Abstract
N-(4-Chloro-3-(trifluoromethyl)phenyl)-2-ethoxybenzamide (CTB) is a small molecule that functions by altering the chromatin architecture to modulate gene expression. We report a new CTB derivative with increased solubility and demonstrate CTB's functionality by conjugating it on the recendy established NanoScript platform to enhance gene expression and induce stem cell differentiation. NanoScript is a nanoparticle-based artificial transcription factor that emulates the structure and function of transcription factor proteins (TFs) to effectively regulate endogenous gene expression. Modifying NanoScript with CTB will more closely replicate the TF structure and enhance CTB functionality and gene expression. To this end, we first conjugated CTB onto NanoScript and initiated a time-dependent increase in histone acetyltransferase activity. Next, because CTB is known to trigger the pathway involved in regulating Sox9, a master regulator of chondrogenic differentiation, we modifed a Sox9-specific NanoScript with CTB to enhance chondrogenic gene activity and differentiation. Because NanoScript is a tunable and robust platform, it has potential for various gene-regulating applications, such as stem cell differentiation.
Stem-cell-based therapies and cellular reprogramming hold immense potential for regenerative medicine. Significant effort has been invested in developing methods for effectively controlling the differentiation of stem cells into specific lineages. One promising approach for enhancing gene expression and inducing stem cell differentiation involves epigenetic modification.1 Epigenetic modification refers to changes in gene activity and expression by modifying chromatin structure rather than altering the DNA sequence.2 The eukaryotic genome is packed into chromatin, in which the DNA wraps around proteins called histones, and gene expression is regulated in part by the dynamics of chromatin structure.3 Moreover, epigenetic modification plays a critical role in enabling transcription factor proteins (TFs)— master regulators of gene expression—to regulate chromatin structure for inducing transcriptional activity.4
Specifically, epigenetic modification involves DNA histone deacetylation and acetylation, which regulate the chromatin structure. One class of epigenetic modification molecules, histone acetyltransferases (HATs), function by weakening the association of DNA with histones, making the DNA more accessible for transcription and gene activation (Figure 1a).5 Of the many FLAT activator small molecules, N-(4-chloro-3-(trifluoromethyl)phenyl)-2-ethoxybenzamide (CTB), is known to be effective in regulating HAT activity.6 CTB functions by triggering the p300 signaling pathway, which influences chromatin structure by facilitating the assembly of chromatin remodeling proteins to DNA binding sites (Figure 1a).7 As a result, CTB enhances p300 HAT-dependent transcriptional activation and regulates expression of p300-related genes.8 One gene regulated by the p300 signaling pathway is Sox9,9 which plays a critical role for inducing stem cell chondrogenesis.10
Figure 1.
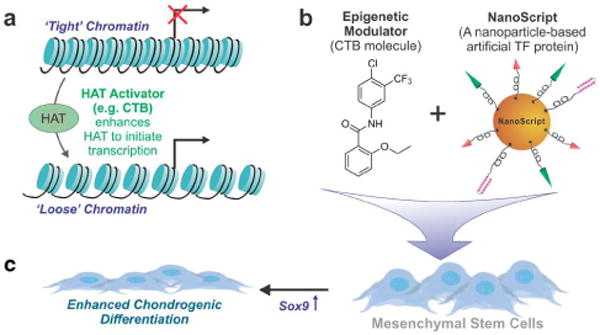
Schematic representation of CTB's function and role in initiating chondrogenesis. (a) If DNA (black line) is tightly coiled around histone proteins (blue cylinders), transcriptional activity and subsequent gene expression are inhibited. Activation of histone acetyltransferase (HAT) via the CTB molecule loosens the DNA making target binding sites available for transcription. (b) CTB is functionalized on the NanoScript platform, and the resulting construct is delivered to adipose-derived mesenchymal stem cells (ADMSCs) to enhance overexpression of targeted genes. The NanoScript construct is also modified with additional small molecules that target and overexpress the Sox9 gene. (c) Activation of the Sox9 gene in ADMSCs guides enhanced differentiation into the chondrogenic lineage.
