Abstract
The ancient phylum Actinobacteria is composed of phylogenetically and physiologically diverse bacteria that help Earth’s ecosystems function. As free-living organisms and symbionts of herbivorous animals, Actinobacteria contribute to the global carbon cycle through the breakdown of plant biomass. In addition, they mediate community dynamics as producers of small molecules with diverse biological activities. Together, the evolution of high cellulolytic ability and diverse chemistry, shaped by their ecological roles in nature, make Actinobacteria a promising group for the bioenergy industry. Specifically, their enzymes can contribute to industrial-scale breakdown of cellulosic plant biomass into simple sugars that can then be converted into biofuels. Furthermore, harnessing their ability to biosynthesize a range of small molecules has potential for the production of specialty biofuels.
Keywords: actinomycetes, biotechnology, biofuels, cellulases, symbiosis, Streptomyces
INTRODUCTION
Actinobacteria are ubiquitous and one of the most diverse groups of bacteria in nature. Its members range from anaerobic, unicellular organisms to aerobic, filamentous, and spore-forming lineages. Alone, the genus Streptomyces accounts for almost 5% of the ~16,000 described bacterial species (http://www.bacterio.net/). Actinobacteria have made a substantial positive contribution to human health; they are the producers of many compounds that are used as important drugs, including most antibiotics (58). In addition, the phylum includes important pathogens, such as Mycobacterium tuberculosis, the causal agent of one of the most devastating diseases in human history, tuberculosis. Largely because of these biomedical impacts, Actinobacteria have avoided the obscurity relegated to most bacterial phyla, receiving substantial scientific study of their genetics, molecular biology, and biochemistry.
In addition to their important influence on human health, Actinobacteria have key ecological roles. Before focusing on antibiotic discovery, Nobel laureate Selman Waksman’s work on soil bacteria and their impact on agricultural productivity was among the first to implicate Actinobacteria as important contributors to the process of plant biomass decomposition (58, 146). More recently, Actinobacteria have been revealed as widespread symbionts of eukaryotes, helping herbivores gain access to plant biomass as nutritional mutualists and producing natural products as defensive mutualists (e.g., 18, 33, 38, 65). Studies of Actinobacteria as defensive mutualists have led to the discovery of new antibiotics with potential pharmaceutical applications (97, 106, 117), renewing recognition of the value in understanding the ecology of Actinobacteria for drug discovery (e.g., 12, 122) and hearkening back to Waksman’s journey to the discovery of streptomycin.
We suggest, much like in drug discovery, the ecology and evolution of Actinobacteria position this phylum to contribute to the development of a sustainable bioenergy industry. Specifically, as Actinobacteria are implicated as important decomposers of plant material in nature, their cellulolytic enzymes can be used to more efficiently break down plant biomass into simple sugars, which can then be used for the production of biofuels. In addition, the diverse biosynthetic capacity of Actinobacteria, which evolved to mediate their environmental interactions, could be leveraged to produce a range of bioproducts, including specialty biofuel compounds. Here, we review our current understanding of the ecology and evolution of Actinobacteria. Then, within this framework, we discuss the potential application of Actinobacteria in bioenergy.
EVOLUTION AND GENOMICS OF ACTINOBACTERIA
The evolutionary origin of the Actinobacteria is ancient, dating back to ~2,700 million years ago, predating oxygenation of the atmosphere (~2,300 million years ago) (Figure 1) (8, 150). 16S rRNA gene sequencing, multilocus phylogenetics, and phylogenomics support a shared common ancestor for Actinobacteria, Cyanobacteria, and Deinococcus (8, 52). Together, these three lineages share a most recent common ancestor with the phylum Firmicutes, with divergence estimated at more than 3,000 million years ago (8). Consistent with evolving in an anoxic atmosphere, the earliest Actinobacteria were obligate anaerobes, were not yet filamentous or able to form spores, and instead had simple rod/coccus morphology (26). The lineage Actinobacteria subsequently diverged into at least six classes and 53 families (52). The ability to grow as filaments and produce spores evolved in the more recently derived lineages but has been secondarily lost in a few groups (26).
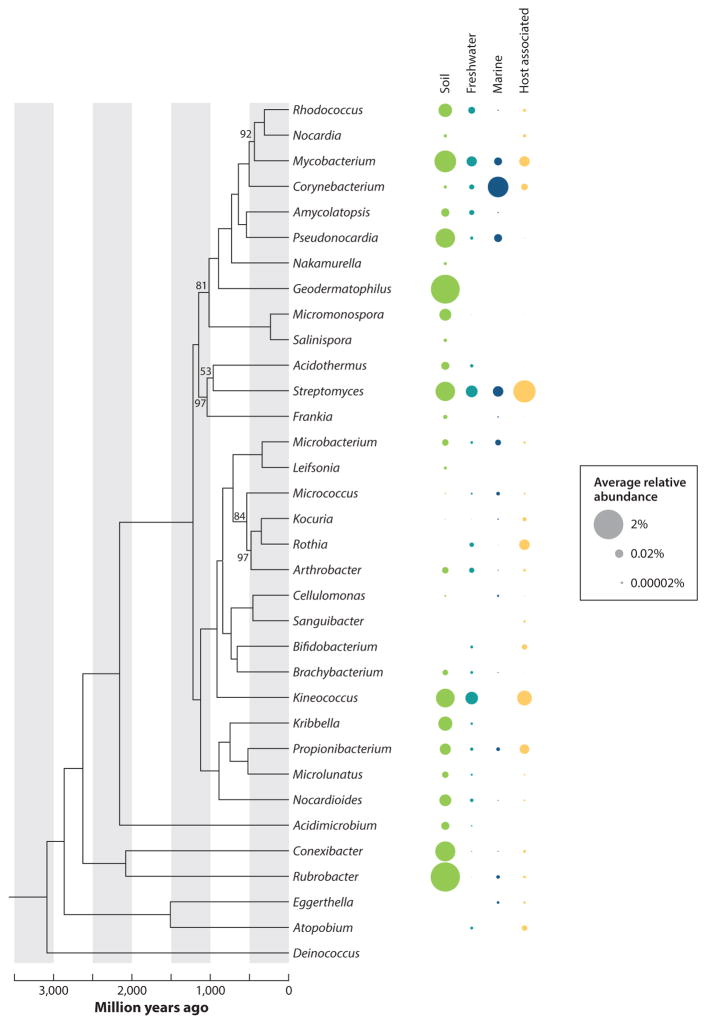
Figure 1.
Actinobacterial phylogenomics and relative abundance across ecosystems. Forty-six publicly available 16S rRNA gene datasets, spanning four ecosystems, were analyzed to determine the average relative percentage of reads assigned to each actinobacterial genus (93) (Supplemental Table 1; follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org). Phylogenetic analysis was performed using concatenated alignments from 94 conserved TIGRFAM protein families in representative complete actinobacterial genomes (Supplemental Table 2), with the outgroup Deinococcus. Sequences were aligned with MAFFT 7.221, and the phylogeny was generated using RAxML 8.1.24 (67, 124). Molecular clock calculations were performed with RelTime using the origin of life (3,500–3,800 million years ago), the origin of Cyanobacteria (2,500–3,500 million years ago), and the Escherichia-Salmonella divergence time (50–150 million years ago) as calibration points (133). Statistical support for the phylogeny, based on a sequence resampling bootstrap analysis, is shown on nodes with less than 100% support.
In general, the genomes of Actinobacteria are large, relative to other phyla of bacteria, with an average genome size of over 5 megabases (Mb). However, genome sizes vary widely, from less than 1 Mb in some Tropheryma species to over 12 Mb in a few Streptomyces species. In general, the genomes have a high guanine-plus-cytosine (G+C) content, with the G+C content of some genera, such as Streptomyces and Pseudonocardia, more than 70%. However, G+C content is less than 50% in Gardnerella vaginalis and Tropheryma whipplei (2, 6). Of those analyzed, all Actinobacteria have a single chromosome, but the presence of large plasmids is not uncommon. The ancestral state is a circular chromosome, but the genera Gordonibacter, Kineococcus, Rhodococcus, and Streptomyces all have linear chromosomes, which appear to have independently evolved in each genus.
Genome content in bacteria is shaped by the interplay between vertical inheritance (i.e., clonal reproduction) with gene loss and acquisition, the latter either involving the genesis of new genes within a lineage or horizontal gene transfer (HGT) from other lineages. Our current understanding of the evolutionary forces shaping gene loss and gene origin in Actinobacteria is limited. Recent genomic analyses of 120 Streptomyces genomes indicate that, at least within this genus, HGT is relatively rare and that long-term retention of horizontally transferred genes occurs several orders of magnitude less frequently than the origin of point mutations (B.R. McDonald & C.R. Currie, unpublished information). Nevertheless, HGT enables adaptive divergence of populations through the introduction of novel genetic diversity. For example, virulence operons in M. tuberculosis and plant pathogenic Streptomyces species appear to have been acquired through HGT (69, 109). Furthermore, HGT has been shown as a major driver of secondary metabolite diversity in the marine actinobacterial genus Salinispora (161).
Coevolution is another evolutionary force involved in shaping the biology of Actinobacteria. Coevolution inherently recognizes the importance of species interactions in the evolution and diversity of lineages (134). Thus, given that Actinobacteria typically occur either within complex communities of interacting microbes or as symbionts of eukaryotic hosts (see “Ecology of Actinobacteria,” below), reciprocal selection is likely to influence the evolution of these lineages. One of the few studied examples of coevolution in Actinobacteria is the Pseudonocardia-ant defensive mutualism (see the sidebar, Coevolution of Actinobacteria in the Fungus-Growing Ant System and Figure 2a). In this example, as well as other defensive mutualisms, Actinobacteria provide antimicrobial protection to a host organism, and species interactions are mediated by secondary metabolites that are likely influenced and shaped by coevolution. Further studies are needed, both on how chemical diversification is shaped by coevolutionary forces and on the nutritional interactions between Actinobacteria and their hosts.
