Abstract
Female fertility is determined in part by the size and development of the primordial follicle pool. The current study investigates the role of glial cell-line-derived neurotrophic factor (GDNF) in the regulation of primordial follicle development in the ovary. Ovaries from 4-day-old female rat pups were maintained in organ culture for 10 days in the absence (control) or presence of GDNF or kit ligand (KL)/stem cell factor. Ovaries treated with GDNF contained a significant increase in developing follicles, similar to that observed with KL treatment previously shown to promote follicle development. The actions of GDNF on the ovarian transcriptome were investigated with a microarray analysis. Immunohistochemical studies demonstrated that GDNF is localized to oocyte cytoplasm in follicles of all developmental stages, as well as to cumulus granulosa cells and theca cells in antral follicles. GDNF receptor α1 (GFRα1) staining was localized to oocyte cytoplasm of primordial and primary follicles, and at reduced levels in the oocytes of antral follicles. GFRα1 was present in mural granulosa cells of antral follicles, theca cells, and ovarian surface epithelium. The localization studies were confirmed with molecular analysis. Microarray analysis was used to identify changes in the ovarian transcriptome and further elucidate the signaling network regulating early follicle development. Observations indicate that GDNF promotes primordial follicle development and mediates autocrine and paracrine cell–cell interactions required during folliculogenesis. In contrast to the testis, ovarian GDNF is predominantly produced by germ cells (oocytes) rather than somatic cells.
Introduction
Female fertility of most mammals is determined by the primordial follicle pool size in the neonatal period and the rate at which primordial follicles leave the arrested pool to begin development. A resting pool of primordial follicles is formed late in embryogenesis or in the first days after birth (Hirshfield 1991, Skinner 2005). A primordial follicle consists of an oocyte arrested in prophase I of meiosis and surrounded by flattened pre-granulosa cells (Parrott & Skinner 1999). Once a primordial follicle begins to develop, it will either continue to develop fully into an ovulatory follicle or undergo atresia via cellular apoptosis at some stages of folliculogenesis. The development process is initiated by the primordial to primary follicle transition, during which the flattened pre-granulosa cells surrounding the oocyte become cuboidal granulosa cells indicative of the primary follicle (Hirshfield 1991, Kezele et al. 2002). The rate at which follicles leave the primordial pool and transition into developing primary follicles determines a female’s future fertility. Elucidation of the processes regulating primordial follicle development is essential in designing therapies for reproductive diseases such as premature ovarian failure in which follicles undergo primordial to primary transition at an abnormally high rate (Richardson et al. 1987, Santoro 2001). Potential therapies could also regulate the transition into menopause or induce menopause for women at risk of developing breast and ovarian cancers. The network of known extracellular signaling factors regulating primordial to primary follicle transition includes kit ligand (KL; Parrott & Skinner 1999, Nilsson & Skinner 2004), leukemia inhibitory factor (LIF; Nilsson et al. 2002), bone morphogenetic protein 4 (BMP4; Nilsson & Skinner 2003), BMP7 (Lee et al. 2004), platelet-derived growth factor (PDGF; Nilsson et al. 2006), and basic fibroblast growth factor (bFGF; Nilsson & Skinner 2004), all of which promote primordial follicle development (Skinner 2005). Anti-Müllerian hormone/Müllerian inhibitory substance (AMH/MIS) and stromal-derived factor-1 (SDF-1/ CXCL12) inhibit primordial to primary follicle transition (Ikeda et al. 2002, Holt et al. 2006, Nilsson et al. 2007). The current research was designed to determine whether glial-derived neurotrophic factor (GDNF) is a part of this network of cell–cell interactions that regulate primordial follicle development.
GDNF has previously been shown to exhibit neurorestorative and neuroprotective actions for dopaminergic neurons in the nigrostriatal pathway of the brain (Gash et al. 1998, Kordower et al. 2000, Kirik et al. 2004, Wissel et al. 2006). It also mediates tube formation in mammary glands via the MAP kinase pathway (Karihaloo et al. 2005). GDNF signaling occurs via a protein complex. The receptor GDNF family receptor α1 (GFRα1) preferentially binds GDNF, and the ligand–receptor complex activates the ubiquitous tyrosine kinase receptor RET (Amoresano et al. 2005, Carmillo et al. 2005, Pozas & Ibanez 2005, Vargas-Leal et al. 2005). Activation of RET by this complex leads to activation of intracellular signaling pathways involved in cell proliferation and differentiation (Naughton et al. 2006).
Testicular GDNF is expressed in the Sertoli cells while its receptors, GFRα1 and RET, are expressed in the spermatogonial cells (Dettin et al. 2003). Signaling from the GDNF–GFRα1–RET complex is necessary for spermatogonial stem cell self-renewal and proliferation in neonatal mice (Widenfalk et al. 2000, Wu et al. 2005, Naughton et al. 2006). GDNF expression in the testis decreases as males reach maturity. By contrast, GDNF expression in the ovary increases with maturity especially in follicles close to ovulation (Golden et al. 1999). Gdnf mRNA is expressed at lower levels in developing follicles (Golden et al. 1999, Widenfalk et al. 2000) and localized to oocytes (Aravindakshan et al. 2006). Microarray experiments by the current investigators suggested that RET receptor expression in ovaries changed with AMH treatment (Nilsson et al. 2007). This observation, coupled with the results of the previous studies on GDNF, led to the hypothesis that GDNF signaling may regulate the primordial to primary follicle transition. The current study investigates the role of GDNF in promoting primordial to primary follicle transition and characterizes GDNF and GFRα1 expression in neonatal and adult ovaries.
Results
Ovaries from 4-day-old female rat pups were maintained in organ culture for 10 days in the absence (control) or presence of 50 ng/ml GDNF, 50 ng/ml KL/stem cell factor (KL), or GDNF and KL combined. Following culture follicles in ovarian cross sections were classified as primordial or developing. Follicles containing an oocyte partially or fully encapsulated by flattened pre-granulosa cells were designated primordial (Fig. 1A), whereas follicles containing an oocyte accompanied by two or more cuboidal granulosa cells were designated developing (Fig. 1B). Developing follicles can be further classified into separate stages (transitional, primary, secondary, pre-antral); however, for the purpose of analyzing primordial to primary follicle transition, a comparison of the percentage of primordial follicles to the percentage of total developing follicles per section was made. At 200× magnification, untreated control ovaries (Fig. 1C) contain notably fewer large developing follicles than the ovaries treated with GDNF (Fig. 1D). Cultured control untreated ovary cross sections contained 41.3 ± 3.4% (mean ± S.E.M.) developing follicles (stages 1–4) due to spontaneous primordial to primary follicle transition (Fig. 2). By contrast, ovary sections treated with GDNF contained an increased proportion of developing follicles (P<0.01) at 54.3 ± 1.9%. Ovarian sections treated with both GDNF and KL also had an increased percentage of developing follicles (P<0.01; 56.9 ± 2.6%) compared with controls. As a positive control, ovary sections treated with KL contained 49.7 ± 0.21% developing follicles (P<0.05). Total follicle pool size did not significantly change with varying treatments (Fig. 3). Although no statistically significant change was observed, a slight reduction in total follicle number was found after treatment. The decrease observed could not account for the stimulation after treatment. The higher percentage of developing follicles in GDNF-treated ovaries indicates that more follicles have undergone primordial to primary follicle transition. Therefore, GDNF promotes primordial follicle development in rat ovaries.
Figure 1.
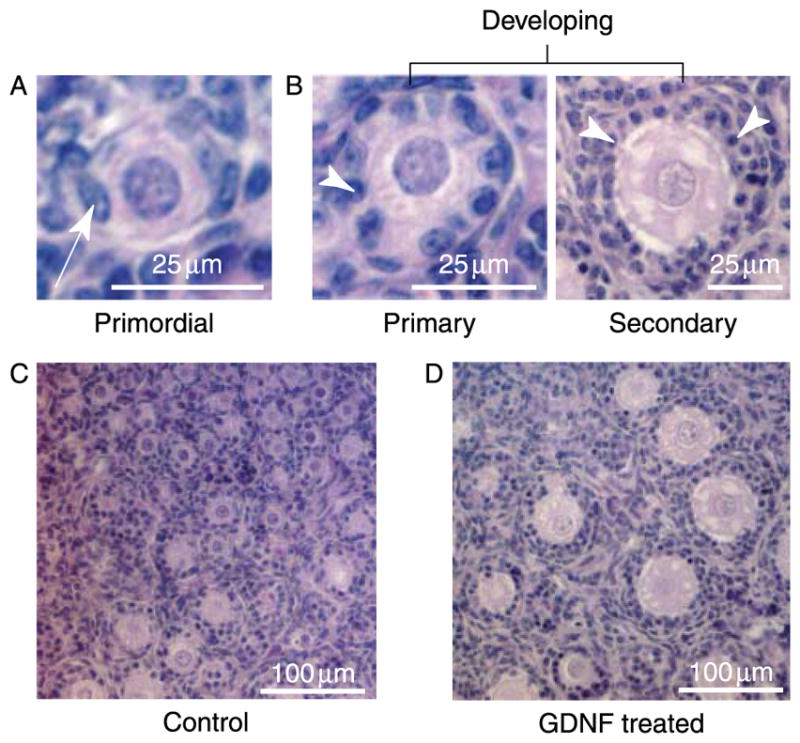
(A) Histology of a primordial follicle with an oocyte surrounded partially or completely by flattened (squamous) pre-granulosa cells (arrow). (B) Histology of primary and pre-antral secondary follicles (both classified as developing) with an oocyte surrounded by one or more cuboidal granulosa cell layers (arrowhead). (C) Postnatal day 4 rat ovary cultured for 10 days with no treatment. (D) Postnatal day 4 rat ovary cultured for 10 days with GDNF treatment.
Figure 2.
