Abstract
The influence of unicellular eukaryotic microorganisms on human gut health and disease is still largely unexplored. Blastocystis spp. commonly colonize the gut, but its clinical significance and ecological role are currently unsettled. We have developed a high-sensitivity bioinformatic pipeline to detect Blastocystis subtypes (STs) from shotgun metagenomics, and applied it to 12 large data sets, comprising 1689 subjects of different geographic origin, disease status and lifestyle. We confirmed and extended previous observations on the high prevalence the microrganism in the population (14.9%), its non-random and ST-specific distribution, and its ability to cause persistent (asymptomatic) colonization. These findings, along with the higher prevalence observed in non-westernized individuals, the lack of positive association with any of the disease considered, and decreased presence in individuals with dysbiosis associated with colorectal cancer and Crohn’s disease, strongly suggest that Blastocystis is a component of the healthy gut microbiome. Further, we found an inverse association between body mass index and Blastocystis, and strong co-occurrence with archaeal organisms (Methanobrevibacter smithii) and several bacterial species. The association of specific microbial community structures with Blastocystis was confirmed by the high predictability (up to 0.91 area under the curve) of the microorganism colonization based on the species-level composition of the microbiome. Finally, we reconstructed and functionally profiled 43 new draft Blastocystis genomes and discovered a higher intra subtype variability of ST1 and ST2 compared with ST3 and ST4. Altogether, we provide an in-depth epidemiologic, ecological, and genomic analysis of Blastocystis, and show how metagenomics can be crucial to advance population genomics of human parasites.
Introduction
Blastocystis spp. (referred to as Blastocystis in the manuscript) is a unicellular eukaryotic microorganism that belongs to the Stramenopile phylum. This phylum encompasses an extremely large diversity of organisms including free-living flagellates, parasites of plants (for example, Peronospora) and animals (for example, Phytium insidiosum), organisms resembling fungi in terms of cytology and ecology, and a myriad of photosynthetic lineages that range from single-cell diatoms to giant multicellular brown algae (Derelle et al., 2016). Blastocystis is a common inhabitant of the gut of humans and other animals (Clark et al., 2013). Its prevalence in humans varies within and between populations, but it is higher in underdeveloped countries, where it can reach 100% (El Safadi et al., 2014). This is likely the result of poor hygiene conditions, contact with animal reservoirs, and consumption of contaminated water or food (Tan, 2008). Isolates of Blastocystis from different hosts are morphologically very similar, yet display substantial genetic variability: based on nucleotide differences in the small subunit ribosomal DNA gene, 17 different subtypes (STs) are recognized, nine of which (ST1 to ST9) are associated with human colonization (Tan, 2008; Alfellani et al., 2013b). Previous studies reported the presence of Blastocystis in all continents (Alfellani et al., 2013a), attesting its ubiquitous distribution, but the overall epidemiological picture is still incomplete.
Whether Blastocystis is to be considered a pathogen, a commensal or even a beneficial member of the human gut microbiome is still unclear (Lukeš et al., 2015). Indeed, some studies have implicated it in intestinal diseases, including inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS), thus supporting a pathogenic potential (Tan et al., 2010). Further, genome analysis of an ST7 isolate revealed the presence of genes encoding potential virulence factors, notably hydrolases and serine and cysteine proteases (Denoeud et al., 2011). On the other hand, studies of healthy, randomly sampled individuals have shown a high carriage of Blastocystis and a prolonged colonization of the gut (Scanlan and Marchesi, 2008; Scanlan et al., 2014). Therefore, unbiased large-scale investigations are needed to clarify its role as an etiological agent of disease, but targeted epidemiological investigations of Blastocystis at a global scale are impractical.
Cultivation-free, sequencing-based metagenomic technologies (Tyson et al., 2004; Venter et al., 2004; Morgan et al., 2013; Segata et al., 2013a) can potentially overcome some of these issues. Many large-scale metagenomic studies have been performed to characterize the complex consortium of organisms constituting the human gut microbiome, and recent strain-level analyses started to unravel the population structure of bacterial species (Scholz et al., 2016; Truong et al., 2017) but little attention has been devoted to intestinal parasites (Andersen et al., 2013). Indeed, until now, only one investigation (Andersen et al., 2015) used a metagenomic approach to study Blastocystis within 316 samples of the MetaHIT data set (Qin et al., 2010a) and there is thus the unmet opportunity to exploit larger sets of metagenomes for parasite profiling and epidemiology.
In order to expand the size, genetic depth, and host population diversity of epidemiologic investigation, we developed a bioinformatic pipeline to detect the presence of Blastocystis from metagenomes, and applied it on 12 published large metagenomic data sets of the human gut microbiome. Overall, 1689 subjects from 18 different countries and 4 continents (Europe, Africa, Asia and North/South America) were studied, allowing us to survey the prevalence, ST distribution, and genome characteristics of the microorganism, and to investigate its association with disease conditions and the structure of the resident gut bacterial population.
Materials and methods
Metagenomic data sets and data pre-processing
We analyzed 2154 publicly available gut metagenomic samples from twelve studies. We considered the nine largest metagenomics studies we were aware of and were available as of July 2015 to which we added three additional studies to expand the geographical span of our analysis (Table 1). The selected raw metagenomes were processed with FastqMcf (Aronesty, 2013) by trimming positions with quality <15, removing low-quality reads (mean quality <25), and discarding reads shorter than 90 nt. Human DNA and Illumina spike-in DNA (Bacteriophage phiX174) were then removed by using BowTie2 (Langmead and Salzberg, 2012) to map the reads against the reference genomes.
Table 1. List and characteristics of the metagenomic data sets used in this study.
| Data set name | Condition | Country | # Subjects | # Total samples | # Samples with conditiona | # Total reads (109) | # Reads per sample (106) mean±std | Age (yrs) median (interquartile range) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Candela | Healthy | Italy, Tanzania | 38 | 38 | — | 0.85 | 22.3±19.3 | 30 (23–38) | Rampelli et al., 2015 |
| HMP | Healthy | USA | 111 | 191 | — | 20.50 | 108.5±31.7 | 26 (23–28) | Huttenhower et al., 2012 |
| Karlsson | T2D | Denmark, 10 EU countries | 145 | 145 | 53 | 4.49 | 31.0±17.6 | 70 (69–71) | Karlsson et al., 2013 |
| Le Chatelier | Obesity | Denmark | 292 | 292 | 169 | 20.14 | 69.0±23.2 | 56 (50–61) | Le Chatelier et al., 2013 |
| Liu | Healthy | China, Mongolia | 110 | 110 | — | 6.41 | 58.2±26.8 | — | Liu et al., 2016 |
| Loman | STEC infection | Germany | 37 | 44 | 44 | 0.39 | 9.0±12.0 | — | Loman et al., 2013 |
| MetaHIT | CD, UC | Denmark, Spain | 124 | 124 | 25 | 5.60 | 45.1±18.4 | 54 (49–60) | Qin et al., 2010a |
| Nielsen | CD, UC | Denmark, Spain | 318 | 396 | 148 | 21.40 | 53.9±20.2 | 49 (40–59) | Nielsen et al., 2014 |
| Obregon-Tito | Healthy | Peru, USA | 58 | 58 | — | 2.73 | 47.1±20.8 | 26 (17–35) | Obregon-Tito et al., 2015 |
| Qin | Liver cirrhosis | China | 237 | 237 | 123 | 12.24 | 51.6±30.9 | 45 (38–54) | Qin et al., 2014 |
| T2D | T2D | China | 363 | 363 | 170 | 14.60 | 40.2±11.8 | 48 (38–57) | Qin et al., 2012 |
| Zeller | Colorectal cancer | France | 156 | 156 | 53 | 9.37 | 60.0±25.4 | 63 (58–70) | Zeller et al., 2014 |
| Total | 1689 | 2154 | 785 | 118.72 | 55.12±29.0 | 49 (36–62) |
Abbreviations: CD, Crohn’s disease; STEC, Shiga-toxigenic Escherichia coli; T2D, type 2 diabetes; UC, ulcerative colitis.
Except for condition ‘healthy’.
Eight of the considered studies aimed at characterizing the human gut in different health conditions whereas four considered subjects not affected by documented medical conditions (Table 1). We collected and manually curated the main available metadata associated with the samples (Pasolli et al., 2016). The metadata fields considered here are body mass index (BMI), age, gender and disease status. We made the complete metadata table associated with the samples publicly available at https://bitbucket.org/CibioCM/metaml/src and inside the curatedMetagenomicData package (Pasolli et al., In press).
