Abstract
Long noncoding RNAs (lncRNAs) are a new class of regulatory noncoding RNAs. Emerging evidences indicate that lncRNAs play a critical role in the development of hepatocellular carcinoma (HCC). Although several lncRNAs have been annotated, the association of most lncRNAs with HCC is unknown. In this study, we investigated lncRNA alterations in HCC by performing lncRNA microarray analysis. We identified a novel lncRNA called HCC-associated lncRNA (HCAL) that was highly expressed in HCC tissues. HCAL upregulation was clinically associated with poor differentiation, intravascular cancer embolus, and decreased survival of patients with HCC. HCAL silencing significantly inhibited the growth and metastasis of HCC cells both in vitro and in vivo. Interestingly, transcriptome-sequencing analysis of HCAL-knockdown cells showed alterations in some cancer-related pathways. Mechanistically, HCAL directly interacted with and functioned as a sponge for microRNAs such as miR-15a, miR-196a, and miR-196b to modulate LAPTM4B expression. Taken together, our findings suggest the presence of a novel lncRNA-miRNA-mRNA regulatory network, i.e., the HCAL-miR-15a/miR-196a/miR-196b-LAPTM4B network, in HCC and indicate that HCAL may be a potential target for treating HCC.
Keywords: long noncoding RNA, competing endogenous RNA, LAPTM4B, heptatocellular carcinoma, microRNA
Introduction
Hepatocellular carcinoma (HCC) is the fifth most prevalent cancer worldwide, with an increasing morbidity rate.1 Despite recent therapeutic advances in HCC prevention, including surgical techniques and medical therapy, the prognosis of patients with HCC remains poor; moreover, approximately 600,000 patients with HCC die each year.2 Therefore, determination of the molecular mechanisms underlying HCC occurrence and progression is important to identify novel diagnostic and therapeutic markers for developing HCC treatment.
The human transcriptome comprises many types of RNAs, including protein-coding mRNAs and noncoding RNAs (ncRNAs).3 Intensive investigations over the last few decades have focused on the role of protein-coding genes in the pathogenesis of HCC. MicroRNAs (miRNAs) are a class of small ncRNAs that induce mRNA degradation or inhibit mRNA translation by binding to the 3ʹ UTR of mRNAs. NcRNAs containing more than 200 nucleotides are called long ncRNAs (lncRNAs).4, 5 Previous studies have shown that miRNAs play important roles in the regulation of cell proliferation, differentiation, invasion, and metabolism.6 Evidences obtained thus far indicate that lncRNAs perform multiple physiological and pathological biological functions.7 Aberrant lncRNA expression is observed in many cancers, including lung cancer, gastric cancer, colon cancer, and HCC.8, 9, 10 LncRNAs perform their functions through various mechanisms, including epigenetic silencing, lncRNA-miRNA interaction, lncRNA-protein interaction, and lncRNA-mRNA interaction.11 For example, lncTCF7, which is highly expressed in HCC and liver cancer stem cells (CSCs), promotes CSC self-renewal and HCC progression by interacting with the switching/sucrose non-fermenting (SWI/SNF) complex and by activating the WNT pathway.12 LncRNA-ATB, an important regulator of the invasion-metastasis cascade, promotes cell invasion by acting as a competing endogenous RNA (ceRNA) of ZEB and facilitates the colonization of disseminated HCC cells in distant organs by binding to interleukin-11 (IL-11) mRNA.13 Although several lncRNAs have been annotated, the association of most lncRNAs with HCC development and progression remains unclear.
In the present study, we identified a novel lncRNA termed HCC-associated lncRNA (HCAL; gene symbol, RP11-21I10.2) in HCC. We found that HCAL was overexpressed in HCC tissues, and its expression was correlated with the poor prognosis of patients with HCC. Results of in vivo and in vitro assays showed that HCAL plays an oncogenic role in regulating the malignant phenotypes of HCC cells, including proliferation, apoptosis, migration, and invasion. Additional mechanistic investigations showed that HCAL functions as a ceRNA to regulate LAPTM4B expression by competitively binding to common miRNAs, such as miR-15a, miR-196a, and miR-196b.
Results
LncRNA Expression Profiles in HCC
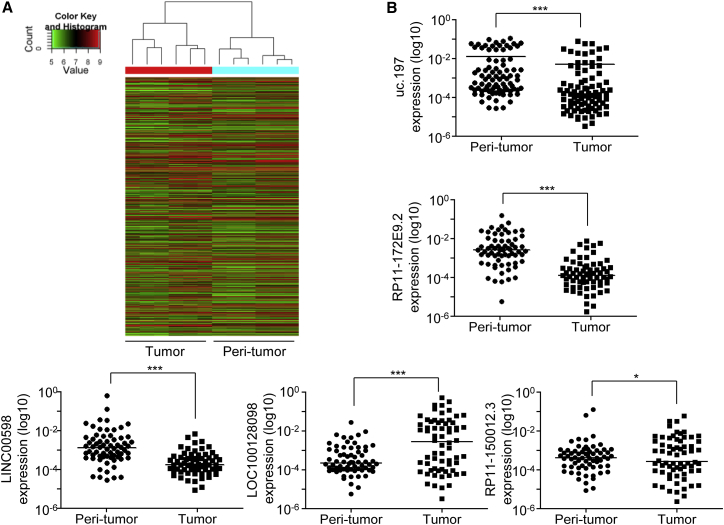
To identify differentially expressed lncRNAs in HCC, we examined lncRNA and mRNA expression in six pairs of HCC and corresponding peritumor tissues by performing microarray analysis. Microarray analysis identified 2,665 and 3,885 lncRNAs and mRNAs, respectively, that were significantly differentially expressed in HCC tissues (fold change >2, p < 0.05) compared with those in corresponding peritumor tissues (Figures 1A and S1). Results of microarray analysis were validated by analyzing the expression of five random differentially expressed lncRNAs by performing qPCR in 66 pairs of HCC and corresponding peritumor tissues (Figure 1B). We observed that the expression of lncRNAs uc.197, RP11-172E9.2, and LINC00598 was downregulated, and the expression of lncRNAs LOC100128098 and RP11-150012.3 was upregulated in HCC tissues. Thus, our results indicate that the phenotypic shift from the normal state to malignant transformation is reflected in not only the expression of protein-coding genes, but also by relative large amount of lncRNAs in HCC.
Figure 1.
Expression Profiles of lncRNA in HCC
(A) Hierarchical clustering analysis of differentially expressed lncRNAs (fold change > 2; p < 0.05) between HCC and paired peritumor tissue samples obtained from six patients with HCC. (B) Differential expression of five randomly selected lncRNAs was validated in 66 HCC and corresponding peritumor tissues by performing qPCR. Horizontal line indicates the mean of indicated lncRNA expression level; *p < 0.05 and ***p < 0.001 (paired Student’s t test).
