Abstract
The cAMP response element binding protein/activating transcription factor (CREB/ATF) family of transcription factors is hormone responsive and critical for nearly all mammalian cell types. The basic helix-loop-helix (bHLH) family of transcription factors is important during the development and differentiation of a wide variety of cell types. Independent studies of the role of the bHLH protein scleraxis in testicular Sertoli cells and paraxis in muscle development using yeast-2-hybrid screens provided the novel observation that bHLH proteins can directly interact with ATF/CREB family members. Analysis of the interactions demonstrated the helix-loop-helix domain of bHLH proteins directly interacts with the leucine zipper (ZIP) region of CREB2/ATF4 to form heterodimers. The direct bHLH–CREB2 binding interactions were supported using co-immunoprecipitation of recombinant proteins. Structural analysis of bHLH and ATF4 heterodimer using previous crystal structures demonstrated the heterodimer likely involves the HLH and Zip domains and has the potential capacity to bind DNA. Transfection assays demonstrated CREB2/ATF4 over-expression blocked stimulatory actions of scleraxis or paraxis. CREB1 inhibited MyoD induced myogenic conversion of C3H10T1/2 cells. CREB2/ATF4 and scleraxis are expressed throughout embryonic and postnatal testis development, with scleraxis specifically expressed in Sertoli cells. ATF4 and scleraxis null mutant mice both had similar adult testis phenotypes of reduced spermatogenic capacity. In summary, bHLH and CREB family members were found to directly heterodimerize and inhibit the actions of bHLH dimers on Sertoli cells and myogenic precursor cells. The observations suggest a mechanism for direct cross-talk between cAMP induced and bHLH controlled cellular differentiation.
Keywords: CREB, bHLH, ATF4, scleraxis, paraxis, transcriptional control, testis, muscle, Sertoli cell, cAMP signal transduction, knockout, spermatogenesis
INTRODUCTION
Distinct signal transduction pathways have increasingly been shown to cross-talk to allow for greater complexity and specificity in signaling. The targets for many signal transduction pathways are specific transcription factor families that act at unique DNA response elements to regulate transcription. In addition to combinatorial binding to specific response elements, direct interactions between members of different transcription factor families has been observed to integrate different pathways. The current study demonstrates that CREB and bHLH transcription factors can physically interact and link these two important transcriptional signaling pathways.
One of the best characterized signal transduction and transcriptional regulation pathways involves cAMP, protein kinase A (PKA) and the CREB transcription factor family (Montminy, 1997). Binding of ligands, such as follicle stimulating hormone (FSH), to G-protein coupled receptors leads to the production of cAMP by activated adenyl cylase and activation of PKA. The catalytic domain of PKA then translocates to the nucleus where it activates members of the CREB/ATF transcription factor family through phosphorylation. These factors in turn direct transcription by binding as dimers to DNA at cyclic AMP response elements (CRE). An important ATF/CREB family member is CREB2, also known as ATF4, which has been shown to regulate hematopoiesis, lens and skeletal development, cellular proliferation, differentiation, and long-term memory (Ameri and Harris, 2007). Further, aberrant over-expression of ATF4 is associated with cancer progression (Ameri et al., 2004; Bi et al., 2005). Consistent with the diversity of biological functions, ATF4 has been reported to interact with a broad set of signaling factors. ATF4 can form homodimers and heterodimers through a basic-leucine zipper (bZip) domain with members of the AP-1 and C/EBP family of proteins (Hai and Curran, 1991). Other factors predicted to interact with ATF4 through a non-bZip mechanism include, kinases (RSK2, NIPK ZIP, and SKIP3), transcription factors (p300, Runx2, Tax, Nrf2, RNA polymerase II subunit RPB3, and GPE1 binding protein), nuclear matrix protein Satb2, and βTrCP, the receptor component of the SCF E3 ubiquitin ligase (Ameri and Harris, 2007). This underscores the potential for integrating signaling pathways through the physical interaction of different regulatory factors of distinct signaling pathways.
The superfamily of basic helix-loop-helix (bHLH) proteins are a well characterized set of transcription factors that can promote cell lineage determination and differentiation in a variety of cell types. These proteins are defined by the inclusion of a helix-loop-helix domain essential for dimerization and a basic domain that mediates DNA binding to a hexanucleotide sequence known as an E-box (CANNTG). Members of the superfamily have been functionally subdivided based on expression pattern and dimer partners. Class A bHLH members, including Drosophila daughterless, mammalian E12/E47, and HEB (rat homolog REBα) have relatively ubiquitous expression and form heterodimers with cell lineage restricted class B bHLH proteins. Members of the Class B proteins are expressed in almost every tissue in the organism. Better characterized members of this class include, the Twist subfamily of bHLH factors (Twist1, Twist2, scleraxis, and paraxis; Quertermous et al., 1994; Li et al., 1995; Olson et al., 1996), the myogenic bHLH genes myf5, MyoD, myogenin and MRF4 for muscle (Davis et al., 1987; Braun et al., 1989; Edmondson and Olson, 1989), HAND1 and HAND2 for heart (Srivastava et al., 1995; Firulli et al., 1998), Sgn 1 for salivary gland (Yoshida et al., 2001), Math1 for intestine (Yang et al., 2001), and NeuroD and neurogenin for the neural cell lineage (Ma et al., 1996). Additional complexity of bHLH regulation occurs through homodimeric and heterodimeric interactions between specific class B bHLH proteins (Firulli et al., 2000). For example, heterodimers of Twist1 and HAND2 are essential for normal limb development. A stoichiometric shift in the Twist1:HAND2 dimer leads to limb abnormalities associated with Saethre–Chotzen syndrome (Firulli et al., 2005). Further regulation of the activity of bHLH proteins occurs through the formation of complexes with other transcriptional regulators such as MEF2 (Molkentin et al., 1995) and GATA4 (Dai et al., 2002). These complexes are able to direct transcriptional activation through either MEF2 or GATA sites or E-boxes.
