Abstract
Internally transcribed spacer (ITS) rRNA profiling is a novel tool for detailed analysis of microbial populations at low taxonomic ranks. Here we exploited this approach to explore species-level biogeography of the Bifidobacterium genus across 291 adult mammals. These include humans and 13 other primates, domesticated animals, such as dogs, cats, cows, sheep, goats, horses and pigs, and 46 additional species. The collected profiles revealed the presence of 89 putative novel bifidobacterial taxa in addition to 45 previously described species. Remarkably, in contrast to what is currently known for many gut commensals, we did not observe host-specialization among bifidobacterial species but rather their widespread distribution across mammals. Moreover, ITS rRNA profiling of wild relatives of domesticated dogs, rabbits and pigs clearly indicates that domestication and close contact with humans have impacted on the composition of the fecal bifidobacterial population. These data were complemented by analysis of bifidobacterial communities in milk of eight mammalian families, showing that bifidobacteria represent prototypical early gut microbiota members which are inherited by newborns from their lactating mother. Thus this study highlights the role of bifidobacteria as pioneering gut colonizers of a wide range of mammals.
Introduction
The development of the mammalian gut microbiota starts at birth, with colonization of the sterile gastro-intestinal tract of the newborn by bacteria that are harbored by the mother or present in the surrounding environment (Yatsunenko et al., 2012). In this regard, recent studies have highlighted a combined effect of bacterial acquisition from the surrounding environment, including that of the mother, and of gut commensal selection based on human genetic heritage (van Opstal and Bordenstein, 2015). In the first 6–12 months following birth, the gut microbiota is in a continuous compositional flux and its biodiversity progressively expands as the diet of the infant evolves from solely milk based to that typical of its parents or carers (De Filippo et al., 2010; Yatsunenko et al., 2012). Despite its biological relevance, the high level of complexity and the absence of cost-effective tools to assess the gut microbiota at strain level have so far prevented a detailed profiling of its transmission route (s).
Being among the first gut colonizers of newborns, bifidobacteria are known to be an abundant component of the gut microbiota of various animals (including humans) that provide parental care to their offspring (Turroni et al., 2009a; Bunesova et al., 2014; Milani et al., 2014a; Avershina et al., 2016). Moreover, bifidobacteria have been shown to have crucial roles in a variety of biological processes, such as development of the gastro-intestinal tract, induction of mucus layer production, protection against pathogens, maturation of the immune system, as well as expansion of the gut glycobiome and participation in the processing of indigestible food components (Ventura et al., 2012; Milani et al., 2015c; Arboleya et al., 2016; Milani et al., 2016; O’Callaghan and van Sinderen, 2016). We have recently developed an internal transcribed spacer (ITS)-based metagenomic tool for bifidobacterial community profiling (Milani et al., 2014b), allowing (i) accurate taxon identification down to phylotype level, (ii) tracking vertical transmission events, and (iii) discovery of putative novel bifidobacterial species (Milani et al., 2014b, 2015b).
Here we describe the composition of the bifidobacterial DNA present in fecal samples of a wide spectrum of mammalian species and provide clues as to how these gut commensals establish themselves in the newborn gut through a vertical transmission route involving maternal milk as both a vehicle and a biological reservoir of early colonizing bacteria.
Materials and methods
Sample collection
Human fecal samples were collected at the Parma University Hospital (Parma, Italy) while stool of other mammals was retrieved at several Italian zoological parks. Samples consisted of 10 g of fresh fecal material, which is considered to be a sufficient amount to represent the overall biodiversity of the fecal microbiota as outlined in previously published reports (Ley et al., 2008). In order to be sure that the fecal material was fresh and of a certified origin, it was collected immediately following defecation. Furthermore, this amount of stool sample is sufficient for both bacterial DNA extraction and isolation purposes (see below for protocol details). Only samples of verifiable origin were retained. In case of aquatic mammals, sampling was performed during a veterinary examination routine and fecal samples were collected with rectal swabs to avoid contamination (Harper et al., 2003). Conservation and shipment were performed following the ‘Stool Packaging Instructions’ of the Core Microbiome Sampling Protocol A reported in the Manual of Procedures of the Human Microbiome Project (http://www.hmpdacc.org/doc/HMP_MOP_Version12_0_072910.pdf). In detail, all samples were cooled to 4 °C in a fridge under anaerobic conditions immediately after collection and within 24 h packed with ice and sent under anaerobic conditions to the laboratory where they were processed. To ensure refrigeration during transfer, samples were shipped using the World Courier Cold Chain Logistics. Notably, it has previously been established that refrigeration at 4 °C for up to 72 h does not impact on microbial composition, as has also been demonstrated for long-term storage at −80 °C (Choo et al., 2015). Upon arrival, approximately 4 g were immediately processed by the DNA isolation protocol, while the remaining ~6 g were maintained at −80 °C until DNA extraction. These storage conditions were chosen to avoid alterations of the relative abundance of taxa caused by bacterial growth or DNA degradation, while also retaining viability for isolation attempts (Cardona et al., 2012; Fouhy et al., 2015). Samples were subjected to DNA extraction using the QIAamp DNA Stool Mini Kit following the manufacturer’s instructions (Qiagen, Manchester, UK).
Ethical statement
This study was carried out in accordance with the recommendations of the ethical committee of the University of Parma (prot. nos. 11201 and 161/OPBA/2016). The protocols were approved by the ‘Comitato di Etica Università degli Studi di Parma’, Italy. All animal procedures were performed according to national guidelines (Decreto legislativo 26/2014). Informed consent was obtained from all participants. All investigations were carried out following the principles of the Declaration of Helsinki.
Bifidobacterial isolation
The existence of putative novel bifidobacterial taxa and vertical transmission of bifidobacterial strains were verified through strain isolations using mupirocin-based medium, which has previously been described to be selective for bifidobacteria (Turroni et al., 2009b; Serafini et al., 2011; for details, see Supplementary Text).
Fermentation profiles on milk glycans
Details regarding growth conditions and determination of growth profiles are provided in Supplementary Text.
Evaluation of the bifidobacterial cell density by quantitative PCR (qPCR)
Evaluation of the presence of bifidobacteria in animal fecal samples was performed by real-time qPCR as described in Supplementary Text.
Bifidobacterial ITS profiling
Bifidobacterial ITS profiling was performed as described previously (Milani et al., 2014b). Additional details are reported in Supplementary Text.
Statistical analyses, network representations and hierarchical clustering
Co-variance statistical analyses were performed with the SPSS software v. 22 (IBM SPSS Statistics for Windows, Version 22.0. IBM Corp., Armonk, NY, USA) and force-driven network representations were obtained with the Gephi software (Bastian and Jacomy, 2009). Network clusters were predicted using the Modularity statistical function of Gephi. Hierarchical clustering was performed using the software MeV v.4.9.0 (http://mev.tm4.org/) and the Pearson correlation distance metric.