We recently demonstrated a new gene-regulating platform called NanoScript, which mimics the fundamental structure and function of TFs.11 NanoScript effectively induced gene expression of both a reporter plasmid and endogenous genes on native DNA in a nonviral manner, establishing it as an attractive platform for applications requiring gene regulation.11 NanoScript was designed to mimic the two fundamental domains on TFs: the activation domain (AD) and the DNA binding domain (DBD). The DBD seeks and binds to target gene sequences on DNA, while the AD recruits the transcriptional basal machinery to initiate transcriptional activity. These two domains function synergistically to induce targeted gene expression. Through a simple modification of the DBD, NanoScript can be designed to target and activate the Sox9 gene. Moreover, studies have shown CTB to trigger the p300 pathway, and the p300 pathway to recruit the transcriptional basal machinery to initiate transcriptional activity.13 Thus, we hypothesize that by modifying NanoScript with a CTB molecule, an AD molecule, and a Sox9-targeting molecule, we can utilize the NanoScript platform to achieve enhanced Sox9 expression in stem cells to accelerate their chondrocyte differentiation.
We combined the NanoScript platform with an epigenetic modulator (i.e., CTB) and a Sox9-targeting molecule to activate Sox9 expression in adipose-derived mesenchymal stem cells (ADMSCs) to induce enhanced chondrogenic differentiation (Figure 1b). First, we developed a new functional CTB derivative modified with a thiol-PEG linker molecule to increase solubility and enable conjugation on NanoScript. Second, the NanoScript platform was modified with CTB, whereupon it more closely mimicked TF protein structure because TFs contain an epigenetic modification domain. Finally, when the CTB-modified NanoScript specific for Sox9 (termed NanoScript-Sox9) was delivered to ADMSCs, enhanced gene expression of Sox9 and subsequent chondrogenic differentiation were observed.
The NanoScript platform was designed and constructed to replicate the fundamental structure of multidomain TFs. Specifically, to replicate the structure of TFs, we designed NanoScript with six components: (1) the CTB molecule that mimics the epigenetic modification domain to initiate HAT activity for enhanced gene expression,6 (2) a hairpin polyamide molecule that mimics the DBD to target and bind to the Sox9 gene,12,14 (3) a transactivation peptide that mimics the AD to initiate transcriptional activity,15 (4) a membrane-penetrating peptide,16 (5) polyethleyne glycol (PEG)-based molecules to increase stability and solubility,17 and (6) a gold nanoparticle (AuNP) to tether these molecules onto a single construct.18 Assembling these individual small molecules on an AuNP enables the resulting NanoScript platform to emulate TF protein structure and function.
The CTB molecule interacts with the p300 protein, which in turn interacts with lysine amino acids on histone proteins. This results in reduced electrostatic attraction between the histone and the DNA, causing the DNA to decrease its affinity, thus exposing more binding sites for transcription. This CTB binds to the target through hydrogen-bonding and hydrophobic interactions, with the -Cl and -CF3 positions having the strongest influence on binding affinity (Figure 2a).8 Moreover, the ortho position on the aromatic hydrocarbon does not play a critical role in overall binding.19 Thus, we modified the ortho position with a PEG-thiol chain to increase solubility in physiological environments and enable conjugation onto the AuNPs (Figure 2a). In doing so, we synthesized a CTB analogue.
Figure 2.