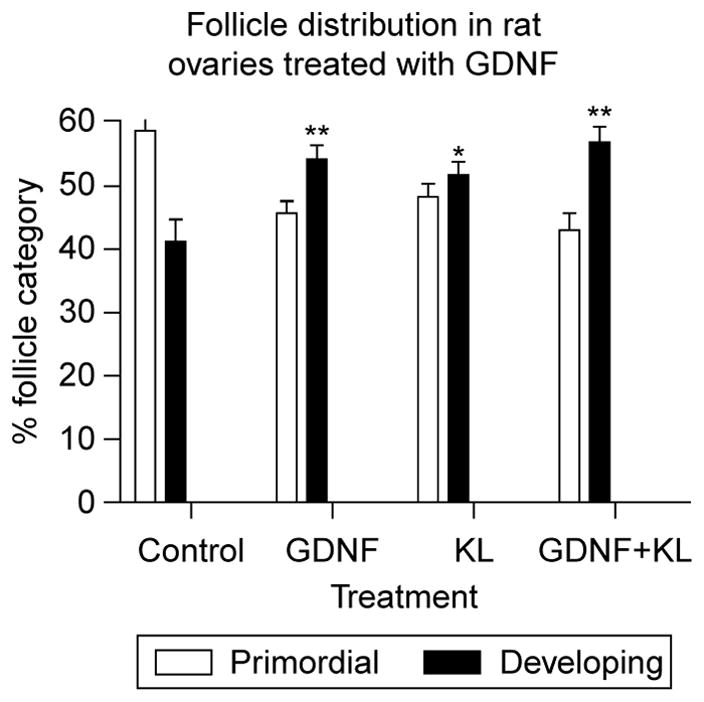
Follicle distribution in control and GDNF-treated rat ovary sections. Postnatal day 4 rat ovaries were maintained for 10 days in organ culture with no treatment (control), GDNF treatment, KL treatment, or GDNF and KL combined. KL is a known promoter of primordial to primary follicle transition. The percent follicle category (control or developing) is presented (mean ± S.E.M.) from a minimum of three experiments in replicate with (**) indicating P<0.01 compared with control, and (*) indicating P<0.05 compared with control.
Figure 3.
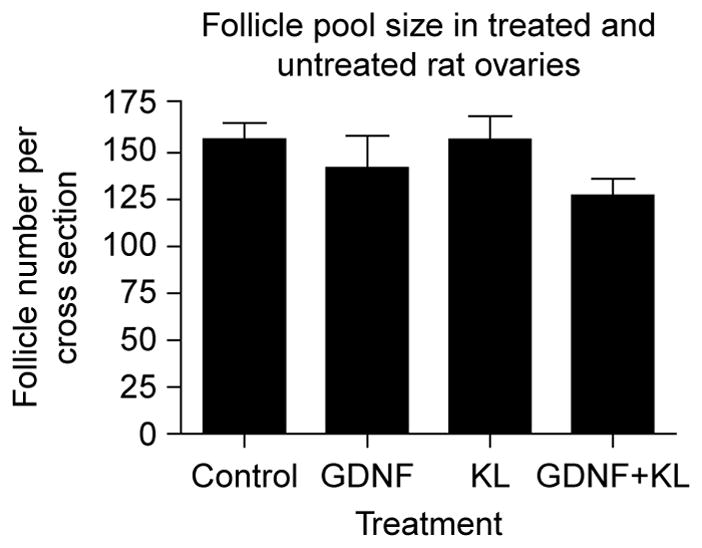
Follicle pool size in treated and untreated rat ovaries. Postnatal day 4 rat ovaries were maintained for 10 days in organ culture with no treatment (control), GDNF treatment, KL treatment, or GDNF and KL combined. Two cross sections from the center of each ovary were analyzed and the total follicle number per cross section averaged. The mean ± S.E.M. from a minimum of three different experiments in replicate are presented with no statistical difference (P>0.05) between treatments.
Immunohistochemistry was used to localize the proteins for GDNF and its receptor GFRα1 in postnatal day 4 (P4) rat ovaries after 10 days of organ culture. Dark-colored GDNF-specific staining was localized to oocyte cytoplasm in primordial and primary follicles of P4 sections (Fig. 4A and B). GFRα1 protein in neonatal ovaries was also localized to the oocyte cytoplasm of primordial and primary follicles (Fig. 4C and D). Non-specific staining throughout the tissue is determined by comparison with sections stained with a non-specific primary antibody (Fig. 4E and F).
Figure 4.
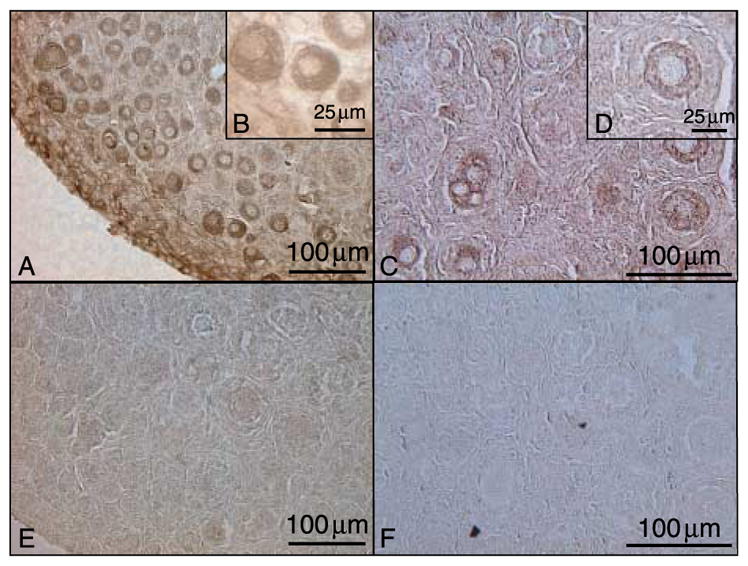
GDNF and GFRα1 immunohistochemical localization in postnatal day 4 (P4) rat ovaries. (A–B) GDNF, (C–D) GFRα1, (E) non-specific IgG developed according to GDNF protocol, and (F) non-specific IgG developed according to GFRα1 protocol. Representative micrograph of a minimum of three different experiments in replicate.
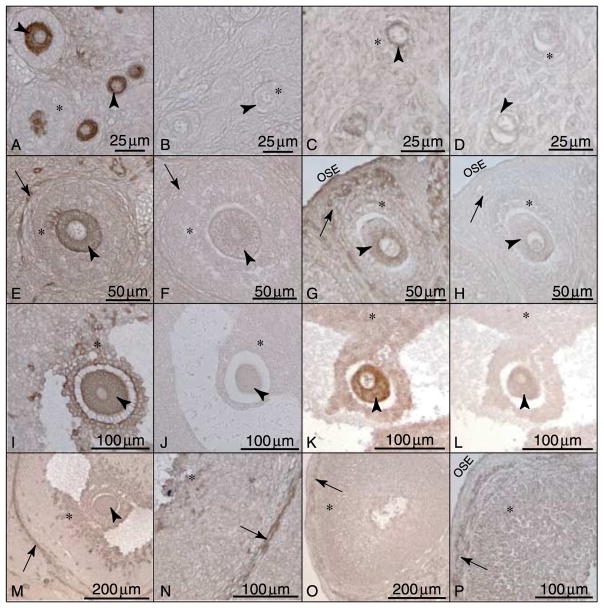
Immunohistochemistry was also used to localize GDNF and GFRα1 in freshly isolated adult female ovaries. In primordial and primary follicles, GDNF and GFRα1 are both present in the oocyte (Fig. 5A and C). This confirms the results seen in cultured ovaries. In pre-antral follicles, GDNF localization expands to include oocytes, nearby granulosa cells, and theca (Fig. 5E). GFRα1 localization in pre-antral follicles remains at low levels in the oocyte and expands to the granulosa cells and theca (Fig. 5G). GDNF localization in antral follicles remains in the oocytes and nearby cumulus granulosa cells (Fig. 5I). GDNF is also present in the theca throughout adult sections (Fig. 5M and N). In antral follicles, GFRα1 is present at low levels in the oocytes and mural granulosa cells (Fig. 5K). In some antral follicle oocytes, GFRα1 is detectable at levels no higher than those of the background staining seen in the negative controls (data not shown), suggesting that GFRα1 protein may not be present in these oocytes. GFRα1 is also present in the theca and ovarian surface epithelium of adult rat ovaries (Fig. 5O and P). These findings are summarized in Table 1. Corresponding negative control sections developed according to matched GDNF and GFRα1 protocols represent non-specific background staining at each follicular stage (Fig. 5B, D, F, H, J and L).
Figure 5.
GDNF and GFRα1 immunohistochemical localization in adult rat ovaries. (A–D) Localization in primordial and primary follicles: (A) GDNF and (C) GFRα1. (E–H) Localization in pre-antral follicles: (E) GDNF and (G) GFRα1. (I–L) Localization in antral follicles: (I) GDNF and (K) GFRα1. (M–P) Localization in theca and granulosa cells: (M and N) GDNF and (O and P) GFRα1. Sections stained with non-specific IgG corresponding to protocol-matched GDNF and GFRα1-stained sections (B, D, F, H, J and L). Arrowheads point to oocyte cytoplasm, arrows point to thecal cells, (*) indicates granulosa cells, and OSE indicates ovarian surface epithelium. Representative micrographs from a minimum of three different experiments in replicate.
Table 1.
Ovarian localization of glial cell-line-derived neurotrophic factor (GDNF) and GDNF receptor α1 (GFRα1) expression.
| Primordial | Primary | Pre-antrala | Antralb | |
|---|---|---|---|---|
| GDNF | ||||
| Oocyte | + + | + + | + + | + + |
| Granulosa | − | − | + | + + |
| Theca | − | − | + | + + |
| GFRα1 | ||||
| Oocyte | + + | + + | + | + /− |
| Granulosa | − | − | + | + |
| Theca | − | − | + + | + + |
Pre-antral follicles have three or more layers of granulosa cell.