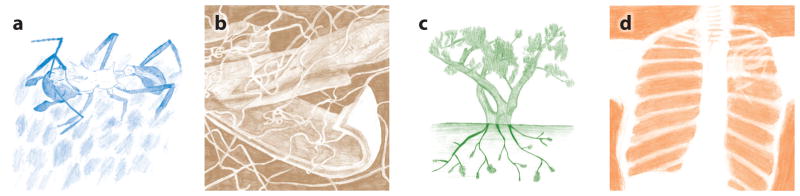
Figure 2.
Actinobacteria in nature. (a) An Acromyrmex leaf-cutter ant in her fungus garden covered in white Pseudonocardia bacteria. (b) A Streptomyces strain growing on decaying plant biomass. (c) Frankia nodules on the roots of a Russian olive tree. (d) X-ray showing a healthy lung on the left and Mycobacterium tuberculosis growth on the right. Illustrations courtesy of Gina Lewin.
COEVOLUTION OF ACTINOBACTERIA IN THE FUNGUS-GROWING ANT SYSTEM.
Some lineages of Pseudonocardia are associated with fungus-growing ants (tribe Attini), with the bacterium residing on the ants’ exoskeleton (see Figure 2a) (38). These Pseudonocardia produce antimicrobial compounds that help defend the ants’ food source, a fungal cultivar, from specific pathogens (38, 97, 152). The bacterium is vertically inherited (38), and there is a critical window in which the ants must be exposed to the bacterium for acquisition to occur (87). Specialized glands and structures on the ant exoskeleton supply nutrients to the bacterium, indicating evolutionary selection on the ant for hosting Pseudonocardia (37, 126). Likewise, molecular phylogenetic studies of symbiotic Pseudonocardia reveal congruence with the host ant phylogeny (22, 23). Additional complexity is seen in the population dynamics of Pseudonocardia, which are correlated to that of a system-specific pathogen, Escovopsis (24, 36, 38). Indeed, studies examining pair-wise interactions between Pseudonocardia and Escovopsis strains show varying degrees of pathogen inhibition, indicating diversity in secondary metabolites produced by Pseudonocardia as well as variation in pathogen resistance (22, 23, 38). These findings support coevolution shaping the dynamics and biology of this system, which likely has generated and influenced the diversity of secondary metabolism of the bacterial symbiont. Similar dynamics are likely common across defensive mutualisms, such as in that of solitary digger wasps (tribe Philanthini), which engage in a defensive mutualism with Streptomyces strains to offer antimicrobial protection during larval development (66, 72). Potentially, similar dynamics also occur in nutritional mutualisms.
ECOLOGY OF ACTINOBACTERIA
Historically, Actinobacteria were largely viewed as soil bacteria but are now recognized as being cosmopolitan; they are found in virtually all ecosystems, with a distribution covering most of the planet. As free-living microbes, they are abundant in soil environments, especially those with a higher pH (77), and in marine and freshwater ecosystems (Figures 1 and 2). Actinobacteria are also associated with eukaryotic hosts in diverse niches, such as the exoskeleton of some tropical ants, the lungs and skin of mammals, and the roots and inner tissues of plants (Figures 1 and 2). Some genera, including Streptomyces, Kineococcus, and Mycobacterium, span diverse ecosystems from soil to marine to freshwater environments. Others, such as Atopobium, Bifidobacterium, Kocuria, and Rothia, are mostly found in host associations (Figure 1) (Supplemental Table 1; follow the Supplemental Materials link from the Annual Reviews home page at http://www.annualreviews.org). The diversity of niche utilization in Actinobacteria mirrors their physiological capability to grow in a range of conditions, as supported by culture- and genome-based analyses. Nevertheless, our understanding of the ecological roles of Actinobacteria in many of these systems remains poorly resolved. Here, we explore their ecology, focusing on cases where the functional role of Actinobacteria has been elucidated with the understanding that Actinobacteria are critical for a wide range of ecosystem services and host interactions.
Role of Free-Living Actinobacteria in Carbon Cycling
The largest terrestrial source of organic carbon is the network of energy-rich but recalcitrant polymers in the plant cell wall (156). The primary polymer in the plant cell wall is cellulose, an insoluble crystal of β-1,4-linked glucose molecules, which is surrounded by a complex mesh of hemicellulose and lignin (99). Hemicellulose is a heterogeneous polymer composed of five- and six-carbon sugars, such as xylose, arabinose, mannose, and uronic acids. Lignin is an amorphous polymer formed from variable, highly cross-linked phenolic alcohols. In contrast to most other phyla of bacteria, Actinobacteria are enriched in the full suite of enzymes necessary to deconstruct plant biomass (14). This enrichment in enzymes, combined with the abundance of Actinobacteria in soil and the ability of some isolates to grow on plant biomass or its constituent polymers, supports their contribution to plant biomass degradation in soil in conjunction with fungi and other organisms (41, 77, 90). Several recent in vivo studies provide further validation (54); for example, in Arctic peat soil, Tveit and colleagues (138) found that most cellulase and hemicellulase RNA transcripts were assigned to Actinobacteria.
Bacteria in the genus Streptomyces are among the most abundant organisms in soil (2a, 62), and the ability of some isolates to grow on cellulose was first identified ~100 years ago (145). Subsequently, a variety of Streptomyces strains isolated from soil have been shown to grow on cellulose (16, 120, 144), hemicellulose (45, 61), and potentially even lignin (34, 163) (Figure 2b). However, the majority of strains tested cannot grow on crystalline cellulose or hemicellulose, suggesting that most Streptomyces species in the soil only use the breakdown products of plant cell wall polymers (18). Other genera of Actinobacteria have also been implicated in plant biomass degradation in the soil, including Cellulomonas, Nocardia, and Micromonospora, again on the basis of their high prevalence in soil and the ability of some isolates to grow on plant biomass-derived substrates (57, 90, 163).
Free-living Actinobacteria contribute to carbon cycling in other environments enriched in decaying plant material, including leaf litter, compost, and manure (91, 116). In many composting systems, genera of Actinobacteria, including Thermobifida, Saccharomonospora, and Thermobispora, are abundant community members during the thermophilic stage (56, 127, 159). For example, metagenomic analysis of rice straw within composting cow manure showed that Actinobacteria were abundant and encoded more cellulases than any other phylum (149). Similar to compost, leaf litter is also host to a high abundance of Actinobacteria, and metagenomic sequencing identified cellulases from Streptomyces and Thermobispora genera, as well as from many other Actinobacteria (13, 70, 89, 162). Micromonospora strains are common in lake sediment, another environment rich in decaying plant material, and isolates in this genus have been shown to degrade cellulose and hemicellulose (35, 48).
Actinobacteria are also implicated in the cycling of a variety of other carbon sources. This includes chitin, the second most abundant organic carbon source in nature, which is a structural polymer in the cell wall of fungi and in the exoskeletons of invertebrates. As with cellulose, few organisms can degrade chitin, but some Actinobacteria, including most Streptomyces and Arthrobacter species, have this ability (68, 92). The Ac1 Actinobacteria, dominant organisms in freshwater, are also thought to degrade chitin or uptake degradation by-products of chitin from fungi, crustaceans, and diatoms (9, 46). Additionally, some Actinobacteria are able to degrade hydrocarbons and organic contaminants (5, 85). For example, Rhodococcus can degrade diverse natural and synthetic molecules, including chlorinated or aromatic hydrocarbons, nitroaromatics, sulfonated azo dyes, and pesticides (10).
Role of Actinobacteria in Host Associations
Although Actinobacteria are still generally viewed as free-living bacteria, the recent increased focus on symbiotic associations has revealed their common association with eukaryotic hosts (18, 49, 84). They have broadly been identified in association with animals, both in their guts and on external surfaces. For example, the genera Bifidobacterium and Corynebacterium are associated with animals ranging from insects to mammals, including humans (137, 143). Additionally, Actinobacteria are associated with plants, including in the rhizosphere and as endophytes (32, 84, 115). Efforts are underway to determine the ecological roles of these bacteria, from aiding hosts with nutrient acquisition to serving as defensive mutualists.
A range of animals and plants rely on Actinobacteria to supplement their diets or help digest complex food sources. Surveys of insect guts, using 16S rRNA gene sequencing, indicate that Actinobacteria are more abundant in the guts of insects that consume detritus or decaying wood, especially termites (31, 64). Strains that can degrade plant biomass have been isolated from termites and from a range of other insects associated with wood niches, including Sirex woodwasps and mountain pine beetles (Figure 2b) (1, 18, 98, 151). Other insects, especially some true bugs, rely on Actinobacteria to provide nutritional supplementation (65). For example, Rhodococcus rhodnii provides B vitamins to Rhodnius prolixus, the insect vector of Chagas disease (20), and likewise, Coriobacteriaceae strains provide B vitamins to Pyrrhocoridae (Hemiptera), such as firebugs (110). In the mammalian gut, Bifidobacterium species aid in digestion of complex carbohydrates, including plant-derived oligosaccharides and gastric mucin (94, 105). The most clearly defined nutritional association between plants and Actinobacteria is the association between nitrogen-fixing Frankia strains and actinorhizal plants (Figure 2c) (30), which enables plants to colonize early during primary succession (78, 118, 141).