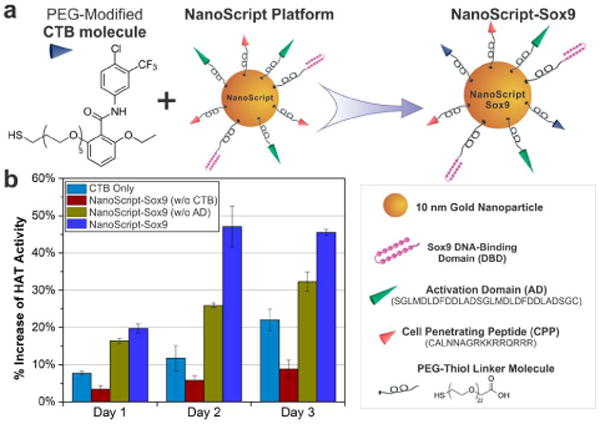
CTB's function is enhanced when conjugated on NanoScript. (a) The CTB molecule is modified with a thiol-PEG chain and functionalized on NanoScript. The remaining components on NanoScript include the AD, which recruits endogenous factors to initiate transcription, and the DBD (refer to Figure S1 for structure details), which targets and binds to the target Sox9 sequence on DNA (b) The HAT activity of this NanoScript-Sox9 construct was shown to increase across three time points (days 1–3). Controls with only the AD (without CTB), only CTB (without AD), or unconjugated CTB in media showed comparatively less HAT activity. All conditions and controls were transfected with 1 nM of the constructs. The increase of HAT activity for all samples was determined by normalizing to the untreated control. Standard error is from three independent trials.
We next synthesized the hairpin polyamide specific for the Sox9 gene. The hairpin polyamide, which acts as the DBD, consists of imidazole and pyrrole motifs that bind to the G-C and A-T base pairs on the DNA respectively, with nanomolar affinity.20 The Sox9 gene has a consensus promoter sequence of 5′-ACAATGG-3′,21 so we designed a hairpin polyamide to target this sequence (Figure S1a). Reported protocols using solid-phase synthesis enabled us to build the hairpin with a sequence of PyPyPy-β-PyImPy-γ-PyPyPy-β-PyImIm-β-Dp-NH2, complementary to the target Sox9 gene.22 In vitro binding association studies were performed using surface plasmon resonance to verify the binding affinity of the Sox9 polyamide. Results showed a high sequence-specific binding affinity of the Sox9 polyamide to the target sequence as compared to controls, including scrambled polyamides and scrambled DNA sequences (Figure S1b).
The third molecule is the transactivation peptide, which emulates the AD and recruits the transcriptional basal machinery required to initiate transcription.23 This peptide is a potent inducer of transcriptional activity.15 The fourth molecule is the cell-penetrating peptide that enables plasma and nuclear membrane penetration. Specifically, we used a modified TAT peptide to shuttle AuNPs into the nucleus.16
The hairpin polyamide, transactivation peptide, and TAT peptide were conjugated to a PEG-thiol molecule (21-chain PEG) via EDC/NHS coupling, which enables functionalization onto AuNPs. AuNPs were chosen because they are highly biocompatible and nontoxic in cells and, most importantly, because their multifunctional surface enables conjugation of multiple molecules on a single NP.18 After 10 nm AuNPs were functionalized with these PEG-terminated molecules, along with the CTB, the resulting NanoScript platform (NanoScript-Sox9) targeted the Sox9 gene (Figure 2a). Characterization of NanoScript-Sox9 revealed a successive shift in the UV absorbance, indicative of functionalization (Figure S2), and the hydrodynamic diameter was found to be 57.9 nm (Figure S3).
After constructing the NanoScript-Sox9, we first wanted to ensure successful nuclear localization, which is essential because transcriptional activity occurs only in the nucleus. Hence, the NanoScript-Sox9 was labeled with a fluorescent dye and transfected into ADMSCs. After 24 h, fluorescence imaging revealed that NanoScript-Sox9 was able to enter and localize within the nucleus of 51.6% of the total cells (Figure S4a,b).
Next, we evaluated the function of the CTB molecule on the NanoScript-Sox9. It is well known that CTB initiates HAT activity.6 The effect of PEG modification on CTB was tested, and both the unmodified CTB and PEG-modified CTB showed similar HAT activity (Figure S5), indicating that the modified CTB retained its functionality. Moreover, the function of CTB conjugated to the NP was tested and revealed that increasing CTB amounts on the NP resulted in increasing HAT activity (Figure S6). We then tested various concentrations of Nano-Script-Sox9 and observed a concentration-dependent HAT activity increase (Figure S7). Next, NanoScript-Sox9-induced HAT activity was evaluated at various time points. Remarkably, the NanoScript-Sox9 with both the AD and CTB induced maximal HAT activity at all the time points compared to the controls, which included a free CTB molecule and NanoScript-Sox9 that lacked either the CTB or AD (Figure 2b). While this result shows induction of HAT activity when CTB or AD alone is on NanoScript, the combined effect of CTB and AD on a single NanoScript shows maximum increase in HAT activity.