A Novel lncRNA, HCAL, Is Overexpressed in HCC Tissues and Is Clinically Correlated with Poor Prognosis of Patients with HCC
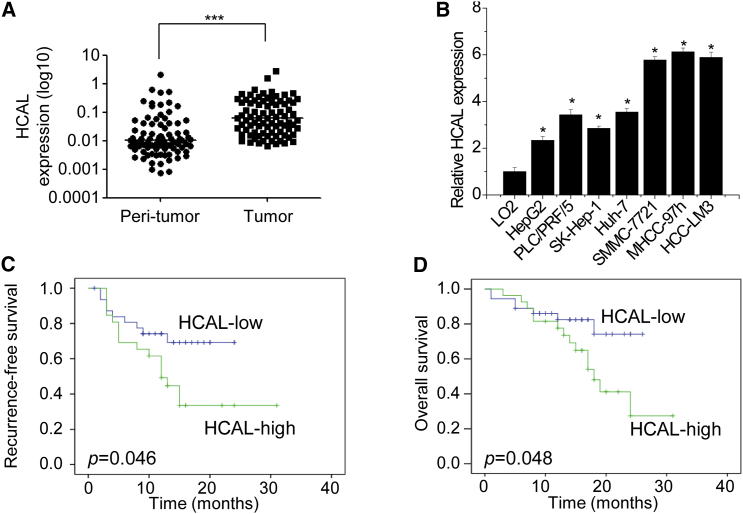
We used gene coexpression networks to identify interactions among different transcripts. Because coexpression modules may correspond to biological pathways,14 we focused on coexpression modules that have a high rate of protein-coding RNAs in the HCC coexpression. HCAL was screened out by this way. In the coexpression network, HCAL was associated with many protein-coding genes involved in tumor progression, such as STMN1, FOS, and IL-3 (Figure S2). The gene encoding HCAL is located on chromosome 4 (119990479–119991452 [−] strand) between two protein-coding genes, namely, SYNPO2 and MYOZ. We examined HCAL expression in 84 pairs of HCC and corresponding peritumor tissues by performing qPCR (Figure 2A). HCAL expression was significantly upregulated in HCC tissues compared with that in the matched peritumor tissues. We also detected HCAL expression in immortalized normal liver cells (LO2) and seven HCC cell lines, namely, HepG2, PLC/PRF/5, SK-Hep-1, Huh-7, SMMC-7721, MHCC-97h, and HCC-LM3 (Figure 2B). We observed that all the HCC cell lines showed higher HCAL expression than LO2 cells.
Figure 2.
HCAL Is Overexpressed in HCC and Is Clinically Correlated with the Poor Prognosis of Patients with HCC
(A) HCAL expression in 84 pairs of HCC and corresponding peritumor tissues was analyzed by performing qPCR. ***p < 0.001 (paired Student’s t test). (B) HCAL expression in the immortalized normal liver cells (LO2 cells) and seven HCC cell lines was analyzed by performing qPCR. LO2 cells were used as controls. *p < 0.05 (Student’s t test). (C and D) Kaplan-Meier survival curves and log rank tests were used to evaluate the association of HCAL expression with (C) recurrence-free and (D) overall survival of patients with HCC. The patients were divided into high- and low-HCAL groups based on median HCAL expression in HCC tissues.
Next, we investigated the clinical significance of HCAL expression in patients with HCC. HCAL overexpression was more frequently observed in HCC tissues obtained from patients showing low-level tumor differentiation and intravascular cancer embolus than in HCC tissues obtained from patients showing high-level tumor differentiation and no intravascular cancer embolus. However, no significant correlation was observed between HCAL expression and the surface antigen of the hepatitis B virus (HBsAg) and serum alpha fetoprotein (AFP) levels, tumor size, liver cirrhosis, or portal vein tumor thrombus (PVTT) (Table 1). Female patients showed lower HCAL expression levels than male patients. Furthermore, the duration of recurrence-free and overall survival was shorter in patients with HCC showing high HCAL expression than in patients showing low HCAL expression (Figures 2C and 2D). Together, these results indicate that HCAL is overexpressed in HCC and that HCAL expression is correlated with HCC progression.
Table 1.
Correlation Analysis between HCAL Expression and Clinicopathological Features of Patients with HCC
| Clinicopathological Variables | HCAL Expression |
p Value |
|
|---|---|---|---|
| Low | High | ||
| Gender | |||
| Male | 29 | 37 | 0.033 |
| Female | 13 | 5 | |
| HBsAg | |||
| Present | 39 | 34 | 0.106 |
| Absent | 3 | 8 | |
| Serum AFP (IU) | |||
| <400 | 19 | 14 | 0.264 |
| >400 | 23 | 28 | |
| Tumor Size | |||
| <5 cm | 19 | 14 | 0.264 |
| ≥5 cm | 23 | 28 | |
| Liver Cirrhosis | |||
| Present | 23 | 24 | 0.826 |
| Absent | 19 | 18 | |
| Tumor Differentiation | |||
| Low | 5 | 17 | 0.01 |
| Intermediate | 34 | 24 | |
| High | 3 | 1 | |
| Intravascular Cancer Embolus | |||
| Absent | 25 | 15 | 0.029 |
| Present | 17 | 27 | |
| PVTT | |||
| Present | 39 | 40 | 0.645 |
| Absent | 3 | 2 | |
Patients with HCC were divided into high- and low-HCAL groups based on median HCAL expression. AFP, alpha fetoprotein; PVTT, portal vein tumor thrombus.
HCAL Regulates HCC Cell Proliferation, Apoptosis, Migration, and Invasion Both In Vitro and In Vivo
Analysis of the sequences using the ORF Finder available on the NCBI website could not predict a protein containing >92 amino acids (Figure S3A). Moreover, we confirmed that HCAL did not have any protein-coding capacity (Figure S3B).