Actinobacteria also function as defensive mutualists for a number of eukaryotic hosts. A well-studied example of this relationship is in fungus-growing ants (see the sidebar, Coevolution of Actinobacteria in the Fungus-Growing Ant System and Figure 2). Similarly, solitary digger wasps, commonly known as beewolves, inoculate Streptomyces symbionts on their larval brood cells to protect cocoons from microbial infection (66), and southern pine beetles harbor a symbiotic Streptomyces species to protect their fungal food source (117). Plants are protected by the genera Streptomyces and Micromonospora, as well as by other Actinobacteria in the rhizosphere, which can help inhibit plant pathogens through the production of secondary metabolites (57, 81, 115). Also, endophytic Actinobacteria suppress pathogens and activate the plant immune system (32, 50). In addition to the nutritional roles discussed above, Bifidobacterium species in the human gut function as defensive mutualists through antimicrobial activity against pathogens and potentially through aiding in immune system development and function (86, 121, 131). Finally, in marine environments, Actinobacteria with diverse secondary metabolism potential are found in association with sponges, but their ecological functions in these environments have not been fully elucidated (119, 153). Although the diversity of Actinobacteria-host defensive relationships has only been explored in a small subset of organisms, it is likely that host defense by Actinobacteria is widespread.
In contrast to these beneficial symbioses, Actinobacteria can also be virulent pathogens of plants and animals. The diverse genus Mycobacterium causes tuberculosis (M. tuberculosis), leprosy (M. leprae), and additional diseases in humans and other mammals (Figure 2d) (111). Actinobacteria also cause a range of other diseases in humans, including diphtheria (Corynebacterium diphtheria), Whipple’s disease (Tropheryma whipplei), and bacterial vaginosis (Gardnerella). In plants, Streptomyces scabies and other plant pathogenic Streptomyces strains share a pathogenicity island, allowing them to infect higher plants and cause diseases including, most famously, potato scab (69, 82). Other common plant pathogens fall within the Microbacteriaceae family, including Leifsonia xyli, which is the cause of ratoon stunting disease in sugarcane, and Clavibacter michiganensis, which infects xylem and causes plant wilting in alfalfa, corn, tomato, and potato (21, 47). Finally, leafy gall syndrome, which infects a wide range of dicotyledonous herbaceous plants, is caused by Rhodococcus fascians (128, 129).
ACTINOBACTERIA IN BIOENERGY
As of 2013, fossil fuels (petroleum, coal, and natural gas) supplied 80% of the world’s energy (60). The negative climate impacts of burning fossil fuels has driven worldwide interest in sustainable, economical production of liquid fuels and other chemicals from renewable sources (40). Use of renewable electric sources, such as solar, wind, and hydroelectric, is expanding, but high-energy density liquid fuels are needed to move ourselves and our goods across continents and oceans (29, 40). Plants and some microbes capture the energy of the sun via photosynthetic carbon dioxide fixation. Efforts to engineer photosynthetic plants, algae, or cyanobacteria to produce fuel compounds are underway and have long-term potential (79, 108, 142, 148). However, near-term advances require tapping into stored energy in the plant cell wall. To develop a sustainable biofuel industry, microbes are needed to convert plant biomass into soluble sugars and to transform these sugars into a wide range of desirable compounds (51). Here, we argue, in large part because of their ecology and evolutionary history, Actinobacteria have potential to contribute to both of these challenges.
Actinobacteria as a Source of Enzymes in Biofuels
Industrial production of fuels from plant biomass represented less than 5% of energy used for transportation in the United States in 2014 (139). This energy is largely derived from the starch in corn kernels, which is readily converted into ethanol to supplement gasoline (154). However, the use of edible carbohydrates for fuel directly competes with their use as food (135). In contrast, using the abundant, nonedible polysaccharide and lignin fractions of corn plants, other grasses, and trees to make fuels and chemicals is a promising alternative and could remove more carbon from the atmosphere than is released during production and use of the fuel (53). Currently, industrial lignocellulose hydrolysis is carried out using enzyme mixtures from engineered fungi, such as Trichoderma reesei and Aspergillus niger (101). However, the enzymatic release of intact clean sugar from plant cell walls remains an important bottleneck to efficient biofuel production (55). As detailed below, the diversity and biochemistry of actinobacterial cellulases, evolved within plant-degradation niches, have promise for integration into a successful cellulosic biofuels industry (also see the sidebar, Integrating Actinobacterial Enzymes in Industrial Cellulosic Biofuel Production).
INTEGRATING ACTINOBACTERIAL ENZYMES IN INDUSTRIAL CELLULOSIC BIOFUEL PRODUCTION.
A number of aspects of the evolution, ecology, and physiology of Actinobacteria make them well suited for integration with cellulosic biofuel production. First, Actinobacteria evolved to break down plant biomass under diverse environmental conditions, including under high temperatures, varying pH levels, and anoxic or aerobic states. Similarly, industrial biofuel production requires enzymes that function under diverse conditions, depending on factors such as the biomass composition and pretreatment protocol. The identification of Actinobacteria that have evolved within particular niches can lead to cellulases suitable for specific industrial settings. For example, thermostable enzymes are found in Thermobifida fusca and Thermomonospora curvata from heated composting environments and in Acidothermus cellulolyticus from acidic hot springs (107, 130, 136, 155). Furthermore, given that Actinobacteria freely secrete their enzymes and have evolved in some niches in close association with fungi, actinobacterial cellulases are promising candidates to complement fungal enzymes for plant biomass deconstruction. Finally, Actinobacteria are attractive for industrial use because of their history as successful workhorse organisms in industrial processes (42), the ability to engineer strains that constitutively secrete cellulases at high levels (18, 88), and the resistance of their cell walls to processes, such as milling, that may be important in sustainable cellulosic biofuel production (100).
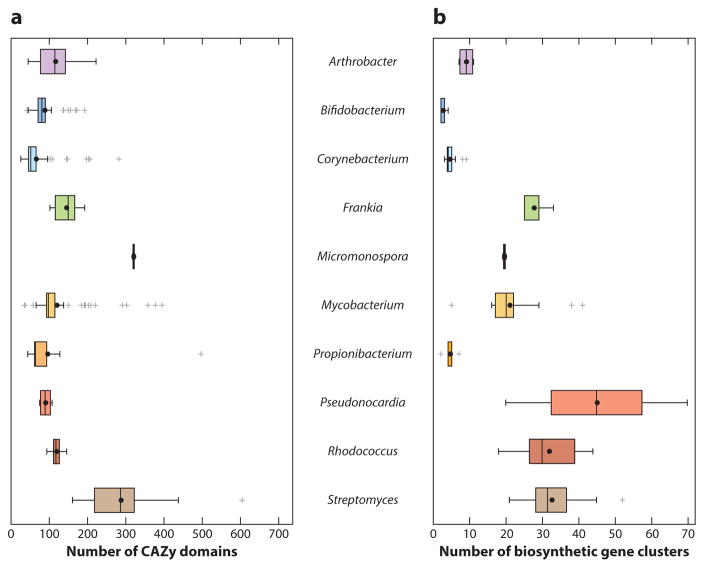
The ecological roles of Actinobacteria in plant biomass degradation, in soil, in compost, and in association with herbivorous animals have selected for the evolution of high cellulolytic abilities (18). Indeed, a number of model cellulolytic bacteria occur in this phylum, such as Cellulomonas spp. and Thermobifida fusca, whose study has helped advance our understanding of the enzymology of cellulose degradation (28, 59, 155). More broadly, the cellulolytic abilities of this phylum are supported by genomic and proteomic analyses, which indicate that actinobacterial genomes have an amazing diversity of enzyme fold families and organizational architectures used to hydrolyze plant biomass compared to most other phyla of bacteria (14). In particular, of the genera analyzed for this review, the genomes of Streptomyces and Micromonospora, some strains of Mycobacterium, and one strain of Propionibacterium are particularly enriched in carbohydrate-active enzyme (CAZy) genes (Figure 3a) (80). Within these enriched genera, the diversity of CAZy classes varies: Eighty-six glycoside hydrolase (GH) classes are found across 42 Streptomyces genomes, whereas only 35 GH classes are found across 149 Mycobacterium genomes (Supplemental Table 3).
Figure 3.
Enrichment of (a) CAZy (carbohydrate-active enzyme) domains and (b) biosynthetic gene clusters in select actinobacterial genera (Supplemental Table 3). Each median value is indicated by the line within the box, each mean value is indicated by the black dot, and outliers are indicated with gray plus signs. CAZy domains were identified using the CAZy database (80). Putative biosynthetic gene clusters were identified through genome searches for conserved elements of polyketide, terpene, and nonribosomal peptide biosynthesis enzymes. Hits <30 kb apart were called the same cluster. The number of clusters may be overestimated in draft genomes.
Here, we focus on the properties of Streptomyces cellulases. The cellulolytic system of Streptomyces was defined in S. reticuli by Schrempf and colleagues (112–114, 144, 147), whose investigations included secreted cellulolytic enzymes, carbohydrate binding proteins, sugar transporters, and gene regulation. Recent studies have expanded on this work by studying host-associated Streptomyces, such as Streptomyces sp. SirexAA-E, isolated from a Sirex woodwasp, which secretes enzymes with comparable specific activity to an enzyme cocktail from Trichoderma reesei (18, 132).
