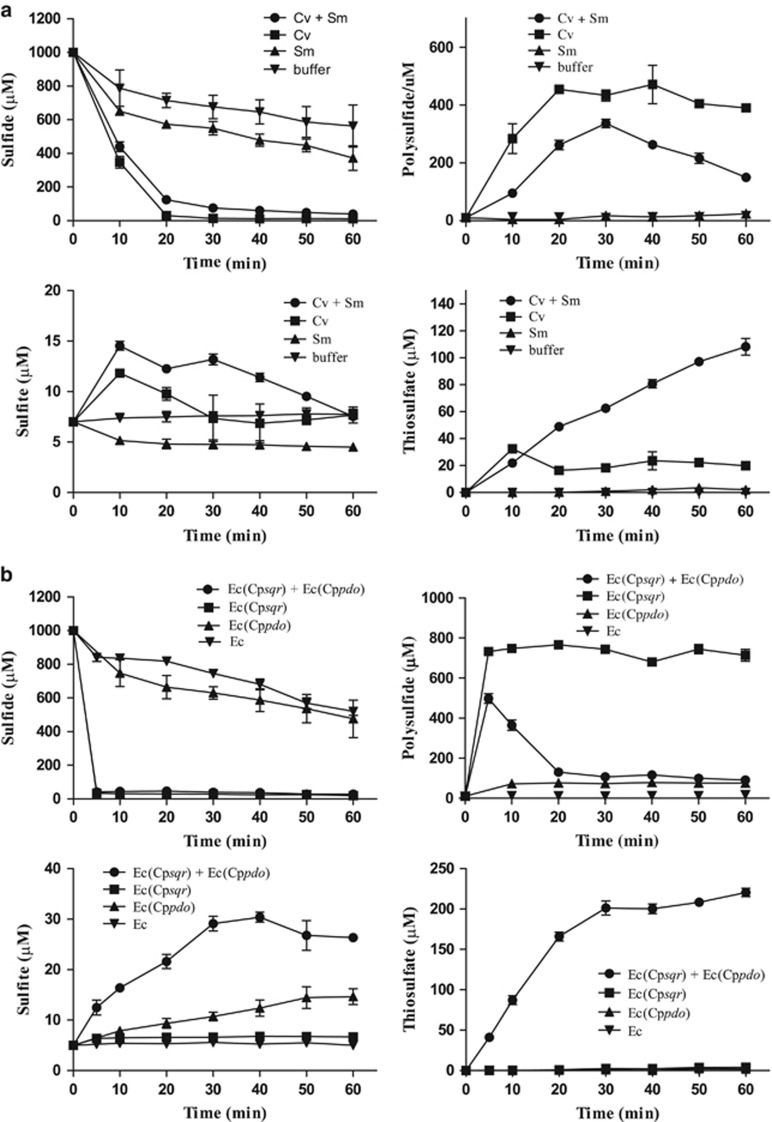
Rapid sulfide oxidation was observed in samples containing sulfide-induced C. vitaeruminis DSM20294 either alone or mixed with S. meliloti 1021 (Figure 5a). C. vitaeruminis DSM20294 with only sqr oxidized sulfide mainly to polysulfide. S. meliloti 1021 with only pdo did not oxidize sulfide to polysulfide, sulfite or thiosulfate. When combined, the two bacteria oxidized sulfide to polysulfide and then to thiosulfate (Figure 5a). The results suggested that in mixed cultures, cells with sqr and cells with pdo can collectively oxidize sulfide to polysulfide and then to sulfite, which reacts with polysulfide to produce thiosulfate (Xin et al., 2016). The collaboration of E. coli BL21(DE3) expressing either sqr or pdo for sulfide oxidation to thiosulfate was also confirmed (Figure 5b). The results indicate that polysulfide or persulfide can be transferred from one cell to another during sulfide oxidation by the combined cells.
Figure 5.
Sulfide oxidation by mixed bacterial cells with SQR or PDO. (a) Combination of C. vitaeruminis DSM20294 (Cv) (with sqr) and S. meliloti 1021 (Sm) (with pdo). (b) Combination of E. coli BL21(DE3) (Ec) with Cpsqr [Ec(Cpsqr)] or Cppdo2 [(Ec(Cppdo2)]. Cv and Sm were induced with sulfide before harvesting, and the recombinant E. coli cells were induced with IPTG before harvesting. Cells were suspended in 50 mm Tris buffer, pH 8.0, with 50 μm DTPA at OD600nm of 2 for individual strains or 4 for mixed strains and 1 mm NaHS was added to initiate the reaction. Controls were done with individual bacterial strains. Sulfide oxidation and the production of polysulfide, sulfite and thiosulfate were detected. All data are average of three samples with standard deviation (error bar).