Genome sequencing and analysis
All genomes used for this study were determined by GenProbio srl (Parma, Italy) using a MiSeq Illumina (Illumina, San Diego, CA, USA) platform and the MEGAnnotator pipeline (Lugli et al., 2016) following protocols described in Supplementary Text.
Data deposition
Bifidobacterial ITS profiling data were deposited in SRA database under accession number PRJNA350611. Genome sequences of Bifidobacterium sp. new_taxa_70, Bifidobacterium sp. new_taxa_71, Bifidobacterium breve 7E and Bifidobacterium pseudocatenulatum 1E were deposited under accession numbers: MVOG00000000, MVOH00000000, MNLA00000000, and MNLB00000000, respectively.
Results and Discussion
Distribution of bifidobacteria across the mammalian tree of life
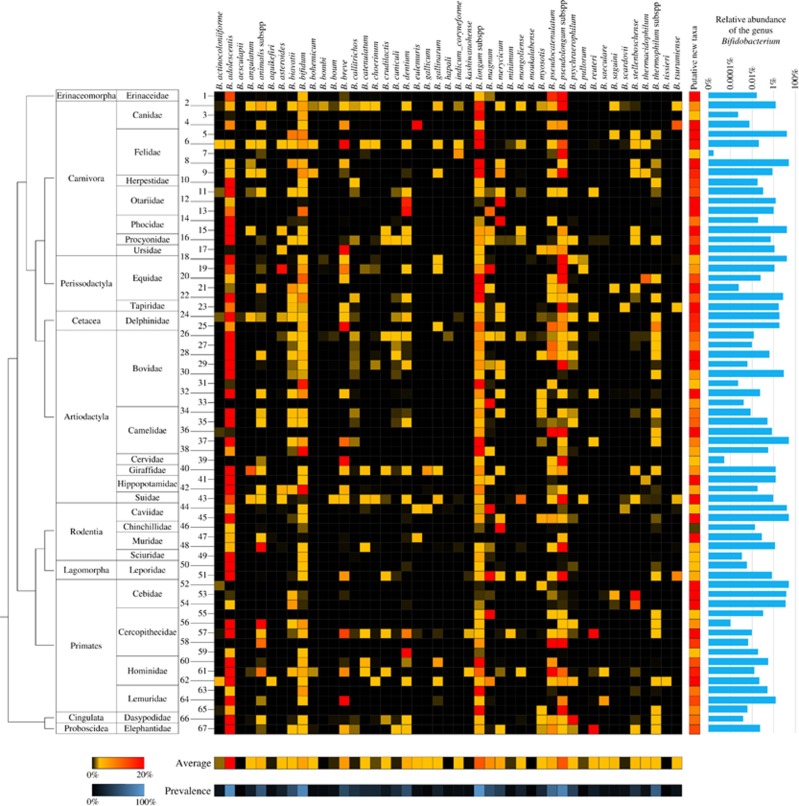
To precisely map the presence of DNA corresponding to bifidobacterial species in complex bacterial communities that reside in the gastro-intestinal tract of mammals, we collected 291 fecal samples of adult animals, corresponding to 67 different mammalian species and 28 mammalian families, and representing a major branch on the phylogenetic tree of life (Supplementary Table S1). Bacterial DNA extracted from these samples was then profiled by means of a recently developed pipeline based on genus-specific primers targeting the hypervariable ITS region and a customized bifidobacterial ITS database representing all sequenced and currently available bifidobacterial strains (Milani et al., 2014b, 2015b). Moreover, the ITS sequences of all sequenced members of other Bifidobacteriaceae genera were also included in the database in order to avoid misclassification of possible non-bifidobacterial Probio_bif_uni/Probio_bif_rev amplicons. Illumina sequencing of the obtained amplicons generated >3.75 million quality-filtered reads that were grouped in clusters of identical sequences (operational taxonomic units (OTUs)) and then taxonomically classified. These data sets were employed to obtain distribution patterns and relative abundance profiles of all currently known bifidobacterial species among the examined mammals. In addition, we also evaluated the proportion of the 67 mammalian species that harbor each bifidobacterial taxa, defined as prevalence. Interestingly, Bifidobacterium longum and Bifidobacterium adolescentis were shown to be present at a prevalence of 95.5% and 91% in the profiled mammalian species, respectively, followed by Bifidobacterium pseudolongum and Bifidobacterium bifidum with a prevalence of 85% (Figure 1). These ubiquitous species thus appear to have evolved an ability to colonize a wide range of mammalian hosts. In contrast, 15 bifidobacterial species, despite showing a very limited distribution with a prevalence in mammalian species <10%, were nonetheless found to be present in an average of 3 mammalian orders and 4 mammalian families (Figure 1). Interestingly, particular species, which were previously thought to be highly specialized to colonize the insect gut, for example, Bifidobacterium actinocoloniiforme, Bifidobacterium asteroides, Bifidobacterium bohemicum, Bifidobacterium bombi and Bifidobacterium indicum (Milani et al., 2014a), were shown to be widely distributed among various mammalian hosts (Figure 1). Notably, despite the low average relative abundance at which the DNA of these bifidobacterial species were detected (ranging from 0.01% to 1.72%), they were not solely identified in mammals that eat insects, demonstrating that their broad distribution is not due to accidental and possibly transient colonization. These data indicate that, even if these species do not possess the same adaptive abilities of bifidobacterial taxa with a high prevalence level, the absence of a strict host-specific specialization behavior is a common feature of (currently known) bifidobacterial species and may indicate the existence of strain-specific adaptation. This hypothesis is further corroborated by the ubiquitous presence of bifidobacterial DNA in fecal samples of Carnivora, that is, mammals with a diet poor in fermentable oligosaccharides/polysaccharides, although we cannot completely exclude transient colonization events. The apparent widespread distribution of DNA corresponding to bifidobacterial species in different mammalian hosts may be a common characteristic of other members of the gut microbiota, as has been suggested by previous genus-level overviews performed by 16S rRNA-based profiling of feces collected from animals kept in captivity (Ley et al., 2008).
Figure 1.
Taxonomic profile of the currently known 45 bifidobacterial species in the mammalian tree of life. The heat map shows the observed relative abundance of 45 known bifidobacterial species in the 291 analyzed samples of adult mammals numbered according to Supplementary Table S9. On the left-hand side, the taxonomy of the samples at Order and Family level, as well as a phylogenetic tree of the mammalian Orders, covered by this study is reported. On the right-hand side of the image, a heat map shows data regarding the relative abundance of putative novel bifidobacterial species with respect to the total bifidobacterial population, while in the lower part two heat maps present the average abundance of the profiled bifidobacterial species with regards to the total bifidobacterial population and their prevalence in the 291 mammalian samples. Bar plots display qPCR data on total bacterial load and bifidobacterial load for each sample in logarithmic scale.