Antral follicles have a fluid-filled antrum.
Observations suggest GDNF has an autocrine role in the primordial follicle since it is produced by the oocyte and has receptors on the oocyte (Figs 4–6). In the pre-antral follicle GDNF continues to have an autocrine role, but the receptor is also present on developing theca cells (Fig. 5). By contrast, antral follicle GDNF is expressed by the oocyte and adjacent cumulus granulosa cells (Figs 5I and M and 6), but the receptor GFRα1 is expressed by the oocyte and mural granulosa (Fig. 5K and O). GDNF expression in the antral follicle is in the oocyte and in cells most proximal to the oocyte (Fig. 6A and B). GDNF expression is also observed in the theca cells (Figs 5M and N and 6). Therefore, GDNF transitions from an autocrine factor in the primordial follicle to a factor with autocrine and paracrine abilities in the antral follicle (Fig. 6C and D; Table 1).
Figure 6.
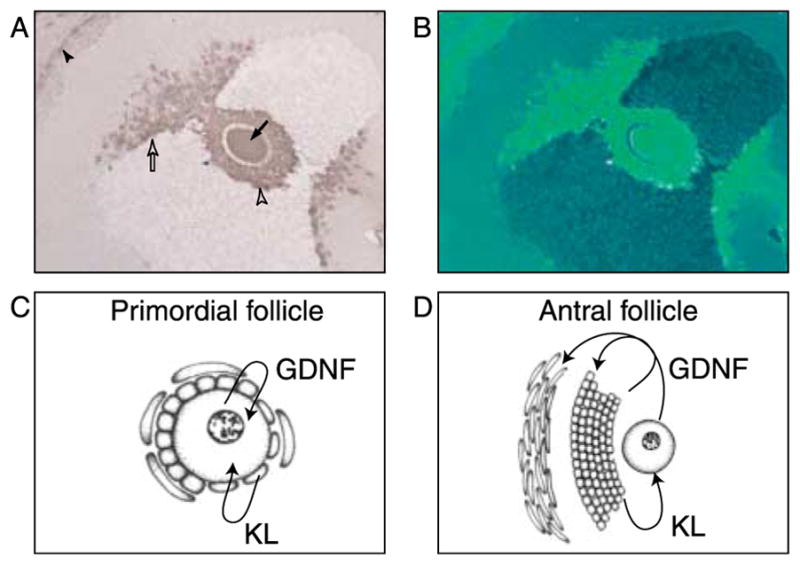
(A and B) GDNF immunohistochemistry in a large antral follicle. (A) The arrow indicates the oocyte, the open arrowhead the cumulus granulosa, the open arrow the oocyte proximal granulosa, and the closed arrowhead the theca interna cells. (B) is an artificial color contrast of (A). Representative micrograph from three different experiments in replicate. (C) Schematic of primordial follicle expression and action of GDNF and KL and (D) schematic of large antral follicle expression and action of GDNF and KL.
To confirm the immunohistochemical analysis, RT-PCR was used to determine which ovarian cell types expressed Gdnf and Gfra1 (GFRα1) mRNA. RNA was extracted separately from isolated oocytes and granulosa cells of rat antral follicles. Negative control samples (created by omitting MMLV enzyme from the RT reaction) did not produce a significant PCR product (Fig. 7). Gfra1 mRNA was localized to granulosa cells of antral follicles, whereas Gdnf mRNA was present in the oocytes and granulosa cells of antral follicles (Fig. 7). Ribosomal gene S2 was amplified as a constitutively expressed gene reference standard. Observations confirm the expression of Gdnf by the oocyte and demonstrated decreased Gfra1 expression in antral follicle oocytes.
Figure 7.
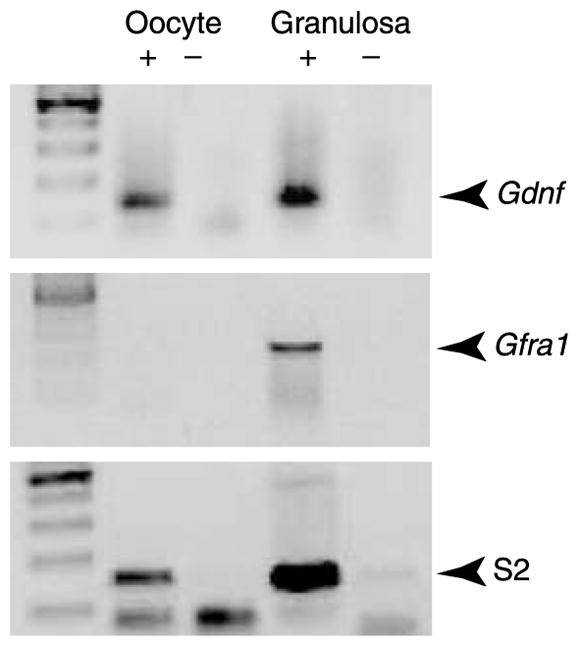
RT-PCR showing expression of Gdnf and mRNA in oocytes and granulosa cells from antral follicles. Gdnf and Gfra1 bands shown were imaged after two rounds of PCR. Products from reactions with (+) and without (−) MMLV reverse transcriptase enzyme are shown. S2 was amplified as a constitutively expressed reference gene. Data are representative of a minimum of three different experiments.
A real-time PCR procedure was used to measure the potential regulation of KL (Kitl) expression in cultured rat ovaries treated with and without GDNF. The ribosomal gene S2 was amplified as a constitutively expressed gene reference standard and used to normalize data. There was no significant difference in KL expression between control and GDNF-treated ovaries (Fig. 8). Therefore, GDNF does not appear to regulate KL expression during primordial follicle development. The level of Gdnf and Gfra1 mRNA expression in the whole ovary was found to be negligible, such that KL regulation of GDNFexpression in whole ovary studies were not useful (data not shown).
Figure 8.
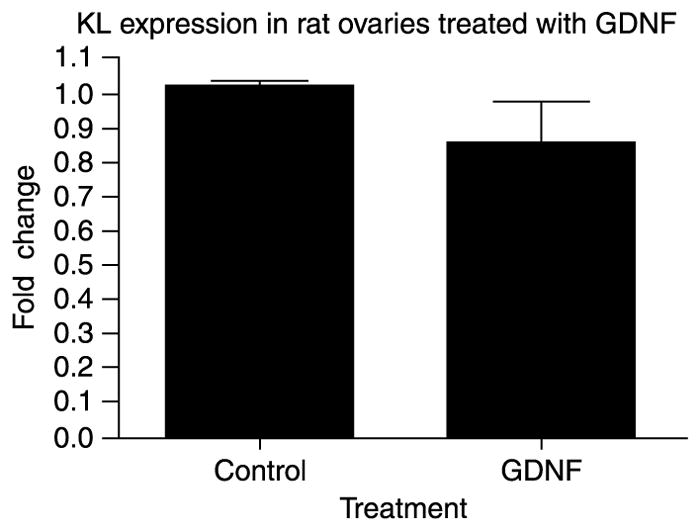
KL expression in GDNF-treated ovaries. Real-time PCR amplification of KL (Kitl) mRNA derived from postnatal day 4 rat ovaries cultured for 2 days in the presence or absence of GDNF treatment. The mean ± S.E.M. from three different experiments is presented as relative expression and normalized with S2 mRNA.
The actions of GDNF on the neonatal rat ovary were further investigated by determining the effects of GDNF on the ovarian transcriptome with a microarray analysis. P4 rat ovaries were cultured for 2 days in the absence or presence of GDNF and then RNA was collected for microarray analysis. GDNF was found to alter the expression of 28 genes (above 1.5-fold change cut-off) with 17 stimulated and 11 decreased in expression (Table 2; Fig. 9). A list of specific genes is provided in Table 2 and categories of altered genes are presented. GDNF was found to affect several growth factors and secreted cytokines including AMH, connective tissue growth factor (CTGF), fibroblast growth factor-8 (FGF8), growth differentiation factor-9 (GDF9), and stanniocalcin-1 (STC1). Observations demonstrate that GDNF participates with previously described members of the signaling network regulating follicle development. This analysis has also identified candidate genes for further analysis.
Table 2.