Both sulfide oxidizers and non-oxidizers were isolated from environmental samples
Various soil and water samples were collected and inoculated in LB; mixed cultures grew rapidly, but none released any detectable H2S (Supplementary Figure 7). The samples were also diluted and spread on LB agar plates. Without exception, both H2S-releasing and H2S non-releasing bacteria were isolated from these samples (Supplementary Table 6). When the H2S-releasing bacteria and H2S-non-releasing bacteria were mixed at different ratios and incubated in LB, H2S was not accumulated in the cultures of the farm, forest and lake isolates (Supplementary Figure 7); the mixed marine isolates released some H2S, but much lower than the control with only the marine H2S-releasing bacterium (Supplementary Figure 7). Thus, the sulfide oxidizers did not release H2S, while the non-oxidizers released H2S in rich medium. The selected bacteria were identified to their closest relatives according to 16 S rRNA gene sequences (GenBank: KT443871-KT443878). Sequence search of the genomes of these close relatives revealed that the sulfide oxidizers were closely related to bacteria containing both sqr and pdo genes and the non-oxidizers were related to bacteria without the genes (Supplementary Figure 7 legend).
The abundance and diversity of PDOs and SQRs in the metagenomes from the GOS Expedition
175 PDOs and 82 SQRs were identified from 124 genomes of the 177 marine bacterial genomes of the Gordon and Betty Moore Foundation Marine Microbial Genome Sequencing project (http://www.jcvi.org/cms/research/past-projects/microgenome/overview/) (Supplementary Figure 8). SQRs and PDOs are common in the Roseobacter clade, but rare in the SAR11 clade (Supplementary Table 7). The marine bacterial PDOs and SQRs were used to search the metagenomic database of the GOS Expedition (Rusch et al., 2007). 1895 PDOs and 439 SQRs were found in the GOS data set. The type I PDOs and type II SQRs were dominant (Supplementary Table 8). The average percentages of bacterial genomes with sqr and pdo were 4 and 34.9%, respectively, in the GOS data set (Supplementary Table 9). The percentages of genomes with SQR and PDO from each GOS sampling site ranged from 0% to 53.9% and 10.7% to 74.9%, respectively (Table 3). The numbers of sqr in hypersaline waters were significantly higher than that in other sites (Table 3). The pdo genes were detected at all sampling sites, and the sqr genes were detectable from 42 of the 58 sampling sites. Thus, the sqr genes were also commonly occurring in marine surface waters, but its abundance was lower than that of pdo (Table 3).
Table 3. The distribution of pdo and sqr homologs in the GOS data set per sample sites.
| Site | Site names | Types | % genomes with pdo | % genomes with sqr |
|---|---|---|---|---|
| 0 | Sargasso Station | Open Ocean | 35.2 | 7.7 |
| 1 | Hydrostation S | Open Ocean | 35.3 | 0.9 |
| 2 | Gulf of Maine | Coastal | 64.9 | 4.7 |
| 3 | Browns Bank, Gulf of Maine | Coastal | 74.9 | 2.4 |
| 4 | Outside Halifax, Nova Scotia | Coastal | 25.7 | 5.4 |
| 5 | Bedford Basin, Nova Scotia | Embayment | 32.7 | 20.5 |
| 6 | Bay of Fundy, Nova Scotia | Estuary | 52.2 | 4.2 |
| 7 | Northern Gulf of Maine | Coastal | 32.1 | 0.0 |
| 8 | Newport Harbor, RI | Coastal | 46.3 | 12.9 |
| 9 | Block Island, NY | Coastal | 53.8 | 0.0 |
| 10 | Cape May, NJ | Coastal | 59.4 | 6.0 |
| 11 | Delaware Bay, NJ | Estuary | 30.3 | 12.3 |
| 12 | Chesapeake Bay, MD | Estuary | 23.0 | 1.3 |
| 13 | Off Nags Head, NC | Coastal | 58.6 | 12.2 |
| 14 | South of Charleston, SC | Coastal | 42.0 | 6.5 |