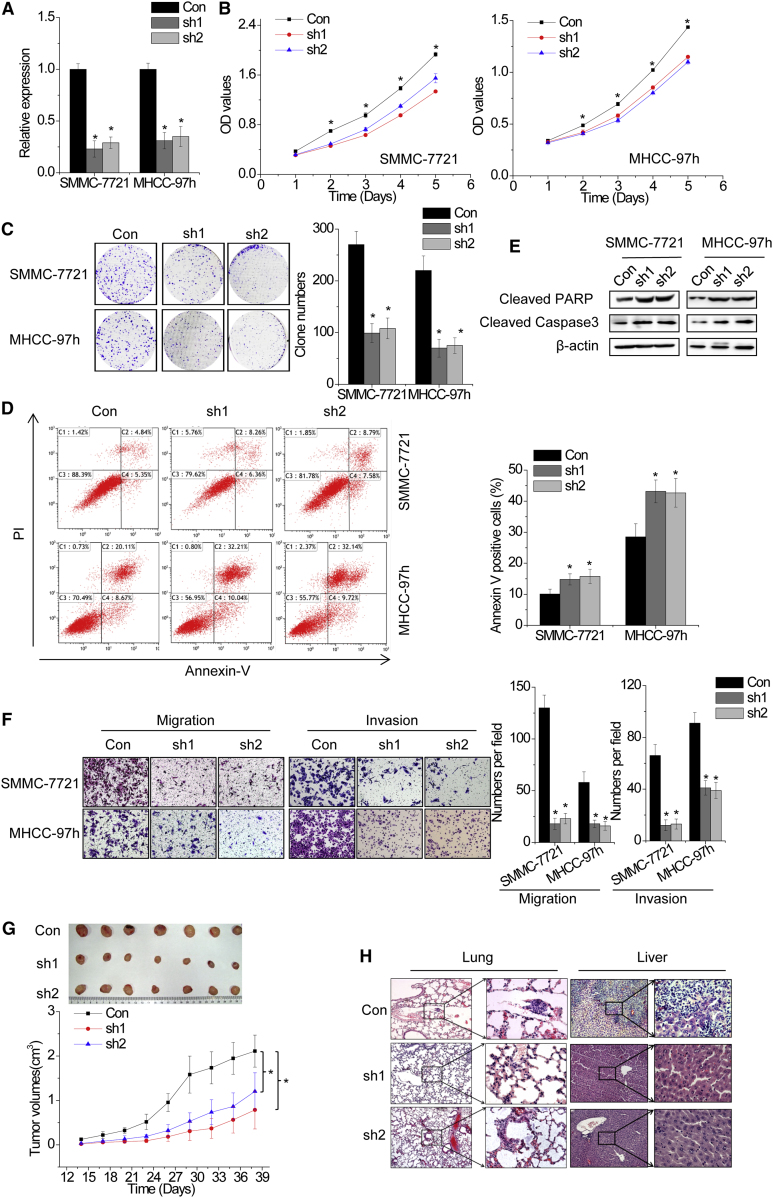
We observed that HCAL was more obviously overexpressed in the seven HCC cell lines, especially in SMMC-7721 and MHCC-97h cells, than in LO2 cells (Figure 2B). Therefore, we selected SMMC-7721 and MHCC-97h cells as representative HCC cells for performing subsequent experiments. We developed stable HCAL-knockdown SMMC-7721 and MHCC-97h cells. To eliminate off-target effects, we designed two independent short hairpin RNAs (shRNAs) against HCAL (HCAL shRNA1 and shRNA2). Both the shRNAs significantly reduced HCAL transcription (Figure 3A). To assess the potential effects of HCAL silencing on cell proliferation, we performed Cell Counting Kit-8 (CCK-8) and colony formation assays. HCAL knockdown significantly decreased the proliferation of HCC cells (Figures 3B and 3C). To determine mechanisms underlying the HCAL-induced increase in HCC cell proliferation, we determined the effect of HCAL silencing on cell cycle distribution by performing flow cytometry. The results show that HCAL depletion did not affect cell cycle progression or expression of cell cycle checkpoint proteins, including CDK4 and CDK6 (Figures S3C and S3D).
Figure 3.
HCAL Regulates HCC Cell Proliferation, Apoptosis, Migration, and Invasion In Vitro and In Vivo
(A) Relative HCAL expression in SMMC-7721 and MHCC-97h cells transduced with lentiviruses expressing control shRNA (Con), HCAL shRNA1 (sh1), or shRNA2 (sh2) was determined by performing qPCR. (B) Proliferation of control and HCAL-knockdown cells was assessed by performing the CCK-8 assay. HCAL knockdown suppressed the proliferation of SMMC-7721 and MHCC-97h cells. (C) Colony-formation assays were performed using control and HCAL-knockdown cells. Left, crystal violet staining; right, number of colonies from three independent experiments. (D) HCAL-knockdown cells were treated with 5-fluorouracil (100 mg/mL) for 48 hr, stained with annexin V and PI, and analyzed by flow cytometry. Annexin V-positive cells were designated as apoptotic cells. Percentage of apoptotic cells is shown. (E) Cleaved PARP1 and caspase-3 levels after HCAL silencing were determined by performing western blot analysis. (F) Left, representative images of the migration and invasion of SMMC-7721 and MHCC-97h cells expressing control and HCAL shRNAs. Right, statistical results obtained from three independent experiments. (G) Effects of HCAL knockdown on HCC growth in vivo. Upper panel, representative images of tumors formed in nude mice subcutaneously injected with HCAL-knockdown SMMC-7721 cells; lower panel, tumor growth curves measured after injecting SMMC7721 cells expressing control and HCAL shRNAs. (H) Representative images of pulmonary and intrahepatic metastases in nude mice subcutaneously injected with control and HCAL-knockdown SMMC-7721 cells obtained by performing HE staining. Data are expressed as mean ± SD; *p < 0.05.
Because HCAL exerted an oncogenic effect in HCC cells, we speculated that HCAL was critical for cell apoptosis. To test this hypothesis, we analyzed the apoptosis of HCC cells treated with 5-fluorouracil by performing flow cytometry with annexin V and propidium iodide (PI) staining. Results of flow cytometry showed that HCAL knockdown increased the percentage of annexin V-positive cells in both SMMC-7721 and MHCC-97H cells compared with control cells (Figure 3D). Moreover, HCAL knockdown increased the levels of apoptosis markers, including cleaved PARP1 and cleaved caspase-3, in SMMC-7721 and MHCC-97H cells, which was consistent with the results of flow cytometry (Figure 3E). Collectively, these results indicate that HCAL affects HCC cell proliferation and apoptosis.
Next, we investigated the role of HCAL in HCC cell migration and invasion. As shown in Figure 3F, HCAL knockdown markedly suppressed the migration and invasion of SMMC-7721 and MHCC-97h cells.
Based on this observation, we determined the effects of HCAL on HCC growth and metastasis in vivo. We used a xenograft mouse model that was generated by subcutaneously injecting control and HCAL-knockdown SMMC-7721 cells into nude mice. Xenografted tumors derived from HCAL-knockdown SMMC-7721 cells had smaller volumes and grew slower than tumors derived from control cells (Figure 3G). Mice injected with HCAL-knockdown cells showed decreased intrahepatic and pulmonary metastases compared with mice injected with control cells, which was further confirmed by performing histological analysis (Figures 3H and S3E). Together, the results of in vitro and in vivo assays indicate that HCAL affects the proliferation, apoptosis, migration, and invasion of HCC cells.
LAPTM4B Is the Target Gene of HCAL
LncRNAs act in cis or trans to regulate the expression of neighboring genes.15 We examined whether HCAL affected the expression of its neighboring genes SYNPO2 and MYOZ. We observed that HCAL knockdown did not affect the transcription of SYNPO2 or MYOZ (Figure S4A). Previous studies have reported that approximately 20% of lncRNAs physically interact with polycomb repressive complex 2 (PRC2) to regulate the expression of their target genes.16 We performed an RNA immunoprecipitation (RIP) assay with anti-EZH2 (an important subunit of PRC2) antibody using nuclear extracts of SMMC-7721 and MHCC-97h cells. Interestingly, HCAL was significantly enriched by the EZH2 antibody, compared with the nonspecific immunoglobulin G (IgG) control antibody (Figure S4B), suggesting that HCAL regulated gene expression by interacting with EZH2.