After determining that both CTB and AD are required on a single NP to induce maximal HAT activity, we focused on overexpressing the Sox9 gene in ADMSCs to enhance chondrogenic differentiation. ADMSCs were chosen as a model cell line because they are excellent multipotent stem cells with a capacity to differentiate into muscle, cartilage, bone, and neuronal cells.24 Moreover, Sox9 was chosen as a target gene because it is known to control chondrogenic differentiation and its expression is regulated by both the p300 pathway and conventional ADs.9 Thus, we expected to observe an enhancement of Sox9 expression and other chondrogenic-related genes when NanoSdpt-Sox9 was delivered to ADMSCs.
NanoScript-Sox9 and chondrogenic differentiation media were incubated with ADMSCs on days 0 and 2. After 7 days, the ADMSCs were evaluated for expression of distinct chondrogenic factors, Aggrecan and Collagen II, which are prominent chondrogenic markers that are expressed based on the progression of chondrogenic differentiation. After the ADMSCs were fixed and stained, fluorescence imaging revealed that the sample treated with NanoScript-Sox9 (having both CTB and AD) showed enhanced expression of Aggrecan and Collagen II as compared to controls (Figure 3a,b). Furthermore, the fluorescence intensity of Aggrecan and Collagen II expression from these images showed a remarkable trend that confirmed maximal expression when both CTB and AD are present on NanoScript-Sox9 (Figure 3c,d), with high cell viability (Figure S8). Although all conditions and controls were treated identically with differentiation media, maximum expression was observed when CTB and AD were both present on NanoScript-Sox9, thus suggesting enhanced chondrogenic differentiation.
Figure 3.
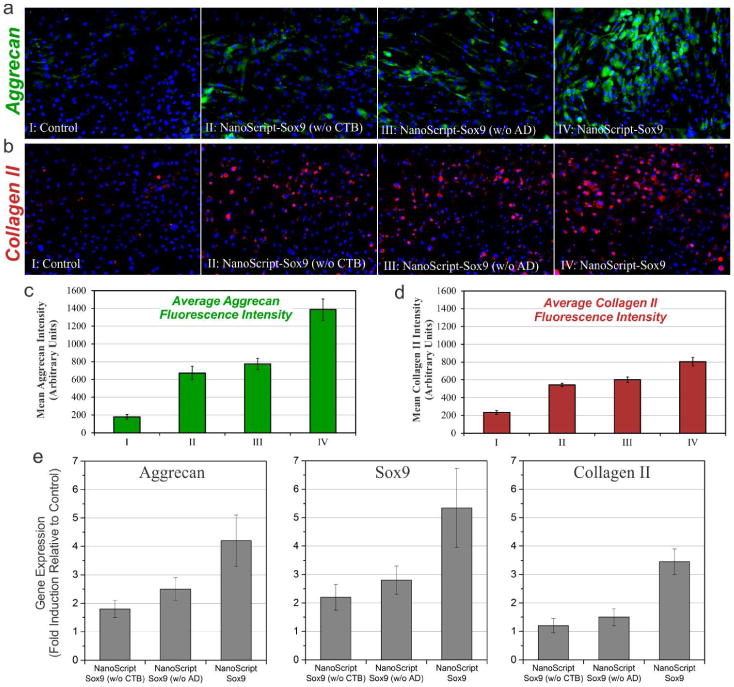
Synergistic effect of CTB and AD on NanoScript enhances expression of chondrogenic markers. After 7 days post-transfection with NanoScript-Sox9, ADMSCs were evaluated for their expression of distinct chondrogenic markers, (a) Aggrecan (green) and (b) Collagen II (red) (scale bar = 50 μm). The intensity levels of both chondrogenic markers, (c) Aggrecan and (d) Collagen II, in the fluorescence images were quantified and revealed a similar trend. (e) After 7 days post-treatment with NanoScript-Sox9, qPCR analysis revealed expression of distinct chondrogenic genes, as compared to the untreated control. Gene expression levels were normalized to GAPDH. Error bars are from three independent experiments.