| 15 | Off Key West, FL | Coastal | 31.9 | 0.0 |
| 16 | Gulf of Mexico | Coastal Sea | 38.3 | 1.1 |
| 17 | Yucatan Channel | Open Ocean | 34.8 | 2.6 |
| 18 | Rosario Bank | Open Ocean | 33.0 | 0.8 |
| 19 | Northeast of Colon | Coastal | 21.3 | 0.0 |
| 20 | Lake Gatun | Fresh Water | 18.9 | 5.3 |
| 21 | Gulf of Panama | Coastal | 51.7 | 0.0 |
| 22 | 250 miles from Panama City | Open Ocean | 40.2 | 0.0 |
| 23 | 30 miles from Cocos Island | Open Ocean | 25.9 | 0.9 |
| 25 | Dirty Rock, Cocos Island | Fringing Reef | 11.5 | 0.0 |
| 26 | 134 miles NE of Galapagos | Open Ocean | 42.6 | 1.4 |
| 27 | Devil's Crown, Floreana Island | Coastal | 37.6 | 1.1 |
| 28 | Coastal Floreana | Coastal | 40.3 | 1.3 |
| 29 | North James Bay, Santigo Island | Coastal | 31.1 | 1.3 |
| 30 | Warm seep, Roca Redonda | Warm Seep | 26.4 | 1.6 |
| 31 | Upwelling, Fernandina Island | Coastal upwelling | 43.4 | 0.9 |
| 32 | Mangrove on Isabella Island | Mangrove | 36.7 | 3.8 |
| 33 | Punta Cormorant, Hypersaline Lagoon, Floreana Island | Hypersaline | 56.9 | 53.9 |
| 34 | North Seamore Island | Coastal | 26.7 | 1.1 |
| 35 | Wolf Island | Coastal | 49.5 | 1.1 |
| 36 | Cabo Marshall, Isabella Island | Coastal | 34.6 | 0.0 |
| 37 | Equatorial Pacific TAO Buoy | Open Ocean | 17.6 | 2.6 |
| 47 | 201 miles from F. Polynesia | Open Ocean | 46.4 | 0.0 |
| 48 | Moorea, Cooks Bay | Coral Reef | 39.8 | 6.3 |
| 49 | Moorea, Outside Cooks Bay | Coastal | 29.2 | 0.0 |
| 51 | Rangirora Atoll | Coral Reef Atoll | 62.0 | 1.2 |
| 108 | Coccos Keeling, Inside Lagoon | Lagoon Reef | 39.8 | 14.5 |
| 109 | Indian Ocean | Open Ocean | 21.6 | 0.0 |
| 110 | Indian Ocean | Open Ocean | 23.4 | 0.6 |
| 111 | Indian Ocean | Open Ocean | 37.8 | 1.8 |
| 112 | Indian Ocean | Open Ocean | 19.9 | 0.6 |
| 113 | Indian Ocean | Open Ocean | 25.4 | 1.9 |
| 114 | 500 Miles west of the Seychelles in the Indian Ocean | Open Ocean | 25.9 | 0.0 |
| 115 | Indian Ocean | Open Ocean | 20.1 | 0.0 |
| 116 | Outside Seychelles, Indian Ocean | Open Ocean | 37.7 | 0.0 |
| 117 | St. Anne Island, Seychelles | Coastal | 20.6 | 0.5 |
| 119 | International Water Outside of Reunion Island | Open Ocean | 24.8 | 0.0 |
| 120 | Madagascar Waters | Open Ocean | 30.3 | 4.4 |
| 121 | International water between Madagascar and South Africa | Open Ocean | 26.8 | 5.3 |
| 122 | International waters between Madagascar and South Africa | Open Ocean | 21.7 | 2.8 |
| 123 | International water between Madagascar and South Africa | Open Ocean | 33.2 | 3.0 |
| 148 | East coast Zanzibar (Tanzania), offshore Paje lagoon | Fringing Reef | 29.7 | 1.0 |
| 149 | West coast Zanzibar (Tanzania), harbor region | Harbor | 33.2 | 0.0 |
| Average | 34.9 | 4.0 |
Identification of pdo and sqr transcripts in marine metatranscriptomic data
The transcripts of sqr and pdo were further detected in three metatranscriptomic data sets of marine samples, and the ratios of pdo and sqr transcripts over total transcripts were 7.6 × 10−6 and 5.5 × 10−7 from station ALOHA (NCBI accession: PRJNA244754), 2.2 × 10−5 and 4.5 × 10−6 from Monterey Bay (PRJNA183166), 5.1 × 10−6 and 6.8 × 10−7 from a coastal California water (PRJNA268385), respectively. Waters from different locations vary in chemical and physical characteristics, being nutrient-rich from the Monterey Bay and coastal waters and oligotrophic at station ALOHA in the North Pacific Gyre (Amin et al., 2015). The sqr transcripts doubled in the samples taken at night, but the pdo transcripts were essentially the same (Supplementary Figure 9). The data suggest that bacterial pdo and sqr are present and expressed in these natural habitats.