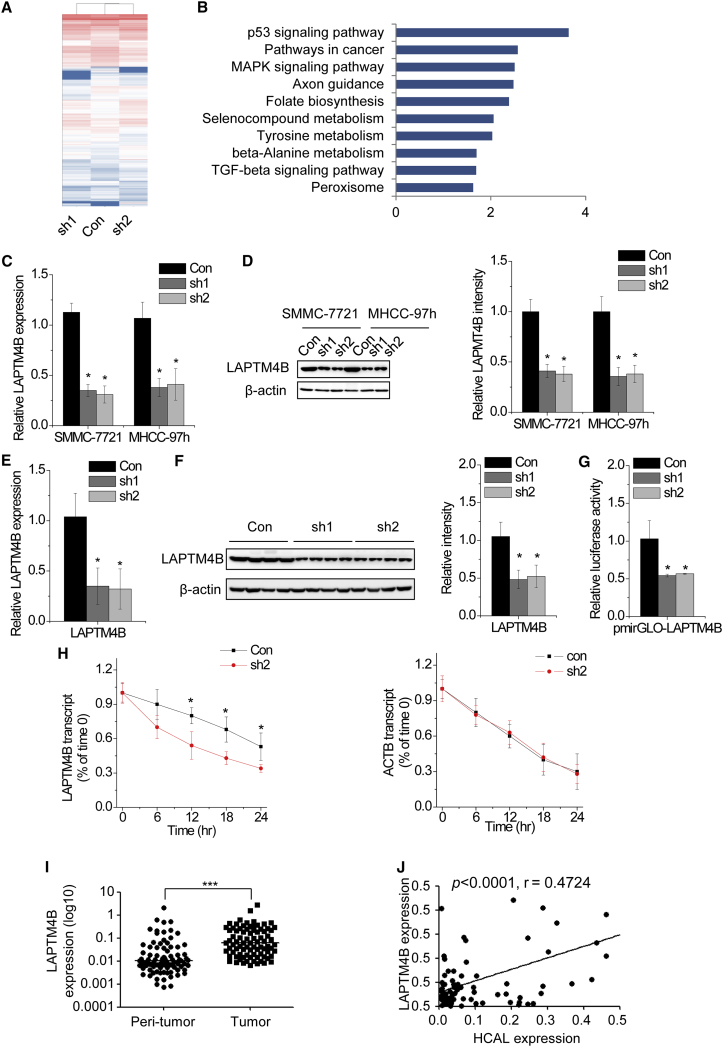
We performed transcriptome-sequencing analysis of HCAL-knockdown SMMC-7721 cells to determine the target genes of HCAL. HCAL knockdown resulted in the differential expression of 204 genes (fold change > 2; p < 0.05; Figure 4A). Next, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the differentially expressed genes to identify key pathways affected by HCAL-mediated transcriptional regulation. HCAL regulated several genes associated with important cancer-related signaling pathways, including the p53, cancer, MAPK, and TGF-beta pathways (Figure 4B).
Figure 4.
LAPTM4B Is the Target Gene of HCAL
(A) Expression heatmap of transcripts regulated by HCAL (fold change > 2; p < 0.05). Red and blue indicate upregulation and downregulation, respectively. (B) The top 10 pathways affected by HCAL downregulation according to the KEGG analysis. (C) Relative LAPTM4B mRNA expression in control and HCAL-knockdown cells was determined by performing qPCR. (D) Left, LAPTM4B protein levels in control and HCAL-knockdown cells were determined by performing western blotting. Right, quantification of LAPTM4B protein levels. (E) LAPTM4B mRNA expression in the xenografts of nude mice injected with HCAL-knockdown cells analyzed by qPCR. (F) Left, LAPTM4B protein levels in the xenografts of nude mice injected with HCAL-knockdown cells was analyzed by performing western blotting. Right, quantification of LAPTM4B protein levels. (G) Relative luciferase activity of LAPTM4B 3′ UTR in control and HCAL-knockdown SMMC-7721 cells. Data are presented as the relative ratio of firefly luciferase activity to Renilla luciferase activity. (H) Stability of LAPTM4B and ACTB mRNA over time was measured by performing qPCR relative to time 0 after blocking new RNA synthesis with α-amanitin (50 mM) in SMMC-7721 cells expressing control and HCAL shRNAs and was normalized to that of 18S rRNA (a product of RNA polymerase I that is unaffected by α-amanitin). (I) LAPTM4B expression in 84 pairs of HCC and corresponding peritumor tissues was analyzed by qPCR. (J) Correlation between HCAL and LAPTM4B mRNA levels in the same set of 84 HCC tissues was determined using Pearson’s correlation analysis. Data are expressed as mean ± SD; *p < 0.05 and ***p < 0.001.
Emerging evidences indicate that many transcripts function as ceRNAs by competitively binding to common miRNAs.13, 17 To examine whether HCAL performed a similar role in HCC, we performed a ceRNA prediction using a bioinformatics method (Table S2). Among the predicted mRNAs, we focused on LAPTM4B mRNA because of its oncogenic activity in various cancers.18 The HCAL sequence showed 96% similarity to the LAPTM4B 3′ UTR sequence, strongly suggesting that HCAL functioned as a ceRNA of LAPTM4B (Figure S4C). We examined changes in LAPTM4B mRNA and protein expression in HCAL-knockdown cells and found that HCAL knockdown significantly suppressed both the mRNA and protein expression of LAPTM4B (Figures 4C and 4D). Moreover, xenografts of nude mice injected with HCAL-knockdown cells showed lower LAPTM4B expression than xenografts of nude mice injected with control cells (Figures 4E and 4F). We also examined the luciferase reporter activity of the LAPTM4B 3′ UTR in HCAL-knockdown SMMC-7721 cells. HCAL depletion significantly reduced the luciferase activity of the LAPTM4B 3′ UTR (Figure 4G). To examine whether HCAL regulated the stability of LAPTM4B mRNA, we treated control and HCAL-knockdown SMMC-7721 cells with α-amanitin to block new RNA synthesis and measured the decrease in the mRNA expression of LAPTM4B, ACTB, and 18S rRNA over a 24-hr period. We observed that HCAL downregulation markedly shortened the half-life of LAPTM4B mRNA (Figure 4H).
To determine the pathological correlation between HCAL and LAPTM4B expression in human HCC samples, we measured LAPTM4B mRNA level in the same 84 pairs of HCC and corresponding peritumor tissues by performing qPCR. LAPTM4B mRNA expression was significantly upregulated in HCC tissues compared with that in corresponding peritumor tissues (Figure 4I). In addition, results of qPCR showed a positive correlation between HCAL and LAPTM4B mRNA expression in the 84 HCC tissues (r = 0.4724, p < 0.0001; Figure 4J). Together, these results indicate that LAPTM4B is the target gene of HCAL.
