Abstract
Phaeoviruses are latent double-stranded DNA viruses that insert their genomes into those of their brown algal (Phaeophyceae) hosts. So far these viruses are known only from members of the Ectocarpales, which are small and short-lived macroalgae. Here we report molecular and morphological evidence for a new Phaeovirus cluster, referred to as sub-group C, infecting kelps (Laminariales) of the genera Laminaria and Saccharina, which are ecologically and commercially important seaweeds. Epifluorescence and TEM observations indicate that the Laminaria digitata Virus (LdigV), the type species of sub-group C, targets the host nucleus for its genome replication, followed by gradual degradation of the chloroplast and assembly of virions in the cytoplasm of both vegetative and reproductive cells. This study is the first to describe phaeoviruses in kelp. In the field, these viruses infected two thirds of their host populations; however, their biological impact remains unknown.
Kelps (brown algae of the order Laminariales) are the largest marine photosynthetic organisms, engineering temperate rocky coastlines into complex habitats comparable to terrestrial forests, that support extensive marine ecosystems (Dayton, 1985). Ectocarpoids (order Ectocarpales) are small filamentous brown algae (Cock et al., 2010) sharing habitat and close evolutionary relationships with kelps (Kawai et al., 2015). Ectocarpoids are host to the only fully characterised seaweed viruses (genus Phaeovirus), which are comprised of nine virus species infecting seven ectocarpoid species. Phaeoviruses are eukaryotic algal viruses (family Phycodnaviridae) with large (150–350 kb), complex double-stranded DNA genomes (Schroeder, 2011), and are Nucleo-Cytoplasmic Large DNA Viruses alongside Poxviridae, Asfarviridae, Iridoviridae, Ascoviridae, and Mimiviridae. The well-studied type species of Phaeovirus sub-group A is Ectocarpus siliculosus virus 1 (EsV-1), which infects Ectocarpus siliculosus using a persistent strategy, integrating its genome into the genome of hosts infected during their short term as motile spores or gametes, that is, the only wall-less life-cycle stages (Maier et al., 2002). Each host cell inherits the phaeoviral genome, but symptoms appear only in reproductive organs (sporangia or gametangia), which are reprogrammed to produce virus particles instead of host zoids (Müller, 1996). Phaeoviral diversity and host range are largely unknown (Park et al., 2011; Schroeder, 2011). For example, Stevens et al. (2014) provided evidence that members of the Phaeovirus sub-group B (mainly viruses that infect the ectocapoid Feldmannia) evolved from sub-group A, through genome reduction and accompanying loss of DNA proofreading capability. This has led to Phaeovirus increasing its host range and changing from a K- to an r- strategist (Stevens et al., 2014).
There are approximately 13.5 thousand described seaweed species (Guiry, 2012), with 13% belonging to the brown algal class Phaeophyceae. Seaweeds are possibly host to a vast, unexplored diversity of viruses. There have been microscopic observations of virus-like particles (VLPs) in some seaweeds other than Ectocarpales, but none are described in any greater detail (Schroeder, 2011). We have examined three ecologically and commercially important European kelp species, Laminaria digitata, L. hyperborea and Saccharina latissima, targeting the phaeovirus-encoded major capsid protein (MCP) gene using a standard PCR methodology (Stevens et al., 2014). Samples were taken on both sides of the English Channel (Supplementary Figure S1); they included 34 field sporophyte tissue samples collected from epiphyte-free meristematic zones, mixtures of gametophytes isolated from 82 fertile field sporophytes, and 28 clonal gametophyte cultures isolated from gametophyte mixtures that were MCP positive in PCR. In total, 64.7% of sporophytes and 23.2% of gametophyte mixes were phaeoviral MCP positive (Supplementary Table S1).