GDNF-regulated gene expression in primordial follicles.
| Gene symbol and category | Control raw | GDNF raw | GDNF/Cont ratio | GenBank | Gene title |
|---|---|---|---|---|---|
| Cell cycle | |||||
| Cdc14a_predicted | 165 | 104 | 0.63 | AW915083 | CDC14 cell division cycle 14 homolog A |
| Cytoskeleton | |||||
| Jam2 | 126 | 204 | 1.62 | AA849471 | Junction adhesion molecule 2 |
| Spire1_predicted | 80 | 125 | 1.58 | H31747 | Spire homolog 1 (Drosophila) (predicted) |
| Growth factors | |||||
| Amh | 146 | 219 | 1.50 | AI059285 | Anti-Müllerian hormone |
| Ctgf | 505 | 716 | 1.42 | NM_022266 | Connective tissue growth factor |
| Fgf8 | 22 | 40 | 1.80 | NM_133286 | Fibroblast growth factor-8 |
| Gdf9 | 626 | 951 | 1.52 | NM_021672 | Growth differentiation factor-9 |
| Stc1 | 644 | 392 | 0.61 | BF552244 | Stanniocalcin-1 |
| Development | |||||
| Lrrn3 | 66 | 107 | 1.62 | NM_030856 | Leucine-rich repeat protein-3, neuronal |
| Metabolism and transport | |||||
| Arg2 | 239 | 396 | 1.66 | NM_019168 | Arginase-2 |
| Aqp9 | 185 | 115 | 0.62 | NM_022960 | Aquaporin-9 |
| Ldhc | 62 | 99 | 1.61 | NM_017266 | Lactate dehydrogenase C |
| Sardh | 92 | 57 | 0.63 | AI103641 | Sarcosine dehydrogenase |
| LOC684802///LOC685152 | 76 | 49 | 0.65 | BI273855 | Similar to probable phospholipid transporting |
| RGD1565367_predicted | 124 | 66 | 0.53 | C07059 | Similar to solute carrier family 23, member 2 |
| Receptor and binding Pr | |||||
| Il17re | 127 | 83 | 0.65 | BM383766 | Interleukin 17 receptor E |
| Trpc3 | 248 | 153 | 0.62 | NM_021771 | Transient receptor potential cation channel |
| Transcription/epigenetic | |||||
| Arid1b | 136 | 86 | 0.63 | AI556402 | AT-rich interactive domain 1B (Swi1 like) |
| Irx3_predicted | 333 | 213 | 0.64 | AI713965 | Iroquois-related homeobox 3 (Drosophila) |
| ESTs | |||||
| RGD1308734 | 84 | 140 | 1.66 | BI285951 | Similar to RIKEN cDNA 1100001H23 |
| – | 103 | 61 | 0.59 | BM391972 | CDNA clone IMAGE:7366335 |
| RGD1310110_predicted | 116 | 176 | 1.52 | AI501165 | Similar to 3632451O06Rik protein (predicted) |
| RGD1307390_predicted | 80 | 123 | 1.54 | BI295093 | Similar to BC282485_1 (predicted) |
| RGD1309888 | 164 | 107 | 0.65 | BG371863 | Similar to RIKEN cDNA 1500002O20 |
| – | 159 | 101 | 0.64 | BF396928 | Transcribed locus |
| – | 122 | 79 | 0.65 | BE109251 | Transcribed locus |
| – | 104 | 61 | 0.59 | BF283840 | Transcribed locus |
| – | 123 | 201 | 1.64 | BM386842 | Transcribed locus, strongly similar to XP_001086149 |
| – | 165 | 96 | 0.58 | BE117343 | – |
| – | 457 | 257 | 0.56 | AI058976 | – |
Microarray of control and GDNF-treated postnatal day 4 rat ovaries after 2 days of culture.
Figure 9.

Microarray of control and GDNF-treated postnatal day 4 rat ovaries after 2 days of culture. Dendrogram shows transcripts that increased (brown) or decreased (green) in GDNF-treated versus control ovaries. Gene symbols provided for each altered expression with (−) indicating an EST.
Discussion
Ovaries from 4-day-old rats were cultured in vitro with and without exogenous GDNF. After culture, morphometric analysis showed that ovaries treated with GDNF displayed higher percentages of developing follicles, indicating that the rate at which arrested primordial follicles left the primordial pool and underwent primordial to primary follicle transition was stimulated by exogenous GDNF. This increased percentage of developing follicles in GDNF-treated ovaries was not due to changes in overall follicle pool size, as there was no significant difference in total follicle number per section between control and GDNF-treated ovaries. The slight decrease in follicle number observed after treatment could not account for the stimulation in primordial follicle development. This observation is similar to those previously observed with several other stimulating growth factors (Parrott & Skinner 1999, Nilsson et al. 2001, 2006, 2007, Kezele et al. 2005a). GDNF treatments stimulated primordial follicle development to the same extent as KL, a known promoter of primordial to primary follicle transition (Parrott & Skinner 1999, Nilsson & Skinner 2004). Localization of GDNF in neonatal rat ovaries further supported the evidence from organ culture experiments that GDNF promotes early follicle development. Immunohistochemical localization revealed that GDNF was present in the oocyte cytoplasm of primordial follicles and was in the oocytes, cumulus granulosa cells, and theca cells of antral (late-stage developing) follicles. This localization in granulosa cells near the oocyte suggests that factors produced and secreted by the oocyte may be differentially regulating GDNF expression in the cumulus granulosa cells of the antral follicle. GDNF expression in the theca cells tended to be limited to the theca externa layers. The presence of GDNF protein in antral follicles supports previous research localizing GDNF message to preovulatory follicles (Golden et al. 1999, Aravindakshan et al. 2006, Linher et al. 2007). Immunohistochemistry localized GFRα1 to oocyte cytoplasm of primordial and primary follicles, and to oocytes and mural granulosa cells of antral follicles. However, in some antral follicles, non-specific oocyte staining was quite prominent, raising the possibility that GFRα1 levels are low in some oocytes. GFRα1 was also present in the theca and ovarian surface epithelium. GFRα1 expression was the strongest in the mural granulosa cells (Fig. 5O and P) and theca interna layer of cells (Fig. 5G, O and P). Additional studies are needed to confirm the localization of GFRα1 to subpopulations of granulosa and theca cells. Western blot analysis was used to verify the specificity of the GFRα1 antibody (data not shown), which confirms observations from previous studies (Wang et al. 2004, Serra et al. 2005). Localization of GDNF and GFRα1 in primordial and primary follicles was the same in both cultured neonatal ovaries and adult uncultured ovaries. This suggests that culture conditions did not affect Gdnf and GFRα1 expression patterns in neonatal ovaries. To confirm the immunohistochemical observations, RT-PCR procedure localized Gdnf mRNA to antral oocytes and granulosa cells, and GFRα1 (Gfra1) mRNA to antral granulosa cells, but not to antral follicle oocytes. Oocytes from antral follicles have some non-specific immunohistochemical staining, so it is possible some antral follicle oocytes have very low levels of GFRα1 protein. Therefore, the immunohistochemical studies suggest that GFRα1 is still expressed in antral oocytes, although possibly decreasing in the largest preovulatory follicles. This expression decrease may also explain why GFRα1 is not detected in the antral oocyte samples by RT-PCR. Faint GFRα1 bands are amplified from antral oocyte samples, although it is not clear whether these bands reflect low oocyte expression or granulosa cell contamination. Another possibility is that antral oocytes do not themselves produce GFRα1 protein, but instead GFRα1 expressed by surrounding granulosa cells is cleaved free of its GPI membrane anchor and taken up by oocytes, so no oocyte Gfra1 mRNA is detectable. Further studies are needed to confirm the cellular expression pattern for GFRα1.
Combined observations suggest endogenous ovarian GDNF promotes the primordial to primary follicle transition through autocrine cell–cell signaling to GFRα1 receptors on oocytes. In late-stage antral follicles, the expression of GDNF changes to include cumulus granulosa cells, while the receptor GFRα1 expression decreases in the oocytes. This shift in ligand and receptor expression alters the physiological function of GDNF to a signaling factor with both autocrine and paracrine roles in developing antral follicles (Fig. 6D; Table 1). GDNF may also have an autocrine function for the antral follicle theca cells. Localization of GDNFand its receptor GFRα1 to oocyte cytoplasm presents one of the first examples of autocrine signaling to regulate primordial follicle development (Fig. 6C). The mechanism by which GDNF promotes follicle development remains to be fully elucidated.
Interestingly, GDNF expression in the ovary is the opposite of GDNF expression in analogous tissues in the testes. Often paracrine or endocrine signaling ligands have similar localization patterns in the gonads of males and females, such that a particular growth factor is expressed by the germ cells (oocytes and spermatogonia) in both sexes, or in the epithelial cells (Sertoli and granulosa) of both sexes. However, there is a difference between male and female GDNF localization. In males, GDNF is expressed by the Sertoli cells and GFRα1 is in the germ cells. GDNF signaling to spermatogonia stimulates proliferation and self-renewal after division (Widenfalk et al. 2000, Wu et al. 2005, Naughton et al. 2006). A similar function in females is not possible, as postnatal oocytes are arrested in meiosis and do not undergo cell division. This may explain why the site of GDNF production is different in females. Ovarian GDNF and GFRα1 are both present in the germ cell (i.e., oocyte). It is possible that over the course of evolution, GDNF’s role diverged from spermatogonial self-renewal in the male to autocrine signaling that promotes the primordial to primary follicle transition in the female. Autocrine signaling loops have been shown to promote cell viability in neural, immune, and tumor cells, and to regulate estradiol production by granulosa cells (Meerschaert et al. 1999, Ireland et al. 2004, Schumacher et al. 2004, Datta & Datta 2006). Preserving oocyte viability may be an important aspect of autocrine GDNF signaling in the oocytes of early-stage follicles. Therefore, GDNF is likely essential for germ cell survival in both the testis and ovary, but the sites of expression are distinct.
Other possible mechanisms for GDNF action in the neonatal rat ovary include interactions with other extracellular signaling factors, thus indirectly promoting follicle development. The known network of extracellular signaling factors involved in primordial to primary follicle transition includes LIF (Nilsson et al. 2002), BMP4 (Nilsson & Skinner 2003), PDGF (Nilsson et al. 2006), nerve growth factor (Dissen et al. 2002), bFGF (Nilsson & Skinner 2004), and KL (Parrott & Skinner 1999, Nilsson & Skinner 2004), all of which promote development (Skinner 2005). KL may be expressed as either free (KL-1) or membrane bound (KL-2) isoforms. There is evidence that KL-2 is the necessary isoform for oocyte growth and formation of germ cells (Flanagan et al. 1991, Thomas et al. 2007). However, soluble KL-1 can promote primordial follicle transition (Parrott & Skinner 1999). AMH and SDF1 inhibit primordial to primary follicle transition (Ikeda et al. 2002, Skinner 2005, Holt et al. 2006, Nilsson et al. 2006). It was hypothesized that GDNF participated in this network by up-regulating KL or other factors that promote primordial to primary follicle transition. However, data from real-time PCR indicate that exogenous GDNF does not influence KL expression. This observation indicates that GDNF acts in a signaling pathway that is not dependent on changing KL expression.