HCAL Regulates LAPTM4B Expression by Competitively Binding to Common miRNAs
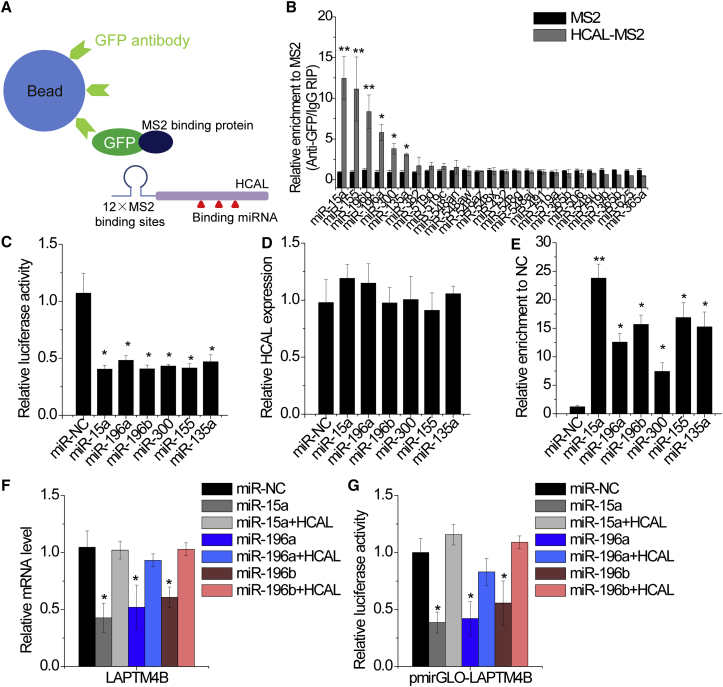
To further confirm the exact mechanisms through which HCAL regulated LAPTM4B expression, we identified miRNAs that potentially interacted with HCAL by using the TargetScan prediction algorithm (Table S3). Direct interaction between miRNAs and HCAL was validated by performing RIP assays (Figure 5A). We detected 25 miRNAs involved in HCC tumorigenesis and progression by performing qPCR. HCAL immunoprecipitates obtained from SMMC-7721 cells were significantly enriched with six miRNAs, namely, miR-15a, miR-155, miR-196b, miR-196a, miR-300, and miR-135a, compared with those obtained from cells transfected with an empty vector (MS2) and treated with control IgG (Figure 5B). For further confirmation, we constructed a luciferase reporter vector containing HCAL and found that ectopic expression of the above six miRNAs dramatically decreased the luciferase activity of the HCAL reporter vector (Figure 5C). However, these miRNAs did not affect HCAL expression (Figure 5D). In addition, we performed anti-AGO2 RIP in SMMC-7721 cells transiently overexpressing these miRNAs. HCAL was significantly pulled down by AGO2 antibodies in SMMC-7721 cells overexpressing the specified miRNAs (Figure 5E). Together, these results indicate that these miRNAs interact with HCAL but do not lead to degradation.
Figure 5.
HCAL Regulates LAPTM4B Expression by Competitively Binding to Common miRNAs
(A) Schematic representation of an RNA immunoprecipitation assay. (B) MS2-RIP followed by miRNA qPCR to detect endogenous miRNAs associated with HCAL. (C) Relative luciferase activity in SMMC-7721 cells cotransfected with the indicated miRNAs and luciferase reporter vectors containing HCAL. (D) SMMC-7721 cells were transfected with the indicated miRNAs. After 48 hr, relative expression of HCAL was analyzed by qPCR. (E) RIP assay using anti-AGO2 antibody and qPCR were performed with SMMC-7721 cells transiently overexpressing the indicated miRNAs to detect the association of HCAL with AGO2. (F) Changes in LAPTM4B mRNA expression levels in the indicated SMMC-7721 cells. (G) Changes in the luciferase activity of LAPTM4B 3′ UTR in the indicated SMMC-7721 cells. Data are presented as the relative ratio of firefly luciferase activity to Renilla luciferase activity. Data are expressed as mean ± SD. *p < 0.05 and **p < 0.01.
The TargetScan prediction algorithm indicated that miR-135a, miR-155, and miR-300 could not potentially bind to the LAPTM4B 3′ UTR. Moreover, overexpression of miR-135a, miR-155, or miR-300 did not affect LAPTM4B expression or luciferase activity of the vector containing the LAPTM4B 3′ UTR (Figures S5A and S5B). Ectopic expression of miR-15a, miR-196a, or miR-196b significantly decreased LAPTM4B expression, while upregulation of HCAL abrogated this suppression (Figure 5F). To confirm whether this effect was mediated by regulating LAPTM4B 3′ UTR activity, we cotransfected SMMC-7721 cells with luciferase vectors containing the LAPTM4B 3′ UTR (pmirGLO-LAPTM4B) and overexpressed miR-15a, miR-196a, or miR-196b. Upregulation of any of these three miRNAs decreased the luciferase activity of pmirGLO-LAPTM4B, whereas ectopic expression of HCAL abolished this effect (Figure 5G). Altogether, these results indicate that HCAL regulates LAPTM4B expression by acting as a ceRNA and by competitively binding to common miRNAs.
HCAL Facilitates HCC Cell Proliferation, Migration, and Invasion by Regulating LAPTM4B
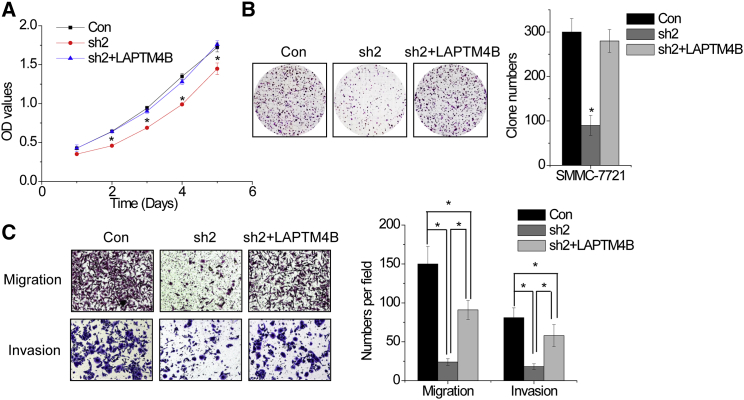
To determine whether HCAL functions upstream of LAPTM4B to regulate the malignant phenotypes of HCC cells, we overexpressed LAPTM4B in HCAL-knockdown SMMC-7721 cells. LAPTM4B overexpression abolished the suppression of HCC cell growth induced by HCAL knockdown (Figures 6A and 6B). Moreover, LAPTM4B overexpression partially rescued the HCAL knockdown-induced inhibition of cell migration and invasion (Figures 6C and 6D). These results indicate that HCAL regulates the malignant phenotypes of HCC cells by regulating LAPTM4B expression.
Figure 6.
HCAL Facilitates HCC Cell Proliferation, Migration, and Invasion by Regulating LAPTM4B
(A) HCAL-knockdown SMMC-7721 cells were transfected with LAPTM4B. Cell proliferation rates were determined by CCK-8 assay. (B) Colony formation of HCAL-knockdown SMMC-7721 cells transiently overexpressing LAPTM4B. (C) Migration and invasion assays were performed using HCAL-knockdown SMMC-7721 cells transiently overexpressing LAPTM4B. Data are expressed as mean ± SD. *p < 0.05.