Furthermore, feces collected from members of the mammalian families Cercopithecidae, Hominidae, Bovidae, Suidae, Equidae and Canidae were shown to carry DNA of a relatively high number of bifidobacterial species, harboring on average at least 29 of the 45 currently known members of the genus Bifidobacterium (Figure 1). Interestingly, these families encompass primates and domesticated mammalian species such as cattle, pigs, horses and dogs that live in close contact with humans (Figure 1). This finding highlights that changes in gut microbiota composition may be triggered by a shift from a wild toward a domesticated lifestyle, thus supporting previous studies on Suidae (Ushida et al., 2016) and birds (Wang et al., 2016). Moreover, the families Hominidae and Canidae were shown to harbor the highest number of known bifidobacterial species, that is, 31 and 30, respectively, of which they had 22 in common (Figure 1), suggesting that co-habitation of these hosts may have favored the spread and exchange of bifidobacterial microbiota members colonizing these animals. This idea has previously been put forward to explain the finding of shared microbiota members between companion animals and their owners (Song et al., 2013; Lax et al., 2014; Misic et al., 2015).
Effect of domestication and artificial selection on the bifidobacterial population
To further investigate the impact of domestication on the biogeography of bifidobacterial communities, we profiled the bifidobacterial population of additional undomesticated species, that is, hares, boars and wolves, and compared the obtained profiles to those retrieved from their domesticated relatives living in close contact with humans, that is, rabbits, pigs and dogs, respectively. (Supplementary Table S1 and Supplementary Figure S1). Notably, the obtained data revealed a higher bifidobacterial biodiversity in the domesticated species as compared with wild animals (Supplementary Figure S1), thus supporting the hypothesis that frequent interactions with humans and a domesticated lifestyle favor acquisition of additional bifidobacterial taxa by these mammals through niche shift.
Furthermore, we evaluated whether artificial selection may have impacted on gut microbiota composition. For this purpose, fecal samples were collected from 10 wolves living in the same nature reserve and from 7 dog breeds living in the same kennel and with an identical diet (Supplementary Table S1). The particular canine hosts were selected to represent the main phylogenetic cluster of breeds (Asian/Ancient, Hunting and Mastiff) as identified by Parker et al. (2004) based on microsatellite analysis. Hierarchical clustering based on the average bifidobacterial profiles obtained from the fecal samples of the included wolves and dog breeds reflected that of host phylogeny (Parker et al. 2004; Supplementary Figure S2). This finding therefore indicates that domestication and artificial selection of the wild wolf has gradually shaped the gut microbiota composition of the mammalian species Canis lupus. During domestication, modulation of the gut microbiota may also have been influenced by close human contact.
Quantitative profiling of bifidobacteria among mammals
ITS profiling of members of the Bifidobacterium genus in the 291 analyzed mammalian fecal samples also generated information regarding the abundance of DNA corresponding to known members of the genus Bifidobacterium relative to the total bifidobacterial population (Figure 1). In order to avoid bias of our analyses due to the variable rRNA locus copy number in bifidobacterial genomes (Lugli et al., 2014), we estimated the number of rRNA gene clusters based on available bifidobacterial genome data and used this information to normalize the ITS taxonomic profiling results (Supplementary Table S2 and Figure 1).
Notably, the bifidobacterial species that exhibit high prevalence are also those that, where present, show high average relative abundance, for example, B. adolescentis (32%), B. longum (11.6%) and B. pseudolongum (13.1%), though with B. bifidum (3.6%) representing an exception to this observation (Figure 1). These species represent dominant bifidobacterial taxa, which may be characterized by effective and broad-ranging adaptive capabilities to colonize the gut of these mammalian hosts compared with other (co-colonizing) bifidobacteria. In contrast, of the remaining 42 bifidobacterial species, 26 exhibited, where present, an average relative abundance <1%, with only Bifidobacterium eulemuris, Bifidobacterium magnum, Bifidobacterium breve and Bifidobacterium merycicum representing on average >5% of the whole bifidobacterial population (Figure 1). The observed low average relative abundance of bifidobacteria reflects the general presence of multiple taxa in each mammalian species analyzed. In fact, on average 9.6 (currently recognized) bifidobacterial taxa were detected at a relative abundance of >0.01% in the analyzed mammalian species (Figure 1).
The above results confirm previous genus-level overviews (Ley et al., 2008) and reinforce the view that bifidobacterial development is characterized by extensive selective pressure toward the colonization of a wider range of different hosts and the development of extensive microbe–microbe cooperation (Milani et al., 2015a; Turroni et al., 2016). In order to assess the quantitative contribution of bifidobacteria to the overall microbiota diversity of each of the analyzed ecological niches, the total bacterial load, as well as bifidobacterial cell counts, were evaluated by means of qPCR. Such analyses showed that in adult mammals the gut microbiota concentration ranges between 1E+06 and 4.7E+10 genome copy number per gram of feces, with bifidobacteria representing on average 3.5% of this microbiota population. Notably, this variability reflects the different environmental conditions that characterize the gastro-intestinal tract of the wide range of sampled mammals. Furthermore, bifidobacteria were detected in all profiled mammalian species, of which 53.7% were shown to exhibit a relative abundance of the genus Bifidobacterium of >0.1% (Figure 1). Thus, while bifidobacteria do not represent one of the dominant genera in the gut environment of mammals, they appear to be widespread and ubiquitous across the entire mammalian branch within the tree of life.