We performed the analysis using the 9 available genomes of Blastocystis subtypes as reference. These include the complete genome sequence of one isolate from ST7 (Denoeud et al., 2011) and one from ST4 (accession codes CABX01000000 and JPUL02000000, respectively). Additionally, we used the draft genomes of other STs (ST1, ST2, ST3, ST4, ST6, ST8 and ST9) isolated from humans that have been recently deposited in public databases (accession codes LXWW00000000, JZRJ00000000, JZRK00000000, JZRL00000000, JZRM00000000, JZRN00000000, JZRO00000000). Before using these genomes in our analysis, and because Blastocystis sequencing projects are likely to contain DNA from other organisms, we screened all contigs of all assemblies for potential bacterial and archaeal contamination. We did this by mapping with BLASTN the Blastocystis assemblies against the set of ~55 000 publicly available archaeal and bacterial genomes. By considering matches over at least 500 nucleotides and a nucleotide identity of at least 90%, we removed all contigs with bacterial or archaeal matches over more than 3% of the length of the contig. Overall, we removed 613 contigs after screening out a minimum of 246 384 nucleotides for ST4 and a maximum of over 4.5 M nucleotides for ST6 (Supplementary Table 1). We notice that our procedure was set to be quite aggressive in avoiding potential contamination, but this is a safe strategy for our investigation as more than 10 M nucleotides remained available for all ST and these are largely sufficient to assess the presence of Blastocystis in metagenomes as reported below.
Detection of Blastocystis STs from metagenomes
Metagenomic reads were mapped to reference genomes using the Bowtie2 aligner (Langmead and Salzberg, 2012) and an end-to-end alignment for paired ends reads. The Bowtie2 output was processed by Samtools (Li et al., 2009) and the sorted and indexed BAM file was processed with BEDtools (Quinlan, 2014) to compute the breadth of coverage for each subtype (‘genomecov –bg’ parameter), which represents the fraction of the target genome covered by at least one metagenomic read (Molnar and Ilie, 2015). The relative abundance in subjects colonized over two timepoints was estimated by counting the number of reads mapped to the Blastocystis reference genome normalized by the total number of reads in the sample.
In this work, we define a sample as positive for a Blastocystis ST if its genome has a breadth of coverage of at least 10%. This value was chosen based both on (i) the similarity between the genomes of different Blastocystis STs and (ii) on the false positive detection rate for the presence of a second Blastocystis ST when another one is present. For the first criteria, we quantified the average fraction of the genome of a Blastocystis ST shared at a sequence similarity higher than 80% with a distinct Blastocystis ST genome using LAST (Kiełbasa et al., 2011) (‘-l 100 -f BlastTab’ parameters). The maximum fraction of matching genome was 3%, with the only exceptions of ST4–ST8 and ST6–ST9 which share more than 15% of the genome. However, this value substantially decreases at percentage identity thresholds >80% which is a very conservative threshold considering that the maximum identity at which a read of 100 nt can be mapped against a reference genome is 95%. Additionally, at the 10% breadth of coverage threshold, we did not find any co-occurrence of ST4 and ST8 in the samples, and for the cases in which ST6 and ST9 co-occurred we manually confirmed that most of the reads outside the shared genomic regions mapped only against the ST with the highest breadth of coverage. For the second criteria, we looked at the distribution of the number of additional STs in addition to the one with the largest breadth of coverage detected when varying the threshold (Supplementary Figure 9). This distribution goes from seven (all the STs in addition to the dominant one) to one (only the dominant ST detected), but it is already plateauing at 10% breadth of coverage confirming that such value does not produce false positives. Multiple lines of evidence thus support the 10% breadth of coverage value to be safe in avoiding false positives. False negatives would be minimized at lower threshold value, but false negatives are arguably less problematic than false positives, and false negatives are an intrinsic and unavoidable problem in metagenomics.
Assessing the limit of detection for Blastocystis in metagenomes
To assess the sensitivity of our procedure in detecting Blastocystis, we performed semi-synthetic experiments by spiking-in known amounts of synthetic reads from known Blastocystis genomes into real Blastocystis-negative gut metagenomes. For each ST, the synthetic reads were obtained with an Illumina-based sequencing simulator with typical sequencing error rates and noise (McElroy et al., 2012). As real Blastocystis-negative gut metagenomes we considered metagenomes from the HMP, Karlsson, LeChatelier, and Obregon-Tito data sets subsampled after QC to the typical metagenome size of 50M reads. The procedure was repeated at multiple fractions of Blastocystis relative abundance from 0.001 to 1% (for a total of 30 abundance values) and considering seven distinct real gut microbiomes for each simulation and ST. With this analysis (Supplementary Figure 1), we empirically found that the chosen detection threshold (10% breadth of coverage) corresponds to a limit of detection slightly below 0.03% abundance. ST7 has an even lower limit of detection which is due to the length of its genome (about 50% larger than the other STs). As mentioned above, our Blastocystis detection pipeline aims at minimizing the false positive rate, so even though thresholds lower than 10% breadth of coverage would positively impact the limit of detection, we again preferred to avoid calling the presence of Blastocystis without strong quantitative evidence. The limit of the detection of our procedure is higher than what can be achieved with PCR-based approaches, that are however limited in the amount of genomic information that they can provide.
Metagenomic assembly and Blastocystis contig binning
The 43 metagenomic samples in which we detected a breadth of coverage higher than 66.6% for at least one Blastocystis genome, were selected for de novo metagenomic assembly. This was performed using SPAdes version 3.9.0 (Bankevich et al., 2012). Contigs shorter than 1000 nt were discarded, and contigs from Blastocystis identified by mapping with BLASTN the screened contigs against the Blastocystis reference genomes. Specifically, we assigned a contig to a Blastocystis subtype if it had at least 90% identity over at least half of its length against the available reference genome.
Whole-genome phylogenetic analysis
To infer the phylogeny of the newly assembled genomes we adopted a core gene based strategy (Segata et al., 2013b; Page et al., 2015). The core gene set was generated by aligning all the annotated genes of the Blastocystis ST4 WR1 genome against all 8 available reference genomes and the 43 genomes we newly assembled using BLASTN (Evalue: 1e-50, word size:9). To be included in the core gene set, a gene was required to be present in at least 75% of the analyzed genomes with an identity higher than 65% over at least 600 bp. The identified core gene sequences were then aligned using MUSCLE (Edgar, 2004), concatenated in a single alignment, and processed with trimAL (‘-gappyout’ parameter) (Capella-Gutiérrez et al., 2009) to remove excessively gapped sub alignments and poorly aligned regions. The phylogeny was then built using RAxML version 8.1.15 (Stamatakis, 2014) with the GTRGAMMA model and 100 bootstrap steps.
Using this approach, we identified a core gene set of 9 genes (average alignment length of each gene of 2443 bp and standard deviation of 1374 bp) for a total concatenated alignment length of 21 984 bp. To improve the resolution at a lower phylogenetic level, we repeated the process within the genomes of ST1, ST2, ST3 and ST4 separately and reconstructed their intra subtype phylogeny on which a larger shared core genome can be identified. Subtype-specific trees were generated by fragmenting each genome in portions of 2000 bp and treating them as genes because no genome annotation was available and de novo annotation would have introduced biases. Criteria for the inclusion in the core gene adapted to the intra-ST case included the requirement that a sequence was present in all the genomes with an identity higher than 95%. Single-nucleotide variant distribution within every subtype was calculated using nucmer (Kurtz et al., 2004) pipeline for computing pairwise alignment and SNV reporting. For ST1 the average pairwise alignment was 3 431 933 bp (s.d. 2 323 310 bp), for ST2 3 876 563 bp (s.d. 1 985 719 bp), for ST3 2 318 013 bp (s.d. 2 917 467 bp) and for ST4 3 432 010 bp (s.d. 3 131 603 bp).
Functional prediction and annotation
We considered the 19 reconstructed genomes accounting >5 Mbp for gene prediction and annotation. Ad initio gene prediction was performed using SNAP (Korf, 2004) to generate HMM models for all the STs using the available annotations to build the HMM reference profile. Genome annotation was performed using MAKER (Cantarel et al., 2008) with default parameters using the HMM models previously generated. Newly predicted proteins were then functionally annotated with eggnog-mapper (Huerta-Cepas et al., 2017) using the eggNOG (Huerta-Cepas et al., 2016) Eukaryotic data set. ST-specific KOG functions were determined by performing a Fisher's exact test between the genomes of a particular ST and the other ones. Adjusted P-values were computed through the false-discovery rate correction.