Discussion
Sulfide production and benefits
During heterotrophic growth, microorganisms are likely released H2S from sulfur-containing amino acids by using a variety of enzymes, including cysteine desulfhydrase, 3-mercaptopyruvate sulfurtransferase, cystathionine β-synthase and cystathionine γ-lyase (Morra and Dick, 1991; Shatalin et al., 2011; Oguri et al., 2012). In agreement with these well documented mechanisms, we have observed H2S releasing from all tested bacteria without SQR and PDO as well as the mutants without SQR and PDO activities in rich media (Table 2). The sulfur in LB is from yeast extract and tryptone, and the determined sulfur contents are similar to related products, in which most sulfur is in cysteine and methionine (Sugata and Koch, 1926; Kassell and Brand, 1938; McManus et al., 1950). The observation highlights the common production of sulfide from organic sulfur by heterotrophic bacteria. We suggest that aerobic sulfide production by heterotrophic bacteria can be significant in soils with high organic contents and in water bodies with eutrophication, during algal bloom, or on organic particulate matter.
The self-produced sulfide may benefit heterotrophic bacteria. It can protect bacteria from antibiotics through reacting and removing reactive oxygen species (Shatalin et al., 2011). Sulfide may have special functions, as the human pathogen Mycoplasma pneumoniae produces sulfide from cysteine for erythrocyte lysis (Großhennig et al., 2016). Further, sulfide and its oxidation intermediate sulfane sulfur have been shown to function as signals by causing protein S-sulfhydration (Ida et al., 2014; Miranda and Wink, 2014). The signaling role of H2S in bacteria has been suggested (Lloyd, 2006), and it will likely join a growing number of volatile molecules produced by bacteria as signaling molecules for interactions among bacteria and with eukaryotic hosts or predators (Schmidt et al., 2015; Schulz-Bohm et al., 2016).
We have demonstrated that bacteria with SQR and PDO oxidized self-produced sulfide and did not release H2S (Figure 3). In the assay of 2-ml bacterial cultures, as low as 5 μm H2S, produced by bacteria without SQR and PDO during the course of incubation, was released into the headspace and clearly detected by the lead-acetate paper strip (Figure 3). Our incubation with shaking may also facilitate the volatilization of H2S. Thus, the sulfide-oxidizing heterotrophic bacteria are able to oxidize sulfide at low levels to prevent the accumulation and volatilization of H2S. We only tested 24 bacteria with sequenced genomes, and the result cannot predict that all bacteria with SQR and PDO will not release H2S when growing in rich media, but can certainly expect most will follow the trend and oxidize sulfide as they produce it. This oxidation of sulfide at low concentrations may have ecological significance as abiotic oxidation of sulfide is slow with a half-life of 55 days in a trace metal-free solution at pH 12 and 25 °C (Luther et al., 2011b) or a half-life of 26 h in seawater at 25 °C (Millero et al., 1987). Without the oxidation, sulfide may accumulate (Figure 3).
Heterotrophic bacteria may get energy from sulfide oxidation. SQR oxidizes sulfide to polysulfide and passes the electrons to the electron transport chain of aerobic respiration, which will generate the proton motive force for ATP production (Marcia et al., 2010). However, this system is not energy efficient, as PDO and rhodanese do not participate in energy conservation. Interestingly, the common marine Roseobacter members often carry the SQR-PDO system as well as the energy-generating sulfite oxidase and Sox systems that further oxidize sulfite and thiosulfate to sulfate (Lenk et al., 2012). In the dark ocean bacteria are rich with sulfur oxidizing genes from metagenomic analysis, suggesting that these bacteria may use both organic and inorganic compounds to cope with the nutrient-poor environment (Swan et al., 2011). However, the source of sulfide is unlikely from the anaerobic sediment, which is farther down below, and sulfide produced in the sediment hardly escapes the interphase of the water-sediment boundary. Our results suggest a possible source of the sulfide from the metabolism by heterotrophic bacteria, especially on sulfur rich compounds, for example, dimethylsulfoniopropionate (DMSP).