Identification of putative novel members of the genus Bifidobacterium
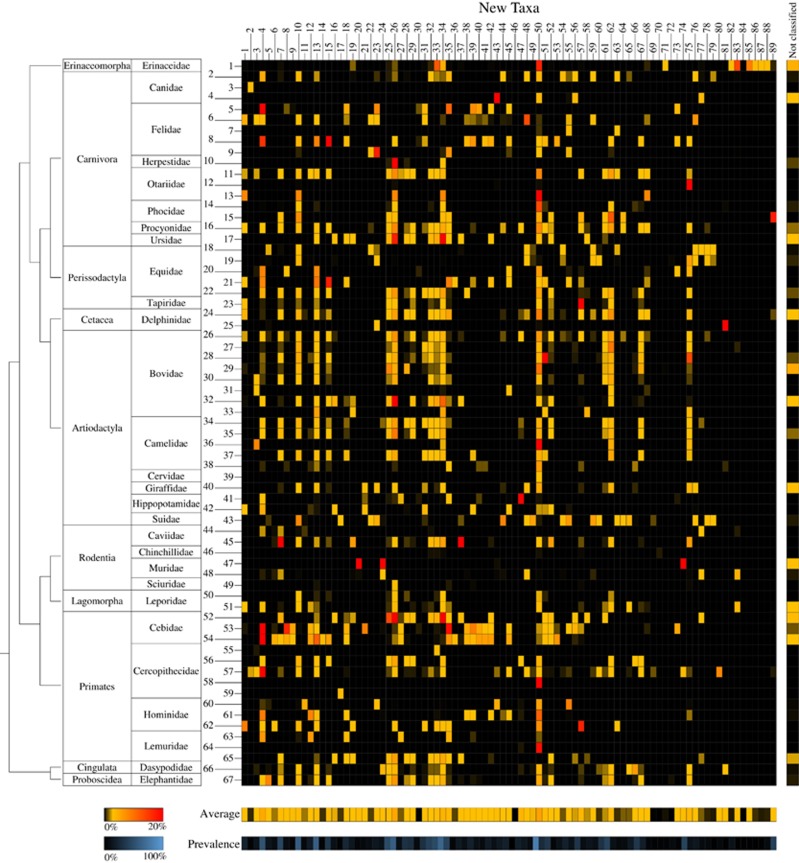
ITS profiling revealed the presence of OTUs with an identity level of <93% with respect to any of the currently recognized bifidobacterial species. This identity cutoff represents the lowest identity level observed between subspecies when comparing the 213 publicly available ITS sequences of known bifidobacterial taxa, focusing on the hypervariable region corresponding to the Probio-bif_uni/Probio-bif_rev amplicon used for bifidobacterial ITS rRNA profiling (Supplementary Table S3). Interestingly, these unclassified bifidobacterial OTUs correspond on average to 21.5% of the bifidobacterial population in the 67 mammalian species analyzed, ranging from a minimum of 0.02% to a maximum of 99.2% (Figures 1 and 2). Notably, to avoid misclassification of ITS sequences corresponding to other Bifidobacteriaceae genera amplified by the Probio_bif_uni/Probio_bif_rev primer pair, we also included in the profiling database the ITS sequences of all the sequenced non-bifidobacteiral members of this family. Alignment and manual evaluation of all identified unclassified OTUs resulted in the identification of 89 putative new (sub)species (named new_taxa_1 through to new_taxa_89) with OTU sequence identity ⩽92% with respect to all currently known bifidobacterial species. These putative novel taxa were added to the bifidobacterial ITS database and profiled in all 291 samples included in this study (Supplementary Table S4 and Figure 2). Profiling data sets were normalized assuming that the putative novel bifidobacterial taxa possess two ribosomal loci, which is the average observed for the genus based on data of currently known bifidobacterial species (Supplementary Table S2). Remarkably, each of the analyzed mammalian species appear to harbor, on average, the DNA of 16 novel bifidobacterial species with an average abundance, when present, of 1.66% with respect to the overall bifidobacterial population (Figure 2). Evaluation of the occurrence of the putative novel taxa among the analyzed mammals failed to reveal specific distribution patterns linked to host taxonomy or diet, yet again highlighting that Primates, Cercopithecidae and Hominidae, as well as mammalian families encompassing domesticated species, harbor the highest number of (novel) bifidobacterial species (Figure 2).
Figure 2.
Taxonomic profile of 89 putative novel bifidobacterial species in the mammalian branch of the tree of life. The heat map displays the relative abundance with respect to the total bifidobacterial population observed for the 89 putative bifidobacterial species, numbered progressively from 1 to 89, in the 291 analyzed samples of adult mammals numbered according to Supplementary Table S9. On the left-hand side, the taxonomy of the samples at Order and Family level is indicated, as well as a phylogenetic tree of the mammalian Orders, covered by this study. The vertical heat map on the right-hand side shows data regarding the relative abundance of OTUs not classified as new bifidobacterial species but displaying similarity with other members of the Bifidobacteriaceae family, while in the lower part of the image two horizontal heat maps present the average abundance with respect to the total bifidobacterial population of the profiled putative bifidobacterial species and their prevalence in the 291 mammalian samples.
Moreover, while six novel taxa (new_taxa_4, 10, 13, 26, 34 and 50) were shown to be prevalent at an average of 63.9% of the mammalian species (ranging from 53.7% to 82.1%), none of these exhibit a relative abundance >4.7% with respect to the overall bifidobacterial population. In fact, evaluation of the average relative abundance of these 89 putative novel bifidobacterial taxa, calculated including only data of the mammalian species where they are detected, revealed that only new_taxa_74 and new_taxa_20 were shown to elicit an average relative abundance >8% (17.9% and 12.4%, respectively) (Figure 2).
These findings indicate that these suspected, and as yet undescribed, novel members of the genus Bifidobacterium are widespread among mammals, though their presence has until now remained undetected, probably owing to their low abundance (Figure 2).
To validate the in silico identification of novel taxa, we isolated and genomically characterized two of these novel bifidobacterial species by means of a previously described cultivation-based approach (Turroni et al., 2009a; Turroni et al., 2012; Supplementary Table S5; Supplementary Text).
Co-variance of bifidobacterial species among mammals
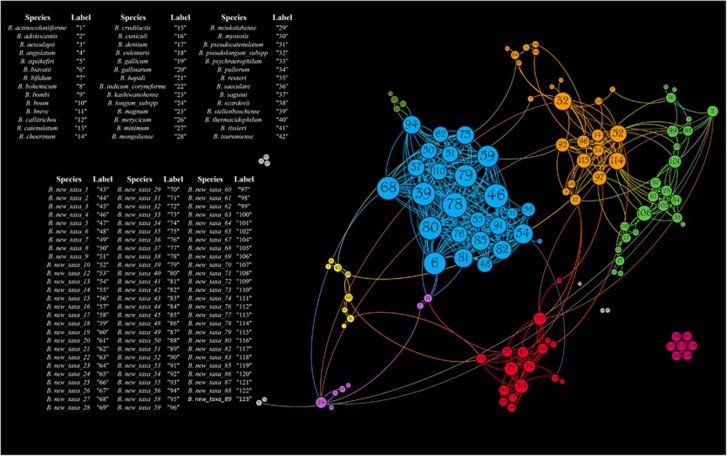
Availability of profiling data for all known and putative novel bifidobacterial species allowed the evaluation of their co-occurrence or co-exclusion across the mammalian tree of life by means of Pearson correlation index (Supplementary Table S6). Statistically significant (P-value<0.05) co-variance data were then used to construct a force-driven network (Figure 3). Interestingly, significant (P-value<0.05) co-exclusion appears to be very limited among members of the Bifidobacterium genus and was only observed among the ubiquitous B. adolescentis, B. longum and B. pseudolongum taxa, probably owing to their high prevalence in the mammalian gut environment, as underlined by the collected profiling data (see above) (Supplementary Table S6). In contrast, all other co-variance interactions with P-value<0.05 were observed to be co-occurrences, which supports the idea of extensive co-evolution among bifidobacterial members that are present in the mammalian gut microbiota (Supplementary Table S6).
Figure 3.