Microbiome profiling and co-occurrence analysis
All samples were processed with MetaPhlAn2 (Segata et al., 2012; Truong et al., 2015) to quantitatively profile the whole microbial population exploiting the properties of species-specific markers (Huang et al., 2014). We used the obtained abundance profiles to investigate the co-occurrence or co-exclusion (Faust et al., 2012) of Blastocystis with other members of the microbiome. In particular, the Wilcoxon rank-sum test was used to identify the microbial features that were associated with the presence or absence of Blastocystis. In computing this test, duplicates from the same subject were discarded and a threshold of 0.05 was considered as significance level. Additional analysis for finding bacterial clades associated with Blastocystis presence was performed using the LEfSe (LDA effect size) tool (Segata et al., 2011). Finally, a machine learning-based approach was applied to further investigate if the microbiome signature is predictive for the presence of Blastocystis. The species abundances generated by MetaPhlAn2 were used to discriminate between Blastocystis-positive and negative samples. For this purpose we considered a random forest (RF) classifier (Breiman 2001) implemented in the MetAML tool (Pasolli et al., 2016). First, prediction accuracies were assessed by an unbiased 10-fold cross-validation procedure, repeated and averaged over 20 independent runs. Then, we applied a leave-one-data set-out approach, in which the presence of Blastocystis in a given data set is predicted by training the model on the samples from the other independent studies. Prediction accuracies were evaluated in terms of area under the ROC curve (AUC) statistics, which can be interpreted as the probability that the classifier ranks a randomly chosen positive sample higher than a randomly chosen negative one, assuming that the positive sample ranks higher than the negative one. The free parameters of the classifiers were set as follows: (i) the number of decision trees was equal to 500; (ii) the number of features to consider when looking for the best split was equal to the root of the number of original features; (iii) the quality of a decision tree split was measured using the Gini impurity criterion. The software framework used for this experiment is open-source and available online at http://segatalab.cibio.unitn.it/tools/metaml. Alpha diversity was computed for each data set by considering Gini-Simpson and Shannon indexes under the condition of presence or absence of Blastocystis and the Student’s t-test (significance level set to 0.05) was used to test significance between the two conditions.
Results
Meta-analysis for Blastocystis in large metagenomic data sets
We screened large-scale intestinal metagenomic data sets to assess the prevalence of Blastocystis and its STs, infer epidemiologic characteristics, and examine the characteristics of their genomes. Overall, we processed 2154 fecal microbiome samples from 1,689 subjects from 12 data sets (Table 1). These data sets span diverse disease conditions including colorectal cancer (Zeller et al., 2014), type 2 diabetes (Qin et al., 2012; Karlsson et al., 2013), liver cirrhosis (Qin et al., 2010a), obesity (Le Chatelier et al., 2013) and IBD (Qin et al., 2010a; Nielsen et al., 2014). All these studies almost exclusively focused on the bacterial components of the microbiome and did not report the presence of microbial Eukaryotes, with the above mentioned exception that focused on a single metagenomic data set (Andersen et al., 2015). The wide range of distinct health conditions and geographic origins of the hosts we considered here are thus a key factor in this study.
To assess the presence of Blastocystis, we used a sequence mapping based approach aided by the availability of draft genome sequences from eight subtypes (ST1, ST2, ST3, ST4, ST6, ST7, ST8 and ST9), all known to be associated with human colonization. After removing potential bacterial sequences contaminating these genomes (see Materials and methods and Supplementary Table 1), we estimated the fraction of each target genome covered by metagenomic reads (that is, the breadth of coverage) and we considered samples positive for Blastocystis when the breadth of coverage was higher than 10% (see Materials and methods). Using this approach, Blastocystis is detected when present at a concentration as low as 0.03% in typical metagenomic samples of 50M reads (Supplementary Figure 1). Downstream analyses detailed in the rest of the work are based on this detection threshold.
Blastocystis prevalence and subtype dominance is biogeographically variable
We first determined the prevalence of Blastocystis in the overall data set, which included 2154 fecal samples from 1689 subjects. The microorganism was detected in 321 samples, originating from subjects in ten countries (China, Denmark, France, Mongolia, Norway, Peru, Spain, Sweden, Tanzania and USA) from four continents, with an overall prevalence of 14.9%. The prevalence was higher in European subjects (243 of 1084 samples, 22.4%) and lower in Chinese ones (24 of 600, 4.0%). Despite the relatively small size of the data set, 15 (55.6%) of the 27 Tanzanian subjects (Rampelli et al., 2015) were positive for Blastocystis, whereas all the Italian subjects (n=11) from the same study were negative. Blastocystis was not detected in the Shiga toxin-producing Escherichia coli (STEC)-infection data set (Loman et al., 2013).
The prevalence of Blastocystis appears to be influenced by the DNA extraction procedure used in the different studies, being higher when methods combining mechanical and chemical lysis steps are used (Supplementary Figure 2). This suggests that efficient DNA extraction from lysis-resistant microorganism cysts requires appropriate procedures, and that comparison across studies should consider this factor (Yoshikawa et al., 2011). On the other hand, cohort differences may have a larger impact on prevalence than methodological aspects, as exemplified by large differences in prevalence between three European data sets (LeChatelier, MetaHIT and Nielsen) and the Chinese T2D data set, despite the use of the same DNA extraction procedure.
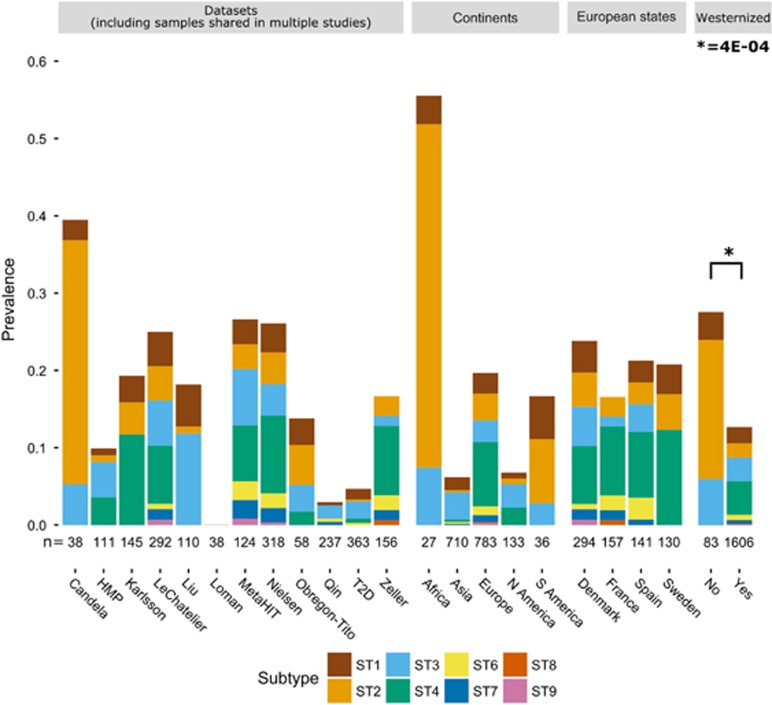
We then examined the prevalence of the different Blastocystis STs among individuals colonized with single STs (Figure 1; Supplementary Table 2). While some aspects such as the wide geographic distribution of ST3 (detected in 10 of 12 data sets), and the overall low prevalence of ST6, ST7 and ST9, are in agreement with the current global epidemiologic information (Clark et al., 2013), two new points of particular relevance emerged. First, ST2 appears to predominate in the non-industrialized cohorts analyzed, which are hunter-gatherer populations from Tanzania (Rampelli et al., 2015) and Peru (Obregon-Tito et al., 2015; Figure 1). The difference in ST2 prevalence between non-westernized (including data from (Liu et al., 2016)) and westernized individuals is highly statistically significant (P=6E−10). This raises the hypothesis that ST2 is one of the members of the gut microbiome that have been affected by westernization processes (Segata, 2015). Second, the prevalence of ST4 is very high among European subjects (Figure 1), which is in sharp contrast with the absence, or extreme rarity, of this ST in other regions of the world (for example, P=7E−16 for the difference in prevalence between Europe and Asia, Supplementary Figure 3), except the US. The difference in ST4 prevalence between westernized and non-westernized individuals is also statistically significant (P=0.046). These data confirm and extend previous observations on the peculiar geographical distribution of ST4 (Forsell et al., 2012). Overall, ST2 and ST4 thus appear to be the Blastocystis subtypes most influenced by geography and lifestyles.
Figure 1.