DMSP is mainly produced by marine macroalgae and single-cell phytoplankton, and it has major physiological roles in phytoplankton, heterotrophic bacteria and zooplankton (Yoch, 2002). Its annual production is estimated around 109 metric tons. The produced DMSP is rapidly converted to dimethylsulfide (DMS) by both phytoplankton and heterotrophic bacteria with DMPS lyases or demethylated to produce methanethiol by bacteria (Alcolombri et al., 2015; Sun et al., 2016). Only a small fraction of the produced DMS is released into the atmosphere, which is chemically oxidized to sulfite and sulfate to serve as nuclei for cloud formation (Vila-Costa et al., 2006). DMS is either oxidized to dimethylsulfoxide by marine bacteria with trimethylamine monooxygenase (Lidbury et al., 2016) or converted to MeSH by bacteria with dimethylsulfide monooxygenase (Boden et al., 2011). MeSH can be metabolized to release sulfide or assimilated to produced methionine (Kiene et al., 1999). Methionine is converted to homocysteine and cysteine, both of which can be further metabolized to release H2S. Thus, it is speculated that the rapid turnover of the huge amount of DMSP may generate significant amounts of sulfide in marine waters, especially during algal bloom, and the heterotrophic bacteria with sqr and pdo present in the community are likely responsible for sulfide oxidation.
Another physiological role of sulfide oxidation by heterotrophic bacteria is likely detoxification when sulfide level is high. Baker’s yeast without SQR and PDO accumulates sulfide, which then slows down respiration and growth due to sulfide inhibition of cytochrome C oxidase (Sohn et al., 2000). Bacterial respiration with cytochrome bo3 oxidase is also inhibited by sulfide with a half-maximal inhibitory concentration of 1.1 μm sulfide for the E. coli oxidase; however, E. coli cytochrome bd oxidase is less sensitive to sulfide (Forte et al., 2016). The data we presented in Figure 3 and Supplementary Figure 7 show that bacteria with SQR and PDO oxidize sulfide and prevent its accumulation in pure and mixed cultures. Further, exogenous sulfide can also be oxidized and detoxified by bacteria with SQR and PDO (Figures 4 and 5). The detoxification role of sulfide oxidation has been reported, as Staphylococcus aureus with SQR and PDO is more resistant to added sulfide than the deletion mutant (Luebke et al., 2014).
Bacteria may also use SQR or SQR and PDO to prevent the loss of sulfur through H2S volatilization. Sulfide can be lost via H2S volatilization (Figure 3; Supplementary Figure 7; Table 2), and bacteria with SQR or SQR and PDO can prevent the loss (Figure 3). The use of SQR for sulfur conservation is inferred from the genome of Candidatus Evansia muelleri Xc1, an endosymbiotic bacterium of the moss insect Xenophyes cascus. The endosymbiont has a very small genome but contains sqr that is proposed for sulfur conservation as the insect is on a sulfur-poor diet (Santos-Garcia et al., 2014). Bacteria with sqr and pdo genes, including some Pseudomonads, Bacilli and Roseobacter, are common in soil, freshwater and seawater (Table 1; Supplementary Table 7), and they oxidize biogenic sulfide to sulfite and thiosulfate (Xin et al., 2016). Sulfite and thiosulfate can be directly used by plants and microorganisms as the sulfur source, or further oxidized by bacteria to sulfate (Anandham et al., 2008; Marshall and Morris, 2013).
The pdo genes are more abundant than the sqr genes in marine waters (Table 3) and sequenced marine bacterial genomes (Supplementary Figure 8). Higher plants possess only PDO and its function is likely to prevent the accumulation of polysulfide and persulfide generated from sulfur-containing amino acids (Höfler et al., 2016). Bacteria with PDO may have a similar role. However, bacteria with only SQR or PDO are able to collaborate in oxidizing sulfide to thiosulfate (Figure 5), suggesting the interspecies transfer of polysulfide or persulfide.
PDO classification
The PDO was initially classified into three groups, SdoA, ETHE1 and Blh (Liu et al., 2014). Structure analysis grouped SdoA and Blh into the type II and ETHE1 related proteins as type I (Sattler et al., 2015), and both used GSSH as the substrate (Liu et al., 2014). The type I and II PDOs, present in Gram-negative bacteria, share about 20–30% sequence identities. The type III PDOs are further distanced from the type I and II PDOs with less than 20% sequence identity to them, and they are present in both Gram-positive and Gram-negative bacteria as well as in Archaea (Figure 1; Supplementary Figure 2; Table 1; Supplementary Table 3).