To further quantify the genetic expression patterns of chondrogenic genes in NanoScript-Sox9-treated ADMSCs, we performed qPCR Analysis showed that NanoScript-Sox9 enhanced expression of all three distinct genes, Sox9, Aggrecan, and Collagen II, as compared to controls and NanoScript lacking either the AD or CTB (Figure 3e). Another control experiment, incubating NanoScript-Sox9 with free CTB, suggested that CTB has maximal function when attached to the AuNP (Figure S9). Furthermore, expression levels of housekeeping genes remained unchanged as compared to the control (Figure S10). Even though NanoScript-Sox9 was designed to target the Sox9 gene, which showed the highest expression among the three genes, Sox9 is sufficient to trigger the chondrogenic pathway, including the Aggrecan and Collagen II genes.
Taken together, the results from our three experiments (HAT assay, immunofluorescence, and qPCR) indicate that the presence of both CTB and AD on NanoScript initiates maximal HAT activity and expression of chondrogenic genes. One possible hypothesis for this is that the CTB and AD initiate HAT activity through different pathways: previous reports showed that CTB initiates HAT activity through the p300 pathway,6 while the AD (specifically the transactivation peptide) initiates HAT activity by recruiting factors such as SAGA and NuA4.23,25 Hence, it is possible that because two gene activation pathways were induced, we observed an enhanced synergistic gene expression when both CTB and AD were present on NanoScript. However, the exact interplay between these signaling pathways is unknown, and we will attempt to elucidate the mechanisms in future studies.
In summary, the recently developed NanoScript platform was modified with an epigenetic modification molecule, CTB, which regulates histone proteins and enhances gene expression through the HAT-dependent p300 pathway. Addition of CTB not only moved NanoScript a step closer toward mimicking natural transcription factors but also enhanced the gene expression capabilities of NanoScript. The role and function of CTB on NanoScript were evaluated and revealed that CTB is effective in inducing HAT activity. Moreover, we observed that NanoScript with both AD and CTB showed the greatest HAT activity, possibly because the AD and CTB triggered two distinct pathways for regulating HAT activity. When NanoScript-Sox9 was incubated with ADMSCs, we confirmed through fluorescence imaging and qPCR that NanoScript-Sox9 activates expression of chondrogenic markers.
Our NanoScript-based transcriptional activator can have a significant impact in the fields of stem cell biology and cellular reprogramming. Through this proof-of-concept demonstration, we have established that NanoScript is an effective and tunable platform that can manipulate targeted genes that are critical for stem cell differentiation in a nonviral manner. Moreover, by modifying NanoScript with epigenetic modification molecules such as HAT activators (e.g., CTB), we optimized it to be more effective in regulating gene expression. As a result, we are confident that NanoScript can be utilized for other applications requiring nonviral and effective gene regulation in stem cell biology and cellular reprogramming.
Supplementary Material
Acknowledgments
We thank Anthony Dudzinski, Ruth Goldman, and Mike Moeller (American CryoStem Corp.) for stem cells and media. K.-B.L. acknowledges the NIH Director's Innovator Award (1DP20D006462-01), NIH R21 (1R21NS085569-01), New Jersey Commission on Spinal Cord (CSR13ERG005), NSF CHE-1429062, CBET-1236508, Busch Biomedical Grant Program, and the Collaborative Research Travel Grant (CRTG) from the Burroughs Wellcome Fund for funding.
Footnotes
The authors declare no competing financial interest.
