Abstract
A wealth of human studies have demonstrated the importance of gut microbiota to health. Research on non-human animal gut microbiota is now increasing, but what insight does it provide? We reviewed 650 publications from this burgeoning field (2009–2016) and determined that animals driving this research were predominantly ‘domestic’ (48.2%), followed by ‘model’ (37.5%), with least studies on ‘wild’ (14.3%) animals. Domestic studies largely experimentally perturbed microbiota (81.8%) and studied mammals (47.9%), often to improve animal productivity. Perturbation was also frequently applied to model animals (87.7%), mainly mammals (88.1%), for forward translation of outcomes to human health. In contrast, wild animals largely characterised natural, unperturbed microbiota (79.6%), particularly in pest or pathogen vectoring insects (42.5%). We used network analyses to compare the research foci of each animal group: ‘diet’ was the main focus in all three, but to different ends: to enhance animal production (domestic), to study non-infectious diseases (model), or to understand microbiota composition (wild). Network metrics quantified model animal studies as the most interdisciplinary, while wild animals incorporated the fewest disciplines. Overall, animal studies, especially model and domestic, cover a broad array of research. Wild animals, however, are the least investigated, but offer under-exploited opportunities to study ‘real-life’ microbiota.
The dawn of modern microbiota research
Technological advances in multi-‘omic platforms such as metataxonomics and metagenomics, have helped fuel the recent expansion of microbiota research (Marchesi and Ravel, 2015), especially on humans, as exemplified by large-scale efforts such as The Human Microbiome Project, started in 2007 (Peterson et al., 2009). Research on microbiota from non-human habitats has followed: in 2010 the Earth Microbiome Project (www.earthmicrobiome.org) was initiated to document microbial diversity across multiple biomes (Gilbert et al., 2014). Studies focusing on microbiota of the gut have especially captivated scientific interest; it is the most dense and diverse microbial community of the body, is influenced by a range of intrinsic and extrinsic variables including diet, genetics and environmental factors (Khachatryan et al., 2008; Phillips, 2009; Bright and Bulgheresi, 2010; Claesson et al., 2012), and is vital to health and development (Round and Mazmanian, 2009; Lozupone et al., 2012). In recent years non-human animal gut microbiota studies have started to appear, for example, characterising the microbiota of giant pandas, Ailuropoda melanoleuca, to make microbial comparisons across age groups (Tun et al., 2014), or of the European honey bee, Apis mellifera, to understand the role of bacteria in nutrition (Engel et al., 2012). But, what other species have been studied, and why? Given this field is burgeoning, it is timely to take stock of the non-human animal gut microbiota literature and examine the research landscape thus far.
Here, we ask ‘what drives research in animal gut microbiota?’ by quantifying the subject of each study as a domestic, model or wild animal. Within these three animal groups we determine whether data collection is purely observational or instead, the result of experimentation; which animal taxa are used, and which research questions are addressed. In addition, we use network analyses to determine unique and overlapping research foci for each animal group. Finally, we determine the extent that animal groups consider microbiota-host-environment interactions, by calculating the interdisciplinarity of studies within each group.
Data-mining the literature
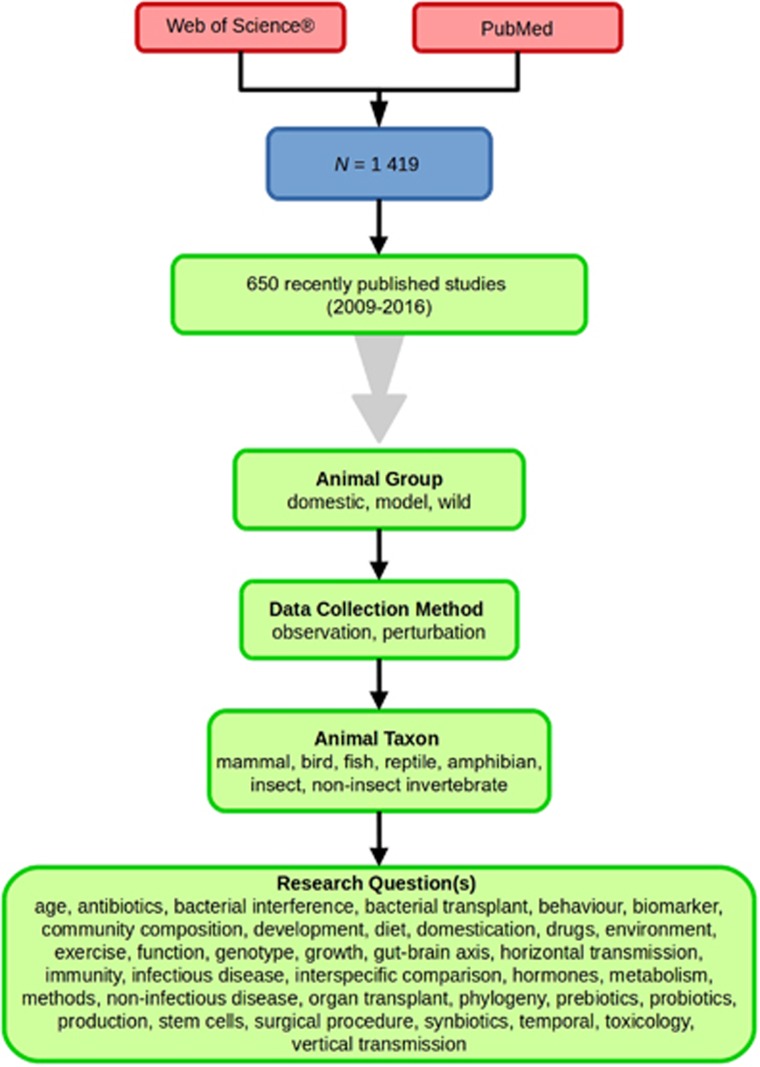