The distribution and transcription of sqr was affected by environmental conditions
The distribution and transcription of sqr in marine surface waters are possibly influenced by salt concentrations and light. The sqr genes were more abundant in hypersaline waters (Punta Cormorant, Hypersaline Lagoon, Floreana Island) (Table 3). This is possibly due to increased availability of sulfide in hypersaline waters because the dissolved oxygen content decreases with the increased salt contents (Wetzel, 2001), making sulfide more stable (Kolluru et al., 2013). Further, in shallow hypersaline lagoons, sulfide may also come from dissimilatory sulfate reduction in the sediment (Cotner et al., 2004). The transcription of sqr during the night was about 2 folds higher than that during the day (Supplementary Figure 9). We speculate that there is likely more sulfide during the night because phytoplankton respiration in the dark may contribute to additional sulfide and the lack of photosynthesis leads to reduced oxygen levels (Watt, 2000). The expression of sqr and pdo can be induced with increased sulfide as shown in this study (Supplementary Figure 6A) and as previously reported (Guimaraes et al., 2011; Luebke et al., 2014; Shimizu et al., 2017).
In conclusion, bacterial production of sulfide from organic sulfur is common during aerobic growth. Due to the wide presence of sulfide-oxidizing heterotrophic bacteria and their ability to oxidize sulfide at low levels, the sulfide production by heterotrophic bacteria is not readily observed. Without the oxidation, sulfide accumulates and volatilizes as the H2S gas. Since heterotrophic bacteria do not solely rely on sulfide to supply energy for growth, they can oxidize it at low levels and when the supply is intermittent. In the ocean, heterotrophic bacteria metabolize the large quantities of DMSP to release sulfide, and they are also likely to consume the released sulfide. Considering the scale of their presence on Earth, heterotrophic bacteria may significantly contribute to sulfide production and oxidation, a missed aspect of the sulfur cycle.
Acknowledgments
The work was supported by grants from the National Key Research and Development Program of China (Grant no. 2016YFA0601103), the National Natural Science Foundation of China (21477062, 31500047), the Natural Science foundation of Shandong Province (ZR2014CM003, 2015BSE27054) and the State Key Laboratory of Microbial Technology at Shandong University. We thank Yuzhong Zhang for stimulating discussion and for Z. profunda SM-A87, Qilong Qin, Yue Wu, Hangyuan Cui, Guanhua Xuan for help with bioinformatics, and Feifei Cui for help with chemical analysis.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Alcolombri U, Ben-Dor S, Feldmesser E, Levin Y, Tawfik DS, Vardi A. (2015). MARINE SULFUR CYCLE. Identification of the algal dimethyl sulfide-releasing enzyme: a missing link in the marine sulfur cycle. Science 348: 1466–1469. [DOI] [PubMed] [Google Scholar]
- Amin S, Hmelo L, van Tol H, Durham B, Carlson L, Heal K et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522: 98–101. [DOI] [PubMed] [Google Scholar]
- Anandham R, Indiragandhi P, Madhaiyan M, Ryu KY, Jee HJ, Sa TM. (2008). Chemolithoautotrophic oxidation of thiosulfate and phylogenetic distribution of sulfur oxidation gene (soxB) in rhizobacteria isolated from crop plants. Res Microbiol 159: 579–589. [DOI] [PubMed] [Google Scholar]
- Boden R, Borodina E, Wood AP, Kelly DP, Murrell JC, Schafer H. (2011). Purification and characterization of dimethylsulfide monooxygenase from Hyphomicrobium sulfonivorans. J Bacteriol 193: 1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PH. (1953). Hydrogen sulphide production by bacteria. J Gen Microbiol 8: 397–407. [DOI] [PubMed] [Google Scholar]
- Cotner JB, Suplee MW, Chen NW, Shormann DE. (2004). Nutrient, sulfur and carbon dynamics in a hypersaline lagoon. Estuar Coast Shelf Sci 59: 639–652. [Google Scholar]
- Forte E, Borisov VB, Falabella M, Colaco HG, Tinajero-Trejo M, Poole RK et al. (2016). The terminal oxidase cytochrome bd promotes sulfide-resistant bacterial respiration and growth. Sci Rep 6: 23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AE, Rogers PL, Skotnicki ML. (1982). Minimal medium for isolation of auxotrophic Zymomonas mutants. Appl Env Microbiol 44: 496–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Großhennig S, Ischebeck T, Gibhardt J, Busse J, Feussner I, Stulke J. (2016). Hydrogen sulfide is a novel potential virulence factor of Mycoplasma pneumoniae: characterization of the unusual cysteine desulfurase/desulfhydrase HapE. Mol Microbiol 100: 42–54. [DOI] [PubMed] [Google Scholar]