Prevalence of Blastocystis and Blastocystis subtypes in the different data sets, different continents, different European states, and between westernized and non-westernized subjects (see Supplementary Table 2 for more details). Stacked barplots show the prevalence in each category; numbers below the bars refer to the number of samples in the corresponding category, where duplicates from the same subject are eventually discarded. Statistical significance was assessed by Fisher's exact test.
Blastocystis prevalence is higher in subjects with low BMI and in healthy controls for Crohn’s disease and colorectal cancer
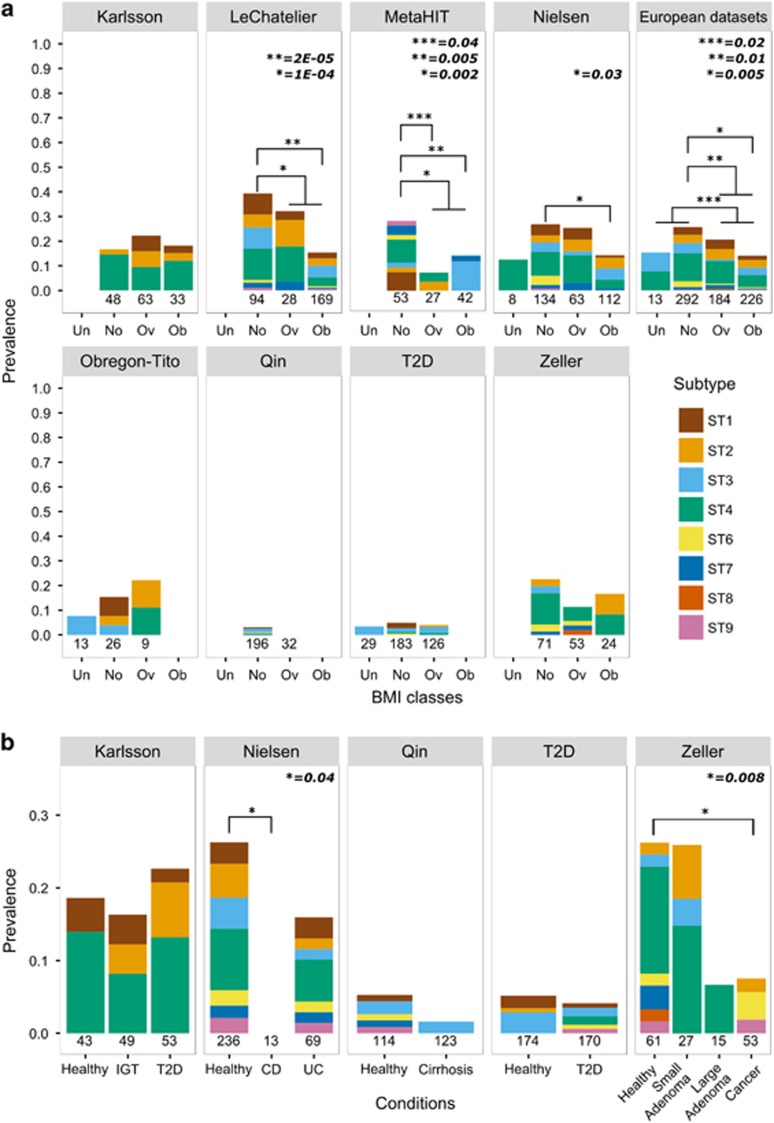
We tested the association between the presence of Blastocystis and available parameters of interest (see Materials and methods), and found that BMI is strongly negatively correlated with Blastocystis prevalence. In the metagenomic study that specifically targeted the obesity phenotype (Le Chatelier et al., 2013), we detected Blastocystis in 39.4% normal weight individuals, compared with 15.4% obese subjects (P=2E−05, Figure 2a). This is consistent with findings from a study of Danish subjects (Andersen et al., 2015). The other data sets include a smaller number of obese subjects, thus providing less statistical power to test the association. Nonetheless, a higher Blastocystis prevalence in normal weight individuals compared with overweight and obese ones was evident in six of the eight data sets, two of which supported by statistical significance (Figure 2a).
Figure 2.
Blastocystis prevalence in BMI classes (a) and different health conditions (b) for the considered data sets. Barplots show the prevalence of Blastocystis in different health conditions reported in the analyzed data sets. BMI classes considered were underweight (Un), normal (No), overweight (Ov) and obese (Ob). The total number of samples in each class and data set is reported below the bars. Bars associated with a total number of samples less than four are not shown. Note that scales in panels A and B are different. Abbreviations: CD, Crohn’s disease; IGT, impaired glucose tolerance; T2D, type 2 diabetes; UC, ulcerative colitis. Fisher's exact test was used as statistical significance test.
Interestingly, when considering all the European data sets that used the same collection and processing protocols (n=715, 126% more samples than (Andersen et al., 2015)), the difference in Blastocystis prevalence between normal weight and obese subjects was again strongly significant (P=5E−03), as it was between normal weight and overweight (P=0.01), and between non-overweight and overweight (P=0.02). At the level of specific subtypes, only ST4 reached statistical significance (P=0.03 between normal weight and obese), suggesting that association between Blastocystis and BMI is probably not subtype-specific.
We did not find an increased prevalence of Blastocystis in subjects affected by any of the considered diseases (lowest one-side P=0.4 for T2D in the Karlsson data set). Conversely, Blastocystis was positively associated (P=0.008) with the control group in the colorectal cancer data set (Zeller et al., 2014), with only 3 of the 53 (5.7%) patients positive for the microorganism compared with 15 of the 61 (24.6%) healthy controls (Figure 2b). This trend was also confirmed in the same data set when considering patients with large adenomas (n=14), as only one was positive for Blastocystis. Crohn’s disease, but not ulcerative colitis, was also negatively associated with the presence of Blastocystis (P=0.04). Our findings seem to contrast other reports especially for colorectal cancer (Kumarasamy et al., 2014), whereas for IBD existing data already associated ulcerative colitis rather than Crohn’s disease with decreased Blastocystis prevalence (Petersen et al., 2013) although different conclusions were reached in other reports (Cekin et al., 2012). Previous data on this association are however sparse, debated in clinical settings, and potentially affected by publication bias. More independent investigations are needed to elucidate these relations, but our results suggest that the ecological niche of Blastocystis is independent from disease-associated microbiome dysbiosis features. A further hypothesis supported by the above associations and the absence of Blastocystis in STEC-positive subjects, is that Blastocystis is actually less common in individuals with gastro-intestinal symptoms and other microbiome-associated disease conditions (Scanlan et al., 2014).
Stable Blastocystis colonization is subtype-independent
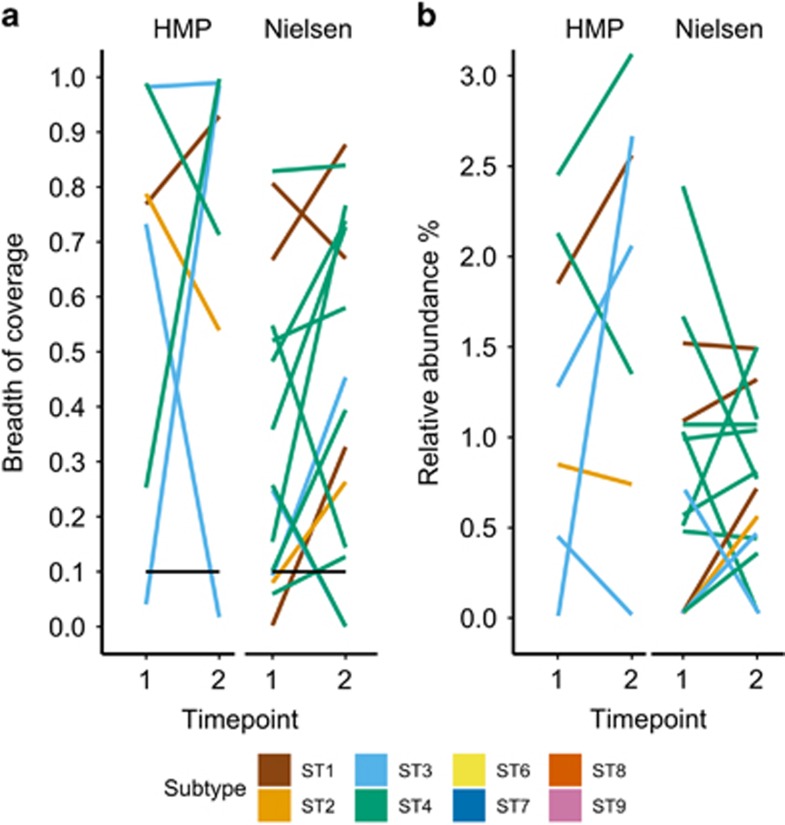
To study the persistence of Blastocystis colonization and determine the subtypes involved in chronic colonization, we analyzed the metagenomic data set of subjects who provided stool samples at multiple timepoints. A total of 121 subjects, 43 from the HMP data set (Huttenhower et al., 2012) and 78 from the Nielsen data set (Nielsen et al., 2014), were sampled at two timepoints (mean 219 and 163 days after first sampling, respectively). Blastocystis was identified above the detection threshold in 22 subjects (7 from HMP and 15 from Nielsen) in at least one of the timepoints considered (Supplementary Table 3). Of the 22 positive subjects, 14 (64%) maintained the colonization over the two timepoints, whereas five subjects acquired and three subjects lost the colonization between the two timepoints (Figure 3a; Supplementary Table 3). For the cases of colonization acquisition/loss and accordingly with our detection limit of 0.03% relative abundance, Blastocystis is indeed absent in the subject or may be present at very low abundance which is still indicative of variations in the ecological relation of Blastocystis with the resident microbiome. In subjects with stable colonization, the relative abundance of Blastocystis changed only slightly in the majority of the cases (Figure 3b) and we did not observe variations higher than three folds.