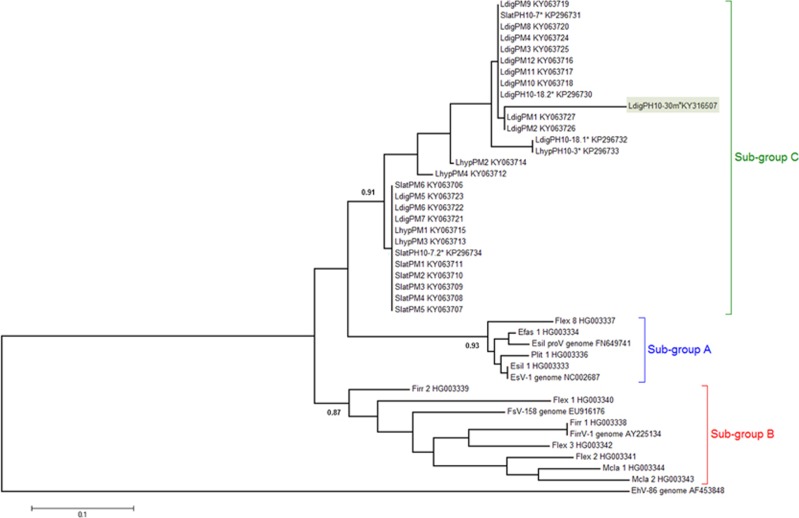
Phylogenetic analysis of 28 sequences of MCP fragments from kelps (160 bp in length; Figure 1), obtained by Sanger sequencing of cloned PCR products showed that the viral sequences from Laminariales (sporophyte MCPs: KY063706-KY063723 and gametophyte MCPs: HG003317-HG003355, LdigPH10-30 m: KY316507) formed a cluster distinct from all known phaeoviruses, which we name sub-group C (Figure 1; posterior probability 0.91). Sub-group C appears to share common ancestry with sub-group A (posterior probability 0.93) and sub-group B (posterior probability 0.87). Similar phaeoviral variants in sub-group C were found in Laminaria and Saccharina, suggesting a host range including multiple genera. The gametophytes LdigPH10-18 and SlatPH10-7 each had 2 different viral MCP sequences, which suggests multiple infection in a single host individual (Figure 1). Given that a combined MCP-DNA polymerase phylogeny of ectocarpoid phaeoviruses showed similar sub-group distinctions (Stevens et al., 2014; Schroeder, 2015), it is unknown if sub-group C diverged at the same time as sub-group B, that is, during or after the speciation of the Ectocarpales, or during or after the divergence of Ectocarpales and Laminariales 90.5 Ma (Kawai et al., 2015). If algal viruses are ancient, then ancestral phaeoviruses may have expanded their host range into all brown algal orders. Many brown algal groups need to be screened for viruses, followed by phylogenetic analyses of any new viral sequences. This would allow the common descent and lateral transfer of brown algal viruses to be disentangled.
Figure 1.
Phylogenetic analysis of sub-group C phaeoviral MCP from Laminariales and Ectocarpales phaeoviral sub-groups A and B. The Coccolithovirus EhV-86 (Schroeder et al., 2002) was used as an outgroup. Topology based on maximum likelihood and decimals are Bayesian posterior probabilities for each sub-group. * denotes sequence variants from gametophyte isolates. Accession numbers are given for each sequence. Scale bar denotes number of nucleotide substitutions per site. Highlighted in green is the strain used for microscopy observations.