- Guimaraes BG, Barbosa RL, Soprano AS, Campos BM, de Souza TA, Tonoli CC et al. (2011). Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J Biol Chem 286: 26148–26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfler S, Lorenz C, Busch T, Brinkkötter M, Tohge T, Fernie AR et al. (2016). Dealing with the sulfur part of cysteine: four enzymatic steps degrade l‐cysteine to pyruvate and thiosulfate in Arabidopsis mitochondria. Physiol Plant 157: 352–66. [DOI] [PubMed] [Google Scholar]
- Harighi B. (2009). Genetic evidence for CheB- and CheR-dependent chemotaxis system in A. tumefaciens toward acetosyringone. Microbiol Res 164: 634–641. [DOI] [PubMed] [Google Scholar]
- Hildebrandt TM, Grieshaber MK. (2008). Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J 275: 3352–3361. [DOI] [PubMed] [Google Scholar]
- Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y et al. (2014). Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc Natl Acad Sci USA 111: 7606–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamyshny A Jr. (2009). Improved cyanolysis protocol for detection of zero-valent sulfur in natural aquatic systems. Limnol Oceanogr Methods 7: 442–448. [Google Scholar]
- Kassell B, Brand E. (1938). The distribution of the sulphur in casein, lactalbumin, edestin, and papain. J Biol Chem 125: 435–443. [Google Scholar]
- Kiene RP, Linn LJ, Gonzalez J, Moran MA, Bruton JA. (1999). Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl Environ Microbiol 65: 4549–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolluru GK, Shen X, Bir SC, Kevil CG. (2013). Hydrogen sulfide chemical biology: pathophysiological roles and detection. Nitric Oxide 35: 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM 2nd et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176. [DOI] [PubMed] [Google Scholar]
- Lenk S, Moraru C, Hahnke S, Arnds J, Richter M, Kube M et al. (2012). Roseobacter clade bacteria are abundant in coastal sediments and encode a novel combination of sulfur oxidation genes. ISME J 6: 2178–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidbury I, Krober E, Zhang Z, Zhu Y, Murrell JC, Chen Y et al. (2016). A mechanism for bacterial transformation of dimethylsulfide to dimethylsulfoxide: a missing link in the marine organic sulfur cycle. Environ Microbiol 18: 2754–2766. [DOI] [PubMed] [Google Scholar]
- Liu H, Xin Y, Xun L. (2014). Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl Env Microbiol 80: 1799–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd D. (2006). Hydrogen sulfide: clandestine microbial messenger? Trends Microbiol 14: 456–462. [DOI] [PubMed] [Google Scholar]
- Luebke JL, Shen J, Bruce KE, Kehl-Fie TE, Peng H, Skaar EP et al. (2014). The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol Microbiol 94: 1343–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther GW, Findlay AJ, MacDonald DJ, Owings SM, Hanson TE, Beinart RA et al. (2011. a). Thermodynamics and kinetics of sulfide oxidation by oxygen: a look at inorganically controlled reactions and biologically mediated processes in the environment. Front Microbiol 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther GW 3rd, Findlay AJ, Macdonald DJ, Owings SM, Hanson TE, Beinart RA et al. (2011. b). Thermodynamics and kinetics of sulfide oxidation by oxygen: a look at inorganically controlled reactions and biologically mediated processes in the environment. Front Microbiol 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Ermler U, Peng G, Michel H. (2010). A new structure-based classification of sulfide:quinone oxidoreductases. Proteins 78: 1073–1083. [DOI] [PubMed] [Google Scholar]
- Marshall KT, Morris RM. (2013). Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD-19 clade. ISME J 7: 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus D, Schultz A, Maynard W. (1950). Microbiological determination of sulfur in yeast. Anal Chem 22: 1187–1190. [Google Scholar]
- Millero FJ, Hubinger S, Fernandez M, Garnett S. (1987). Oxidation of H2S in seawater as a function of temperature, pH, and ionic strength. Environ Sci Technol 21: 439–443. [DOI] [PubMed] [Google Scholar]