Figure 3.
Breadth of coverage (a) and relative abundance (b) of Blastocystis in subjects colonized over two timepoints (see Supplementary Table 3 for more details). In the breadth of coverage plots, samples below the threshold of detection are also indicated. The breadth of coverage represents the fraction of the reference genome covered by at least one metagenomic read. The relative abundance is estimated by dividing the number of reads mapped to the Blastocystis reference genome with the total number of reads in the sample.
In the 14 subjects with stable colonization, we always found the same ST at the two timepoints, suggesting that ST replacement is not a frequent event in the healthy human gut, at least over the relatively short timeframes considered in the data sets (Figure 3). The subtypes commonly found in humans (ST1–ST4) all appeared as stable colonizers, suggesting that this phenomenon is not subtype dependent.
Whole-genome genetic analysis of Blastocystis subtypes
Isolates belonging to the same Blastocystis subtype display some genetic variability, as highlighted by studies of ribosomal markers (Yoshikawa et al., 2016) and a few housekeeping genes (Stensvold et al., 2012; Yoshikawa et al., 2016). However, the extent of polymorphism at the genome level and variability in gene content within different STs is unknown. To this end, we reconstructed draft Blastocystis genomes from the metagenomes and performed comparative genomic analysis. In total, 43 assemblies were obtained using metagenomic assembly with SPAdes (Bankevich et al., 2012) followed by binning and taxonomic assignment (see Materials and methods, Supplementary Table 4) from the samples with very high Blastocystis abundance. Specifically, 16 new genomes were very closely related to the available genome of ST4, 7 to ST2 and ST1, 9 to ST3, whereas only 4 genomes were assembled from the phylogenetically related ST6, ST8 and ST9. A simple genetic feature such as the average GC content (Supplementary Figure 4) was already distinctive across STs, in that ST1, ST2 and ST3 that have a genome much richer in GC (average 52.6%, 52.0%, 51.5% respectively) than ST4, ST8 and ST9 (average 40.0%, 42.3% and 41.5% respectively), whereas ST6 is in between these two groups (average 44.9%).
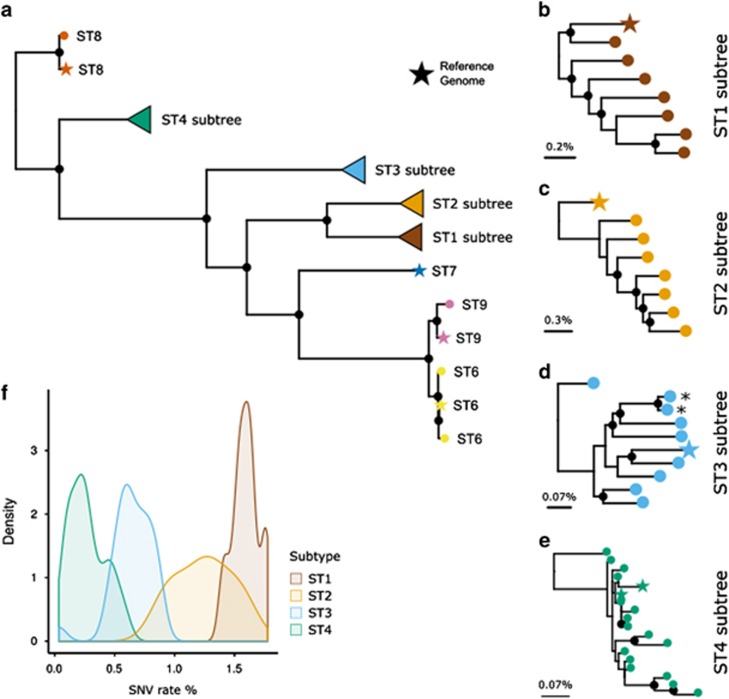
We then integrated the nine available genomes with the 43 new assemblies to reconstruct the genome-scale phylogeny of the Blastocystis genus using the concatenation of aligned core genomic fragments. This reconstruction relies upon the substantial fraction of the genome that is conserved across strains and have been performed for assemblies with an average of 4.4 Mb of reconstructed genome (Supplementary Table 4). While the overall structure of the tree (Figure 4a) confirms previous phylogenetic analyses based on single marker genes (Yoshikawa et al., 2016) and ST-specific phylogenies consistently place the reconstructed genomes from multiple sample of the same patient (Figure 4d), substantial genetic diversity is detected within each ST (Figures 4b and e). Strains belonging to ST1 show the highest genetic diversity with, on average, 1.5% (s.d. 0.10%) single-nucleotide substitutions in the genomic regions conserved between pairs of strains (Figure 4f). ST4 shows instead an overall much higher sequence conservation (average 0.27% s.d. 0.14% divergence), in agreement with findings from single marker genes (Stensvold et al., 2012). ST2 and ST3 display intermediate genetic diversity compared with ST1 and ST4.
Figure 4.
Phylogenetic relation between the 9 available Blastocystis reference genomes and 43 newly reconstructed genome assemblies from metagenomes. From the overall phylogenetic tree (a) we also report the subtrees of the four subtypes with more than 3 genomes (b–e) and compare the sequence diversity they span (f). Maximum likelihood phylogenetic trees were inferred using concatenated aligned shared genomic regions identified in reference genomes and assemblies (see Materials and methods). The asterisk highlights samples acquired from the same subject at two different timepoints. Black filled circles denote bootstrap support greater than 80%. The scale bar represents the average SNV rate calculated on the pairwise alignment.