A search for peer-reviewed articles on non-human gut microbiota published between the years 1911 and 2016 was performed in Web of Science and PubMed. Search terms were ‘microbi*’ AND ‘gut’ OR other gut-related terms (‘anal’ OR ‘anus’ OR ‘caec*’ OR ‘cec*’ OR ‘cloac*’ OR ‘colon’ OR ‘duoden*’ OR ‘faec*’ OR ‘fec*’ OR ‘gastro*’ OR ‘ile*’ OR ‘intest*’ OR ‘jejun*’ OR ‘rect*’ OR ‘rum*’ OR ‘stomach’). The search excluded common irrelevant terms (‘ferment*’, ‘microbiol*’, ‘reactor*’, ‘review*’, ‘vitro’), and those related to humans (‘child*’, ‘human*’, ‘infan*’, ‘men’, ‘paedi*’, ‘patient*’). All abstracts of the resulting 3 095 articles were reviewed manually and 1419 were found to characterise the microbiota of the non-human animal gut (either the entire digestive tract, one or more sections, and/or faeces). A sub-set of 650 studies (November 2009 to July 2016) were randomly selected for analysis based on corresponding randomly generated numbers from all studies (Figure 1, Supplementary Information 1). Firstly, we categorised each study as focussing on animal species that were: ‘domestic’ (livestock and companion animals), ‘model’ (studied to provide insight into the microbiota of other organisms), or ‘wild’ (free-living or undomesticated animal species studied in their natural habitat or captivity). For each publication we noted whether data were ‘observational’, that is, purely descriptive, or the result of a ‘perturbation’, that is, a treatment was applied, such as a probiotic. We categorised the focal taxon for each study as mammal, bird, fish, reptile, amphibian, insect or non-insect invertebrate. Finally, 36 broad lines of enquiry (‘research questions’) were identified and quantified within each of the three animal groups (Figure 1, Supplementary Information 1).
Figure 1.
Work flow for categorising gut microbiota studies on non-human animals following searches in Web of Science and PubMed. Of the 1419 relevant articles identified, 650 recently published studies (2009–2016) were categorised into one of three animal groups (domestic, model or wild animals). Data collection method, animal taxon and research question(s) addressed were determined for each study.
What is driving animal microbiota studies?
The 650 publications reviewed here were dominated by studies on domestic animals (48.2%) followed by model animals (37.5%), while wild animal studies were comparatively few (14.3% Table 1). Perturbation is crucial to understand how a system functions, as exemplified by classic ecological experiments (Paine, 1966), and we found that it was used heavily, as opposed to observational data, in domestic studies (81.1% Table 1). Likewise, perturbation was frequent in model studies (87.7%), but was rarely used in wild animals (20.4%), where instead observational data (79.6%) were favoured. All of the reviewed studies focussed on the bacterial communities of the microbiota, and of these, 12.5% studies also characterised at least one other microbial community: archaea (8.8%), fungi (4.3%), protozoa (2.8%) and/or viruses (0.6% Supplementary Information 1). Just over half (54.3%) of studies that investigated the non-bacterial microbiota used perturbation, the remaining half being observational; in addition, about half investigated domestic animals (53.1%), followed by wild (32.1%) and model (14.8%) organisms.
Table 1. The number of studies categorised into three animal study groups: domestic, model or wild, from 650 non-human animal gut microbiota studies, showing data collection methods (observation or perturbation) and network indices of three network graphs investigating research question interdisciplinarity and overlap.
| Animal group |
Data collection method |
Number of nodes (N) | Maximum node size (s) | Maximum node degreea(k) | Maximum node strengthb (NS) | Network densityc (D) | Mean betweenness centralityd (± SEM) (BC) | |
|---|---|---|---|---|---|---|---|---|
| Perturbation | Observation | |||||||
| Domestic (48.2%) | 256 (81.8%) | 57 (18.2%) | 27 | Diet (158) | Diet (20) | Diet (175) | 0.17 | 15.99 (±3.41) |
| Model (37.5%) | 214 (87.7%) | 30 (12.3%) | 34 | Diet (95) | Immunity (23) | Immunity (164) | 0.23 | 19.09 (±3.99) |
| Wild (14.3%) | 19 (20.4%) | 74 (79.6%) | 22 | Community composition (39) | Diet (13) | Community composition (41) | 0.08 | 12.19 (±3.41) |
Node degree (k): The number of edges connected to a node, that is the number of research questions that co-occur.