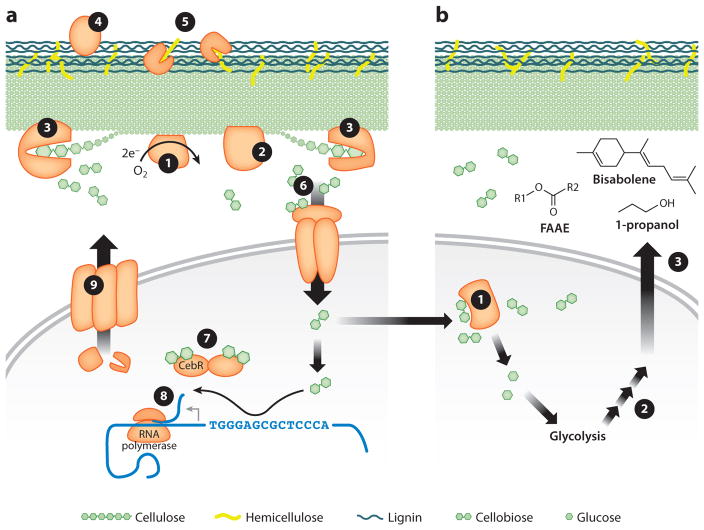
As in many Actinobacteria, cellulolytic Streptomyces species rely on a simple, freely secreted catalytic assembly to efficiently deconstruct cellulose, including endoglucanases (GH5, GH9, and GH12) and reducing- and nonreducing-end exoglucanases (GH48 and GH6, respectively). The cellobiose produced by these enzymes is cleaved into glucose by β-glucosidases (GH1 or GH3) in the cytoplasm (Figure 4a) (17, 18, 132). In addition, the dominant carbohydrate-binding module in these organisms is carbohydrate-binding module family 2. Parallel minimal catalytic assemblies deconstruct other plant cell wall polysaccharides, e.g., xylan (GH10 and GH11), mannan (GH5), and chitin (GH18, GH19, and GH46) (132). Carbohydrate esterases (CEs) that remove sugar modifications and break linkages between hemicellulose and lignin include families CE4 and CE7 in Streptomyces sp. SirexAA-E (132).
Figure 4.
Molecular details of the proposed role of Actinobacteria in biofuels. (a) Use of Actinobacteria for the deconstruction of plant biomass. ❶ Lytic polysaccharide monooxygenases and ❷ endoglucanases create internal cuts in the cellulose structure, creating free ends. ❸ Reducing- and nonreducing-end exoglucanases break cellulose chains into glucose and cellobiose. Additionally, ❹ peroxidases may break apart lignin, and ❺ hemicellulases hydrolyze xylan, mannan, and other hemicelluloses into constituent sugars. ❻ Cellobiose is imported into the cell, where it ❼ binds to the CebR repressor, releasing the repressor from the DNA and ❽ allowing RNA polymerase to transcribe cellulase genes. ❾ These cellulases are secreted from the cell using the Tat or Sec pathways. (b) Use of Actinobacteria for the production of biofuels. ❶ Cellobiose also is broken by β-glucosidases into glucose, which enters central metabolism. ❷ These sugars are processed in the cell and ❸ can be used for production of secreted fuel compounds, such as bisabolene, 1-propanol, or various fatty acid alkyl ester (FAAE) species where R1 is a small alkyl group and R2 is an aliphatic hydrocarbon. Currently, there are limited examples of cells that can both degrade plant biomass and produce fuel compounds, but we propose this scheme as one possible future direction in actinobacterial biofuels research.
Cellulose-oxidizing lytic polysaccharide monooxygenases from the auxiliary activity (AA) family AA10 enzymes primarily are found in Actinobacteria (19). These enzymes likely contribute to the high cellulolytic ability of Actinobacteria and provide an alternative to hydrolytic endoglucanases (Figure 4a) (63, 140). Interestingly, although Streptomyces are enriched in AA10 enzymes, Streptomyces do not appear to secrete an electron donor (e.g., cellobiose dehydrogenase or exogenous donors, such as ascorbate, gallate, or quinol), which is required for lytic polysaccharide monooxygenase function (17, 18, 132). Possibly instead, Streptomyces species depend on community association with fungal partners to provide this electron donor (104).
Nonreducing-end exoglucanase (GH6) genes are also expanded in Actinobacteria relative to many other groups of bacteria and fungi (14). Genetic and biochemical analyses have indicated that GH6 enzymes are necessary but not sufficient for cellulose degradation (7, 18, 157). Evolutionary analyses indicate that cellulolytic Streptomyces strains evolved to degrade cellulose by maintaining GH6 enzymes, which are highly expressed and secreted during growth on plant biomass (17, 18, 132).
Actinobacteria, Specialty Biofuels, and Bioproducts
The enzymatic capabilities described above allow Actinobacteria to deconstruct plant biomass into simple sugars that can be converted into biofuels and other bioproducts. Here, we focus on how Actinobacteria, having evolved to produce diverse chemicals, may contribute to the production of these bioproducts.
Although bioethanol from corn and cane sugars and biodiesel from plant oils have already seen widespread adoption, increasing their use as transportation fuels faces substantial hurdles, including storage, engine infrastructure, hygroscopy, and land-use concerns (3, 4, 25, 102). New bioproducts, including microbially produced specialty biofuels, more closely match the properties of traditional fuels and thus are highly desirable (4). Specialty biofuels include higher-alcohol biofuels, fatty acid alkyl esters (FAAEs or biodiesel), triacylglycerols (TAGs) that can serve as fuel precursors, and a diverse family of energy-dense isoprenoid compounds that can be used as diesel or jet-fuel replacements (11, 27, 39, 73, 102). Also, bio-based production of fuel additives and commodity chemicals, especially those currently obtained from petroleum, would help support a bioenergy industry. Actinobacteria are a particularly promising group to contribute to the development of these classes of chemicals or their precursors, especially those developed through alternative biosynthesis routes, given their well-known capacity to produce and secrete a diversity of compounds, the ability to engineer their biosynthetic pathways, and their long history of use in industrial processes (Figure 4b).
Several promising studies support the potential utility of Actinobacteria for biocommodity production (Supplemental Table 4). For example, Corynebacterium glutamicum has been targeted to produce specialty biofuels because of its innate isobutanol tolerance and ability to produce large amounts of glutamate and various 2-keto acids, which are precursors to branched-chain alcohols. Genetic engineering and optimization of C. glutamicum led to strains that could produce isobutanol at similar yields as in optimized Escherichia coli strains (15, 123, 158). The oleaginous Rhodococcus opacus was shown to accumulate high yields of high-energy density TAG (74). Additionally, native long-chain FAAE (biodiesel) production has been demonstrated in a Streptomyces strain isolated from sheep feces (83), and an isoprenoid biosynthesis pathway in Streptomyces venezuelae was combined with a cyclase gene from Abies grandis (grand fir) to produce bisabolene, another alternative to diesel fuel (103). 1-Propanol, an industrially relevant solvent with good fuel properties, was produced in T. fusca at a titer of 0.5 g/L after the addition of an aldehyde/alcohol dehydrogenase, approaching the yields possible in E. coli (44). Transcriptomic and metabolomic analyses suggested that additional pathway engineering of the intermediates would result in increased yields (43).
These studies represent a few of the many possible options to engineer Actinobacteria to produce bioproducts. The evolution of diverse natural product pathways in Actinobacteria, especially in genera such as Pseudonocardia, Rhodococcus, and Streptomyces (Figure 3b), make these organisms attractive as sources for discovery of biosynthetic enzymes and as hosts for pathway engineering and heterologous expression (45a, 103). The use of terpenoid biosynthetic machinery for bisabolene production in Streptomyces represents just one possible adaptation of natural product pathways for biofuel production. Polyketide natural products represent another source of industrially relevant molecules from Actinobacteria (160). Potentially, rational engineering of modular polyketide synthases can lead to molecules with extremely diverse structural and functional characteristics (71, 102).
Combining the natural abilities of Actinobacteria both to efficiently degrade plant biomass and to produce diverse chemicals make them well suited to contribute to the long-term goal of deploying consolidated bioprocessing (CBP) (Figure 4). The potential of Actinobacteria to contribute to CBP was illustrated by the FAAE-producing Streptomyces species, which grew readily on cellulose and xylan and produced higher biodiesel titers on complex carbohydrates than CBP-engineered E. coli (83, 125). The TAG-accumulating R. opacus was engineered with Streptomyces xylose metabolism genes (76) and subjected to directed evolution for high production of TAG with lignocellulosic biomass as a carbon source (75). Use of CBP approaches with Actinobacteria may also be possible for commodity chemicals. For example, the strain of T. fusca engineered for 1-propanol production produced higher titers when grown on a complex biomass, including cellulose and switchgrass, than on refined sugars. Similarly, Streptomyces maritimus produced benzoate, a food preservative and plastics additive, at higher yields when engineered to grow on cellulose as compared to growth on simple sugars (96). Furthermore, Streptomyces lividans modified with a T. fusca endoglucanase (for cellulose consumption) and two biosynthesis genes could produce 4-vinylphenol, a compound used in the plastics and electronics industries (95).
CONCLUSIONS
As our main source of antibiotics, Actinobacteria became famous for helping contribute to one of the greatest achievements of the twentieth century—vastly reducing the impact of infectious diseases. This century, we have an urgent and critical need for bio-based approaches that help meet our energy requirements while reducing carbon emissions. As described in this review, efforts are underway to leverage the diverse ecology of Actinobacteria to identify enzymes, pathways, and biosynthetic products that can contribute to biofuel and chemical production. We believe that, in the twenty-first century, Actinobacteria have potential to further increase in prominence as they help us meet our global needs for renewable biofuels and other bioproducts while mitigating the dangers of climate change.