Co-variance network of the 45 currently recognized and 89 putative novel bifidobacterial species present in mammals. The force-driven network shows the predicted co-variances with P-values<0.05 between the profiled bifidobacterial species. The node size is proportional to the number of co-variances, while the node color indicates the eight clusters observed.
Furthermore, the constructed force-driven co-variance network highlights the presence of eight clusters of bifidobacterial species with a high degree of co-occurrence among the analyzed mammals (Figure 3). Notably, four clusters involved just a small number of species (⩽8), while the four remaining clusters were formed owing to extensive co-occurrence between 16, 23, 24 and 26 species (Figure 3). In contrast, very limited co-occurrence interactions (⩽5) were shown to exist between different clusters (Figure 3), with the exception of two clusters that were linked by 18 connections, of which 3, 4 and 6 involve new_taxa_60, new_taxa_10 and new_taxa_78, respectively (Figure 3). Notably, these data indicate that bifidobacteria have developed specific sets of cooperative behavior between co-colonizers, which act as powerful evolutionary drivers in the mammalian gut microbiota. This assumption is supported by previous studies that have reported the existence of cross-feeding between specific bifidobacterial species (Milani et al., 2015a; Turroni et al., 2016).
In constrast, correlation analyses between the presence/absence of bifidobacterial species and the taxonomy or dietary lifestyles (vegetarian, carnivorous or omnivorous) of the mammalian hosts failed to identify any statistically significant correlation. This finding reinforces the notion that bifidobacterial species did not evolve individually toward adaptation to specific environments but instead relied on microbe–microbe cooperation to achieve colonization of a wider range of different hosts (Belenguer et al., 2006; Rios-Covian et al., 2015; Riviere et al., 2016). Notably, while co-variance analyses clearly pointed out the existence of interactions between members of this genus, bifidobacteria should also be considered as valuable key candidates for intergenus cooperation in the light of the complex bacterial community harbored by the mammalian gut.
Exploring vertical (mother-to-child) transmission of bifidobacteria
Vertical transmission of gut microbiota members from mother to offspring during delivery and subsequent parental care is believed to have a key role in colonization and persistence of bacteria in humans (Milani et al., 2015b). In order to evaluate transmission of specific bacterial phylotypes among mammals, we collected fecal samples of four human mother–child and 12 animal mother–child pairs, together representing seven different mammalian species and five mammalian orders, and encompassing humans and domesticated animals (Supplementary Table S7). In addition, we re-analyzed two datasets from previous work (Milani et al., 2015b), listed in Supplementary Table S7 as Human_5 and Human_6 mother–child dyads. Reanalysis yielded improved results due to the use of the updated version of our extended, customized bifidobacterial ITS database. Where possible, we also sampled the milk of the mother and feces of her multiple newborns (Supplementary Table S7). The resulting qPCR evaluation of the relative abundance of the genus Bifidobacterium showed that, despite considerable variability among samples, in mammals bifidobacteria represent on average 0.7% and 8.9% of the whole fecal microbiota of sampled mothers and corresponding offspring, respectively (Supplementary Figure S3). Interestingly, comparison of data collected from human and non-human species revealed that, on average, human infants harbor a higher abundance of bifidobacteria (32.3% compared with 0.7% of non-human offspring, analysis of variance P-value<0.001; Supplementary Figure S3). This finding reflects previous observations that human milk differs in oligosaccharide composition from that of other mammals. More specifically, human milk contains a higher abundance of type I oligosaccharides, that is, containing lacto-N-biose type I structures, as compared with milk from non-primate mammals (Urashima et al., 2012; Albrecht et al., 2014).
Taxonomic classification of the predicted ITS-based OTUs allowed us to reconstruct the composition of the bifidobacterial population in the analyzed mother–child pairs and to identify phylotypes that they had in common (Supplementary Figure S4). Interestingly, the large majority of such common OTUs in mother–offspring pairs corresponds to a single species constituting on average 82.9% of the profile retrieved for the pool of common OTUs (Supplementary Figure S4). This finding suggests that specific members of the microbiota possess enhanced capabilities supporting vertical transmission, including the ability to reach a high relative abundance in the mother and/or a genetic repertoire that supports transmission to and subsequent colonization of her offspring.
The retrieved species-level profiles of the offspring were also implied in the construction of a principal coordinate analysis plot based on Bray–Curtis dissimilarity (Supplementary Figure S5). This analysis revealed that siblings born in the same litter from the same mother cluster together, thus indicating the development of a similar gut bifidobacterial population, as also confirmed by bar plot representation of the taxonomic profiles (Supplementary Figure S4).
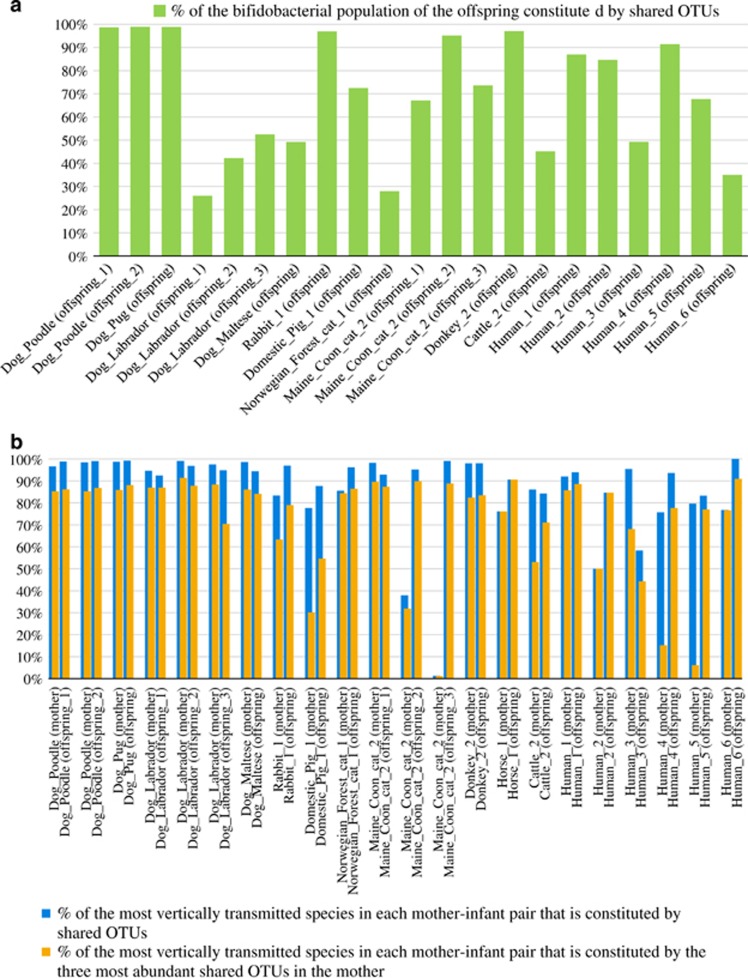
Homing in on the phylotype level, analysis of the retrieved bifidobacterial ITS-based OTUs (as calculated based on 100% identity) revealed the presence of identical phylotypes in fecal samples of mother and corresponding offspring for all analyzed mammalian families (Figure 4). Notably, the OTUs shared by mother–child dyads represent an average of 69.4% of the total bifidobacterial population of the corresponding single or multiple offspring (Figure 4). This data suggests extensive mother–child transmission of gut bifidobacterial phylotypes among mammalian representatives and underlines the key role exerted by vertical transmission in defining the biodiversity and composition of the bifidobacterial population (and perhaps other microbiota components) colonizing the newborn gut.