Node strength (NS): The sum of the weighted edges connected to a node, that is the total number of separate co-occurrences of a research question and all others that it is connected to.
Network density (D): The connections present in a network as a proportion of the total number of possible connections.
Mean betweenness centrality (BC): The mean shortest number of paths required to pass through each research question in the network, that is how well connected research questions are and thus interdisciplinarity of the whole network.
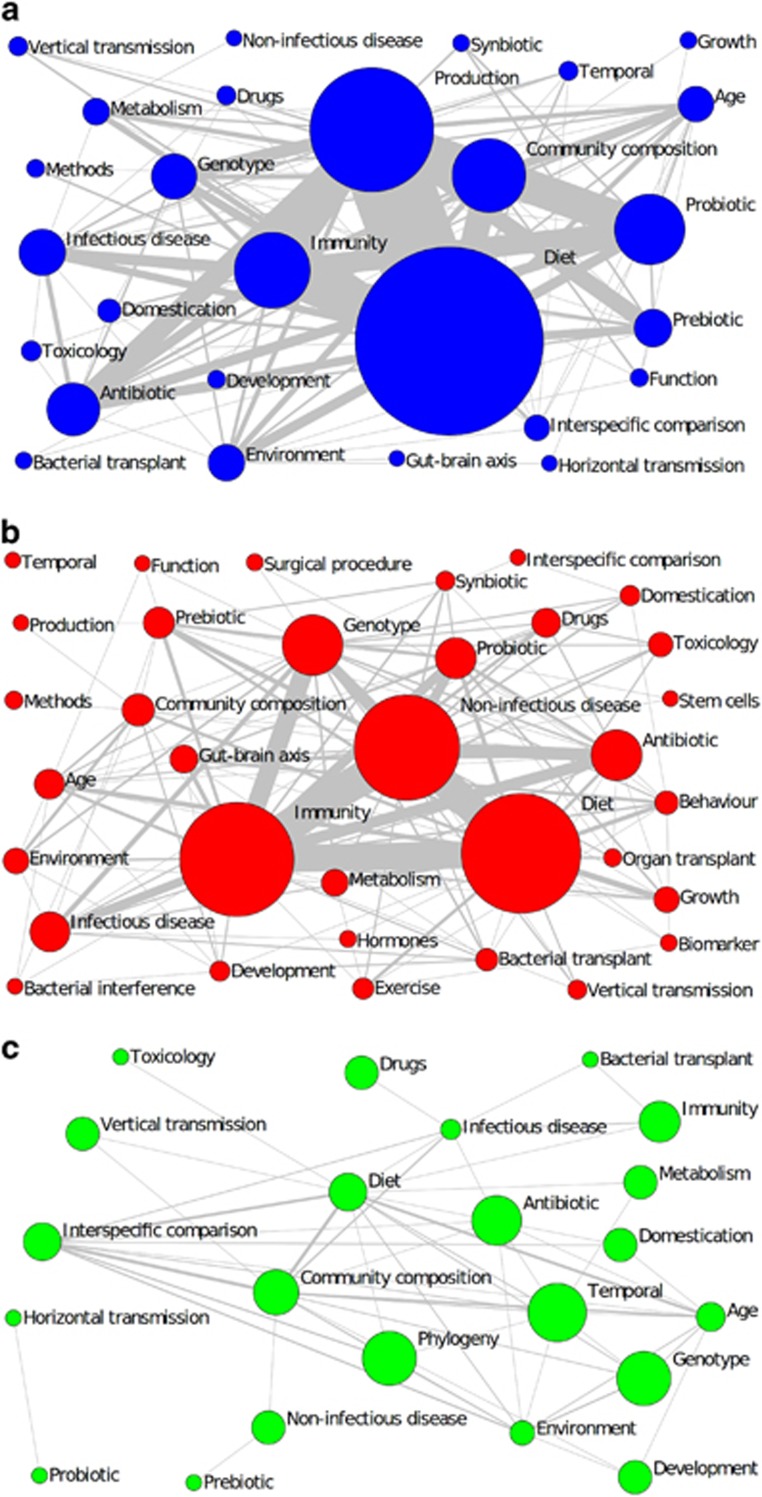
In domestic animals, perturbation was used with the aim of improving animal productivity (29.7%), for example by administering probiotics (16.3%, for example, Ahmed et al., 2014) or prebiotics (6.4%, for example, Hoseinifar et al., 2014; Figure 2a). In model animals perturbation was used to determine interactions between gut microbiota and host health, for example, the role of microbiota in eliciting an immune response (‘immunity’ 36.6% for example, Brinkman et al., 2011) for forward translation to humans. For model animals, perturbation also included therapeutics, such as antibiotics (13.5% for example, Carvalho et al., 2012), and more rarely, organ transplants (1.2% Li et al., 2011) and other surgical procedures (0.8% Devine et al., 2013,Figure 2b). The few wild animal studies to use perturbation did so to understand system functions, for example, by examining the effect of dietary treatments on microbiota of wild-caught giraffes, Giraffa camelopardalis, as a means to understand microbial symbioses (Roggenbuck et al., 2014). Instead, observational data were the norm for wild animals in order to characterise ‘natural’ microbiota structure and function, especially community composition (41.9% Figure 2c).