Discussion
The key finding of the present study is that the expression of the lncRNA HCAL is significantly upregulated in HCC tissues compared with corresponding peritumor tissues. HCAL expression is closely associated with sex, tumor differentiation level, and intravascular cancer embolus. High HCAL expression is a predictor of poor recurrence-free and overall survival. We determined the function of HCAL in HCC cells using loss-of-function approaches. HCAL knockdown inhibited the proliferation, colony formation, migration, and invasion of HCC cells and induced apoptosis. This is the first study to show the clinical and functional significance of HCAL in the malignant progression of HCC. Our results indicate that HCAL functions as an oncogene in HCC and suggest that it can be used as a novel diagnostic and prognostic marker for HCC.
The incidence of HCC is higher in men than in women, with average ratios between 2:1 and 4:1;1 sex hormones and cytokines are suggested to contribute to this gender disparity. Estrogen and estrogen receptors prevent HCC in women, while androgen and androgen receptors increase the risk of HCC in men.19 IL-6 is one of the most important cytokines that promotes HCC development in men. IL-6 ablation partially abolishes diethylnitrosamine-induced development of HCC in male mice.20 In the present study, we observed that HCAL expression was lower in HCC tissues obtained from female patients than in those obtained from male patients, suggesting that HCAL partially contributes to the gender disparity observed in HCC. However, additional studies should be performed to determine whether HCAL function depends on sex hormones.
lncRNAs play critical roles in gene regulation21 and exert their effects through diverse mechanisms, including transcriptional regulation, epigenetic modification, post-transcriptional regulation, and post-translational modulation.13, 22, 23, 24 Accumulating evidence indicates that lncRNAs regulate gene expression by functioning as ceRNAs. The lncRNAs act as miRNA sponges or inhibitors to suppress the interactions between miRNAs and target mRNAs,17, 25 e.g., lncRNA HULC, which is highly upregulated in liver cancer and directly binds to miR-372 to repress its expression and activity. Reduction of miR-372 expression increases the expression of PRKACB mRNA, which is a target of miR-372.26 A recent study showed that HULC functions as a ceRNA to induce epithelial-mesenchymal transition (EMT) by sponging miR-200a and subsequently upregulating ZEB1 expression.27 The present study provides additional evidence that lncRNAs function as miRNA sponges, indicating that mRNA post-transcriptional regulation contributes to HCC pathogenesis and suggesting that the lncRNA-miRNA-mRNA regulatory network is critical for HCC tumorigenesis and progression.
We selected LAPTM4B mRNA because of its oncogenic activity in various cancers. LAPTM4B is highly expressed in various solid tumors and may be a marker of poor prognosis. LAPTM4B plays an important role in promoting tumor cell proliferation, migration, and invasion; autophagy initiation; and apoptosis suppression in different cancers, including HCC.28, 29, 30 However, limited information is available on the regulatory mechanism underlying LAPTM4B expression. To our knowledge, this is the first study to show that HCAL regulates LAPTM4B expression by functioning as a ceRNA. Our data showed that HCAL downregulation significantly suppressed the expression and stability of LAPTM4B mRNA and decreased the luciferase activity of the LAPTM4B 3′ UTR. In addition, we observed a positive correlation between HCAL and LAPTM4B expression in HCC tissues, confirming the regulatory association between HCAL and LAPTM4B expression. Because LAPTM4B is upregulated in other cancers such as breast cancer, glioblastoma, and lung cancer,31, 32, 33 we speculated that HCAL functioned upstream of LATPM4B in these cancers. Results of RIP and luciferase assays showed that HCAL directly interacted with miRNAs. Overexpression of miR-15a, miR-196a, or miR-196b inhibited LAPTM4B expression and the luciferase activity of the LAPTM4B 3′ UTR, and restoration of HCAL expression reversed this inhibition. Finally, we demonstrated that HCAL functioned upstream of LAPTM4B to regulate the malignant phenotypes of HCC cells. However, LATPM4B expression could not completely rescue the suppression of cell migration and invasion induced by HCAL knockdown, suggesting the involvement of other mechanisms in the HCAL-induced malignant phenotypes. Results of ceRNA prediction analysis suggested that HCAL acts as ceRNA of other transcripts. Moreover, results of the RIP assay showed a direct interaction between HCAL and EZH2. Together, these results suggest that HCAL affects the malignant phenotypes of HCC cells through diverse mechanisms.
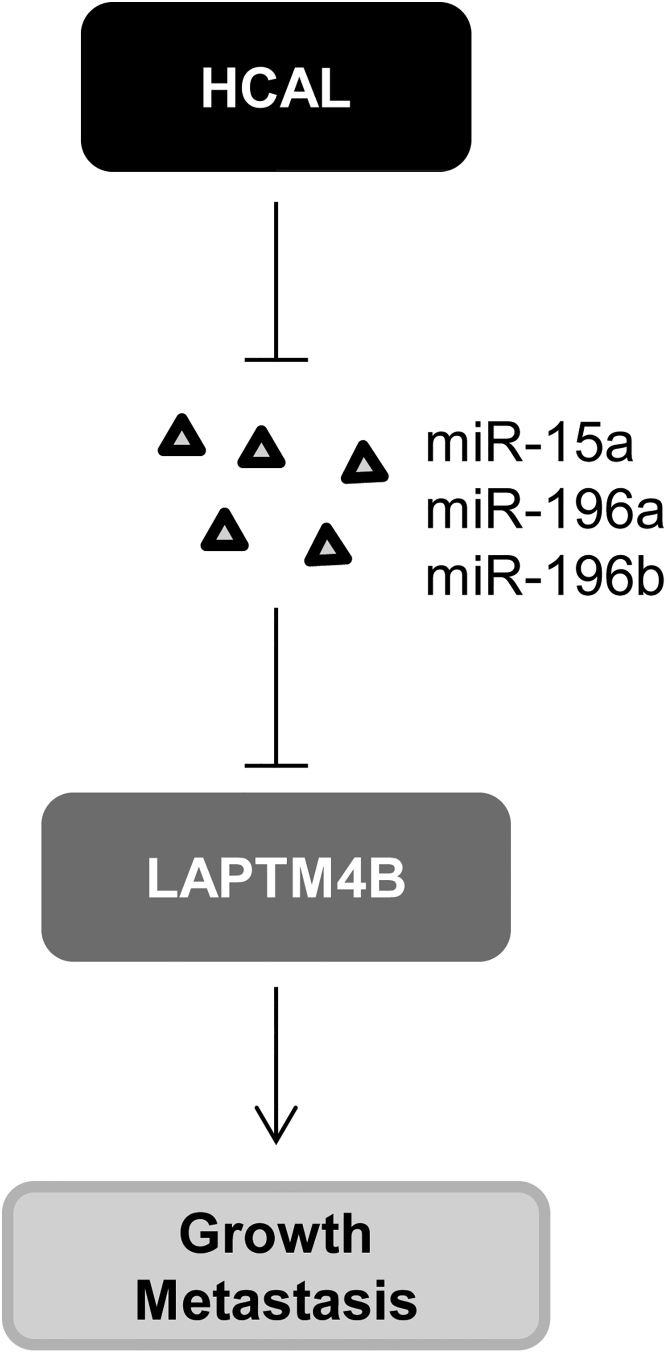
In summary, our results indicate that HCAL is an oncogenic lncRNA that facilitates HCC cell proliferation, migration, and invasion and inhibits HCC cell apoptosis by acting as a ceRNA of LAPTM4B (Figure 7). Thus, these findings indicate that HCAL is important for HCC progression and suggest that it can be used as a potential therapeutic target for HCC treatment.