Figure 4.
Impact of vertical transmission events on the offspring microbiota. Panel (a) shows a bar plot representing for each mother–offspring set the percentage of the total bifidobacterial population of the offspring that is constituted by OTUs also found in the mother. Panel (b) displays a bar plot depicting for each mother–offspring set: in blue the percentage of the most frequently observed vertically transmitted bifidobacterial strains as represented by OTUs that were in common between mother and offspring, and in orange the percentage of the most frequently observed vertically transmitted strains represented by the three most abundant OTUs from the mother that were also found in her offspring.
Furthermore, the most represented bifidobacterial taxa in the shared OTUs pool of each mother–offspring pair were observed to represent, on average, the 81.7% and 92.3% of the total bifidobacterial population in the respective mother and offspring samples (Figure 4).
Moreover, evaluation of the three most abundant OTUs of the mother representing the shared OTU pool revealed that they constitute, on average, 64.7% and 81.1% of the population of their species in the mothers and their children, respectively (Figure 4). Such findings support the hypothesis that competition for vertical transmission does not only occur at the species level but also at the phylotype level, meaning that only a small number of phylotypes for each species, in particular those that are able to reach a high relative abundance, will be successfully transmitted from mother to child. Nevertheless, further experiments involving shotgun metagenomics and/or strain isolation followed by genome reconstruction are needed to verify this hypothesis.
Role of milk as modulator of gut microbiome development
Production of milk as the sole infant nutrition during the first weeks and months following birth is one of the distinctive features of Mammalia. Moreover, mammary glands have been proposed to act as a bacterial reservoir that supports transmission of infant gut microbial colonizers during suckling (Rodriguez, 2014).
The availability of 11 milk samples from representatives of four mammalian orders and six species (Supplementary Table S7) showed the presence of bifidobacterial DNA in all milk samples, thus suggesting a specific adaptation of bifidobacteria to colonize and/or persist in the mammary gland (Supplementary Figure S3; Supplementary Text). Moreover, ITS profiling of milk samples allowed us to assess the ability of milk to act as a vector for the migration of gut bacteria from mother to offspring. Similar to previous observations for human samples (Martin et al., 2012; Milani et al., 2015b), species-level profiling of mother’s milk and fecal samples, as well as of corresponding newborn fecal samples, suggests the presence of the same bifidobacterial strains in each of the analyzed mammalian mother–child dyads. Notably, isolation and genomic characterization of these strains is needed to evaluate their vertical transmission (see below). Our analysis also indicates that the taxonomic profile of the phylotypes that are common between mother–offspring resemble, in terms of composition and species abundance, that of OTUs commonly found in offspring milk and mother milk samples (Supplementary Figure S6). In fact, the OTUs shared by milk and offspring represent on average 74.6% of the bifidobacterial population of the newborn, while 58.0% is also shared by the mother (Supplementary Figure S7). Intriguingly, these data further substantiate the notion that maternal milk (and perhaps the mammary gland) acts as a reservoir of (bifido)bacteria, which may be transmitted to the suckling infant.
Isolation and genomic characterization of two phylotypes corresponding to OTUs shared by Human_2 and Cattle_2 families, that is, identified in both mother and her offspring, confirmed the in silico prediction. In fact, isolates obtained from mother and the respective child, and corresponding milk sample, displayed an average nucleotide identity value >99.99%, as well as 100% ITS identity (Supplementary Text).
Altogether, the data obtained by dissection of the genome of these two isolates (Supplementary Table S8) and growth experiments on human milk oligosaccharide (HMO) and HMO-derived sugars (Supplementary Figure S7) indicated that these vertically transmitted strains have evolved to use carbon sources typically present in milk, thus pointing at mammary glands as an effective reservoir and milk as an optimal vector for vertical transmission of (seemingly specifically adapted) bifidobacterial members of the gut microbiota from mother to offspring.
Conclusions
Bifidobacteria represent a prototype for the investigation of the ecological and functional roles exerted by the mammalian gut microbiota, especially during the crucial developmental phase immediately following birth when gut colonization takes place.
Nevertheless, current ecological knowledge on bifidobacteria is essentially restricted to the human gut, and in particular the period during which the infant’s nutrition is exclusively milk (Roger et al., 2010; Turroni et al., 2012; Avershina et al., 2016; Martin et al., 2016; Nuriel-Ohayon et al., 2016; Yassour et al., 2016). In the current study, we explored the bifidobacterial ecology in 67 mammalian species, representing an exhaustive array of species within this class of animals, by means of bifidobacterial ITS profiling. This culture-independent approach was chosen for its unique ability to allow in-depth phylotype-level community profiling and being cost-effective for screening of a wide range of samples in order to reconstruct a complete overview of, in this case, the bifidobacterial distribution across the mammalian branch of the tree of life. Notably, our analyses demonstrate that bifidobacterial species are universally distributed across the mammalian branch of the tree of life irrespective of host’s taxonomy or diet, thus revealing that these gut commensals have evolved a broad colonization potential, rather than that they engaged in host-specific specialization. Nevertheless, while this seems evident at a species level, bifidobacteria may have developed strain-specific adaptation to specific hosts as a result of long-term co-evolution, as suggested by a recent observation that HMO provided through breast-feeding induces selection of specific bifidobacterial strains in the infant gut environment (LoCascio et al., 2007).