Figure 2.
(a–c) Network graphs illustrating the frequency of 36 research questions addressed by gut microbiota studies on (a) domestic (b) model and (c) wild animals, and how frequently these questions co-occur within the 650 studies. Each node (circle) represents a research question, with diameter weighted by the number of studies. Edges (lines) connecting each node represent the co-occurrence of different research questions, with width weighted by the total number of co-occurrences.
Although perturbation, under controlled conditions, is more straightforward in domestic and model animals, thus facilitating treatment comparisons and reducing confounding factors such as genetic variation and diet, the complex combination of factors that influence microbiota are unlikely to be understood by looking at laboratory animals alone (McGuire et al., 2008; Amato, 2013). Standardisation may appear logical to obtain less noisy data, but it does not reflect the human condition, where such identical factors are not experienced throughout life nor between individuals, and risks, what Ronald Fisher stated as ‘(supplying) direct information only in respect of the narrow range of conditions achieved by standardisation’ (Fisher, 1937). It would appear that wild animals could provide an opportunity not only to examine natural gut microbiota function, but to extend observations to incorporate understanding of complex multidirectional microbiota-host-environment interactions that they are subject to. Already, other areas of traditionally animal-model dominated research, such as immunology, study and sometimes perturb wild model systems, giving rise to ‘wild immunology’ (Pedersen and Babayan, 2011), and it could be timely for microbiota research to follow suit. Consequently, the obvious progression of wild studies is to understand how ‘natural’ microbiota responds to perturbation as a model for humans and other species, and to determine directionality of microbiota-host-environment interactions (Gordon, 2012). However, difficulties in doing so may be imposed by legislation relating to scientific procedures on wild animals in any given country. In the UK, for example, the Animals Scientific Procedures Act 1986, must be complied with under Home Office regulations. In addition, species may be afforded protection from perturbation due to their international conservation status, for example, those appearing on the International Union for Conservation of Nature (IUCN) red list. Movement of samples between collaborators working on protected species may also be complex due to Convention on International Trade in Endangered Species (CITES) regulations; and permits are required for the translocation of samples from given species between countries. In a compromise between studying wild animals and meeting legal and logistical requirements, 40.9% of wild studies examined here used wild-caught (captured for purposes of study) or captive ‘wild’ animals (for example, from a zoo or research facility), with the remaining 59.1% investigating free-living, or a combination of free-living and captive animals. Even this level of compromise may significantly alter research outcomes, as it has consistently been found that wild animals exhibit a loss of natural microbes following captivity (Xenoulis et al., 2010; Nelson et al., 2013; Kohl and Dearing, 2014).
How taxonomically diverse are animal microbiota studies?
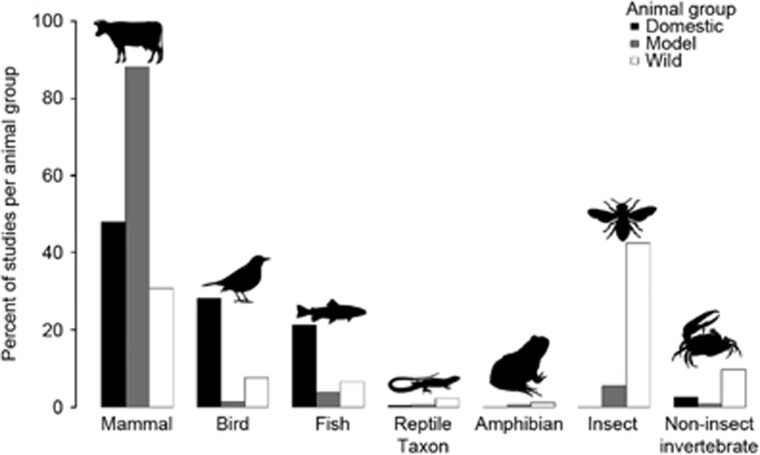
Domestic and model studies were composed of similar taxonomic groups (predominantly vertebrates, that is mammals, birds and fish, within 97.1% and 93.0% of studies respectively), but the opposite was true of wild studies, which predominantly focussed on invertebrates (52.2% Figure 3). Domestic animals that have large farmed populations in economically developed regions were most studied; that is, pigs, cattle (49.7 and 28.7% of mammals respectively), and chickens (80.5% of birds; Figure 3). Species from all seven taxonomic categories have been exploited as models, but model studies mostly focused on laboratory mice (70.2% mammals) or rats (23.3% mammals; Figure 3), in part because the dominant bacterial phyla in the rodent and human gut are similar—Firmicutes, Bacteroidetes and Actinobacteria (Spor et al., 2011).
Figure 3.
The percentage of gut microbiota studies within three animal groups: domestic (black), model (grey) or wild (white), investigating different animal taxa. For each animal group the combined percentage of studies across all taxa equates to 100% of studies for that group.