Supporting Information: Sox9 polyamide structure and characteristics, absorbance, NanoScript-Sox9 diameter, cell viability, effect of CTB modification and ratios, and nuclear localization images. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Spivakov M, Fisher AG. Nat Rev Genet. 2007;8:263. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 2.Portela A, Esteller M. Nat Biotechnol. 2010;28:1057. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 3.Apostolou E, Hochedlinger K. Nature. 2013;502:462. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaret KS, Carroll JS. Gene Dev. 2011;25:2227. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selvi BR, Cassel JC, Kundu TK, Boutillier AL. BBA-Gene Regul Mech. 2010;1799:840. doi: 10.1016/j.bbagrm.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. J Biol Chem. 2003;278:19134. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- 7.Chan HM, La Thangue NB. J Cell Sci. 2001;114:2363. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 8.Mantelingu K, Kishore AH, Balasubramanyam K, Kumar GVP, Altaf M, Swamy SN, Selvi R, Das C, Narayana C, Rangappa KS, Kundu TK. J Phys Chem B. 2007;111:4527. doi: 10.1021/jp067655s. [DOI] [PubMed] [Google Scholar]
- 9.Furumatsu T, Tsuda M, Yoshida K, Taniguchi N, Ito T, Hashimoto M, Ito T, Asahara H. J Biol Chem. 2005;280:35203. doi: 10.1074/jbc.M502409200. [DOI] [PubMed] [Google Scholar]
- 10.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombragghe B. Mol Cell Biol. 1997;17:2336. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel S, Jung D, Yin PT, Carlton P, Yamamoto M, Bando T, Sugiyama H, Lee KB. ACS Nano. 2014;8:8959. doi: 10.1021/nn501589f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez-Martínez JA, Peterson-Kaufman KJ, Ansari AZ. BBA-Gene Regul Mech. 2010;1799:768. doi: 10.1016/j.bbagrm.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh A, Varga J. J Cell Physiol. 2007;213:663. doi: 10.1002/jcp.21162. [DOI] [PubMed] [Google Scholar]
- 14.Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Nature. 1997;387:202. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 15.Nyanguile O, Uesugi M, Austin DJ, Verdine GL. Proc Natl Acad Sci U S A. 1997;94:13402. doi: 10.1073/pnas.94.25.13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krpetić Ž, Saleemi S, Prior IA, Sée V, Qureshi R, Brust M. ACS Nano. 2011;5:5195. doi: 10.1021/nn201369k. [DOI] [PubMed] [Google Scholar]
- 17.Rahme K, Chen L, Hobbs RG, Morris MA, O'Driscoll C, Holmes JD. RSC Adv. 2013;3:6085. [Google Scholar]
- 18.Dykman L, Khlebtsov N. Chem Soc Rev. 2012;41:2256. doi: 10.1039/c1cs15166e. [DOI] [PubMed] [Google Scholar]
- 19.Devipriya B, Parameswari AR, Rajalakshmi G, Palvannan T, Kumardhas P. Indian J Biochem Biol. 2010;47:364. [PubMed] [Google Scholar]
- 20.Dervan PB, Edelson BS. Curr Opin Struct Biol. 2003;13:284. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 21.Mertin S, McDowall SG, Harley VR. Nucleic Acids Res. 1999;27:1359. doi: 10.1093/nar/27.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morinaga H, Bando T, Takagaki T, Yamamoto M, Hashiya K, Sugiyama H. J Am Chem Soc. 2011;133:18924. doi: 10.1021/ja207440p. [DOI] [PubMed] [Google Scholar]
- 23.Mapp AK, Ansari AZ. ACS Chem Biol. 2007;2:62. doi: 10.1021/cb600463w. [DOI] [PubMed] [Google Scholar]
- 24.(a) Gimble JM, Bunnell BA. Circ Res. 2007;100:1249. doi: 10.1161/01.RES.0000265074.83288.09. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Goudenege S, Pisani DF, Wdziekonski B, Di Santo JP, Bagnis C, Dani C, Dechesne CA. Mol Ther. 2009;17:1064. doi: 10.1038/mt.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeda K, Steger DJ, Eberharter A, Workman JL. Mol Cell Biol. 1999;19:855. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