Figure 7.
A Schematic Diagram Showing the Role of HCAL in HCC Progression
HCAL promotes HCC growth and metastasis by competitively binding to miR-15a, miR-196a, or miR-196b and by subsequently increasing LAPTM4B expression.
Materials and Methods
Cell Culture
LO2, HepG2, PLC/PRF/5, SK-Hep-1, Huh-7, and SMMC-7721 cells were obtained from the Cell Bank of Chinese Academy of Sciences, and MHCC-97h and HCC-LM3 cells were obtained from Zhongshan Hospital of Fudan University. The cells were cultured in DMEM (Hyclone) supplemented with 10% fetal bovine serum (FBS; PAN Biotech) at 37°C in 5% CO2.
Patients and Tissue Samples
The 84 pairs of HCC and corresponding peritumor tissues were collected from patients with HCC who initially underwent hepatectomy without any preoperative treatment at the Zhongshan Hospital of Xiamen University from 2011 to 2013. The procedure for sample collection was approved by the ethics committee of the Zhongshan Hospital of Xiamen University, and written informed consent was obtained from all patients.
RNA Isolation and Real-Time qPCR
Total RNA was isolated by TRIzol reagent (Invitrogen) according to standard protocol. cDNA was synthesized using One-Step gDNA Removal and cDNA Synthesis Kit (Transgen, Beijing, China). Real-time PCR was performed in the Lightcycle Real-Time PCR System (Roche) using FastStart Universal SYBR Green Master (Rox) (Roche). The gene-specific primers are shown in Table S1. ACTB was employed as an endogenous control for mRNA and lncRNA. For miRNA detection, cDNA was synthesized using Mir-X miRNA First-Strand Synthesis kit (Clontech), and real-time qPCR was performed as per the manufacturer’s instructions. ACTB was employed as an endogenous control for miRNA. Comparative quantification was determined using the 2−ΔΔCt method.
Coexpression Network
A coexpression network in HCC samples was constructed to identify interactions among mRNA and lncRNAs, as described previously.34, 35
Lentivirus Production and Construction of Stable Cell Lines with HCAL Knockdown
Two different shRNAs (shRNA1 and shRNA2) targeting HCAL were inserted into the pLKO.1 plasmid (Addgene). The target sequences of HCAL shRNA1 and shRNA2 are 5′-TCCCTAGGCAGTGAAACATT-3′ and 5′-CCCCAAATCTGATGGATCTT-3′, respectively. Lentiviral and packaging vectors (pMD2.G and psPAX2) were transfected into 293T cells (ATCC) by using TurboFect Transfection Reagent (Thermo Scientific). The culture medium of the cells was changed 24 hr after transfection, and medium containing the lentivirus was collected after 48 hr. Next, the cells were centrifuged, filtered, and transduced with the lentivirus in the presence of 5 μg/mL polybrene (Sigma). Culture supernatant was replaced with a complete culture medium after 12 hr, and stable cells were selected using puromycin for 1 week.
Cell Proliferation Assay
2.0 × 103 cells per well were seeded in 96-well culture plates. At the indicated time points, CCK-8 (Dojindo) was added to each well and incubated at 37°C for 1.5 hr. The absorbance values (optical density [OD] 450 nm) were measured using a spectrophotometer (Bio-Rad).
Colony Formation
2 × 103 cells per well were seeded in the 6-well culture plates. After 14 days’ culture, cells were fixed with 4% paraformaldehyde and stained with crystal violet.
Apoptosis Analysis
The apoptosis was detected by using Apoptosis Detection Kit (Dojindo) according to the manufacturer’s instructions. The data were analyzed by Kaluza software.
Migration and Invasion Assay
1.0 × 105 MHCC-97h or 5.0 × 104 SMMC-7721 cells were suspended and seeded in serum-free DMEM in the upper chamber of a 24-well transwell migration (Corning) or invasion insert (BD Biosciences). The lower chamber was filled with DMEM containing 10% FBS. After 24 hr of incubation, the cells that had traversed the membrane were fixed in 4% paraformaldehyde and staining by crystal violet.
Animal Studies
A xenograft mouse model was developed using 5- to 6-week-old male BALB/c nude mice. Control and HCAL-knockdown SMMC-7721 cells were trypsinized and harvested in serum-free DMEM, and 0.1 mL serum-free DMEM containing 3 × 106 cells was injected subcutaneously into the right flank of the nude mice. Tumors were detected after approximately 14 days, and tumor size was measured every 3 days. Tumor-bearing mice were killed 38 days after the injection, and tumors were removed for further analysis. All animal experiments were approved by the Animal Care and Use Committee of the Xiamen University. To detect pulmonary and hepatic metastasis, the tissues were fixed in 10% neutral formalin, embedded in paraffin, and cut into 3-μm-thick sections. H&E staining was performed using a standard protocol.
Luciferase Reporter Assay
Luciferase activity was determined using Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer’s instructions. In brief, transfected cells were lysed in culture dishes containing lysis buffer. Relative luciferase activity was determined using Varioskan Lux detection System (Thermo Scientific) and was normalized to Renilla luciferase activity at 48 hr after transfection.
RIP
For the RIP assays, SMMC-7721 cells were cotransfected with pcDNA-MS2 or pcDNA-HCAL-MS2 and pBobi-MS2-GFP. The RIP assay was performed after 48 hr using anti-GFP antibody (Abcam) and a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA), according to the manufacturer’s instructions.
For performing RIP with anti-AGO2 antibody, SMMC-7721 cells were transfected with the pMIR vector expressing a negative control or miR-15a, miR-196a, miR-196b, miR-300, miR-155, or miR-135a (Vigenebio Company, Shandong, China). The RIP assay was performed after 48 hr using anti-AGO2 antibody (Millipore, Bedford, MA) as described above.
Statistical Analysis
Statistical analysis was performed using SPSS 19.0 software. Data are expressed as mean ± standard deviation (SD). Comparison between two groups was performed using Student’s t test. Pearson’s correlation analysis was performed to determine the correlation between two variables. Survival curves were constructed using the Kaplan-Meier method and were compared using a log rank test. A p value of < 0.05 was considered statistically significant. Other experimental procedures are described in the Supplemental Materials and Methods.