Laboratory model rodent studies have been fundamental for progressing our understanding of microbiota function and modulation, for example rats have demonstrated microbiota may be used as a biomarker to predict liver transplant rejection (Ren et al., 2013). However, extrapolating data from laboratory animals to other species (including humans) has limitations, for example, similarities in microbiota between rodents and humans are reduced beyond the phyla level (Spor et al., 2011; Nguyen et al., 2015). In addition, laboratory animals have a highly inbred genetic background (Hufeldt et al., 2010), and are exposed to very different conditions to those experienced by humans and wild animals, but which influence microbiota, for example, captive rearing (Zeng et al., 2012), and constant extrinsic factors such as diet and housing conditions (Le Floc’h et al., 2014). Indeed, the disparity between laboratory animals and humans is believed to be a major contributing factor towards ‘attrition’, whereby drug trials are successful in laboratory animals but later fail in human trials (Garner, 2014), and this same lack of successful forward translation is also likely to occur in microbiota research. As such, there appears to be a niche for utilising wild rodents as model organisms, which are physiologically and genetically similar to those already used and understood in the laboratory (Pedersen and Babayan, 2011), but host an intact and diverse gut microbiota (Amato, 2013). However, microbiota studies on wild mammals are currently relatively uncommon (30.6%) and include species not related to those traditionally used as model organisms for example, Arctic ground squirrels (Urocitellus parryii) have been studied to monitor temporal changes in microbiota composition (Stevenson et al., 2014). Instead, wild studies focussed on insects (42.5%), and although wild insects such as Drosophila, whose simple microbiota has provided insight into host-microbe interactions, could be developed as a model system (Chandler et al., 2011), studies were instead driven by the potential for microbiota manipulation to be used in biocontrol. As such, wild insect studies were mainly focussed on agricultural pests and vectors of pathogens for example, bee (23.4%), termite (22.1%) and mosquito species (13.0% Figure 3). These, and similar studies, have suggested that removal of important symbiotic bacteria responsible for lignocellulose digestion could be used to control crop pests (Schloss et al., 2006), and probiotics may be used to control vector-borne pathogens such as Plasmodium (malaria) in mosquitoes, since bacteria can stimulate an up-regulation of immunity genes that reduce Plasmodium acquisition (Dong et al., 2009; Boissière et al., 2012).
Using network analyses to visualise and quantify the research landscape
To visualise research foci and interdisciplinarity, network graphs were constructed for domestic, model and wild animal studies based on research questions. A network graph consists of nodes linked by edges; in this case, a node represented one of the 36 research questions identified, and the edges the co-occurrence of those questions within a scientific paper(s). Each network was constructed from an n by n symmetrical adjacency matrix; composed of a corresponding row and column for every node, where entries indicated links between two nodes (i, j). Edges were non-directed, that is, a link between the nodes i, j had the same value as j, i. Node size (s) was weighted according to the total number of studies addressing that question, and edge width was weighted by the number of studies in which the two research questions co-occurred (Figures 2a–c).
What are the research foci of animal microbiota?
To quantify and compare the foci of research questions between animal groups, we calculated a series of network metrics. Node size (s), or the number of studies investigating any given question depicts how common a question is; node degree (k) represents the number of edges connected to a question, thus its importance in forging links between disciplines; and node strength (NS) is the sum of weighted connections to a question, hence how core the question is to the research.
‘Diet’ was consistently a question of focus in all three animal groups (Table 1), but its research associations differed. In domestic animals ‘Diet’ was most commonly studied (s=158), created the most links to other questions (k=20) and did so frequently (NS=175, Table 1). Thus, diet was fundamental and at the core of this research; often as a means to manipulate animal health via the microbiota, particularly to increase animal production (38.0% domestic diet studies; Figure 2a). ‘Diet’ was also most frequently studied in model animals (s=95), but with respect to host health and disease: 34.7% of such studies used diet specifically to treat or simulate non-infectious diseases such as obesity (Esposito et al., 2015) and diabetes (Prajapati et al., 2015; Figure 2b). Despite its popularity ‘diet’ was not the most integrated or interdisciplinary question in the network, but ‘immunity’ was (k=23 and NS=164; Table 1), highlighting the importance of the shared relationship between microbiota and immunity, and how it consequently affects many other aspects of health (Round and Mazmanian, 2009). In contrast ‘community composition’ was most studied (s=39) and embedded (NS=41) within wild studies, but ‘diet’ was key to creating research links between questions (k=13, Table 1). This link results from the fact that wild studies focus on microbiota structure (for example, Delsuc et al., 2014), and suggests we are currently acquiring more basal knowledge on wild animal microbiota. In addition, only 25.9% of wild animal ‘diet’ studies used perturbations, with the remaining 74.1% observing microbiota composition under a ‘natural’ diet (33.3% Figure 2c). Given that 72% of emerging zoonotic pathogens are transmitted to humans from wildlife (Jones et al., 2008), and microbiota and immunity are strongly interlinked (Round and Mazmanian, 2009), determining how microbiota interacts with host immunity and/or infectious disease (currently only 17.9 and 9.3% in domestic animals which have frequent contact with humans, and 3.2 and 10.8% of wild studies, respectively) deserves further consideration.