Supplementary Material
Acknowledgments
We thank Adam Book for his insights and assistance with this review. This work was funded in part by the Department of Energy Great Lakes Bioenergy Research Center, Office of Science grant BER DE-FC02-07ER64494, and National Institutes of Health (NIH) grants U19 AI109673 and U19 TW009872. G.R.L. is funded by the NIH National Research Service Award T32 GM07215, and M.G.C. is funded by the NIH National Research Service Award T32 GM008505.
Glossary
- Nutritional mutualist
a symbiont that provides one or more key dietary benefits to another organism
- Natural product
any chemical compound produced by an organism, colloquially used for secondary metabolites that have uses in medicine and biotechnology
- Defensive mutualist
a symbiont that provides protection, usually physical or chemical, to another organism
- Biofuel
fuel produced through biotechnology, as opposed to fossil fuel
- Horizontal gene transfer (HGT)
the acquisition of genetic material into a genome other than through direct vertical inheritance
- Coevolution
reciprocal evolutionary change in interacting species
- Rhizosphere
the interface between plant roots and soil, often colonized by a specific microbial community, mediated by the plant’s chemical secretions
- Endophyte
a symbiont that lives within a plant
- Carbohydrate-active enzyme (CAZy)
an enzyme that acts on polysaccharides, curated at http://www.cazy.org and elsewhere
- Glycoside hydrolase (GH)
a diverse group of enzymes that cleave glycosidic bonds, for example, between glucose molecules in cellulose
- Endoglucanase
a GH enzyme that reacts with internal glycosidic bonds of the cellulose chain
- Exoglucanase
a GH enzyme that acts at the reducing or nonreducing end of the cellulose chain, often releasing cellobiose
- β-Glucosidase
a GH enzyme that cleaves cellobiose into glucose monomers
- Carbohydrate-binding module
proteins that target a specific carbohydrate, often tethered to GH domains to increase catalytic efficiency
- Lytic polysaccharide monooxygenase
a copper-dependent enzyme that hydroxylates the glycosidic bond in cellulose, xylan, starch, and chitin
- Bioproduct
a chemical, fuel, or other goods produced using biotechnology
- Consolidated bioprocessing (CBP)
the combination of enzyme production, biomass degradation, and fuel production in one reactor, often by one organism
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Adams AS, Jordan MS, Adams SM, Suen G, Goodwin LA, et al. Cellulose-degrading bacteria associated with the invasive woodwasp Sirex noctilio. ISME J. 2011;5(8):1323–31. doi: 10.1038/ismej.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed A, Earl J, Retchless A, Hillier SL, Rabe LK, et al. Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J Bacteriol. 2012;194(15):3922–37. doi: 10.1128/JB.00056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Andam CP, Doroghazi JR, Campbell AN, Kelly PJ, Choudoir MJ, Buckley DH. A latitudinal diversity gradient in terrestrial bacteria of the genus Streptomyces. mBio. 2016;7(2):e02200–15. doi: 10.1128/mBio.02200-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atabani AE, Silitonga AS, Badruddin IA, Mahlia TMI, Masjuki HH, Mekhilef S. A comprehensive review on biodiesel as an alternative energy resource and its characteristics. Renew Sustain Energy Rev. 2012;16:2070–93. [Google Scholar]
- 4.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 5.Barabas G, Vargha G, Szabo IM, Penyige A, Damjanovich S, et al. n-Alkane uptake and utilisation by Streptomyces strains. Antonie Van Leeuwenhoek. 2001;79:269–76. doi: 10.1023/a:1012030309817. [DOI] [PubMed] [Google Scholar]
- 6.Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, et al. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev. 2016;80(1):1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barr BK, Hsieh YL, Ganem B, Wilson DB. Identification of two functionally different classes of exocellulases. Biochemistry. 1996;35(2):586–92. doi: 10.1021/bi9520388. [DOI] [PubMed] [Google Scholar]
- 8.Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol. 2004;4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beier S, Bertilsson S. Uncoupling of chitinase activity and uptake of hydrolysis products in freshwater bacterioplankton. Limnol Oceanogr. 2011;56(4):1179–88. [Google Scholar]
- 10.Bell KS, Philp JC, Aw DWJ, Christofi N. A review: the genus Rhodococcus. J Appl Microbiol. 1998;85:195–210. doi: 10.1046/j.1365-2672.1998.00525.x. [DOI] [PubMed] [Google Scholar]
- 11.Beller HR, Lee TS, Katz L. Natural products as biofuels and bio-based chemicals: fatty acids and isoprenoids. Nat Prod Rep. 2015;32:1508–26. doi: 10.1039/c5np00068h. [DOI] [PubMed] [Google Scholar]
- 12.Berenbaum MR, Eisner T. Bugs’ bugs. Science. 2008;322:52–53. doi: 10.1126/science.1164873. [DOI] [PubMed] [Google Scholar]
- 13.Berlemont R, Allison SD, Weihe C, Lu Y, Brodie EL, et al. Cellulolytic potential under environmental changes in microbial communities from grassland litter. Front Microbiol. 2014;5:639. doi: 10.3389/fmicb.2014.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berlemont R, Martiny AC. Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol. 2013;79(5):1545–54. doi: 10.1128/AEM.03305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blombach B, Riester T, Wieschalka S, Ziert C, Youn J-W, et al. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol. 2011;77(10):3300–10. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bontemps C, Toussaint M, Revol P-V, Hotel L, Jeanbille M, et al. Taxonomic and functional diversity of Streptomyces in a forest soil. FEMS Microbiol Lett. 2013;342(2):157–67. doi: 10.1111/1574-6968.12126. [DOI] [PubMed] [Google Scholar]
- 17.Book AJ, Lewin GR, McDonald BR, Takasuka TE, Doering DT, et al. Cellulolytic Streptomyces strains associated with herbivorous insects share a phylogenetically linked capacity to degrade lignocellulose. Appl Environ Microbiol. 2014;80(15):4692–701. doi: 10.1128/AEM.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Book AJ, Lewin GR, McDonald BR, Takasuka TE, Wendt-Pienkowski E, et al. Evolution of high cellulolytic activity in symbiotic Streptomyces through selection of expanded gene content and coordinated gene expression. PLOS Biol. 2016;14(6):e1002475. doi: 10.1371/journal.pbio.1002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Book AJ, Yennamalli RM, Takasuka TE, Currie CR, Phillips GN, Jr, Fox BG. Evolution of substrate specificity in bacterial AA10 lytic polysaccharide monooxygenases. Biotechnol Biofuels. 2014;7:109. doi: 10.1186/1754-6834-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brecher G, Wigglesworth VB. The transmission of Actinomyces rhodnii Erikson in Rhodnius prolixus stål (Hemiptera) and its influence on the growth of the host. Parasitology. 1944;35(04):220–24. [Google Scholar]
- 21.Brumbley SM, Petrasovits LA, Hermann SR, Young AJ, Croft BJ. Recent advances in the molecular biology of Leifsonia xyli subsp xyli, causal organism of ratoon stunting disease. Australas Plant Pathol. 2006;35(6):681–89. [Google Scholar]
- 22.Cafaro MJ, Currie CR. Phylogenetic analysis of mutualistic filamentous bacteria associated with fungus-growing ants. Can J Microbiol. 2005;51(6):441–46. doi: 10.1139/w05-023. [DOI] [PubMed] [Google Scholar]
- 23.Cafaro MJ, Poulsen M, Little AEF, Price SL, Gerardo NM, et al. Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc R Soc B. 2011;278:1814–22. doi: 10.1098/rspb.2010.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caldera EJ, Currie CR. The population structure of antibiotic-producing bacterial symbionts of Apterostigma dentigerum ants: impacts of coevolution and multipartite symbiosis. Am Nat. 2012;180(5):604–17. doi: 10.1086/667886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caspeta L, Nielsen J. Economic and environmental impacts of microbial biodiesel. Nat Biotechnol. 2013;31(9):789–93. doi: 10.1038/nbt.2683. [DOI] [PubMed] [Google Scholar]
- 26.Chandra G, Chater KF. Developmental biology of Streptomyces from the perspective of 100 actinobacterial genome sequences. FEMS Microbiol Rev. 2014;38:345–79. doi: 10.1111/1574-6976.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi YJ, Lee J, Jang Y-S, Lee SY. Metabolic engineering of microorganisms for the production of higher alcohols. mBio. 2014;5(5):e01524–14. doi: 10.1128/mBio.01524-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christopherson MR, Suen G, Bramhacharya S, Jewell KA, Aylward FO, et al. The genome sequences of Cellulomonas fimi and “Cellvibrio gilvus” reveal the cellulolytic strategies of two facultative anaerobes, transfer of “Cellvibrio gilvus” to the genus Cellulomonas, and proposal of Cellulomonas gilvus sp nov. PLOS ONE. 2013;8(1):e53954. doi: 10.1371/journal.pone.0053954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu S, Majumdar A. Opportunities and challenges for a sustainable energy future. Nature. 2012;488:294–303. doi: 10.1038/nature11475. [DOI] [PubMed] [Google Scholar]