The microarray analysis suggests that GDNF activity affects a variety of secreted growth factors, including AMH, CTGF, FGF8, GDF9, and STC1. AMH performs many functions in the ovary, the most relevant being that it inhibits the primordial to primary follicle transition in rat, mouse, and humans (Durlinger et al. 2002, Carlsson et al. 2006, Nilsson et al. 2007). Similarly, GDF9 is also critical for normal follicle development. GDF9 is an oocyte-secreted factor necessary for follicle development from the primary follicle stage through ovulation (Dong et al. 1996, Elvin et al. 1999). The other factors flagged by the microarray analysis have less documented function in early follicle development. FGF8 is expressed in antral follicles in rat and cow. It may also play a role in the development of ovarian and breast tumors (Valve et al. 1997, 2000, Daphna-Iken et al. 1998, Zammit et al. 2002, Buratini et al. 2005). STC1 was originally shown to have high expression levels in the ovary (Varghese et al. 1998). Further studies described an increase in STC1 expression during pregnancy and lactation, and a possible function in luteinization inhibition (Ishibashi & Imai 2002, Luo et al. 2004). STC1 knockout mice develop as phenotypically and reproductively normal (Chang et al. 2005). CTGF is a matrix-associated protein with many possible roles in development and differentiation processes (Leask & Abraham 2003). CTGF is regulated by gonadotropins and is expressed in antral follicles and corpus lutea (Landis 2002, Harlow et al. 2007). Early follicle development is a vital process for female fertility, and GDNF participates in a network of compensatory factors that act as checks and balances. Therefore, GDNF alone only modestly alters the ovarian transcriptome. Manipulation of the primordial to primary follicle transition for potential therapeutic purposes will likely need to target many factors congruently.
In males, GDNF promotes spermatogonial stem cell self-renewal by signaling through src family kinases (Braydich-Stolle et al. 2007, Oatley et al. 2007). Interestingly, src signaling is necessary in females for antral follicle development (Roby et al. 2005), and in primordial germ cells src kinase activation is vital for migration to the gonad and is a part of the KL signaling cascade (De Miguel et al. 2002, Farini et al. 2007). This leads to the speculation that GDNF may signal through src family kinases to promote primordial to primary follicle transition. The study of GDNF and GFRα1 knockout animals is difficult as these animals die prior to the onset of primordial to primary follicle transition due to kidney agenesis (Pichel et al. 1996, Sanchez et al. 1996, Cacalano et al. 1998). Further studies are necessary to fully characterize GDNF interactions in other extracellular signaling pathways in primordial follicle development. Since GDNF appears to act in an autocrine manner on the oocyte, other oocyte factors such as bFGF may be involved.
In summary, experiments using postnatal day 4 cultured rat ovaries were performed to explore how GDNF influences primordial to primary follicle transition. Observations from organ cultures, combined with immunohistochemical and RT-PCR localization, indicate that GDNF does promote the primordial to primary follicle transition, and GDNF and GFRα1 are present specifically in the oocyte cytoplasm of the primordial follicle. Combined data indicate GDNF is a part of the network of extracellular signaling factors that regulate primordial to primary follicle transition. Understanding the regulation of this process may lead to the development of therapies for reproductive ailments such as premature ovarian failure, or to methods to control the transition into menopause. The ability to regulate the primordial follicle pool size will directly influence female reproductive potential and associated endocrine abnormalities.
Materials and Methods
Organ culture
Sprague–Dawley rats were obtained from a Washington State University breeding colony. All animal procedures were approved by the WSU Animal Care and Use Committee. Ovaries dissected from 4-day-old female rat pups were maintained in a whole organ culture system on floating filters (0.4 μm Millicell-CM; Millipore Corp., Billerica, MA, USA) in 0.5 ml DMEM–Ham’s F-12 medium (1:1, vol/vol; Life Technologies Inc.) containing 0.1% BSA (Sigma), 0.1% AlbuMAX (Life Technologies Inc.), 200 ng/ml insulin (rh Insulin; Sigma), 0.05 mg/ml L-ascorbic acid (Sigma), and 27.5 μg/ml transferrin (Sigma) in a 4-well culture plate (Nunc plate; Applied Scientific, South San Francisco, CA, USA). The medium was supplemented with final concentration 5 μg/ml gentamicin, 3.25 μg/ml streptomycin, and 3.25 U/ml penicillin to prevent bacterial contamination. Ovaries were treated with no factor (control), GDNF (r-metHu GDNF, 50 ng/ml; Amgen, Thousand Oaks, CA, USA), KL/stem cell factor (rm-SCF, 50 ng/ml; R&D Systems, Minneapolis, MN, USA), or GDNF and KL combined. The 50 ng/ml GDNF dose was found to be optimal since an increased dose (500 ng/ml) had the same biological response. Two to three ovaries were placed on each filter, and no two ovaries from the same animal were placed into the same treatment group. Culture medium and treatments were replaced every 2 days. After 10 days, ovaries were fixed in Bouin’s fixative (Sigma) for 1 h followed by immersion in 70% ethanol. Tissues were paraffin embedded, sectioned at 3 μm, and hematoxylin/eosin stained. The experiments were repeated so that each treatment group contained five to seven different ovaries.
Morphological analysis
The number of follicles at each developmental stage was counted and averaged in two serial sections from the largest cross section through the center of the ovary and averaged. Studies by Bucci et al. (1997) have shown that ovarian follicle counts from a representative sample of as few as 1% of sections gives the same results as counting every fifth section (i.e., 20%). Previously, the data obtained from this analysis of two mid-diameter cross sections have been shown to provide similar results as the analysis of compiled data from all serial sections (data not shown; Parrott & Skinner 1999, Nilsson & Skinner 2003). In addition, total follicle number per section does not change between treatment groups as shown with a number of previous studies (Nilsson et al. 2001, 2006, 2007, Kezele et al. 2005a). Rather, only the percentage of follicles at each developmental stage changes with treatment (Nilsson et al. 2001, 2002). Follicles in ovarian cross sections were classified as primordial (stage 0) or developing (stages 1–4: early primary, primary, transitional, and pre-antral) as described previously (Oktay et al. 1995). Primordial follicles consist of an oocyte arrested in prophase I of meiosis that is partially or completely encapsulated by flattened squamous pre-granulosa cells. Early primary follicles have initiated development (i.e., undergone primordial to primary follicle transition) and contain at least two cuboidal granulosa cells. Primary, transitional, and pre-antral follicles exhibit one or more complete layers of cuboidal granulosa cells. Four-day-old ovaries contain predominantly primordial follicles (Parrott & Skinner 1999). Hematoxylin/eosin-stained ovarian sections were analyzed at 400× magnification using a light microscope. The populations of follicles were determined by the analysis of follicle morphology, rather than oocyte counts, to assess follicle developmental stages.
Immunohistochemistry
Ovary sections from freshly isolated adult or cultured postnatal day 4 (P4) ovaries (cultured for 10 days) were immunostained as described previously (Nilsson et al. 2002), for the presence of GDNF using anti-GDNF primary antibody (anti-GDNF rabbit IgG, 5 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Briefly, 3 μm sections were deparaffinized, rehydrated through a graded ethanol series, boiled in 10 mM sodium citrate buffer, quenched in 3% hydrogen peroxide/20% methanol and 0.1% Triton-X solution, and then blocked with 10% goat serum (normal goat serum; Vector Laboratories Inc., Burlingame, CA, USA) for 20 min prior to incubation with 5 μg/ml primary antibody for 12 h. The sections were then washed with PBS and incubated with 1:300 diluted biotinylated secondary antibody for 45 min (goat anti-rabbit IgG; Vector Laboratories Inc.), washed again, and incubated with streptavidin peroxidase (Zymed, San Francisco, CA, USA) prior to color development with a DAB peroxidase substrate kit (Vector Laboratories Inc). Following development, the sections were dehydrated, coverslips mounted with xylene-based medium (Cytoseal-XYL; Richard Allan Scientific, Kalamazoo, MI, USA), and analyzed at 200×, 400×, and 1000×magnification using light microscope. Negative control experiments were performed using a non-specific primary antibody at 5 μg/ml (rabbit IgG; Sigma). Non-cultured adult rat ovaries were also stained for the presence of GDNF with the same antibody (anti-GDNF rabbit IgG, 5–10 μg/ml; Santa Cruz Biotechnology) using the protocol described below for GFRα1 localization.
Neonatal cultured and freshly isolated adult rat ovaries were stained for GFRα1 localization using anti-GFRα1 primary antibody (anti-GFRα1 rabbit IgG, 5–10 μg/ml; Santa Cruz Biotechnology). The protocol for GFRα1 immunohistochemistry excluded the boiling step and introduced a second wash with 0.1% Triton-X solution following incubation with primary antibody. Negative control experiments were performed using a non-specific primary antibody at 5–10 μg/ml (rabbit IgG; Sigma). The protocol was otherwise identical to that used for GDNF staining.
Antral follicle cell preparation
Female 24-day-old rats were injected with 10 IU pregnant mares serum gonadotrophin subcutaneously to induce follicle development. Forty-eight-hour post-injection ovaries were extracted, bursae and fat trimmed away, and ovaries placed in the plates of F12/DMEM media (1:1, vol/vol; Life Technologies Inc.) supplemented with a final concentration of 5 μg/ml gentamicin, 3.25 μg/ml streptomycin, and 3.25 U/ml penicillin to prevent bacterial contamination. Ovaries were then incubated at 37 °C for 30 min in EGTA (6 mM; Sigma), transferred to a sucrose solution (0.5 M; Life Technologies Inc.), and incubated for another 30 min (both solutions made in media above). Following incubation, ovaries were again placed in F12/DMEM + antibiotics medium, where large follicles were lanced open to extrude granulosa cells and oocytes. Oocytes were aspirated into a separate tube creating two cell preparations, each enriched for either oocytes or granulosa cells from antral follicles. The oocyte preparation was further subjected to gentle aspiration through a 100 μl pipette tip and the resulting supernatant, containing dislodged granulosa cells, was discarded. This created oocyte preparations in which some granulosa cells would still adhere to many oocytes, and so a small amount of granulosa cell contamination is expected. All other cell types remain associated with the ovarian connective tissue. The isolated cell populations were placed in Trizol reagent at −75° (Sigma) for storage until RNA isolation.