Collection and analysis of fecal samples from wild relatives of common domesticated animals was performed to explore the effect of domestication and close contact with humans. The obtained data clearly indicate that domestication of animals has promoted a genetic diversification that followed or accompanied such human interventions. Notably, comparison of the bifidobacterial biogeography between domesticated vs wild animals revealed an increase in the number of colonizing species following domestication. Moreover, further exploration of the impact of artificial selection and interaction with humans was also performed by means of profiling of seven dog breeds. These data revealed that the composition of the bifidobacterial gut population of dog breeds differs from that of wolves and corresponds to their phylogenetic distance (as based on microsatellite typing. Even if the profiling data of bifidobacteria across mammals appears to exclude a major role of host genetics, data retrieved from wild specimens and dog breeds indicates that close contact with humans has progressively shaped the bifidobacterial population. In particular, the increase in species richness may have resulted from prolonged co-existence with humans (characterized by high bifidobacterial biodiversity) suggestive of a niche shift. Another key ecological feature of bifidobacteria is that they are numerically relevant during the first weeks and months following birth in the human gut (Avershina et al., 2016), a feat that appears to be a general trait of bifidobacteria across the mammalian tree of life. Altogether, these data, as well as the presence of bifidobacteria in milk, suggest a genetic specialization of bifidobacteria to the mammalian gut, possibly re-enforced by direct transmission of bifidobacterial consortia from mother to offspring. Previous studies have shown that certain human-derived bifidobacterial species are shared between mothers and their children (Milani et al., 2015b; Avershina et al., 2016), and our findings imply that this scenario extends to many other mammals. Moreover, we show that the gut bifidobacterial population of siblings is rather similar, implying a key role exerted by the mother in defining the gut bifidobacterial community. The establishment of the gut microbiota by means of vertical transmission points to a key functional role exploited by the transmitted microorganisms in the gut of the host. In this context, the role exerted by the gut microbiota in priming and maturing the immune system and energy harvesting in human neonates is well documented, while experimental proof is also emerging regarding its role in nervous system development (Diaz Heijtz et al., 2011; Cox et al., 2014; Matsuki et al., 2016).
Thus bifidobacteria represent ideal prototypes to investigate gut microbiota establishment and development. The developmental trajectory of bifidobacteria in humans includes a decrease in the relative abundance during adult and elderly stages of life, but their maintenance in the core microbiota, even at very low levels, has been noticed in many metagenomic studies involving healthy individuals (Claesson et al., 2012; Rodriguez et al., 2015; Odamaki et al., 2016). Persistence of bifidobacteria in humans across their entire life also reinforces their putative role as key members of the gut microbiota and underlines the need of additional studies for the understanding of the intricate network of interactions that links bacterial genera in the gut environment.
Previous metagenomic analyses have revealed a high prevalence of bifidobacteria in the human milk microbiome (Collado et al., 2009; Martin et al., 2009; Turroni et al., 2011), even though the origin of these microbes is unclear. In this study, we have shown that bifidobacteria are (sometimes highly) prevalent in the milk microbiome not only in humans, as previously described (Collado et al., 2009; Martin et al., 2009), but also in various other mammalian species. These data suggest that milk oligosaccharides represent key bifidogenic compounds that shape the mammalian gut microbiota with strain specificity at an early stage of life confirming and extending previous observations for HMOs in humans (LoCascio et al., 2007).
Furthermore, bifidobacterial phylotypes that appear to be commonly present in stool samples of mother–offspring dyads were sometimes also detected in the corresponding mother’s milk sample, thus reinforcing the notion that milk may serve as an inoculum for breast-feeding infants (Martin et al., 2009; Martin et al., 2012; Jeurink et al., 2013).
The genomic characterization of two vertically transmitted bifidobacterial strains (identified in the milk microbiome, as well as in stool samples, of the corresponding mother–infant pair) revealed a rich repertoire of genes encoding glycosyl hydrolases, as well as transporters (as discussed in detail in Supplementary Text), supporting an active metabolism of lactose and milk oligosaccharides in a strain isolated in humans, whose milk is rich in these glycans. In contrast, limited adaptation has been observed for a strain isolated from cattle milk, where these complex sugars are less abundant. These results indicate that certain bifidobacterial taxa have genetically adapted to specific milk environments, thus resulting in improved fitness for colonization of mammary glands and the gut of breast-fed newborns. Notably, genomic characterization of additional isolates is needed to confirm that genetic adaptation to the milk environment is a distinctive feature of vertically transmitted strains.
ITS profiling data revealed that the genus Bifidobacterium may exist of many more species than the currently 45 taxonomically recognized species, with 89 predicted novel (sub)species identified in this work, 2 of which were isolated and genetically characterized. Notably, and in contrast to what has previously been proposed (Lugli et al., 2014; Milani et al., 2014a), the broad distribution of these 134 bifidobacterial (sub)species in mammals does not reveal any host-specific specialization behavior nor does it seem to correspond to the taxonomy or diet of the host. Arguably, pointed out by a previous study (Oh et al., 2010) concerning diversification of Lactobacillus reuteri strains isolated in different hosts. These data together with the identification of common complex bifidobacterial populations and eight groups of bifidobacterial taxa showing strict co-occurrence demonstrate that bifidobacteria have evolved to achieve extensive co-colonization, which may depend on functional cooperation.
Nevertheless, additional shotgun metagenomics analyses are still required to explore the functional contribution of bifidobacteria in the gut environment of each mammalian species. Moreover, the ITS profiling approach relies on a relatively small genomic region for phylogenetic classification. Thus culturomic approaches will be needed to dissect the microbial composition of this complex ecosystem so as to allow genomic characterization of strains that are adapted to colonize different mammals and to facilitate formal verification of putative novel bifidobacterial taxa. Furthermore, a large majority of the fecal samples from non-human mammals were collected from animals living in zoos, thus additional sampling from wild representatives of these species are required to confirm our observations in a completely natural environment.
Acknowledgments
Part of this research is conducted using the High Performance Computing (HPC) facility of the University of Parma. We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. This work was supported by the EU Joint Programming Initiative—A Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) to DvS (project reference 15/JP/HDHL/3280) and to MV (MIUR Prot. 001988). DvS is a member of The APC Microbiome Institute, which is funded by Science Foundation Ireland (SFI), through the Irish Government’s National Development Plan (Grant number SFI/12/RC/2273). We thank ‘Fondazione Bioparco di Roma’ (Viale del Giardino Zoologico 20, 00197, Rome, Italy), ‘Zoomarine’ (via dei Romagnoli, 00040 Torvaianica, Pomezia, Rimini, Italy), ‘Giardino zoologico di Pistoia’ (Via Pieve a Celle, 160, 51100, Pistoia, Italy) and ‘Centro Tutela e Ricerca Fauna Esotica e Selvatica Monte Adone’ (Via Brento, 9, 40037, Sasso Marconi, Bologna, Italy) for their support in the recruitment of the samples.
Author contributions
CM designed all experiments, performed bioinformatics analyses and wrote the manuscript. GAL and LM performed genomics bioinformatics analyses. MV and DvS conceived the study, revised and approved the manuscript. MM performed sample collection, qPCR experiments and bifidobacterial ITS profiling. SD, FT and CF participated in qPCR experiments and bifidobacterial ITS profiling. KJ performed growth analyses on HMOs. MCO participated in data analysis. All authors reviewed and approved the final version of the manuscript.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Albrecht S, Lane JA, Marino K, Al Busadah KA, Carrington SD, Hickey RM et al. (2014). A comparative study of free oligosaccharides in the milk of domestic animals. Br J Nutr 111: 1313–1328. [DOI] [PubMed] [Google Scholar]
- Arboleya S, Watkins C, Stanton C, Ross RP. (2016). Gut bifidobacteria populations in human health and aging. Front Microbiol 7: 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avershina E, Lundgard K, Sekelja M, Dotterud C, Storro O, Oien T et al. (2016). Transition from infant- to adult-like gut microbiota. Environ Microbiol 18: 2226–2236. [DOI] [PubMed] [Google Scholar]
- Bastian MHS, Jacomy M. (2009). Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media, 7th July 2009, New York, NY, USA.
- Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE et al. (2006). Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72: 3593–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunesova V, Vlkova E, Rada V, Killer J, Musilova S. (2014). Bifidobacteria from the gastrointestinal tract of animals: differences and similarities. Benef Microbes 5: 377–388. [DOI] [PubMed] [Google Scholar]
- Cardona S, Eck A, Cassellas M, Gallart M, Alastrue C, Dore J et al. (2012). Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol 12: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo JM, Leong LE, Rogers GB. (2015). Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep 5: 16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488: 178–184. [DOI] [PubMed] [Google Scholar]
- Collado MC, Delgado S, Maldonado A, Rodriguez JM. (2009). Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett Appl Microbiol 48: 523–528. [DOI] [PubMed] [Google Scholar]
- Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I et al. (2014). Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158: 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 107: 14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A et al. (2011). Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA 108: 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouhy F, Deane J, Rea MC, O’Sullivan O, Ross RP, O’Callaghan G et al. (2015). The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS One 10: e0119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper CG, Whary MT, Feng Y, Rhinehart HL, Wells RS, Xu S et al. (2003). Comparison of diagnostic techniques for Helicobacter cetorum infection in wild Atlantic bottlenose dolphins (Tursiops truncatus. J Clin Microbiol 41: 2842–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeurink PV, van Bergenhenegouwen J, Jimenez E, Knippels LM, Fernandez L, Garssen J et al. (2013). Human milk: a source of more life than we imagine. Benef Microbes 4: 17–30. [DOI] [PubMed] [Google Scholar]
- Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM et al. (2014). Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345: 1048–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS et al. (2008). Evolution of mammals and their gut microbes. Science 320: 1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB et al. (2007). Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem 55: 8914–8919. [DOI] [PubMed] [Google Scholar]
- Lugli GA, Milani C, Turroni F, Duranti S, Ferrario C, Viappiani A et al. (2014). Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl Environ Microbiol 80: 6383–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. (2016). MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363: 1–8. [DOI] [PubMed] [Google Scholar]
- Martin R, Jimenez E, Heilig H, Fernandez L, Marin ML, Zoetendal EG et al. (2009). Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol 75: 965–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E et al. (2016). Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 11: e0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Maldonado-Barragan A, Moles L, Rodriguez-Banos M, Campo RD, Fernandez L et al. (2012). Sharing of bacterial strains between breast milk and infant feces. J Hum Lact 28: 36–44. [DOI] [PubMed] [Google Scholar]
- Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S et al. (2016). A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun 7: 11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M et al. (2014. a). Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 80: 6290–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Lugli GA, Turroni F, Mancabelli L, Duranti S, Viappiani A et al. (2014. b). Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol Ecol 90: 493–503. [DOI] [PubMed] [Google Scholar]
- Milani C, Andrea Lugli G, Duranti S, Turroni F, Mancabelli L, Ferrario C et al. (2015. a). Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep 5: 15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C et al. (2015. b). Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81: 7078–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, Ferrario C et al. (2015. c). Genomics of the genus bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol 82: 980–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Ticinesi A, Gerritsen J, Nouvenne A, Lugli GA, Mancabelli L et al. (2016). Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci Rep 6: 25945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misic AM, Davis MF, Tyldsley AS, Hodkinson BP, Tolomeo P, Hu B et al. (2015). The shared microbiota of humans and companion animals as evaluated from Staphylococcus carriage sites. Microbiome 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuriel-Ohayon M, Neuman H, Koren O. (2016). Microbial changes during pregnancy, birth, and infancy. Front Microbiol 7: 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan A, van Sinderen D. (2016). Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol 7: 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ et al. (2016). Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol 16: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S et al. (2010). Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J 4: 377–387. [DOI] [PubMed] [Google Scholar]
- Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB et al. (2004). Genetic structure of the purebred domestic dog. Science 304: 1160–1164. [DOI] [PubMed] [Google Scholar]
- Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de los Reyes-Gavilan CG. (2015). Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett 362: 20–28. [DOI] [PubMed] [Google Scholar]
- Riviere A, Selak M, Lantin D, Leroy F, De Vuyst L. (2016). Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol 7: 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JM. (2014). The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv Nutr 5: 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N et al. (2015). The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis 26: 26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. (2010). Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156: 3329–3341. [DOI] [PubMed] [Google Scholar]
- Serafini F, Bottacini F, Viappiani A, Baruffini E, Turroni F, Foroni E et al. (2011). Insights into physiological and genetic mupirocin susceptibility in bifidobacteria. Appl Environ Microbiol 77: 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D et al. (2013). Cohabiting family members share microbiota with one another and with their dogs. eLife 2: e00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P et al. (2009. a). Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol 75: 1534–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, Margolles A et al. (2009. b). Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J 3: 745–751. [DOI] [PubMed] [Google Scholar]
- Turroni F, Foroni E, Serafini F, Viappiani A, Montanini B, Bottacini F et al. (2011). Ability of Bifidobacterium breve to grow on different types of milk: exploring the metabolism of milk through genome analysis. Appl Environ Microbiol 77: 7408–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ et al. (2012). Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7: e36957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A et al. (2016). Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J 10: 1656–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urashima T, Asakuma S, Leo F, Fukuda K, Messer M, Oftedal OT. (2012). The predominance of type I oligosaccharides is a feature specific to human breast milk. Adv Nutr 3: 473S–482S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushida K, Tsuchida S, Ogura Y, Toyoda A, Maruyama F. (2016). Domestication and cereal feeding developed domestic pig-type intestinal microbiota in animals of suidae. Anim Sci J 87: 835–841. [DOI] [PubMed] [Google Scholar]
- van Opstal EJ, Bordenstein SR. (2015). MICROBIOME. Rethinking heritability of the microbiome. Science 349: 1172–1173. [DOI] [PubMed] [Google Scholar]
- Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. (2012). Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol 20: 467–476. [DOI] [PubMed] [Google Scholar]
- Wang W, Zheng S, Sharshov K, Cao J, Sun H, Yang F et al. (2016). Distinctive gut microbial community structure in both the wild and farmed Swan goose (Anser cygnoides. J Basic Microbiol 56: 1299–1307. [DOI] [PubMed] [Google Scholar]
- Yassour M, Vatanen T, Siljander H, Hamalainen AM, Harkonen T, Ryhanen SJ et al. (2016). Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med 8: 343ra381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M et al. (2012). Human gut microbiome viewed across age and geography. Nature 486: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.