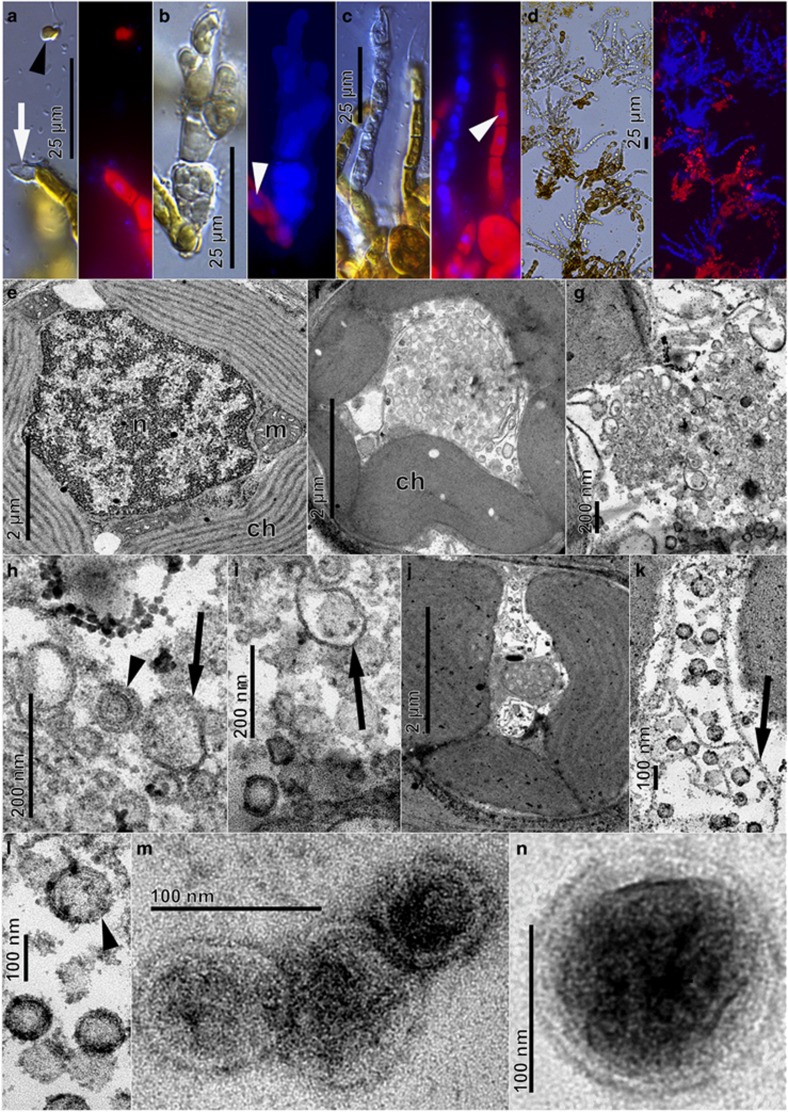
To further describe the sub-group C kelp phaeoviruses, we focused on the L. digitata strain LdigPH10-30 m (Supplementary Table S1), a male gametophyte culture that produced an array of consistent phaeoviral infection-like symptoms (Figures 2a–n), alongside normal growth and gametogenesis (Figure 2a). Gametangia formed preferentially on short side branches (Figure 2a), with one to several spermatozoids developing in each (~5 μm in diameter, arrowhead Figure 2a). The gametes were ejected through a mucilaginous cap, leaving empty translucent gametangia (white arrow, Figure 2a). Female L. digitata gametophyte strains (LdigPH10-31f and LdigPH10-22f) showed similar phaeoviral infection symptoms (Supplementary Figure S2). Healthy gametophyte cells have a large nucleus that can be visualised through DAPI staining and epifluorescence microscopy (discrete and localised blue fluorescence, white arrowheads Figures 2b and c); these are often closely associated with chloroplasts (large irregular red auto-fluorescent structures, Figures 2a–c) distributed around the cell periphery (Figure 2e). Heavily DAPI stained cells were associated with many opaque and not translucent cells (Figures 2b–d). It has been previously reported that similar cells in Ectocarpales were a result of viral infection and that the phaeovirus DNA genomes could be detected through DAPI staining (Müller et al., 1990).
Figure 2.
Light and epifluorescence (a–d, DAPI stained) and transmission electron (e–n) micrographs of Laminaria digitata gametophyte strain LdigPH10-30 m. (a) Spermatozoid (arrowhead) released from antheridium (white arrow), (b,c). Deformed opaque structures with high DAPI blue fluorescence in contrast to normal nuclei (white arrowheads). (d) High prevalence of DAPI-fluorescent filaments. (e) Cross-section of healthy vegetative cell showing chloroplast (ch), nucleus (n), and mitochondria (m). (f–l) VLP formation in vegetative gametophyte cells. Chloroplasts detached from cell periphery, loss of internal structure, appearance of tubular structures (arrows) and various stages of VLP assembly (arrowheads). (m,n) VLPs isolated from extracellular medium and visualised by negative staining.