Do animal microbiota studies take an interdisciplinary approach?
Animal microbiota studies with a single research focus have provided important basal knowledge on microbial composition and function for example, in-depth analyses of microbiota community composition in laboratory mice has revealed that the intestinal crypts, which harbour gut stem cells, also accommodate a niche microbial community (Pédron et al., 2012). Likewise, there is also great value in an interdisciplinary approach in which multiple factors are studied simultaneously, and can aid in progressing knowledge and teasing apart complex and multidirectional host-microbiota-environment interactions (Gordon, 2012). We quantified the ‘interdisciplinarity’ of each group by measuring the mean ‘betweenness centrality’ (BC) of each network: BC indicates how closely associated all questions are in relation to each other, and is the number of shortest paths required to pass through each question to connect it to all other questions; larger values indicate questions are more closely associated (Leydesdorff, 2007). Network density (D), indicates the level at which interdisciplinarity has been exploited in each group, calculated as a proportion of the total number of possible connections, whereby 0=no connections present and 1=all possible connections are present and maximum interdisciplinarity has been reached. Network analyses were conducted using the igraph package in R v. i386 3.0.3 (Csardi and Nepusz, 2006).
Model studies exploited an interdisciplinary approach the most, with the highest proportion of possible links between questions (D=0.23), followed by domestic (D=0.17) and wild (D=0.08) studies (Table 1). In addition, research questions in model studies were more closely associated, directly or indirectly, with one another, (mean BC=19.09 ±3.99), than in domestic (BC=15.99 ±3.41) or wild (BC=12.19 ±3.41) studies (Table 1). The comparatively high interdisciplinarity of model studies reflects the large range of questions addressed (N=34), compared to the domestic (N=27) and wild (N=22) groups, and the motivation of many model studies to improve medical treatments which often requires an interdisciplinary approach to monitor the range of subsequent effects on health (for example, to investigate the associations between organ transplantation, non-infectious disease, immunity and microbiota; Xie et al., 2014). Conversely, wild studies were the least integrated and interdisciplinary, and more questions were addressed independently of one another. However, this group did address a unique research question: ‘phylogeny’ and how phylogeny is driven across species by gut microbiota and diet, and vice versa; for example, myrmecophagous mammals from different evolutionary lineages exhibit striking convergence with respect to gut microbial composition, driven by dietary adaptations (Delsuc et al., 2014).
While the more focussed approach of wild animal research has allowed us to assemble fundamental microbiota knowledge, it has been argued that an interdisciplinary approach is necessary to progress research on basic and applied gut microbiota (Gordon, 2012). We predict that the interdisciplinarity of wild animal studies will increase as they are adopted in microbiota research, particularly if done so as model organisms. Indeed the first interdisciplinary microbiota studies using wild populations provide interesting insight into the interactions between host, microbiota and environment. For example, parasitic helminths infecting the gut have up- and down-stream effects on microbiota composition (Kreisinger et al., 2015) and seasonal variation in wild rodent microbiota is largely driven by changes in food availability (Maurice et al., 2015).
Conclusion and outlooks
Although more than 10% of studies investigated the microbial community of non-bacterial species in addition to the bacterial component of the microbiota, of these only 0.6% studies investigated the virome, despite evidence that viruses bestow a number of functional traits to bacteria (Ogilvie and Jones, 2015). Complementary studies that simultaneously investigate multiple components of the gut biome are likely to shed light on microbiota composition and functionality (see for example, Glendinning et al., 2014). We demonstrate that most animal gut microbiota studies are driven by economic (domestic animals) or human health (model animals) issues, although more microbiota studies on immunity and/or infectious disease in domestic animals could benefit both livestock and humans in close proximity to them. There are, however, well-founded concerns regarding the limitations of laboratory animals as model organisms, as highlighted by attrition (Fisher, 1937; Garner, 2014). In 2013 the former director of the NIH, Prof. Elias Zerhouni, stated that ‘We have moved away from studying human disease in humans’ (NIH Record: http://bit.ly/2f5UpII), arguing that we should ‘….refocus and adapt new methodologies for use in humans to understand disease biology in humans’ raising interesting issues about the use of animal models, including in microbiota research, and whether it is scientifically legitimate to forward translate our findings to humans. This does not mean that we should not use animal models, but rather that we should consider changing the way in which we study them, so that they may more accurately represent human inter-individuality. The intact gut biomes of wild species that experience inter-individual and environmental variation more similar to humans than their laboratory counterparts, rendering the results more ‘realistic’, could form the basis of more relevant models to study microbiota. However, field experiments would need to be carefully designed to provide statistical power in the face of extensive variation (for example, controlling for genetic background, diet, sex and so on). Under some circumstances, manipulation of microbiota in wildlife is not possible (for example, for rare, elusive or protected species). In these cases, development of mathematical and/or statistical models to assign directionality to observational data could be beneficial. Examples of applications in other fields include, identifying interactions between immune components using network theory (Thakar et al., 2012), and determining interspecific interactions among an unperturbed community of gut parasites, using generalised linear mixed models (Fenton et al., 2010). Studies on wild animals are currently comparatively few, and generally aim to characterise natural microbiota, combining few disciplines. However, we expect interdisciplinarity to increase in wild animals should they be developed as model systems.