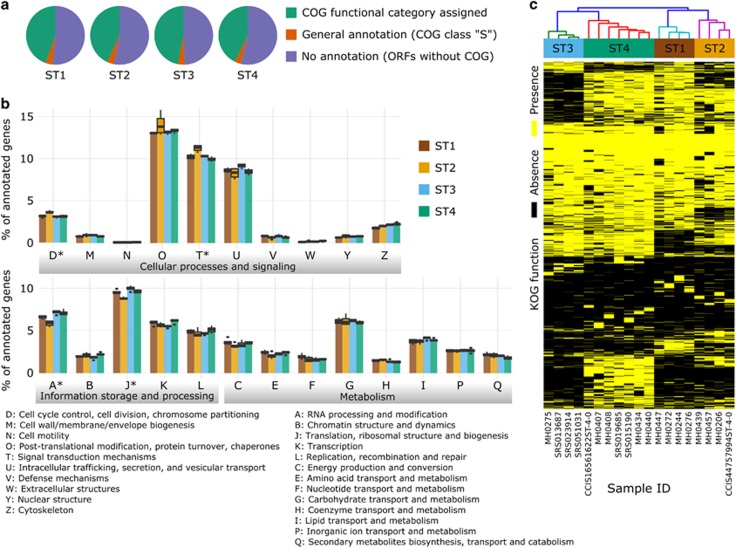
We then restricted the genomic analysis to the 19 genomes for which at least 5 Mb have been reconstructed and performed a functional annotation and characterization of these high-quality assemblies (4 for ST1, 4 for ST2, 4 for ST3, and 7 for ST4, see Supplementary Table 4) by using the eggNOG (Huerta-Cepas et al., 2016) database (see Materials and methods). Unsurprisingly, less than half of the genes identified were assigned to known COG functional categories (from 42.7% of ST4 to 46.7% of ST3, Figure 5a). Only few categories were not represented in Blastocystis (for example, as expected, the cellular machinery for cell motility) and the four STs generally contained a very similar number of proteins in these broad categories (Figure 5b). The only exceptions are category J (Translation, ribosomal structure and biogenesis) and category A (RNA processing and modification) that are overrepresented in the genomes of ST3 and underrepresented in those of ST2 (P<1E−04), as well as categories D (Cell cycle control and mitosis) and T (Signal Transduction, all P<1E−04). More specific functional assignments based on the manually curated Clusters of Orthologous Groups for Eukaryotes (KOG, see Supplementary Table 5; Huerta-Cepas et al., 2016) further highlighted the differences in functional potential between STs and the substantial intra-ST functional consistency. This is clear from the hierarchical clustering analysis of the KOG profiles in each reconstructed genome (Figure 5c), in which the close phylogenetic relationship between strains in the same ST is recapitulated at the level of their functional potential. A total of 795 KOGs were found to be ST-specific (Supplementary Table 5; Supplementary Figure 5) after statistical significance testing with false-discovery rate correction (see Materials and methods). For example, a cystatin (0IZK7 Cystatin B), that in ST7 has a potential role in parasitic cysteine protease and inhibition of host proteases (Denoeud et al., 2011; Wawrzyniak et al., 2012), is present in ST2 but not in ST1, ST3, and ST4. Likewise, we found a glycoside hydrolase (hydrolase family 47) only in ST3, and this may be involved in the attack of the host intestinal epithelial cells (Denoeud et al., 2011). Finally, in ST4 genomes we found heat shock proteins (like 0PHA3 and KOG3047—ubiquitously-expressed, prefoldin-like chaperone) and cytosolic Ca2+-dependent cysteine proteases (like KOG0045—Calpain-like cysteine peptidase) that were not present in other ST genomes, and these may represent virulence factors unique to this ST. Altogether these data indicate that different Blastocystis STs have distinct functional potential niches that are currently only partially characterized. Further, we show for the first time that it is possible to characterize ST-specific functional repertoires that are conserved among strains of the same ST.
Figure 5.
Functional annotation analysis of the 19 reconstructed genomes spanning four Blastocystis STs. Less than half of the genes predicted by MAKER (Cantarel et al., 2008) were assigned to known COG functional categories using the eggNOG (Huerta-Cepas et al., 2016) database (a, see Materials and methods). These annotated genes can be grouped into 23 broad COG categories (b) that are a variable fraction of the total annotated genes. The asterisks denote categories for which one-way ANOVA statistical test gave P<1E−04. Hierarchical clustering performed on the more specific KOG functions show that samples associated with the same ST cluster together (c).
The presence of Blastocystis is highly correlated with gut microbiome composition
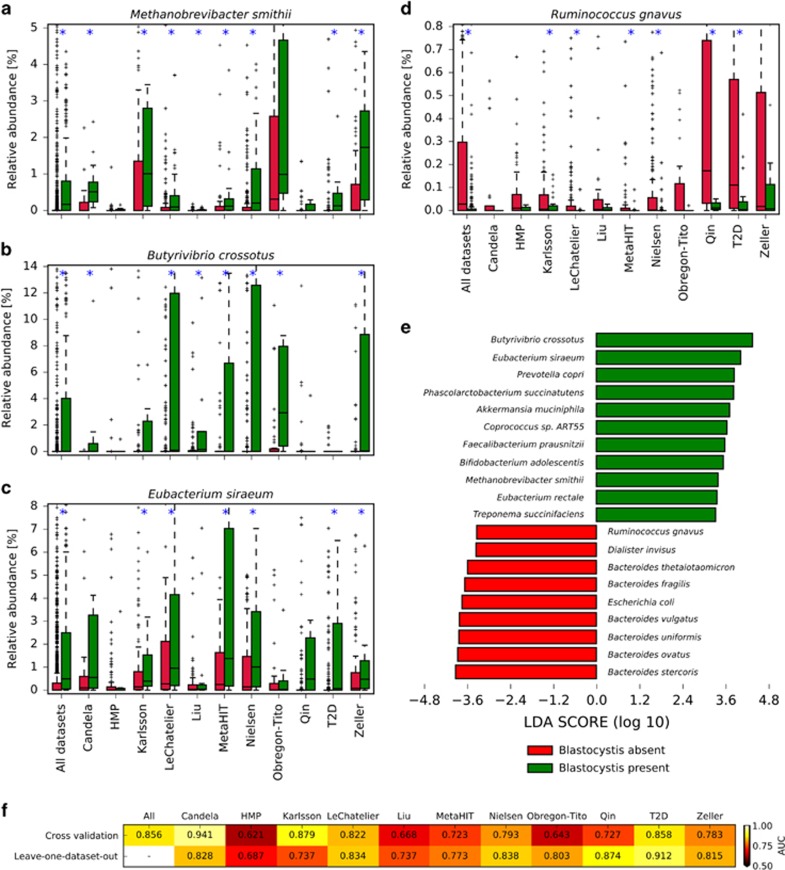
We found a very strong association between the presence of Blastocystis and the abundance of archaeal organisms (P<7E−37). On the overall data set, this association may be inflated because some DNA extraction procedures may favor non-bacterial organisms (Wesolowska-Andersen et al., 2014), but we observed strong statistical significance in all but two single data sets (Figure 6 and Supplementary Table 6). Archaea in the human gut are represented primarily by Methanobrevibacter smithii (Figure 6a) which is in fact strongly associated with the presence of Blastocystis. Interestingly, several archaeal genes, likely acquired horizontally, are present in the Blastocystis genome (Denoeud et al., 2011), suggesting that the common ecological niche favors the interaction and the exchange of genetic material between the microorganism and the Archaea.
Figure 6.
The presence (or absence) of Blastocystis is associated with major differences in the intestinal microbiome. Some species are strongly associated with the presence (a–c) or absence (d) of Blastocystis (plots for additional microbes are reported in Supplementary Figure 7). Boxplots report the distribution of abundances in samples with and without Blastocystis. Blue asterisks denote data sets where significant differences exist between the absence and presence of Blastocystis. LEfSe analysis (e) showed several other microorganisms statistically associated (α=0.05) with Blastocystis presence at high effect size (threshold at 3.3). Machine learning-based approach reveals that the microbiome signature is predictive for the presence of Blastocystis (f). This is valid not only when considering specific data sets with an unbiased cross-validation procedure, but also when predicting the presence of the parasite in a given data set considering only the samples from other independent studies (leave-one-data set-out approach).
Several bacterial clades were also found to be strongly associated with Blastocystis presence, with a total of 68 significant associations with effect size larger than 3.3 as found by LEfSe analysis (Segata et al., 2011) (Figure 6e; Supplementary Figure 6). Bacteria in the Firmicutes phylum and in the Clostridiales order also appeared strongly enriched in samples positive for Blastocystis (Supplementary Figure 7). Species in this order included Butyrivibrio crossotus (significant in 7 data sets, Figure 6b), Eubacterium siraeum (significant in 6 data sets, Figure 6c), and Coprococcus catus (significant in 6 data sets, Supplementary Figure 7). In addition, the overall Clostridiales order is associated with the presence of Blastocystis (Supplementary Table 6). However, some clostridia tend to co-exclude with Blastocystis, such as Ruminococcus gnavus (significant in 6 data sets, Figure 6d) and Clostridium bolteae (significant in 5 data sets, Supplementary Figure 7). Therefore, while there is a general positive association between Firmicutes/Clostridia and Blastocystis, there are negative associations at the species-level, possibly due to competition for resources or different ecological niches.
In contrast, the most abundant intestinal bacterial genus, Bacteroides, is generally more abundant in Blastocystis-negative samples (Supplementary Figure 7), with five data sets in which this trend is significant. This association is also driving the general higher abundance of the Bacteroidetes phylum in Blastocystis-negative samples and possibly contrasting the opposite trend observed for the Firmicutes phylum (Supplementary Figure 7). Proteobacteria and Actinobacteria seem instead generally not influenced by the presence of Blastocystis with only one and two data sets, respectively, in which they appear significant. Specific species in these phyla can however still be strongly associated with Blastocystis presence (e.g., the proteobacterium Oxalobacter formigenes significant in 5 data sets, Supplementary Figure 7) or absence (for example, the actinobacterium Eggerthella significant in 5 data sets, Supplementary Figure 7), suggesting that species-specific functional specialization has a higher ecological connection with Blastocystis than more general phylum-level characteristics.