- 30.Clawson ML, Bourret A, Benson DR. Assessing the phylogeny of Frankia-actinorhizal plant nitrogen-fixing root nodule symbioses with Frankia 16S rRNA and glutamine synthetase gene sequences. Mol Phylogenet Evol. 2004;31:131–38. doi: 10.1016/j.ympev.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Colman DR, Toolson EC, Takacs-Vesbach CD. Do diet and taxonomy influence insect gut bacterial communities? Mol Ecol. 2012;21:5124–37. doi: 10.1111/j.1365-294X.2012.05752.x. [DOI] [PubMed] [Google Scholar]
- 32.Conn VM, Walker AR, Franco CM. Endophytic Actinobacteria induce defense pathways in Arabidopsis thaliana. Mol Plant-Microbe Interact. 2008;21(2):208–18. doi: 10.1094/MPMI-21-2-0208. [DOI] [PubMed] [Google Scholar]
- 33.Coombs JT, Michelsen PP, Franco CMM. Evaluation of endophytic Actinobacteria as antagonists of Gaeumannomyces graminis vartritici in wheat. Biol Control. 2004;29(3):359–66. [Google Scholar]
- 34.Crawford DL. Lignocellulose decomposition by selected Streptomyces strains. Appl Environ Microbiol. 1978;35(6):1041–45. doi: 10.1128/aem.35.6.1041-1045.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cross T. Aquatic actinomycetes: a critical survey of the occurrence, growth and role of actino-mycetes in aquatic habitats. J Appl Bacteriol. 1981;50(3):397–423. doi: 10.1111/j.1365-2672.1981.tb04245.x. [DOI] [PubMed] [Google Scholar]
- 36.Currie CR, Mueller UG, Malloch D. The agricultural pathology of ant fungus gardens. PNAS. 1999;96(14):7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science. 2006;311(5757):81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 38.Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398:701–4. [Google Scholar]
- 39.d’Espaux L, Mendez-Perez D, Li R, Keasling JD. Synthetic biology for microbial production of lipid-based biofuels. Curr Opin Chem Biol. 2015;29:58–65. doi: 10.1016/j.cbpa.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Dale BE, Ong RG. Energy, wealth, and human development: Why and how biomass pretreatment research must improve. Biotechnol Prog. 2012;28(4):893–98. doi: 10.1002/btpr.1575. [DOI] [PubMed] [Google Scholar]
- 41.de Boer W, Folman LB, Summerbell RC, Boddy L. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 42.Demain AL. The business of biotechnology. Ind Biotechnol. 2007;3(3):269–83. [Google Scholar]
- 43.Deng Y, Fisher AB, Fong SS. Systematic analysis of intracellular mechanisms of propanol production in the engineered Thermobifida fusca B6 strain. Appl Microbiol Biotechnol. 2015;99(19):8089–8100. doi: 10.1007/s00253-015-6850-4. [DOI] [PubMed] [Google Scholar]
- 44.Deng Y, Fong SS. Metabolic engineering of Thermobifida fusca for direct aerobic bioconversion of untreated lignocellulosic biomass to 1-propanol. Metab Eng. 2011;13:570–77. doi: 10.1016/j.ymben.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 45.Ding CH, Jiang ZQ, Li XT, Li LT, Kusakabe I. High activity xylanase production by Streptomyces olivaceoviridis E-86. World J Microbiol Biotechnol. 2004;20:7–10. [Google Scholar]
- 45a.Doroghazi JR, Metcalf WW. Comparative genomics of actinomycetes with a focus on natural product biosynthesis genes. BMC Genomics. 2013;14:611. doi: 10.1186/1471-2164-14-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eckert EM, Baumgartner M, Huber IM, Pernthaler J. Grazing resistant freshwater bacteria profit from chitin and cell-wall-derived organic carbon. Environ Microbiol. 2013;15(7):2019–30. doi: 10.1111/1462-2920.12083. [DOI] [PubMed] [Google Scholar]
- 47.Eichenlaub R, Gartemann K-H. The Clavibacter michiganensis subspecies: molecular investigation of gram-positive bacterial plant pathogens. Annu Rev Phytopathol. 2011;49:445–64. doi: 10.1146/annurev-phyto-072910-095258. [DOI] [PubMed] [Google Scholar]
- 48.Erikson D. Some studies on lake-mud strains of Micromonospora. J Bacteriol. 1941;41(3):277–300. doi: 10.1128/jb.41.3.277-300.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flórez LV, Biedermann PHW, Engl T, Kaltenpoth M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat Prod Rep. 2015;32:904–36. doi: 10.1039/c5np00010f. [DOI] [PubMed] [Google Scholar]
- 50.Franco C, Michelsen P, Percy N, Conn V, Listiana E, et al. Actinobacterial endophytes for improved crop performance. Australas Plant Pathol. 2007;36:524–31. [Google Scholar]
- 51.Fulton LM, Lynd LR, Korner A, Greene N, Tonachel LR. The need for biofuels as part of a low carbon energy future. Biofuels Bioprod Biorefining. 2015;9:476–83. [Google Scholar]
- 52.Gao B, Gupta RS. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev. 2012;76(1):66–112. doi: 10.1128/MMBR.05011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gelfand I, Snapp SS, Robertson GP. Energy efficiency of conventional, organic, and alternative cropping systems at a site in the U.S. Midwest. Environ Sci Technol. 2010;44(10):4006–11. doi: 10.1021/es903385g. [DOI] [PubMed] [Google Scholar]
- 54.Gittel A, Bárta J, Kohoutová I, Mikutta R, Owens S, et al. Distinct microbial communities associated with buried soils in the Siberian tundra. ISME J. 2014;8:841–53. doi: 10.1038/ismej.2013.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harris PV, Xu F, Kreel NE, Kang C, Fukuyama S. New enzyme insights drive advances in commercial ethanol production. Curr Opin Chem Biol. 2014;19:162–70. doi: 10.1016/j.cbpa.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 56.Herrmann RF, Shann JF. Microbial community changes during the composting of municipal solid waste. Microb Ecol. 1997;33:78–85. doi: 10.1007/s002489900010. [DOI] [PubMed] [Google Scholar]
- 57.Hirsch AM, Valdés M. Micromonospora: an important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol Biochem. 2010;42:536–42. [Google Scholar]
- 58.Hopwood DA. Nature and Medicine: The Antibiotic Makers. New York: Oxford Univ. Press; 2007. Streptomyces. [Google Scholar]
- 59.Hsing W, Canale-Parola E. Cellobiose chemotaxis by the cellulolytic bacterium Cellulomonas gelida. J Bacteriol. 1992;174(24):7996–8002. doi: 10.1128/jb.174.24.7996-8002.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Int. Energy Agency. 2015 Key World Energy Statistics. Paris: Int. Energy Agency; 2015. [Google Scholar]
- 61.Ishaque M, Kluepfel D. Production of xylanolytic enzymes by Streptomyces flavogriseus. Biotechnol Lett. 1981;3(9):481–86. [Google Scholar]
- 62.Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72(3):1719–28. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johansen KS. Discovery and industrial applications of lytic polysaccharide mono-oxygenases. Biochem Soc Trans. 2016;44:143–49. doi: 10.1042/BST20150204. [DOI] [PubMed] [Google Scholar]
- 64.Jones RT, Sanchez LG, Fierer N. A cross-taxon analysis of insect-associated bacterial diversity. PLOS ONE. 2013;8(4):e61218. doi: 10.1371/journal.pone.0061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaltenpoth M. Actinobacteria as mutualists: general healthcare for insects? Trends Microbiol. 2009;17(12):529–35. doi: 10.1016/j.tim.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 66.Kaltenpoth M, Gottler W, Herzner G, Strohm E. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol. 2005;15:475–79. doi: 10.1016/j.cub.2004.12.084. [DOI] [PubMed] [Google Scholar]
- 67.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawase T, Saito A, Sato T, Kanai R, Fujii T, et al. Distribution and phylogenetic analysis of family 19 chitinases in Actinobacteria. Appl Environ Microbiol. 2004;70(2):1135–44. doi: 10.1128/AEM.70.2.1135-1144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kers JA, Cameron KD, Joshi MV, Bukhalid RA, Morello JE, et al. A large, mobile pathogenicity island confers plant pathogenicity on Streptomyces species. Mol Microbiol. 2005;55(4):1025–33. doi: 10.1111/j.1365-2958.2004.04461.x. [DOI] [PubMed] [Google Scholar]
- 70.Kim M, Kim W-S, Tripathi BM, Adams J. Distinct bacterial communities dominate tropical and temperate zone leaf litter. Microb Ecol. 2014;67(4):837–48. doi: 10.1007/s00248-014-0380-y. [DOI] [PubMed] [Google Scholar]
- 71.Klaus M, Ostrowski MP, Austerjost J, Robbins T, Lowry B, et al. Protein–protein interactions, not substrate recognition, dominates the turnover of chimeric assembly line polyketide synthases. J Biol Chem. 2016 doi: 10.1074/jbc.M116.730531. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kroiss J, Kaltenpoth M, Schneider B, Schwinger M-G, Hertweck C, et al. Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol. 2010;6(4):261–63. doi: 10.1038/nchembio.331. [DOI] [PubMed] [Google Scholar]
- 73.Kung Y, Runguphan W, Keasling JD. From fields to fuels: recent advances in the microbial production of biofuels. ACS Synth Biol. 2012;1:498–513. doi: 10.1021/sb300074k. [DOI] [PubMed] [Google Scholar]