RT-PCR
Granulosa cells and oocytes were extracted from antral rat follicles or microdissected as described above. The cells were immersed in Trizol reagent (Sigma) and RNA extracted according to the manufacturer’s protocols. RNA samples were DNase treated with the TURBO DNA-free kit (Ambion, Austin, TX, USA) prior to reverse transcription of RNA to cDNA according to the standard oligo-dT RT protocol in a reaction volume of 25 μl. Negative controls for each sample were created at this step by omitting MMLV enzyme in the RT reaction. All PCR primer sets span intron–exon boundaries. PCR used the following primers for Gdnf (NCBI: NM_019139), forward: 5′-CTGGAAGATTCCCCGTATGA-3′ and reverse: 5′-TCTTCGGGCATATTGGAGTC-3′. A second PCR for GFRα1 (Gfra1) used 2 μl of the first PCR product as template, and the previous reverse primer was used in conjunction with a nested forward primer: 5′-CTGTCTGCCTGGTGTTGCT-3′. Gdnf thermocycling conditions were as follows: 95 °C for 4 min, 35 cycles of 95 °C for 30 s, 60 °C for 60 s, 72 °C for 30 s, followed by 72 °C for 5 min. Second Gdnf PCR required 20 cycles. Primers for Gfra1 (NCBI: NM_012959.1) were forward: 5′-GGCAGTCCCGTTCATATCAG-3′ and reverse: 5′-AGCA-GAAGAGCATCCCGTAG-3′. Second PCR for Gdnf used 1 μl of the first PCR product as template and identical primers. The protocol for Gfra1 amplification was as follows: 95 °C for 4 min, 35 cycles of 95 °C for 30 s, 60 °C for 60 s, 72 °C for 30 s, followed by 72 °C for 5 min. Second Gfra1 PCR required 20 cycles. Gdnf and Gfra1 PCR products were electrophoretically analyzed and sequenced to confirm identity. Amplification of the ribosomal protein S2 was used as a reference standard. The S2 reference gene primers (NCBI: NM_031838) were rS2-F, 5′-CTGCTCCTGTGCCCAA-GAAG-3′ and rS2-R, 5′-AAGGTG-GCCTTGGCAAAGTT-3′ . Ribosomal S2 mRNA expression does not change in ovarian cells regardless of treatment (Kezele et al. 2005a).
Real-time PCR
Ovaries from 4-day-old female rat pups were maintained in the organ culture system described above for 2 days with GDNF treatment (r-metHu GDNF, 50 ng/ml; Amgen), or were left untreated as controls. Two ovaries from each culture well were combined to make each RNA sample. After 2 days of culture, ovaries were homogenized in Trizol reagent (Sigma) and RNA extracted according to the manufacturer’s protocols. RNA samples were DNase treated with TURBO DNA-free kit (Ambion) prior to reverse transcription of RNA to cDNA according to the standard oligo-dT RT protocol in a reaction volume of 25 μl. Negative controls for each sample were created at this step by omitting MMLV enzyme in the RT reaction. Five microliters of each 1:10 diluted cDNA sample, as well as each 1:10 diluted negative control sample, were used as template for real-time PCR analysis. Each sample was run in triplicate. The Platinum SYBR Green qPCR SuperMix kit (Invitrogen) was used according to the manufacturer’s instructions for detection of KL (Kitl ). The Kitl primers (NCBI: NM_021843) were rKitl-720: 5′-ATTTATGTTACCCCCTGTTG-CAGCC-3′ and rKitl-859: 5′-CAATTACAAGCGAAATGA-GAGCCG-3′ . SYBR Green was also used to detect the ribosomal protein gene S2, which was used as the reference standard for real-time PCR. Real-time PCR was performed on an ABI-7000 real-time machine with the following protocol: 50 °C for 2 min, 95 °C for 2 min, then 40 cycles of 95 °C for 15 s, and 66 °C for 30 s. Fluorescent detection data were analyzed for KL mRNA levels and normalized to S2 mRNA levels, and then GDNF-treated sample KL levels were normalized to untreated control KL mRNA levels.
Microarray and bioinformatics
Postnatal day 4 rat ovaries were cultured for 2 days in the presence or absence of GDNF (r-metHu GDNF, 50 ng/ml; Amgen). Culture conditions were identical to those described for 10-day organ culture experiments. Each sample contained six to ten pooled ovaries, and no two ovaries from the same animal were placed into the same treatment group. For each control and treated sample, two biological replicates were produced using different sets of ovaries. After culture ovaries were placed into Trizol reagent for RNA extraction as per the manufacturer’s protocols.
RNA was hybridized to the Affymetrix (Santa Clara, CA, USA) Rat 230 2.0 gene chip. One chip was used for each biological replicate (i.e., four chips total: two for control and two for treated). The number of chips required for specific experiments has been previously reviewed (Chen et al. 2004). The Genomics Core in the Center for Reproductive Biology at the Washington State University performed the analysis as described previously (McLean et al. 2002, Shima et al. 2004). Briefly, RNA from control and treated cultured ovaries were reverse transcribed into cDNA, which was transcribed into biotin-labeled RNA. Biotin-labeled RNA was then hybridized to the Affymetrix Rat 230 2.0 gene chips. Biotinylated RNA was then visualized by labeling with phycoerythrin-coupled avidin. The microarray chip was scanned on an Affymetrix Gene Chip Scanner 3000 (Affymetrix). The microarray image data were converted to numerical data with GeneChip Operating Software (GCOS version 1.2; Affymetrix) using a probe set target signal of 125. An analysis was performed with GCOS to assess the relative abundance of the transcripts based on signal and detection calls (present, absent, or marginal). The 11 perfect match and 11 mismatch oligonucleotides for a specific gene were used to statistically determine present/absent calls using a one-sided Wilcoxon’s signed-rank test.
In GCOS, the Excel files were generated with expression signals and absent/present calls for each probe set. Using the Excel files, R2 for control or treated sample replicates were calculated (>0.98), indicating negligible total variability between chips, experiments, and samples. The Excel files from GCOS were imported into GeneSpring software (Silicon Genetics, Redwood City, CA, USA) and normalized using the recommended defaults. This includes setting signal values below 0.01 to a value of 0.01, total chip normalization to the 50th percentile, and per gene normalization to the median. Unless otherwise indicated, in order for a transcript to be considered present, it had to be both tagged as present in the GCOS present/absent call, and have an expression level >75. In order for a transcript to be considered changed between treatment groups, it had to exhibit at least a 1.5-fold change between the means of the treatments and have a Student’s t-test P value of <0.05 between control and treatment samples. Therefore, the data presented are for genes that were determined to be statistically present and found to be statistically different from control with a given treatment.
Previous studies have demonstrated that microarray data are validated with quantitative PCR data (Shima et al. 2004, Kezele et al. 2005b). Due to the presence of 11 different oligonucleotide sets for each specific gene being used on the microarray versus only a single primer set for a gene in a quantitative PCR, the microarray is more effective at eliminating false-positive or -negative data and provides a more robust quantitation of changes in gene expression. However, validation of microarray data was performed with the KL gene using a real-time PCR procedure, as described in the Materials and Methods section. As presented in the Results section, similar data were obtained with the real-time PCR analysis as with the microarray analysis.
Statistical analysis
Organ culture treatment groups were compared using one-way ANOVA. Following a significant result with ANOVA, treated groups were compared with the control using Dunnet’s multiple comparison post hoc test. Real-time PCR treatment groups were compared using a one-sample t-test. All statistics were calculated with the help of GraphPad Prism version 4.0b software (GraphPad Software, Inc., San Diego, CA, USA).
Acknowledgments
We acknowledge the technical contributions of Dr Ingrid Sadler-Riggleman and Dr Marina Savenkova, and thank Dr Mary Hunzicker-Dunn for the initial antral follicle cell samples and for critically reviewing the manuscript. We also acknowledge the assistance of Ms Rochelle Pedersen and Ms Jill Griffin in preparation of this manuscript. The authors declare that they have no competing financial interests. This research was supported in part by a grant from the National Institute of Health, NIH/NICHD to M K S. Grant Statement: The research was supported by an NIH (NICHD) grant to M K S. The authors have no conflict of interest to disclose.