- Miranda KM, Wink DA. (2014). Persulfides and the cellular thiol landscape. Proc Natl Acad Sci USA 111: 7505–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra MJ, Dick WA. (1991). Mechanisms of h(2)s production from cysteine and cystine by microorganisms isolated from soil by selective enrichment. Appl Environ Microbiol 57: 1413–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguri T, Schneider B, Reitzer L. (2012). Cysteine catabolism and cysteine desulfhydrase (CdsH/STM0458) in Salmonella enterica serovar typhimurium. J Bacteriol 194: 4366–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Garcia D, Latorre A, Moya A, Gibbs G, Hartung V, Dettner K et al. (2014). Small but powerful, the primary endosymbiont of moss bugs, Candidatus Evansia muelleri, holds a reduced genome with large biosynthetic capabilities. Genome Biol Evol 6: 1875–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SA, Wang X, Lewis KM, DeHan PJ, Park CM, Xin Y et al. (2015). Characterizations of two bacterial persulfide dioxygenases of the metallo-beta-lactamase superfamily. J Biol Chem 230: 18914–18923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Cordovez V, de Boer W, Raaijmakers J, Garbeva P. (2015). Volatile affairs in microbial interactions. ISME J 9: 2329–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz-Bohm K, Geisen S, Wubs EJ, Song C, de Boer W, Garbeva P. (2016). The prey’s scent–Volatile organic compound mediated interactions between soil bacteria and their protist predators. ISME J 11: 817–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatalin K, Shatalina E, Mironov A, Nudler E. (2011). H2S: a universal defense against antibiotics in bacteria. Science 334: 986–990. [DOI] [PubMed] [Google Scholar]
- Shen J, Keithly ME, Armstrong RN, Higgins KA, Edmonds KA, Giedroc DP. (2015). Staphylococcus aureus CstB is a novel multidomain persulfide dioxygenase-sulfurtransferase involved in hydrogen sulfide detoxification. Biochemistry 54: 4542–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Peter EA, Bir S, Wang R, Kevil CG. (2012). Analytical measurement of discrete hydrogen sulfide pools in biological specimens. Free Radic Biol Med 52: 2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Shen J, Fang M, Zhang Y, Hori K, Trinidad JC et al. (2017). Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc Natl Acad Sci USA 114: 2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn HY, Murray DB, Kuriyama H. (2000). Ultradian oscillation of Saccharomyces cerevisiae during aerobic continuous culture: hydrogen sulphide mediates population synchrony. Yeast 16: 1185–1190. [DOI] [PubMed] [Google Scholar]
- Sugata H, Koch FC. (1926). Sulphur metabolism of yeast. Plant Physiol 1: 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Todd JD, Thrash JC, Qian Y, Qian MC, Temperton B et al. (2016). The abundant marine bacterium Pelagibacter simultaneously catabolizes dimethylsulfoniopropionate to the gases dimethyl sulfide and methanethiol. Nat Microbiol 1: 16065. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Vila-Costa M, Simo R, Harada H, Gasol JM, Slezak D, Kiene RP. (2006). Dimethylsulfoniopropionate uptake by marine phytoplankton. Science 314: 652–654. [DOI] [PubMed] [Google Scholar]
- Wagner CA. (2009). Hydrogen sulfide: a new gaseous signal molecule and blood pressure regulator. J Nephrol 22: 173–176. [PubMed] [Google Scholar]
- Watt MK. (2000). A hydrologic primer for New Jersey watershed management. US Geological Survey.
- Wetzel RG. (2001) Limnology: Lake and River Ecosystems. Gulf Professional Publishing: Oxford, UK. [Google Scholar]
- Xin Y, Liu H, Cui F, Liu H, Xun L. (2016). Recombinant Escherichia coli with sulfide: Quinone oxidoreductase and persulfide dioxygenase rapidly oxidizes sulfide to sulfite and thiosulfate via a new pathway. Environ Microbiol 18: 5123–5136. [DOI] [PubMed] [Google Scholar]
- Yang X-P, Wei L-J, Lin J-P, Yin B, Wei D-Z. (2008). Membrane-bound pyrroloquinoline quinone-dependent dehydrogenase in Gluconobacter oxydans M5, responsible for production of 6-(2-hydroxyethyl) amino-6-deoxy-L-sorbose. Appl Environ Microbiol 74: 5250–5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoch DC. (2002). Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl Environ Microbiol 68: 5804–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.