Author Contributions
C.-R.X., X.-M.W., and Z.-Y.Y. conducted the experiments. C.-R.X., F.W., S.Z., F.-Q.W., S.Z., Z.L., J.L., H.-Q.Q., and Q.-L.F. finished all of the experiments. C.-R.X. and Z.-Y.Y. designed the experiments and wrote the paper.
Conflicts of Interest
The authors declare that they do not have any conflict of interest related to this study.
Acknowledgments
We thank Prof. Jian Zhou (Zhongshan Hospital of Fudan University) for providing MHCC-97h and HCC-LM3 cells. This work was supported by the National Natural Science Foundation of China (81372618, 81672418, and 81702351) and the Fujian Youth Key Personnel Research Project (2013-ZQN-ZD-34).
Footnotes
Supplemental Information includes five figures and three tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.10.018.
Supplemental Information
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Maluccio M., Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J. Clin. 2012;62:394–399. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 3.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Fu M., Zou C., Pan L., Liang W., Qian H., Xu W., Jiang P., Zhang X. Long noncoding RNAs in digestive system cancers: Functional roles, molecular mechanisms, and clinical implications (Review) Oncol. Rep. 2016;36:1207–1218. doi: 10.3892/or.2016.4929. [DOI] [PubMed] [Google Scholar]
- 5.Zhang F., Zhang L., Zhang C. Long noncoding RNAs and tumorigenesis: genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol. 2016;37:163–175. doi: 10.1007/s13277-015-4445-4. [DOI] [PubMed] [Google Scholar]
- 6.D’Anzeo M., Faloppi L., Scartozzi M., Giampieri R., Bianconi M., Del Prete M., Silvestris N., Cascinu S. The role of micro-RNAs in hepatocellular carcinoma: from molecular biology to treatment. Molecules. 2014;19:6393–6406. doi: 10.3390/molecules19056393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beermann J., Piccoli M.T., Viereck J., Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 8.Terashima M., Tange S., Ishimura A., Suzuki T. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J. Biol. Chem. 2017;292:82–99. doi: 10.1074/jbc.M116.750950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumura K., Kawasaki Y., Miyamoto M., Kamoshida Y., Nakamura J., Negishi L., Suda S., Akiyama T. The novel G-quadruplex-containing long non-coding RNA GSEC antagonizes DHX36 and modulates colon cancer cell migration. Oncogene. 2017;36:1191–1199. doi: 10.1038/onc.2016.282. [DOI] [PubMed] [Google Scholar]
- 10.Cao C., Sun J., Zhang D., Guo X., Xie L., Li X., Wu D., Liu L. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology. 2015;148:415–426.e18. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Huang J.L., Zheng L., Hu Y.W., Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35:507–514. doi: 10.1093/carcin/bgt405. [DOI] [PubMed] [Google Scholar]
- 12.Wu J., Zhang J., Shen B., Yin K., Xu J., Gao W., Zhang L. Long noncoding RNA lncTCF7, induced by IL-6/STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. J. Exp. Clin. Cancer Res. 2015;34:116. doi: 10.1186/s13046-015-0229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F., Liu F., Pan W., Wang T.T., Zhou C.C. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Nayak R.R., Kearns M., Spielman R.S., Cheung V.G. Coexpression network based on natural variation in human gene expression reveals gene interactions and functions. Genome Res. 2009;19:1953–1962. doi: 10.1101/gr.097600.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng Y., Wang L., Chen D., Chang Y., Zhang M., Xu J.-J., Zhou R., Zhang Q.-Y. LAPTM4B: an oncogene in various solid tumors and its functions. Oncogene. 2016;35:6359–6365. doi: 10.1038/onc.2016.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu I., Kohno N., Tamaki K., Shono M., Huang H.W., He J.H., Yao D.F. Female hepatology: favorable role of estrogen in chronic liver disease with hepatitis B virus infection. World J. Gastroenterol. 2007;13:4295–4305. doi: 10.3748/wjg.v13.i32.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naugler W.E., Sakurai T., Kim S., Maeda S., Kim K., Elsharkawy A.M., Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 21.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu X., Feng Y., Zhang D., Zhao S.D., Hu Z., Greshock J., Zhang Y., Yang L., Zhong X., Wang L.P. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., He L., Du Y., Zhu P., Huang G., Luo J., Yan X., Ye B., Li C., Xia P. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Wang K., Long B., Zhou L.Y., Liu F., Zhou Q.Y., Liu C.Y., Fan Y.Y., Li P.F. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y., Chen N., Sun F., Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S.P., Xu H.X., Yu Y., He J.D., Wang Z., Xu Y.J., Wang C.Y., Zhang H.M., Zhang R.X., Zhang J.J. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431–42446. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., Zhang Q., Tian R., Wang Q., Zhao J.J., Iglehart J.D., Wang Z.C., Richardson A.L. Lysosomal transmembrane protein LAPTM4B promotes autophagy and tolerance to metabolic stress in cancer cells. Cancer Res. 2011;71:7481–7489. doi: 10.1158/0008-5472.CAN-11-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng F., Chen X., Song H., Lou G. LAPTM4B down regulation inhibits the proliferation, invasion and angiogenesis of HeLa cells in vitro. Cell. Physiol. Biochem. 2015;37:890–900. doi: 10.1159/000430216. [DOI] [PubMed] [Google Scholar]
- 30.Yang H., Xiong F., Wei X., Yang Y., McNutt M.A., Zhou R. Overexpression of LAPTM4B-35 promotes growth and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer Lett. 2010;294:236–244. doi: 10.1016/j.canlet.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Li S., Wang L., Meng Y., Chang Y., Xu J., Zhang Q. Increased levels of LAPTM4B, VEGF and survivin are correlated with tumor progression and poor prognosis in breast cancer patients. Oncotarget. 2017;8:41282–41293. doi: 10.18632/oncotarget.17176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong X., Tamura K., Kobayashi D., Ando N., Sumita K., Maehara T. LAPTM4B-35 is a novel prognostic factor for glioblastoma. J. Neurooncol. 2017;132:295–303. doi: 10.1007/s11060-017-2369-0. [DOI] [PubMed] [Google Scholar]
- 33.Kong F., Gao F., Chen J., Sun Y., Zhang Y., Liu H., Li X., Yang P., Zheng R., Liu G., Jia Y. Overexpressed LAPTM4B-35 is a risk factor for cancer recurrence and poor prognosis in non-small-cell lung cancer. Oncotarget. 2016;7:56193–56199. doi: 10.18632/oncotarget.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang F., Zhang L., Huo X.S., Yuan J.H., Xu D., Yuan S.X., Zhu N., Zhou W.P., Yang G.S., Wang Y.Z. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 35.Wang X., Sun W., Shen W., Xia M., Chen C., Xiang D., Ning B., Cui X., Li H., Li X. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J. Hepatol. 2016;64:1283–1294. doi: 10.1016/j.jhep.2016.01.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.