References
- Amoresano A, Incoronato M, Monti G, Pucci P, de Franciscis V, Cerchia L. Direct interactions among Ret, GDNF and GFRalpha1 molecules reveal new insights into the assembly of a functional three-protein complex. Cellular Signalling. 2005;17:717–727. doi: 10.1016/j.cellsig.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Aravindakshan J, Chen XL, Sairam MR. Age-dependent bimodal GDNF regulation during ovarian tumorigenesis in follitropin receptor mutant mice. Biochemical and Biophysical Research Communications. 2006;351:507–513. doi: 10.1016/j.bbrc.2006.10.069. [DOI] [PubMed] [Google Scholar]
- Braydich-Stolle L, Kostereva N, Dym M, Hofmann MC. Role of Src family kinases and N-Myc in spermatogonial stem cell proliferation. Developmental Biology. 2007;304:34–45. doi: 10.1016/j.ydbio.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci TJ, Bolon B, Warbritton AR, Chen JJ, Heindel JJ. Influence of sampling on the reproducibility of ovarian follicle counts in mouse toxicity studies. Reproductive Toxicology. 1997;11:689–696. doi: 10.1016/s0890-6238(97)00034-8. [DOI] [PubMed] [Google Scholar]
- Buratini J, Jr, Teixeira AB, Costa IB, Glapinski VF, Pinto MG, Giometti IC, Barros CM, Cao M, Nicola ES, Price CA. Expression of fibroblast growth factor-8 and regulation of cognate receptors, fibroblast growth factor receptor-3c and -4, in bovine antral follicles. Reproduction. 2005;130:343–350. doi: 10.1530/rep.1.00642. [DOI] [PubMed] [Google Scholar]
- Cacalano G, Farinas I, Wang LC, Hagler K, Forgie A, Moore M, Armanini M, Phillips H, Ryan AM, Reichardt LF, et al. GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron. 1998;21:53–62. doi: 10.1016/s0896-6273(00)80514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson IB, Scott JE, Visser JA, Ritvos O, Themmen AP, Hovatta O. Anti-Mullerian hormone inhibits initiation of growth of human primordial ovarian follicles in vitro. Human Reproduction. 2006;21:2223–2227. doi: 10.1093/humrep/del165. [DOI] [PubMed] [Google Scholar]
- Carmillo P, Dago L, Day ES, Worley DS, Rossomando A, Walus L, Orozco O, Buckley C, Miller S, Tse A, et al. Glial cell line-derived neurotrophic factor (GDNF) receptor alpha-1 (GFR alpha 1) is highly selective for GDNF versus artemin. Biochemistry. 2005;44:2545–2554. doi: 10.1021/bi049247p. [DOI] [PubMed] [Google Scholar]
- Chang AC, Cha J, Koentgen F, Reddel RR. The murine stanniocalcin 1 gene is not essential for growth and development. Molecular and Cellular Biology. 2005;25:10604–10610. doi: 10.1128/MCB.25.23.10604-10610.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Delongchamp RR, Tsai CA, Hsueh HM, Sistare F, Thompson KL, Desai VG, Fuscoe JC. Analysis of variance components in gene expression data. Bioinformatics. 2004;20:1436–1446. doi: 10.1093/bioinformatics/bth118. [DOI] [PubMed] [Google Scholar]
- Daphna-Iken D, Shankar DB, Lawshe A, Ornitz DM, Shackleford GM, MacArthur CA. MMTV-Fgf8 transgenic mice develop mammary and salivary gland neoplasia and ovarian stromal hyperplasia. Oncogene. 1998;17:2711–2717. doi: 10.1038/sj.onc.1202212. [DOI] [PubMed] [Google Scholar]
- Datta S, Datta MW. Sonic Hedgehog signaling in advanced prostate cancer. Cellular and Molecular Life Sciences. 2006;63:435–448. doi: 10.1007/s00018-005-5389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettin L, Ravindranath N, Hofmann MC, Dym M. Morphological characterization of the spermatogonial subtypes in the neonatal mouse testis. Biology of Reproduction. 2003;69:1565–1571. doi: 10.1095/biolreprod.103.016394. [DOI] [PubMed] [Google Scholar]
- Dissen GA, Romero C, Paredes A, Ojeda SR. Neurotrophic control of ovarian development. Microscopy and Research Technique. 2002;59:509–515. doi: 10.1002/jemt.10227. [DOI] [PubMed] [Google Scholar]
- Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383:531–535. doi: 10.1038/383531a0. [DOI] [PubMed] [Google Scholar]
- Durlinger AL, Gruijters MJ, Kramer P, Karels B, Ingraham HA, Nachtigal MW, Uilenbroek JT, Grootegoed JA, Themmen AP. Anti-Mullerian hormone inhibits initiation of primordial follicle growth in the mouse ovary. Endocrinology. 2002;143:1076–1084. doi: 10.1210/endo.143.3.8691. [DOI] [PubMed] [Google Scholar]
- Elvin JA, Yan C, Wang P, Nishimori K, Matzuk MM. Molecular characterization of the follicle defects in the growth differentiation factor 9-deficient ovary. Molecular Endocrinology. 1999;13:1018–1034. doi: 10.1210/mend.13.6.0309. [DOI] [PubMed] [Google Scholar]
- Farini D, La Sala G, Tedesco M, De Felici M. Chemoattractant action and molecular signaling pathways of Kit ligand on mouse primordial germ cells. Developmental Biology. 2007;306:572–583. doi: 10.1016/j.ydbio.2007.03.031. [DOI] [PubMed] [Google Scholar]
- Flanagan JG, Chan DC, Leder P. Transmembrane form of the kit ligand growth factor is determined by alternative splicing and is missing in the Sld mutant. Cell. 1991;64:1025–1035. doi: 10.1016/0092-8674(91)90326-t. [DOI] [PubMed] [Google Scholar]
- Gash DM, Zhang Z, Gerhardt G. Neuroprotective and neurorestorative properties of GDNF. Annals of Neurology. 1998;44:S121–S125. doi: 10.1002/ana.410440718. [DOI] [PubMed] [Google Scholar]
- Golden JP, DeMaro JA, Osborne PA, Milbrandt J, Johnson EM., Jr Expression of neurturin, GDNF, and GDNF family-receptor mRNA in the developing and mature mouse. Experimental Neurology. 1999;158:504–528. doi: 10.1006/exnr.1999.7127. [DOI] [PubMed] [Google Scholar]
- Harlow CR, Bradshaw AC, Rae MT, Shearer KD, Hillier SG. Oestrogen formation and connective tissue growth factor expression in rat granulosa cells. Journal of Endocrinology. 2007;192:41–52. doi: 10.1677/joe.1.06689. [DOI] [PubMed] [Google Scholar]
- Hirshfield AN. Development of follicles in the mammalian ovary. International Review of Cytology. 1991;124:43–101. doi: 10.1016/s0074-7696(08)61524-7. [DOI] [PubMed] [Google Scholar]
- Holt JE, Jackson A, Roman SD, Aitken RJ, Koopman P, McLaughlin EA. CXCR4/SDF1 interaction inhibits the primordial to primary follicle transition in the neonatal mouse ovary. Developmental Biology. 2006;293:449–460. doi: 10.1016/j.ydbio.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Nagai A, Ikeda MA, Hayashi S. Increased expression of Mullerian-inhibiting substance correlates with inhibition of follicular growth in the developing ovary of rats treated with E2 benzoate. Endocrinology. 2002;143:304–312. doi: 10.1210/endo.143.1.8603. [DOI] [PubMed] [Google Scholar]
- Ireland JL, Jimenez-Krassel F, Winn ME, Burns DS, Ireland JJ. Evidence for autocrine or paracrine roles of alpha2-macroglobulin in regulation of estradiol production by granulosa cells and development of dominant follicles. Endocrinology. 2004;145:2784–2794. doi: 10.1210/en.2003-1407. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Imai M. Prospect of a stanniocalcin endocrine/paracrine system in mammals. American Journal of Physiology. Renal Physiology. 2002;282:F367–F375. doi: 10.1152/ajprenal.00364.2000. [DOI] [PubMed] [Google Scholar]
- Karihaloo A, Nickel C, Cantley LG. Signals which build a tubule. Nephron. Experimental Nephrology. 2005;100:e40–e45. doi: 10.1159/000084111. [DOI] [PubMed] [Google Scholar]
- Kezele P, Nilsson E, Skinner MK. Cell–cell interactions in primordial follicle assembly and development. Frontiers in Bioscience. 2002;7:d1990–d1996. doi: 10.2741/kezele. [DOI] [PubMed] [Google Scholar]
- Kezele P, Nilsson EE, Skinner MK. Keratinocyte growth factor acts as a mesenchymal factor that promotes ovarian primordial to primary follicle transition. Biology of Reproduction. 2005a;73:967–973. doi: 10.1095/biolreprod.105.043117. [DOI] [PubMed] [Google Scholar]
- Kezele PR, Ague JM, Nilsson E, Skinner MK. Alterations in the ovarian transcriptome during primordial follicle assembly and development. Biology of Reproduction. 2005b;72:241–255. doi: 10.1095/biolreprod.104.032060. [DOI] [PubMed] [Google Scholar]
- Kirik D, Georgievska B, Bjorklund A. Localized striatal delivery of GDNF as a treatment for Parkinson disease. Nature Neuroscience. 2004;7:105–110. doi: 10.1038/nn1175. [DOI] [PubMed] [Google Scholar]
- Kordower JH, Emborg ME, Bloch J, Ma SY, Chu Y, Leventhal L, McBride J, Chen EY, Palfi S, Roitberg BZ, et al. Neurodegeneration prevented by lentiviral vector delivery of GDNF in primate models of Parkinson’s disease. Science. 2000;290:767–773. doi: 10.1126/science.290.5492.767. [DOI] [PubMed] [Google Scholar]
- Landis SE. Buncombe County Medical Society Project Access. Expanding access to care at the local level. North Carolina Medical Journal. 2002;63:23–29. [PubMed] [Google Scholar]
- Leask A, Abraham DJ. The role of connective tissue growth factor, a multifunctional matricellular protein, in fibroblast biology. Biochemistry and Cell Biology. 2003;81:355–363. doi: 10.1139/o03-069. [DOI] [PubMed] [Google Scholar]