We expanded the analysis on the association between Blastocystis and specific intestinal organisms by searching overall microbiome signatures predictive for the presence of the microorganism. Our previous work on such machine learning signatures (Pasolli et al., 2016) showed that all the diseases considered here can be associated, with a variable degree of accuracy, with their microbiome structures. In the case of Blastocystis, we found that microbiome signatures (Figure 6f) are always statistically significant and are even stronger than for the disease. Importantly, this is true not only when considering specific data sets with an unbiased cross-validation procedure (Pasolli et al., 2016), but also when predicting the presence of Blastocystis in a given data set considering only the samples from the other independent studies. This confirms that Blastocystis-positive microbiomes have distinguishing features that are consistent across populations, geography, and batch effects.
Overall, our analysis suggests that a consistent set of bacterial and archaeal organisms, and the overall composition of the microbiome, are associated with the presence (or absence) of Blastocystis. Interestingly, despite the many ecological associations found, microbiome diversity is instead not associated with the presence of Blastocystis (Supplementary Figure 8). Recent studies addressed the possible correlation between the presence of Blastocystis and other microbiome members that can of course also be influenced by other factors such as intestinal transit time. 16 S rRNA amplicon sequencing revealed a higher abundance of Clostridia, Ruminococcaceae and Prevotellaceae, among Blastocystis-colonized individuals, while Enterobacteriaceae were enriched in Blastocystis-free patients (Audebert et al., 2016). Two other studies found that individuals with an intestinal microbiome dominated by Bacteroides had less Blastocystis than those with Ruminococcus and Prevotella-driven enterotypes (Andersen and Stensvold, 2015; O'Brien Andersen et al., 2016); this was interpreted in terms of a correlation between Blastocystis and species richness, since the Bacteriodes-driven enterotype has a lower species richness compared with the other enterotypes. The same authors, however, pointed out that species richness alone could not explain other observed trends, such as the correlation between Blastocystis carriage and BMI in Danish individuals. They thus argued that the presence of specific microbial species could influence the ability of the microorganism to thrive in the gut, but were unable to identify those species. Here we expand this concept on a much larger cohort size and higher taxonomic resolution, and provide a list of bacterial and archaeal organisms that should be prioritized in future experimental investigations (for example, in vitro) aimed at understanding the ecology of Blastocystis in the human gut and its potential direct interaction with bacterial members of the microbiome.
Discussion
We have developed a computational pipeline to detect Blastocystis in human gut metagenomic samples and applied it to a collection of >2000 metagenomes from subjects representing all continents except Australia and Antarctica. This is the largest investigation on the prevalence of Blastocystis and its subtypes in humans, overcoming in size and geographic diversity the single metagenomic study of an European cohort (Andersen et al., 2015) and the other more traditional investigations (Bart et al., 2013; Ramirez et al., 2014; Scanlan et al., 2014, 2016; Villalobos et al., 2014). Importantly, we also assessed the association between the presence of Blastocystis STs and a number of disease conditions, studied the co-occurrence (or co-exclusion) with other members of the gut microbiome, and reconstructed the genomes of strains belonging to different STs and used them for phylogenetic and functional potential analyses.
We detected Blastocystis in subjects from 11 of the 12 data sets, confirming its global distribution. In agreement with current literature, the geographic distribution of subtypes was not random: ST3 was widely distributed, ST4 was strongly underrepresented outside Europe and USA, and ST2 predominated in the non-industrialized cohorts. These findings illustrate how important epidemiologic aspects can be studied by mining appropriate metagenomics data sets.
We confirmed that the microorganism is able to persist for months (Scanlan et al., 2014), and that all the Blastocystis STs commonly associated with humans are able to stably colonize the gut. The presence of Blastocystis was strongly negatively correlated with BMI, but microbiome diversity was not statistically associated with its presence, suggesting that the low microorganism prevalence in obese subjects is independent from the documented decrease in overall diversity (Pareek et al., 2011). Importantly, Blastocystis was significantly more prevalent in the control groups for the investigations on colorectal cancer and ulcerative colitis, and was absent in individuals with STEC infection (although no controls are available for this study). While these associations require additional follow-up studies, they are consistent with the general trend we observed of higher Blastocystis prevalence in healthy individuals. If we also consider the increased detection rate of the microorganism in non-westernized populations, its stable colonization in healthy subjects, and the high global prevalence, our work provides multiple and robust evidence to consider Blastocystis as a common member of the healthy human gut microbiome and further expands the findings of clinical studies of chronic colonization (Roberts et al., 2014) and carriage among healthy individuals (Scanlan and Marchesi, 2008).
We completed the analysis by showing how the presence and abundance of Blastocystis were strongly correlated with those of Archaea; other bacterial species and phyla were similarly correlated (or anti-correlated) with Blastocystis. These analyses, which raise new hypotheses about potential ecological or direct interactions of Blastocystis with specific bacterial members of the gut microbiome, would have not been possible with purely cultivation-based approaches. Phylogenomic analyses are another essential tool for microbial population genomics, but are almost exclusively performed on genomes obtained by sequencing isolates (Budroni et al., 2011; Klemm and Dougan, 2016). Blastocystis can be cultivated in vitro (Tan, 2008), but establishing a collection of microorganism cultures from individuals of diverse geographic origin is very laborious and time-consuming. Here we show that full Blastocystis genomes can be reconstructed from metagenomes, and provide novel information on the diversity in the genus, the phylogenetic relation within subtypes and functional traits.
With the collections of publicly available metagenomes quickly growing in number and size, there is an unprecedented opportunity to unravel the population genomics of Blastocystis at multiple levels of resolution without the need of targeted isolation work. Importantly, the computational pipeline we developed here is applicable to other parasites and fungi, if genome information is available and the target organism is present at a sufficient abundance. We thus anticipate that metagenomic analysis coupled with the opportunity of mining the vast collections of gut metagenomes will soon become an indispensable tool to explore the epidemiology, genetics and diversity of Eukaryotic microorganisms in the human host.
Acknowledgments
This work was supported in part by the European Union FP7 Marie-Curie grant (PCIG13-618833), MIUR grant FIR RBFR13EWWI, Fondazione Caritro grant Rif.Int.2013.0239, European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (MetaPG project), and Terme di Comano grants to NS, by the European Union H2020 Marie-curie grant (707345) to EP, and by the European Commission H2020 program under contract number 643476 (www.compare-europe.eu) to SMC.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Alfellani Ma, Stensvold CR, Vidal-Lapiedra A, Onuoha ESU, Fagbenro-Beyioku AF, Clark CG. (2013. a). Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop 126: 11–18. [DOI] [PubMed] [Google Scholar]
- Alfellani Ma, Taner-Mulla D, Jacob AS, Imeede CA, Yoshikawa H, Stensvold CR et al. (2013. b). Genetic diversity of Blastocystis in livestock and zoo animals. Protist 164: 497–509. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andersen LOB, Bonde I, Nielsen HB, Stensvold CR. (2015). A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiol Ecol 91: fiv072. [DOI] [PubMed] [Google Scholar]
- Andersen LOB, Nielsen HV, Stensvold CR. (2013). Waiting for the human intestinal Eukaryotome. Int Soc Microb Ecol 7: 1253–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen LOB, Stensvold CR. (2015). Blastocystis in health and disease-Are we moving from a clinical to a public health perspective? J Clin Microbiol 54: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty E. (2013). Comparison of sequencing utility programs. Open Bioinformatics J 7: 1–8. [Google Scholar]
- Audebert C, Even G, Cian A Blastocystis Investigation G Loywick A, Merlin S et al. (2016). Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci Rep 6: 25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart A, Wentink-Bonnema EM, Gilis H, Verhaar N, Wassenaar CJ, van Vugt M et al. (2013). Diagnosis and subtype analysis of Blastocystis sp. in 442 patients in a hospital setting in the Netherlands. BMC Infect Dis 13: 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. (2001). Random forests. Machine Learning 45: 5–32. [Google Scholar]
- Budroni S, Siena E, Dunning Hotopp JC, Seib KL, Serruto D, Nofroni C et al. (2011). Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci USA 108: 4494–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B et al. (2008). MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18: 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekin AH, Cekin Y, Adakan Y, Tasdemir E, Koclar FG, Yolcular BO. (2012). Blastocystosis in patients with gastrointestinal symptoms: a case-control study. BMC Gastroenterol 12: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CG, van der Giezen M, Alfellani Ma, Stensvold CR. (2013). Recent developments in Blastocystis research. Adv Parasitol 82: 1–32. [DOI] [PubMed] [Google Scholar]