Transmission electron microscopy (TEM) of the L. digitata strain LdigPH10-30 m suggests that LdigV-1, similar to phaeovirus infections in Ectocarpales, targets the nucleus resulting in the eventual degeneration (Figures 2f and g) as the cytoplasm fills with long tubular structures (arrows; Figures 2h, i and k), followed by the development of virus-like particles (VLPs) (Figures 2f–l). Simultaneously, the chloroplasts detached from the cell periphery and lost their internal structure and pigmentation (Figure 2f). After nuclear and chloroplast degeneration, more fully formed VLPs were visible in the cytoplasm (Figures 2j–l). VLPs were 80–150 nm in diameter, with a 60–100 nm granular core (Figures 2l and n). The VLPs appeared round to hexagonal and may have icosahedral capsids, as known in other phaeoviruses. Mature VLPs were observed in ultrafiltered gametophyte culture medium (Figures 2m and n) showing a structure similar to intracellular VLPs. Our observations in kelp compare well with the characteristics of EsV-1 in Ectocarpus as described by Müller et al. (1990). However, unlike the ectocarpoid phaeoviruses, the infection in kelp appears to be common in vegetative cells (Figures 2d and e) and we do not know yet how the virions are released. Examination of the sorus of field sporophytes did not reveal any abnormal structures, suggesting that kelp viruses, unlike those in Ectocarpoids, may only be expressed in the gametophytes.
Natural reservoirs of gametophytes stabilise kelp populations by allowing new sporophyte recruitment (Steneck et al., 2002) following natural or anthropogenic deforestation (Dayton, 1985; Dayton et al., 1999; Smale et al., 2013). Sea surface temperature increases of 1.4–5.8 °C over the next century (Cox et al., 2000; Houghton et al., 2001) may cause local extinctions of European kelps (Raybaud et al., 2013). In ectocarpoids, phaeoviral symptoms are temperature sensitive (Müller et al., 1998), but it is unknown how phaeoviruses will affect the biology and ecology of their ectocarpoid and kelp hosts in future climate change scenarios.
Seaweeds are exploitated for human consumption, livestock feed (MacArtain et al., 2007; Evans and Critchley, 2014), unique polysaccharides with many industrial applications, pharmaceuticals (Kraan, 2012; Schiel and Foster, 2015), bioremediation, and biofuel (Kraan, 2013). Knowledge of kelp phaeoviruses may be required to meet challenges to seaweed aquaculture (Cottier-Cook et al., 2016), especially since phaeoviruses are transmitted through the germline and could have unexpected effects in cultivation conditions. Though the effects of viruses on kelps remain to be studied in detail, if phaeoviruses commonly occur in kelps they may be transmitted in the host genome, and could alter host reproduction. The discovery of phaeoviruses in kelps highlights the need to further explore the diversity, biology, and ecology of brown algal viruses.
Acknowledgments
This research was funded by INTERREG programme France (Channel)—England, MARINEXUS (Ref: 1956/4073) and Plymouth University. We thank Di Frank Ehrlich for his help with the kelp DNA extractions, and Matt Hall, Dan Smale, Angela Ward, Glen Wheeler and Andrea Highfield from the Marine Biological Association for their help and advice.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Cottier-Cook EJ, Nagabhatla N, Badis Y, Campbell M, Chopin T, Dai W et al. (2016) Safeguarding the future of the global seaweed aquaculture industry. United Nations University (INWEH) and Scottish Association for Marine Science Policy Brief. p 12.
- Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G et al. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465: 617–621. [DOI] [PubMed] [Google Scholar]
- Cox PM, Betts RA, Jones CD, Spall SA, Totterdell IJ. (2000). Acceleration of global warming due to carbon-cycle feedbacks in a coupled model. Nature 408: 184–187. [DOI] [PubMed] [Google Scholar]
- Dayton PK. (1985). Ecology of kelp communities. Ann Rev Ecol Syst 16: 215–245. [Google Scholar]
- Dayton PK, Tegner MJ, Edwards PB, Riser KL. (1999). Temporal and spatial scales of kelp demography: the role of oceanographic climate. Ecological Monographs 69: 219–250. [Google Scholar]
- Evans F, Critchley A. (2014). Seaweeds for animal production use. J Appl Phycol 26: 891–899. [Google Scholar]
- Guiry MD. (2012). How many species of algae are there? J Phycol 48: 1057–1063. [DOI] [PubMed] [Google Scholar]
- Houghton JT, Ding Y, Griggs DJ, Noguer M, van der Linden PJ, Dai X et al. (2001) Contribution of Working Group I to the Third Assessment Report of Intergovernmental Panel on Climate Change. Cambridge University Press: Cambridge, UK. [Google Scholar]
- Kawai H, Hanyuda T, Draisma SG, Wilce RT, Andersen RA. (2015). Molecular phylogeny of two unusual brown algae, Phaeostrophion irregulare and Platysiphon glacialis, proposal of the Stschapoviales ord. nov. and Platysiphonaceae fam. nov., and a re-examination of divergence times for brown algal orders. J Phycol 51: 918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraan S. (2012). Carbohydrates –Comprehensive Studies on Glycobiology and Glycotechnology. In: Chang C-F (ed). Algal Polysaccharides, Novel Applications and Outlook. INTECH Open Access Publisher: Vienna, Austria, pp 459–532. [Google Scholar]
- Kraan S. (2013). Mass-cultivation of carbohydrate rich macroalgae, a possible solution for sustainable biofuel production. Mitigat Adapt Strateg Global Change 18: 27–46. [Google Scholar]
- MacArtain P, Gill CI, Brooks M, Campbell R, Rowland IR. (2007). Nutritional value of edible seaweeds. Nutr Rev 65: 535–543. [DOI] [PubMed] [Google Scholar]
- Maier I, Müller DG, Katsaros C. (2002). Entry of the DNA virus, Ectocarpus fasciculatus virus type 1 (Phycodnaviridae), into host cell cytosol and nucleus. Phycol Res 50: 227–231. [Google Scholar]
- Müller DG, Kawai H, Stache B, Lanka S. (1990). A virus infection in the marine brown alga Ectocarpus siliculosus (Phaeophyceae). Bot Acta 103: 72–82. [Google Scholar]
- Müller DG. (1996). Host-virus interactions in marine brown algae. Hydrobiologia 326-327: 21–28. [Google Scholar]
- Müller DG, Kapp M, Knippers R, Maramorosch K, Murphy FA, Aaron JS. (1998) Viruses in marine brown algae. In:Maramorosch K, Murphy FA, Shatkin AJ (eds) Advances in Virus Research. Academic Press: New York, NY, USA, pp 49–67. [DOI] [PubMed] [Google Scholar]
- Park Y, Lee K, Seok LY, Kim SW, Choi T-J. (2011). Detection of diverse marine algal viruses in the South Sea regions of Korea by PCR amplification of the DNA polymerase and major capsid protein genes. Virus Res 159: 43–50. [DOI] [PubMed] [Google Scholar]
- Raybaud V, Beaugrand G, Goberville E, Delebecq G, Destombe C, Valero M et al. (2013). Decline in kelp in west Europe and climate. PLos One 8: e66044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel DR, Foster MS. (2015) The Biology and Ecology of Giant Kelp Forests. Univ of California Press: Oakland, CA, USA, p395. [Google Scholar]
- Schroeder D. (2015). More to Phaeovirus infections than first meets the eye. Perspectives in Phycology 2: 105–109. [Google Scholar]
- Schroeder DC, Oke J, Malin G, Wilson WH. (2002). Coccolithovirus (Phycodnaviridae): characterisation of a new large dsDNA algal virus that infects Emiliania huxleyi. Archives of Virology 147: 1685–1698. [DOI] [PubMed] [Google Scholar]
- Schroeder DC. (2011) Viruses of seaweeds. In: Hurst C (ed). Studies in Viral Ecology: Microbial and Botanical Host Systems. Wiley-Blackwell: New Jersey, p 208. [Google Scholar]
- Smale DA, Burrows MT, Moore P, O'Connor N, Hawkins SJ. (2013). Threats and knowledge gaps for ecosystem services provided by kelp forests: a northeast Atlantic perspective. Ecol Evol 3: 4016–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steneck RS, Graham MH, Bourque BJ, Corbett D, Erlandson JM, Estes JA et al. (2002). Kelp forest ecosystems: biodiversity, stability, resilience and future. Environ Conserv 29: 436–459. [Google Scholar]
- Stevens K, Weynberg K, Bellas C, Brown S, Brownlee C, Brown MT et al. (2014). A novel evolutionary strategy revealed in the phaeoviruses. PLos One 9: e86040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.