- Lee WS, Yoon SJ, Yoon TK, Cha KY, Lee SH, Shimasaki S, Lee S, Lee KA. Effects of bone morphogenetic protein-7 (BMP-7) on primordial follicular growth in the mouse ovary. Molecular Reproduction and Development. 2004;69:159–163. doi: 10.1002/mrd.20163. [DOI] [PubMed] [Google Scholar]
- Linher K, Wu D, Li J. Glial cell line-derived neurotrophic factor: an intraovarian factor that enhances oocyte developmental competence in vitro. Endocrinology. 2007;148:4292–4301. doi: 10.1210/en.2007-0021. [DOI] [PubMed] [Google Scholar]
- Luo CW, Kawamura K, Klein C, Hsueh AJ. Paracrine regulation of ovarian granulosa cell differentiation by stanniocalcin (STC) 1: mediation through specific STC1 receptors. Molecular Endocrinology. 2004;18:2085–2096. doi: 10.1210/me.2004-0066. [DOI] [PubMed] [Google Scholar]
- McLean DJ, Friel PJ, Pouchnik D, Griswold MD. Oligonucleotide microarray analysis of gene expression in follicle-stimulating hormone-treated rat Sertoli cells. Molecular Endocrinology. 2002;16:2780–2792. doi: 10.1210/me.2002-0059. [DOI] [PubMed] [Google Scholar]
- Meerschaert J, Vrtis RF, Shikama Y, Sedgwick JB, Busse WW, Mosher DF. Engagement of alpha4beta7 integrins by monoclonal antibodies or ligands enhances survival of human eosinophils in vitro. Journal of Immunology. 1999;163:6217–6227. [PubMed] [Google Scholar]
- De Miguel MP, Cheng L, Holland EC, Federspiel MJ, Donovan PJ. Dissection of the c-Kit signaling pathway in mouse primordial germ cells by retroviral-mediated gene transfer. PNAS. 2002;99:10458–10463. doi: 10.1073/pnas.122249399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton CK, Jain S, Strickland AM, Gupta A, Milbrandt J. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biology of Reproduction. 2006;74:314–321. doi: 10.1095/biolreprod.105.047365. [DOI] [PubMed] [Google Scholar]
- Nilsson EE, Skinner MK. Bone morphogenetic protein-4 acts as an ovarian follicle survival factor and promotes primordial follicle development. Biology of Reproduction. 2003;69:1265–1272. doi: 10.1095/biolreprod.103.018671. [DOI] [PubMed] [Google Scholar]
- Nilsson EE, Skinner MK. Kit ligand and basic fibroblast growth factor interactions in the induction of ovarian primordial to primary follicle transition. Molecular and Cellular Endocrinology. 2004;214:19–25. doi: 10.1016/j.mce.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Nilsson E, Parrott JA, Skinner MK. Basic fibroblast growth factor induces primordial follicle development and initiates folliculogenesis. Molecular and Cellular Endocrinology. 2001;175:123–130. doi: 10.1016/s0303-7207(01)00391-4. [DOI] [PubMed] [Google Scholar]
- Nilsson EE, Kezele P, Skinner MK. Leukemia inhibitory factor (LIF) promotes the primordial to primary follicle transition in rat ovaries. Molecular and Cellular Endocrinology. 2002;188:65–73. doi: 10.1016/s0303-7207(01)00746-8. [DOI] [PubMed] [Google Scholar]
- Nilsson EE, Detzel C, Skinner MK. Platelet-derived growth factor modulates the primordial to primary follicle transition. Reproduction. 2006;131:1007–1015. doi: 10.1530/rep.1.00978. [DOI] [PubMed] [Google Scholar]
- Nilsson E, Rogers N, Skinner MK. Actions of anti-Mullerian hormone on the ovarian transcriptome to inhibit primordial to primary follicle transition. Reproduction. 2007;134:209–221. doi: 10.1530/REP-07-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. Journal of Biological Chemistry. 2007;282:25842–25851. doi: 10.1074/jbc.M703474200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktay K, Schenken RS, Nelson JF. Proliferating cell nuclear antigen marks the initiation of follicular growth in the rat. Biology of Reproduction. 1995;53:295–301. doi: 10.1095/biolreprod53.2.295. [DOI] [PubMed] [Google Scholar]
- Parrott JA, Skinner MK. Kit-ligand/stem cell factor induces primordial follicle development and initiates folliculogenesis. Endocrinology. 1999;140:4262–4271. doi: 10.1210/endo.140.9.6994. [DOI] [PubMed] [Google Scholar]
- Pichel JG, Shen L, Sheng HZ, Granholm AC, Drago J, Grinberg A, Lee EJ, Huang SP, Saarma M, Hoffer BJ, et al. Defects in enteric innervation and kidney development in mice lacking GDNF. Nature. 1996;382:73–76. doi: 10.1038/382073a0. [DOI] [PubMed] [Google Scholar]
- Pozas E, Ibanez CF. GDNF and GFRalpha1 promote differentiation and tangential migration of cortical GABAergic neurons. Neuron. 2005;45:701–713. doi: 10.1016/j.neuron.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Richardson SJ, Senikas V, Nelson JF. Follicular depletion during the menopausal transition: evidence for accelerated loss and ultimate exhaustion. Journal of Clinical Endocrinology and Metabolism. 1987;65:1231–1237. doi: 10.1210/jcem-65-6-1231. [DOI] [PubMed] [Google Scholar]
- Roby KF, Son DS, Taylor CC, Montgomery-Rice V, Kirchoff J, Tang S, Terranova PF. Alterations in reproductive function in SRC tyrosine kinase knockout mice. Endocrine. 2005;26:169–176. doi: 10.1385/ENDO:26:2:169. [DOI] [PubMed] [Google Scholar]
- Sanchez MP, Silos-Santiago I, Frisen J, He B, Lira SA, Barbacid M. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature. 1996;382:70–73. doi: 10.1038/382070a0. [DOI] [PubMed] [Google Scholar]
- Santoro N. Research on the mechanisms of premature ovarian failure. Journal of the Society for Gynecologic Investigation. 2001;8:S10–S12. doi: 10.1016/s1071-5576(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Schumacher M, Guennoun R, Robert F, Carelli C, Gago N, Ghoumari A, Gonzalez Deniselle MC, Gonzalez SL, Ibanez C, Labombarda F, et al. Local synthesis and dual actions of progesterone in the nervous system: neuroprotection and myelination. Growth Hormone and IGF Research. 2004;14(Suppl A):S18–S33. doi: 10.1016/j.ghir.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Serra MP, Quartu M, Mascia F, Manca A, Boi M, Pisu MG, Lai ML, Del Fiacco M. Ret, GFRalpha-1, GFRalpha-2 and GFRalpha-3 receptors in the human hippocampus and fascia dentata. International Journal of Developmental Neuroscience. 2005;23:425–438. doi: 10.1016/j.ijdevneu.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biology of Reproduction. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Skinner MK. Regulation of primordial follicle assembly and development. Human Reproduction Update. 2005;11:461–471. doi: 10.1093/humupd/dmi020. [DOI] [PubMed] [Google Scholar]
- Thomas FH, Ismail RS, Jiang JY, Vanderhyden BC. Kit ligand 2 promotes murine oocyte growth in vitro. Biology of Reproduction. 2007;78:167–175. doi: 10.1095/biolreprod.106.058529. [DOI] [PubMed] [Google Scholar]
- Valve E, Penttila TL, Paranko J, Harkonen P. FGF-8 is expressed during specific phases of rodent oocyte and spermatogonium development. Biochemical and Biophysical Research Communications. 1997;232:173–177. doi: 10.1006/bbrc.1997.6256. [DOI] [PubMed] [Google Scholar]
- Valve E, Martikainen P, Seppanen J, Oksjoki S, Hinkka S, Anttila L, Grenman S, Klemi P, Härkönen P. Expression of fibroblast growth factor (FGF)-8 isoforms and FGF receptors in human ovarian tumors. International Journal of Cancer. 2000;88:718–725. doi: 10.1002/1097-0215(20001201)88:5<718::aid-ijc6>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Vargas-Leal V, Bruno R, Derfuss T, Krumbholz M, Hohlfeld R, Meinl E. Expression and function of glial cell line-derived neurotrophic factor family ligands and their receptors on human immune cells. Journal of Immunology. 2005;175:2301–2308. doi: 10.4049/jimmunol.175.4.2301. [DOI] [PubMed] [Google Scholar]
- Varghese R, Wong CK, Deol H, Wagner GF, DiMattia GE. Comparative analysis of mammalian stanniocalcin genes. Endocrinology. 1998;139:4714–4725. doi: 10.1210/endo.139.11.6313. [DOI] [PubMed] [Google Scholar]
- Wang LM, Zhang Q, Zhang Q, Zhu W, He C, Lu CL, Ding DF, Chen ZY. Identification of the key amino acids of glial cell line-derived neurotrophic factor family receptor alpha1 involved in its biological function. Journal of Biological Chemistry. 2004;279:109–116. doi: 10.1074/jbc.M306287200. [DOI] [PubMed] [Google Scholar]
- Widenfalk J, Parvinen M, Lindqvist E, Olson L. Neurturin, RET, GFRalpha-1 and GFRalpha-2, but not GFRalpha-3, mRNA are expressed in mice gonads. Cell and Tissue Research. 2000;299:409–415. doi: 10.1007/s004419900068. [DOI] [PubMed] [Google Scholar]
- Wissel K, Wefstaedt P, Rieger H, Miller JM, Lenarz T, Stover T. Upregulation of glial cell line-derived neurotrophic factor and artemin mRNA in the auditory nerve of deafened rats. Neuroreport. 2006;17:875–878. doi: 10.1097/01.wnr.0000221836.26093.85. [DOI] [PubMed] [Google Scholar]
- Wu Z, Templeman JL, Smith RA, Mackay S. Effects of glial cell line-derived neurotrophic factor on isolated developing mouse Sertoli cells in vitro. Journal of Anatomy. 2005;206:175–184. doi: 10.1111/j.1469-7580.2005.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit C, Coope R, Gomm JJ, Shousha S, Johnston CL, Coombes RC. Fibroblast growth factor 8 is expressed at higher levels in lactating human breast and in breast cancer. British Journal of Cancer. 2002;86:1097–1103. doi: 10.1038/sj.bjc.6600213. [DOI] [PMC free article] [PubMed] [Google Scholar]