- Denoeud F, Roussel M, Noel B, Wawrzyniak I, Da Silva C, Diogon M et al. (2011). Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol 12: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, López-García P, Timpano H, Moreira D. (2016). A phylogenomic framework to study the diversity and evolution of stramenopiles (= heterokonts). Mol Biol Evol 33: 2890–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Safadi D, Gaayeb L, Meloni D, Cian A, Poirier P, Wawrzyniak I et al. (2014). Children of Senegal river basin show the highest prevalence of Blastocystis sp. ever observed worldwide. BMC Infect Dis 14: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J et al. (2012). Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8: e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsell J, Granlund M, Stensvold CR, Clark CG, Evengard B. (2012). Subtype analysis of Blastocystis isolates in Swedish patients. Eur J Clin Microbiol Infect Dis 31: 1689–1696. [DOI] [PubMed] [Google Scholar]
- Huang K, Brady A, Mahurkar A, White O, Gevers D, Huttenhower C et al. (2014). MetaRef: a pan-genomic database for comparative and community microbial genomics. Nucleic Acids Res 42: D617–D624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Forslund K, Pedro Coelho L, Szklarczyk D, Juhl Jensen L, von Mering C et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34: 2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC et al. (2016). eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44: D286–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT et al. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B et al. (2013). Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498: 99–103. [DOI] [PubMed] [Google Scholar]
- Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. (2011). Adaptive seeds tame genomic sequence comparison. Genome Res 21: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm E, Dougan G. (2016). Advances in understanding bacterial pathogenesis gained from whole-genome sequencing and phylogenetics. Cell Host Microbe 19: 599–610. [DOI] [PubMed] [Google Scholar]
- Korf I. (2004). Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarasamy V, Roslani AC, Rani KU, Kumar Govind S. (2014). Advantage of using colonic washouts for Blastocystis detection in colorectal cancer patients. Parasites Vectors 7: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C et al. (2004). Versatile and open software for comparing large genomes. Genome Biol 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500: 541–546. [DOI] [PubMed] [Google Scholar]
- Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31: 1674–1676. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Zhang J, Wu C, Cai S, Huang W, Chen J et al. (2016). Unique features of ethnic Mongolian gut microbiome revealed by metagenomic analysis. Sci Rep 6: 34826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Constantinidou C, Christner M, Rohde H, JZ-M Chan, Quick J et al. (2013). A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. JAMA 309: 1502–1510. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Stensvold CR, Jirků-Pomajbíková K, Wegener Parfrey L. (2015). Are human intestinal eukaryotes beneficial or commensals? PLoS Pathog 11: e1005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy KE, Luciani F, Thomas T. (2012). GemSIM: general, error-model based simulator of next-generation sequencing data. BMC Genomics 13: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Ilie L. (2015). Correcting Illumina data. Brief Bioinformatics 16: 588–599. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Segata N, Huttenhower C. (2013). Biodiversity and functional genomics in the human microbiome. Trends Genet 29: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S et al. (2014). Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 32: 822–828. [DOI] [PubMed] [Google Scholar]
- O'Brien Andersen L, Karim AB, Roager HM, Vigsnaes LK, Krogfelt KA, Licht TR et al. (2016). Associations between common intestinal parasites and bacteria in humans as revealed by qPCR. Eur J Clin Microbiol Infect Dis 35: 1427–1431. [DOI] [PubMed] [Google Scholar]
- Obregon-Tito AJ, Tito RY, Metcalf J, Sankaranarayanan K, Clemente JC, Ursell LK et al. (2015). Subsistence strategies in traditional societies distinguish gut microbiomes. Nat Commun 6: 6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31: 3691–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek C, Smoczynski R, Tretyn A. (2011). Sequencing technologies and genome sequencing. J Appl Genet 52: 413–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasolli E, Schiffer L, Manghi P, Renson A, Obenchain V, Truong DT et al. (In press). Accessible, curated metagenomic data through ExperimentHub. Nat Methods doi:http://dx.doi.org10.1101/103085. [DOI] [PMC free article] [PubMed]
- Pasolli E, Truong DT, Malik F, Waldron L, Segata N. (2016). Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. PLoS Comput Biol 12: e1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen AM, Stensvold CR, Mirsepasi H, Engberg J, Friis-Moller A, Porsbo LJ et al. (2013). Active ulcerative colitis associated with low prevalence of Blastocystis and Dientamoeba fragilis infection. Scand J Gastroenterol 48: 638–639. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al. (2010. a). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490: 55–60. [DOI] [PubMed] [Google Scholar]
- Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L et al. (2014). Alterations of the human gut microbiome in liver cirrhosis. Nature 513: 59–64. [DOI] [PubMed] [Google Scholar]
- Quinlan AR. (2014). BEDTools: The Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics 47: 11 12 11–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez JD, Sanchez LV, Bautista DC, Corredor AF, Florez AC, Stensvold CR. (2014). Blastocystis subtypes detected in humans and animals from Colombia. Infect Genet Evol 22: 223–228. [DOI] [PubMed] [Google Scholar]
- Rampelli S, Schnorr SL, Consolandi C, Turroni S, Severgnini M, Peano C et al. (2015). Metagenome sequencing of the Hadza Hunter-Gatherer gut microbiota. Curr Biol 25: 1682–1693. [DOI] [PubMed] [Google Scholar]
- Roberts T, Ellis J, Harkness J, Marriott D, Stark D. (2014). Treatment failure in patients with chronic Blastocystis infection. J Med Microbiol 63: 252–257. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Knight R, Song SJ, Ackermann G, Cotter PD. (2016). Prevalence and genetic diversity of Blastocystis in family units living in the United States. Infect Genet Evol 45: 95–97. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Marchesi JR. (2008). Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J 2: 1183–1193. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Stensvold CR, Rajilić-Stojanović M, Heilig HGHJ, De Vos WM, O'Toole PW et al. (2014). The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol Ecol 90: 326–330. [DOI] [PubMed] [Google Scholar]
- Scholz M, Ward DV, Pasolli E, Tolio T, Zolfo M, Asnicar F et al. (2016). Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods 13: 435–438. [DOI] [PubMed] [Google Scholar]
- Segata N. (2015). Gut Microbiome: westernization and the disappearance of intestinal diversity. Curr Biol 25: R611–R613. [DOI] [PubMed] [Google Scholar]
- Segata N, Boernigen D, Tickle TL, Morgan XC, Garrett WS, Huttenhower C. (2013. a). Computational meta'omics for microbial community studies. Mol Syst Biol 9: 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Börnigen D, Morgan XC, Huttenhower C. (2013. b). PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4: 2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9: 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensvold CR, Alfellani M, Clark CG. (2012). Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect Genet Evol 12: 263–273. [DOI] [PubMed] [Google Scholar]
- Tan KS, Mirza H, Teo JD, Wu B, Macary PA. (2010). Current views on the clinical relevance of Blastocystis spp. Curr Infect Dis Rep 12: 28–35. [DOI] [PubMed] [Google Scholar]
- Tan KSW. (2008). New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin Microbiol Rev 21: 639–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E et al. (2015). MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12: 902–903. [DOI] [PubMed] [Google Scholar]
- Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. (2017). Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27: 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaousis AD, Ollagnier de Choudens S, Gentekaki E, Long S, Gaston D, Stechmann A et al. (2012). Evolution of Fe/S cluster biogenesis in the anaerobic parasite. Proc Natl Acad Sci USA 109: 10426–10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74. [DOI] [PubMed] [Google Scholar]
- Villalobos G, Orozco-Mosqueda GE, Lopez-Perez M, Lopez-Escamilla E, Cordoba-Aguilar A, Rangel-Gamboa L et al. (2014). Suitability of internal transcribed spacers (ITS) as markers for the population genetic structure of Blastocystis spp. Parasites Vectors 7: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzyniak I, Texier C, Poirier P, Viscogliosi E, Tan KS, Delbac F et al. (2012). Characterization of two cysteine proteases secreted by Blastocystis ST7, a human intestinal parasite. Parasitol Int 61: 437–442. [DOI] [PubMed] [Google Scholar]
- Wesolowska-Andersen A, Bahl MI, Carvalho V, Kristiansen K, Sicheritz-Ponten T, Gupta R et al. (2014). Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H, Dogruman-AI F, Turk S, Kustimur S, Balaban N, Sultan N. (2011). Evaluation of DNA extraction kits for molecular diagnosis of human Blastocystis subtypes from fecal samples. Parasitol Res 109: 1045–1050. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H, Koyama Y, Tsuchiya E, Takami K. (2016). Blastocystis phylogeny among various isolates from humans to insects. Parasitol Int 65: 750–759. [DOI] [PubMed] [Google Scholar]
- Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI et al. (2014). Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol 10: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.