Acknowledgments
This research is supported by the Autonomous Province of Trento under the ‘Trentino programme of research, training and mobility of post-doctoral researchers’ Incoming Team project ECOBIOME (EU FP7 Marie Curie actions COFUND: 2011 Call). We thank the Fondazione E Mach and Cardiff University for facilities, and J Lello and the anonymous reviewers for useful comments on the manuscript.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Ahmed HA, Sirohi SK, Dagar SS, Puniya AK, Singh N. (2014). Effect of supplementation of Selenomonas ruminantium NDRI-PAPB 4 as direct fed microbial on rumen microbial population in Karan Fries male calves. Indian J Anim Nutr 31: 20–26. [Google Scholar]
- Amato KR. (2013). Co-evolution in context: The importance of studying gut microbiomes in wild animals. Microbiome Sci Med 1: 10–29. [Google Scholar]
- Boissière A, Tchioffo MT, Bachar D, Abate L, Marie A, Nsango SE et al. (2012). Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog 8: e1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright M, Bulgheresi S. (2010). A complex journey: transmission of microbial symbionts. Nat Rev Microbiol 8: 218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman BM, Hildebrand F, Kubica M, Goosens D, Del Favero J, Declercq W et al. (2011). Caspase deficiency alters the murine gut microbiome. Cell Death Dis 2: e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho BM, Guadagnini D, Tsukumo DML, Schenka AA, Latuf-Filho P, Vassallo J et al. (2012). Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia 55: 2823–2834. [DOI] [PubMed] [Google Scholar]
- Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. (2011). Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet 7: e1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488: 178–184. [DOI] [PubMed] [Google Scholar]
- Csardi G, Nepusz T. (2006). The igraph software package for complex network research. Int J Complex Syst 1695: 1–9. [Google Scholar]
- Delsuc F, Metcalf JL, Wegener Parfrey L, Song SJ, González A, Knight R. (2014). Convergence of gut microbiomes in myrmecophagous mammals. Mol Ecol 23: 1301–1317. [DOI] [PubMed] [Google Scholar]
- Devine AA, Gonzalez A, Speck KE, Knight R, Helmrath M, Lund PK et al. (2013). Impact of ileocecal resection and concomitant antibiotics on the microbiome of the murine jejunum and colon. PLoS ONE 8: e73140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. (2009). Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog 5: e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P, Martinson VG, Moran NA. (2012). Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci USA 109: 11002–11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito D, Damsud T, Wilson M, Grace MH, Strauch R, Li X et al. (2015). Black currant anthocyanins attenuate weight gain and improve glucose metabolism in diet-induced obese mice with intact, but not disrupted, gut microbiome. J Agric Food Chem 63: 6172–6180. [DOI] [PubMed] [Google Scholar]
- Fenton A, Viney ME, Lello J. (2010). Detecting interspecific macroparasite interactions from ecological data: patterns and process. Ecol Lett 13: 606–615. [DOI] [PubMed] [Google Scholar]
- Fisher RA. (1937) The Design of Experiments. 2nd ednOliver and Boyd: London, UK. [Google Scholar]
- Garner JP. (2014). The significance of meaning: why do over 90% of behavioral neuroscience results fail to translate to humans, and what can we do to fix it? ILAR J 55: 438–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Jansson JK, Knight R. (2014). The Earth Microbiome project: successes and aspirations. BMC Biol 12: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JI. (2012). Honor thy gut symbionts redux. Science 336: 1251–1253. [DOI] [PubMed] [Google Scholar]
- Glendinning L, Nausch N, Free A, Taylor DW, Mutapi F. (2014). The microbiota and helminths: sharing the same niche in the human host. Parasitology 141: 1255–1271. [DOI] [PubMed] [Google Scholar]
- Hoseinifar SH, Sharifian M, Vesaghi MJ, Khalili M, Esteban MÁ. (2014). The effects of dietary xylooligosaccharide on mucosal parameters, intestinal microbiota and morphology and growth performance of Caspian white fish (Rutilus frisii kutum fry. Fish Shellfish Immunol 39: 231–236. [DOI] [PubMed] [Google Scholar]
- Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK. (2010). Variation in the gut microbiota of laboratory mice is related to both genetic and environmental factors. Comp Med 60: 336–347. [PMC free article] [PubMed] [Google Scholar]
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL et al. (2008). Global trends in emerging infectious diseases. Nature 451: 990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI. (2008). Predominant role of host genetics in controlling the composition of gut microbiota. PLoS ONE 3: e3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl KD, Dearing MD. (2014). Wild-caught rodents retain a majority of their natural gut microbiota upon entrance into captivity. Environ Microbiol Rep 6: 191–195. [DOI] [PubMed] [Google Scholar]