- 74.Kurosawa K, Boccazzi P, de Almeida NM, Sinskey AJ. High-cell-density batch fermentation of Rhodococcus opacus PD630 using a high glucose concentration for triacylglycerol production. J Biotechnol. 2010;147:212–18. doi: 10.1016/j.jbiotec.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 75.Kurosawa K, Laser J, Sinskey AJ. Tolerance and adaptive evolution of triacylglycerol-producing Rhodococcus opacus to lignocellulose-derived inhibitors. Biotechnol Biofuels. 2015;8:76. doi: 10.1186/s13068-015-0258-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kurosawa K, Wewetzer SJ, Sinskey AJ. Engineering xylose metabolism in triacylglycerol-producing Rhodococcus opacus for lignocellulosic fuel production. Biotechnol Biofuels. 2013;6:134. doi: 10.1186/1754-6834-6-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75(15):5111–20. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lawrence DB, Schoenike RE, Quispel A, Bond G. The role of Dryas drummondii in vegetation development following ice recession at Glacier Bay, Alaska, with special reference to its nitrogen fixation by root nodules. J Ecol. 1967;55(3):793–813. [Google Scholar]
- 79.Liu J, Rice A, Mcglew K, Shaw V, Park H, et al. Metabolic engineering of oilseed crops to produce high levels of novel acetyl glyceride oils with reduced viscosity, freezing point and calorific value. Plant Biotechnol J. 2015;13:858–65. doi: 10.1111/pbi.12325. [DOI] [PubMed] [Google Scholar]
- 80.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(1):D490–95. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Loqman S, Barka EA, Clément C, Ouhdouch Y. Antagonistic actinomycetes from Moroccan soil to control the grapevine gray mold. World J Microbiol Biotechnol. 2009;25:81–91. [Google Scholar]
- 82.Loria R, Kers J, Joshi M. Evolution of plant pathogenicity in Streptomyces. Annu Rev Phytopathol. 2006;44:469–87. doi: 10.1146/annurev.phyto.44.032905.091147. [DOI] [PubMed] [Google Scholar]
- 83.Lu Y, Wang J, Deng Z, Wu H, Deng Q, et al. Isolation and characterization of fatty acid methyl ester (FAME)-producing Streptomyces sp S161 from sheep (Ovis aries) faeces. Lett Appl Microbiol. 2013;57:200–5. doi: 10.1111/lam.12096. [DOI] [PubMed] [Google Scholar]
- 84.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Margesin R, Mörtelmaier C, Mair J. Low-temperature biodegradation of petroleum hydrocarbons (n-alkanes, phenol, anthracene, pyrene) by four actinobacterial strains. Int Biodeterior Biodegrad. 2013;84:185–91. [Google Scholar]
- 86.Marin ML, Lee JH, Murtha J, Ustunol Z, Pestka JJ. Differential cytokine production in clonal macrophage and T-cell lines cultured with Bifidobacteria. J Dairy Sci. 1997;80(11):2713–20. doi: 10.3168/jds.S0022-0302(97)76232-5. [DOI] [PubMed] [Google Scholar]
- 87.Marsh SE, Poulsen M, Pinto-Tomás A, Currie CR. Interaction between workers during a short time window is required for bacterial symbiont transmission in Acromyrmex leaf-cutting ants. PLOS ONE. 2014;9(7):e103269. doi: 10.1371/journal.pone.0103269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marushima K, Ohnishi Y, Horinouchi S. CebR as a master regulator for cellulose/cellooligosaccharide catabolism affects morphological development in Streptomyces griseus. J Bacteriol. 2009;191(19):5930–40. doi: 10.1128/JB.00703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matulich KL, Weihe C, Allison SD, Amend AS, Berlemont R, et al. Temporal variation overshadows the response of leaf litter microbial communities to simulated global change. ISME J. 2015;9(11):2477–89. doi: 10.1038/ismej.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCarthy AJ. Lignocellulose-degrading actinomycetes. FEMS Microbiol Rev. 1987;46:145–63. [Google Scholar]
- 91.McCarthy AJ, Williams ST. Actinomycetes as agents of biodegradation in the environment—a review. Gene. 1992;115:189–92. doi: 10.1016/0378-1119(92)90558-7. [DOI] [PubMed] [Google Scholar]
- 92.Metcalfe AC, Krsek M, Gooday GW, Prosser JI, Wellington EMH. Molecular analysis of a bacterial chitinolytic community in an upland pasture. Appl Environ Microbiol. 2002;68(10):5042–50. doi: 10.1128/AEM.68.10.5042-5050.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, et al. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol. 2016;82(4):980–91. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Noda S, Kawai Y, Tanaka T, Kondo A. 4-Vinylphenol biosynthesis from cellulose as the sole carbon source using phenolic acid decarboxylase- and tyrosine ammonia lyase-expressing Streptomyces lividans. Bioresour Technol. 2015;180:59–65. doi: 10.1016/j.biortech.2014.12.064. [DOI] [PubMed] [Google Scholar]
- 96.Noda S, Kitazono E, Tanaka T, Ogino C, Kondo A. Benzoic acid fermentation from starch and cellulose via a plant-like β-oxidation pathway in Streptomyces maritimus. Microb Cell Fact. 2012;11:49. doi: 10.1186/1475-2859-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oh D-C, Poulsen M, Currie CR, Clardy J. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol. 2009;5(6):391–93. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pasti MB, Pometto AL, III, Nuti MP, Crawford DL. Lignin-solubilizing ability of actinomycetes isolated from termite (Termitidae) gut. Appl Environ Microbiol. 1990;56(7):2213–18. doi: 10.1128/aem.56.7.2213-2218.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;54:559–68. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- 100.Paye JMD, Guseva A, Hammer SK, Gjersing E, Davis MF, et al. Biological lignocellulose solubilization: comparative evaluation of biocatalysts and enhancement via cotreatment. Biotechnol Biofuels. 2016;9:8. doi: 10.1186/s13068-015-0412-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, et al. Fungal cellulases. Chem Rev. 2015;115:1308–448. doi: 10.1021/cr500351c. [DOI] [PubMed] [Google Scholar]
- 102.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. Microbial engineering for the production of advanced biofuels. Nature. 2012;488(7411):320–28. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 103.Phelan RM, Sekurova ON, Keasling JD, Zotchev SB. Engineering terpene biosynthesis in Streptomyces for production of the advanced biofuel precursor bisabolene. ACS Synth Biol. 2014;4:393–99. doi: 10.1021/sb5002517. [DOI] [PubMed] [Google Scholar]
- 104.Phillips CM, Beeson WT, IV, Cate JH, Marletta MA. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol. 2011;6:1399–406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 105.Pokusaeva K, Fitzgerald GF, van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011;6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Poulsen M, Oh D-C, Clardy J, Currie CR. Chemical analyses of wasp-associated Streptomyces bacteria reveal a prolific potential for natural products discovery. PLOS ONE. 2011;6(2):e16763. doi: 10.1371/journal.pone.0016763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ransom C, Balan V, Biswas G, Dale B, Crockett E, Sticklen M. Heterologous Acidothermus cellulolyticus 1,4-β-endoglucanase E1 produced within the corn biomass converts corn stover into glucose. Appl Biochem Biotechnol. 2007;136–140:207–19. doi: 10.1007/s12010-007-9053-3. [DOI] [PubMed] [Google Scholar]
- 108.Razeghifard R. Algal biofuels. Photosynth Res. 2013;117:207–19. doi: 10.1007/s11120-013-9828-z. [DOI] [PubMed] [Google Scholar]
- 109.Rosas-Magallanes V, Deschavanne P, Quintana-Murci L, Brosch R, Gicquel B, Neyrolles O. Horizontal transfer of a virulence operon to the ancestor of Mycobacterium tuberculosis. Mol Biol Evol. 2006;23(6):1129–35. doi: 10.1093/molbev/msj120. [DOI] [PubMed] [Google Scholar]
- 110.Salem H, Bauer E, Strauss AS, Vogel H, Marz M, Kaltenpoth M. Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc R Soc B. 2014;281:20141838. doi: 10.1098/rspb.2014.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Saviola B, Bishai W. The genus Mycobacterium—medical. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes. 3 Vol. 3. New York: Springer; 2006. pp. 919–33. [Google Scholar]
- 112.Schlosser A, Jantos J, Hackmann K, Schrempf H. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl Environ Microbiol. 1999;65(6):2636–43. doi: 10.1128/aem.65.6.2636-2643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schlosser A, Kampers T, Schrempf H. The Streptomyces ATP-binding component MsiK assists in cellobiose and maltose transport. J Bacteriol. 1997;179(6):2092–95. doi: 10.1128/jb.179.6.2092-2095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schrempf H, Walter S. The cellulolytic system of Streptomyces reticuli. Int J Biol Macromol. 1995;17(6):353–55. doi: 10.1016/0141-8130(96)81845-9. [DOI] [PubMed] [Google Scholar]
- 115.Schrey SD, Erkenbrack E, Früh E, Fengler S, Hommel K, et al. Production of fungal and bacterial growth modulating secondary metabolites is widespread among mycorrhiza-associated streptomycetes. BMC Microbiol. 2012;12:164. doi: 10.1186/1471-2180-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Scott JJ, Budsberg KJ, Suen G, Wixon DL, Balser TC, Currie CR. Microbial community structure of leaf-cutter ant fungus gardens and refuse dumps. PLOS ONE. 2010;5(3):e9922. doi: 10.1371/journal.pone.0009922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scott JJ, Oh DC, Yuceer MC, Klepzig KD, Clardy J, Currie CR. Bacterial protection of beetle-fungus mutualism. Science. 2008;322:63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Seeds JD, Bishop JG. Low Frankia inoculation potentials in primary successional sites at Mount St. Helens, Washington, USA. Plant Soil. 2009;323:225–33. [Google Scholar]