- Kreisinger J, Bastien G, Hauffe HC, Marchesi J, Perkins SE. (2015). Interactions between multiple helminths and the gut microbiota in wild rodents. Philos Trans R Soc Lond B Biol Sci 370: 20140295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Floc’h N, Knudsen C, Gidenne T, Montagne L, Merlot E, Zemb O. (2014). Impact of feed restriction on health, digestion and faecal microbiota of growing pigs housed in good or poor hygiene conditions. Animal 8: 1632–1642. [DOI] [PubMed] [Google Scholar]
- Leydesdorff L. (2007). Betweenness centrality as an indicator of the interdisciplinarity of scientific journals. J Assoc Inf Sci Technol 58: 1303–1319. [Google Scholar]
- Li Q, Zhang Q, Wang C, Tang C, Zhang Y, Li N et al. (2011). Fish oil enhances recovery of intestinal microbiota and epithelial integrity in chronic rejection of intestinal transplant. PLoS ONE 6: e20460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Ravel J. (2015). The vocabulary of microbiome research: a proposal. Microbiome 3: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice CF, Knowles SC, Ladau J, Pollard KS, Fenton A, Pedersen AB et al. (2015). Marked seasonal variation in the wild mouse gut microbiota. ISME J 9: 2423–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AL, Colgrove J, Whitney SN, Diaz CM, Bustillos D, Versalovic J. (2008). Ethical, legal, and social considerations in conducting the Human Microbiome Project. Genome Res 18: 1861–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson TM, Rogers TL, Carlini AR, Brown MV. (2013). Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ Microbiol 15: 1132–1145. [DOI] [PubMed] [Google Scholar]
- Nguyen TLA, Vieira-Silva S, Liston A, Raes J. (2015). How informative is the mouse for human gut microbiota research? Dis Model Mech 8: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie LA, Jones BV. (2015). The human gut virome: a multifaceted majority. Front Microbiol 6: 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine RT. (1966). Food web complexity and species diversity. Am Nat 100: 65–75. [Google Scholar]
- Pedersen AB, Babayan SA. (2011). Wild immunology. Mol Ecol 20: 872–880. [DOI] [PubMed] [Google Scholar]
- Pédron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G et al. (2012). A crypt-specific core microbiota resides in the mouse colon. mBio 3: e00116–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA et al. (2009). The NIH Human Microbiome Project. Genome Res 19: 2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips ML. (2009). Gut reaction: environmental effects on the human microbiota. Environ Health Perspect 117: A198–A205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati B, Rajput P, Jena PK, Seshadri S. (2015). Investigation of chitosan for prevention of diabetic progression through gut microbiota alteration in sugar rich diet induced diabetic rats. Curr Pharm Biotechnol 17: 173–184. [DOI] [PubMed] [Google Scholar]
- Ren Z, Cui G, Lu H, Chen X, Jiang J, Liu H et al. (2013). Liver ischemic preconditioning (IPC) improves intestinal microbiota following liver transplantation in rats through 16 s rDNA-based analysis of microbial structure shift. PLoS ONE 8: e75950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggenbuck M, Sauer C, Poulsen M, Bertelsen MF, Sørensen SJ. (2014). The giraffe (Giraffa camelopardalis rumen microbiome. FEMS Microbiol Ecol 90: 237–246. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Delalibera I, Handelsman J, Raffa KF. (2006). Bacteria associated with the guts of two wood-boring beetles: Anoplophora glabripennis and Saperda vestita (Cerambycidae). Environ Entomol 35: 625–629. [Google Scholar]
- Spor A, Koren O, Ley R. (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9: 279–290. [DOI] [PubMed] [Google Scholar]
- Stevenson TJ, Buck CL, Duddleston KN. (2014). Temporal dynamics of the cecal gut microbiota of juvenile arctic ground squirrels: a strong litter effect across the first active season. Appl Environ Microbiol 80: 4260–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakar J, Pathak AK, Murphy L, Albert R, Cattadori IM. (2012). Network model of immune responses reveals key effectors to single and co-infection dynamics by a respiratory bacterium and a gastrointestinal helminth. PLoS Comput Biol 8: e1002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun HM, Mauroo NF, Yuen CS, Ho JCW, Wong MT, Leung FC-C. (2014). Microbial diversity and evidence of novel homoacetogens in the gut of both geriatric and adult giant pandas (Ailuropoda melanoleuca. PLoS ONE 9: e79902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenoulis PG, Gray PL, Brightsmith D, Palculict B, Hoppes S, Steiner JM et al. (2010). Molecular characterization of the cloacal microbiota of wild and captive parrots. Vet Microbiol 146: 320–325. [DOI] [PubMed] [Google Scholar]
- Xie Y, Chen H, Zhu B, Qin N, Chen Y, Li Z et al. (2014). Effect of intestinal microbiota alteration on hepatic damage in rats with acute rejection after liver transplantation. Microb Ecol 68: 871–880. [DOI] [PubMed] [Google Scholar]
- Zeng B, Yuan J, Li W, Tang H, Wei H. (2012). The effect of artificial rearing on gut microbiota in a mouse pup-in-a-cup model. Exp Anim 61: 453–460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.