- 119.Seipke RF, Kaltenpoth M, Hutchings MI. Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol Rev. 2012;36:862–76. doi: 10.1111/j.1574-6976.2011.00313.x. [DOI] [PubMed] [Google Scholar]
- 120.Semedo LTAS, Gomes RC, Linhares AA, Duarte GF, Nascimento RP, et al. Streptomyces drozdowiczii sp nov., a novel cellulolytic streptomycete from soil in Brazil. Int J Syst Evol Microbiol. 2004;54:1323–28. doi: 10.1099/ijs.0.02844-0. [DOI] [PubMed] [Google Scholar]
- 121.Servin AL. Antagonistic activities of lactobacilli and Bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28:405–40. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 122.Smanski MJ, Schlatter DC, Kinkel LL. Leveraging ecological theory to guide natural product discovery. J Ind Microbiol Biotechnol. 2015;43:115–28. doi: 10.1007/s10295-015-1683-9. [DOI] [PubMed] [Google Scholar]
- 123.Smith KM, Cho K-M, Liao JC. Engineering Corynebacterium glutamicum for isobutanol production. Appl Microbiol Biotechnol. 2010;87:1045–55. doi: 10.1007/s00253-010-2522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–13. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–62. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 126.Steffan SA, Chikaraishi Y, Currie CR, Horn H, Gaines-Day HR, et al. Microbes are trophic analogs of animals. PNAS. 2015;112(49):15119–24. doi: 10.1073/pnas.1508782112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Steger K, Jarvis Å, Vasara T, Romantschuk M, Sundh I. Effects of differing temperature management on development of Actinobacteria populations during composting. Res Microbiol. 2007;158(7):617–24. doi: 10.1016/j.resmic.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 128.Stes E, Francis I, Pertry I, Dolzblasz A, Depuydt S, Vereecke D. The leafy gall syndrome induced by Rhodococcus fascians. FEMS Microbiol Lett. 2013;342(2):187–94. doi: 10.1111/1574-6968.12119. [DOI] [PubMed] [Google Scholar]
- 129.Stes E, Vandeputte OM, El Jaziri M, Holsters M, Vereecke D. A successful bacterial coup d’état: how Rhodococcus fascians redirects plant development. Annu Rev Phytopathol. 2011;49:69–86. doi: 10.1146/annurev-phyto-072910-095217. [DOI] [PubMed] [Google Scholar]
- 130.Stutzenberger FJ. Cellulolytic activity of Thermomonospora curvata: nutritional requirements for cellulase production. Appl Microbiol. 1972;24(1):77–82. doi: 10.1128/am.24.1.77-82.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Takahashi T, Oka T, Iwana H, Kuwata T, Yamamoto Y. Immune response of mice to orally administered lactic acid bacteria. Biosci Biotechnol Biochem. 1993;57(9):1557–60. [Google Scholar]
- 132.Takasuka TE, Book AJ, Lewin GR, Currie CR, Fox BG. Aerobic deconstruction of cellulosic biomass by an insect-associated Streptomyces. Sci Rep. 2013;3:1–10. doi: 10.1038/srep01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S. Estimating divergence times in large molecular phylogenies. PNAS. 2012;109(47):19333–38. doi: 10.1073/pnas.1213199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thompson JN. The Coevolutionary Process. Chicago: Univ. Chicago Press; 1994. [Google Scholar]
- 135.Thompson PB. The agricultural ethics of biofuels: climate ethics and mitigation arguments. Poiesis Prax. 2012;8:169–89. doi: 10.1007/s10202-012-0105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tucker MP, Mohagheghi A, Grohmann K, Himmel ME. Ultra-thermostable cellulases from Acidothermus cellulolyticus: comparison of temperature optima with previously reported cellulases. Nat Biotechnol. 1989;7:817–20. [Google Scholar]
- 137.Turroni F, van Sinderen D, Ventura M. Genomics and ecological overview of the genus Bifidobacterium. Int J Food Microbiol. 2011;149:37–44. doi: 10.1016/j.ijfoodmicro.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 138.Tveit AT, Urich T, Svenning MM. Metatranscriptomic analysis of Arctic peat soil microbiota. Appl Environ Microbiol. 2014;80(18):5761–72. doi: 10.1128/AEM.01030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.US Energy Inf. Adm. January 2016 Monthly Energy Review. Washington, DC: US Energy Inf. Adm; 2016. [Google Scholar]
- 140.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330:219–22. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 141.Vitousek PM, Walker LR. Biological invasion by Myrica faya in Hawai’i: plant demography, nitrogen fixation, ecosystem effects. Ecol Monogr. 1989;59(3):247–65. [Google Scholar]
- 142.Vollmann J, Eynck C. Camelina as a sustainable oilseed crop: contributions of plant breeding and genetic engineering. Biotechnol J. 2015;10:525–35. doi: 10.1002/biot.201400200. [DOI] [PubMed] [Google Scholar]
- 143.von Graeventiz A, Bernard K. The genus Corynebacterium—medical. 2006:819–42. See Ref. 111. [Google Scholar]
- 144.Wachinger G, Bronnenmeier K, Staudenbauer WL, Schrempf H. Identification of mycelium-associated cellulase from Streptomyces reticuli. Appl Environ Microbiol. 1989;55(10):2653–57. doi: 10.1128/aem.55.10.2653-2657.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Waksman SA. Studies in the metabolism of actinomycetes II. J Bacteriol. 1918;4:307–30. doi: 10.1128/jb.4.4.307-330.1.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Waksman SA. Decomposition of the various chemical constituents etc of complex plant materials by pure cultures of fungi and bacteria. Arch Mikrobiol. 1931;2(1):136–54. [Google Scholar]
- 147.Walter S, Schrempf H. The synthesis of the Streptomyces reticuli cellulase (avicelase) is regulated by both activation and repression mechanisms. Mol Gen Genet. 1996;251(2):186–95. doi: 10.1007/BF02172917. [DOI] [PubMed] [Google Scholar]
- 148.Wang B, Wang J, Zhang W, Meldrum DR. Application of synthetic biology in cyanobacteria and algae. Front Microbiol. 2012;3:344. doi: 10.3389/fmicb.2012.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang C, Dong D, Wang H, Müller K, Qin Y, et al. Metagenomic analysis of microbial consortia enriched from compost: new insights into the role of Actinobacteria in lignocellulose decomposition. Biotechnol Biofuels. 2016;9:1–17. doi: 10.1186/s13068-016-0440-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Watanabe Y, Martini JE, Ohmoto H. Geochemical evidence for terrestrial ecosystems 2.6 billion years ago. Nature. 2000;408:574–78. doi: 10.1038/35046052. [DOI] [PubMed] [Google Scholar]
- 151.Watanabe Y, Shinzato N, Fukatsu T. Isolation of actinomycetes from termites’ guts. Biosci Biotechnol Biochem. 2003;67(8):1797–801. doi: 10.1271/bbb.67.1797. [DOI] [PubMed] [Google Scholar]
- 152.Weber NA. Fungus-growing ants. Science. 1966;153(3736):587–604. doi: 10.1126/science.153.3736.587. [DOI] [PubMed] [Google Scholar]
- 153.Webster NS, Taylor MW. Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol. 2012;14(2):335–46. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- 154.Weise SE, van Wijk KJ, Sharkey TD. The role of transitory starch in C3, CAM, and C4 metabolism and opportunities for engineering leaf starch accumulation. J Exp Bot. 2011;62(9):3109–18. doi: 10.1093/jxb/err035. [DOI] [PubMed] [Google Scholar]
- 155.Wilson DB. Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem Rec. 2004;4(2):72–82. doi: 10.1002/tcr.20002. [DOI] [PubMed] [Google Scholar]
- 156.Wilson DB. Microbial diversity of cellulose hydrolysis. Curr Opin Microbiol. 2011;14:259–63. doi: 10.1016/j.mib.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 157.Wu M, Bu L, Vuong TV, Wilson DB, Crowley MF, et al. Loop motions important to product expulsion in the Thermobifida fusca glycoside hydrolase family 6 cellobiohydrolase from structural and computational studies. J Biol Chem. 2013;288(46):33107–17. doi: 10.1074/jbc.M113.502765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yamamoto S, Suda M, Niimi S, Inui M, Yukawa H. Strain optimization for efficient isobutanol production using Corynebacterium glutamicum under oxygen deprivation. Biotechnol Bioeng. 2013;110(11):2938–48. doi: 10.1002/bit.24961. [DOI] [PubMed] [Google Scholar]
- 159.Yu H, Zeng G, Huang H, Xi X, Wang R, et al. Microbial community succession and lignocellulose degradation during agricultural waste composting. Biodegradation. 2007;18:793–802. doi: 10.1007/s10532-007-9108-8. [DOI] [PubMed] [Google Scholar]
- 160.Yuzawa S, Keasling JD, Katz L. Insights into polyketide biosynthesis gained from repurposing antibiotic-producing polyketide synthases to produce fuels and chemicals. J Antibiot. 2016 doi: 10.1038/ja.2016.64. In press. [DOI] [PubMed] [Google Scholar]
- 161.Ziemert N, Lechner A, Wietz M, Millán-Aguiñaga N, Chavarria KL, Jensen PR. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. PNAS. 2014;111(12):E1130–39. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Žifčáková L, Větrovský T, Howe A, Baldrian P. Microbial activity in forest soil reflects the changes in ecosystem properties between summer and winter. Environ Microbiol. 2015;18(1):288–301. doi: 10.1111/1462-2920.13026. [DOI] [PubMed] [Google Scholar]
- 163.Zimmermann W. Degradation of lignin by bacteria. J Biotechnol. 1990;13:119–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.