Recently, we have demonstrated that Sertoli cell differentiation is mediated through both the CREB and bHLH signaling pathways (Chaudhary and Skinner, 1999, 2001; Muir et al., 2005). The ability of CREB and bHLH proteins to interact on a transcriptional level in the Sertoli cell was previously shown to involve transcriptional complexes and adapter proteins (e.g., CBP) to bridge and link bHLH dimers binding to E-box elements and CREB dimers binding to CRE (Chaudhary and Skinner, 1999, 2001). Interestingly, many gene promoters are responsive to cAMP, but do not contain CRE elements. Observations have suggested that cAMP regulation of these genes occurred either through indirect actions of cAMP and the CREB family, or unique protein associations between CREB family members and other transcription factors resulting in binding to unique regulatory sites (Hai and Hartman, 2001; Mayr and Montminy, 2001). The current study investigates the integration of the cAMP responsive CREB pathway with the bHLH transcriptional pathway through potential direct interactions of these factors. Observations demonstrate that scleraxis is able to interact with ATF4 through the bHLH and bZip domains of the respective proteins. The ability to bind ATF4 was shared by paraxis, another bHLH transcription factor that has 95% sequence identity with scleraxis within the bHLH domain. Since potential direct interactions between ATF4 and scleraxis were found in the current study, the role of ATF4 and scleraxis in the testis was investigated using respective null mutant mice.
MATERIALS AND METHODS
Yeast-2-Hybrid Screens
The bait used for the yeast-2-hybrid screen contained full-length paraxis or scleraxis fused in-frame with the GAL4 DNA-binding domain (Wilson-Rawls et al., 2004). This encompasses the bHLH domain and was cloned into the pAS yeast expression vector. The PAS-bHLH construct was co-transformed into yeast with an embryonic day 10.5 mouse (i.e., paraxis) or rat Sertoli cell (i.e., scleraxis) cDNA library that contained the GAL4 activation domain fused to random cDNAs. Over 5 million primary library transformations were screened. From each individual colony, the activating plasmid was rescued and the cDNA insert was sequenced. Clones containing cDNA inserts in the antisense orientation or out-of-frame were discarded. The remaining clones were retransformed back into yeast to test binding specificity. The isolated clones had to recapitulate the interaction and could not interact with nonspecific bait. Individual clones or constructs were tested for the ability to auto-activate and if they did provide a false positive signal were discarded. Therefore, none of the bait or prey constructs used provided a false positive signal, even if activation domains were present in the constructs. The degree of bait and prey interaction used the level of activation of β-galactosidase compared to a negative control.
Cell Preparation and Culture
Sertoli cells were isolated from 20-day-old rats by sequential enzymatic digestion (Chaudhary and Skinner, 1999, 2001). All animal procedures and protocols were reviewed and approved by the Washington State University Animal Care and Use Committee. Decapsulated testis fragments were digested first with trypsin (1.5 mg/ml; Life Technologies, Gaithersburg, MD) to remove the interstitial cells and then with collagenase (1 mg/ml type I; Sigma Chemical Co., St. Louis, MO) and hyaluronidase (1 mg/ml; Sigma Chemical Co.). Sertoli cells were plated under serum-free conditions in 24-well Falcon plates (Falcon Plastics, Oxnard, CA) at 1 × 106 cells/well. Cells were maintained in a 5% CO2 atmosphere in Ham’s F-12 medium (Life Technologies) with 0.01% BSA at 32°C. Sertoli cells were left untreated (control) or treated with FSH (100 ng/ml) or dibutryl cAMP (dbcAMP; 100 μM) as previously described (Chaudhary and Skinner, 1999, 2001). Butyrate was previously shown not to influence Sertoli cells. The cells were cultured for variable durations of treatment as indicated or for 72 hr in the absence or presence of treatments.
Transfection Procedure
Sertoli cells cultured for 48 hr were transfected with the mammalian reporter and expression vectors by the calcium phosphate method coupled with hyper osmotic shock (10% glycerol). The luciferase reporter vector contains 581 bp of the mouse transferrin (mTf) promoter. The mTf used in the present study included the transcriptional initiation site of the transferrin gene, which is 54 bp upstream of the start site of translation (Chaudhary and Skinner, 1999, 2001). Within this 581 bp sequence is a functional E-box. Full-length scleraxis was inserted into the EcoRI site of pCMV-HA (Promega, Madison, WI). The pCMV-myc/ATF4 fusion vector was generated (Promega). Each vector (1.5 μg) was added to 150 μl of transfection buffer (250 mM NaCl, 50 mM HEPES, and 1.47 mM Na2HPO4, pH 7.05) and placed in each well of a 24-well plate containing 1 × 106 Sertoli cells. Incubation occurred for 4 hr and was followed by a 3 min 10% glycerol shock. Cells were washed twice with HBSS (Gibco, Gaithersburg, MD) before adding fresh Ham’s F-12. Cells were treated with dibutryl cAMP and incubated for 72 hr. Cells were lysed after 72 hr and luciferase activity determined by measuring luminescence of the cell lysates. C3H10T1/2 mouse fibroblasts were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal calf serum. For CAT assays, 10T1/2 fibroblasts were seeded into 60-mm-diameter dishes and the cells were transfected by the calcium phosphate method. The cells were transfected with plasmids containing Gal4E1b5CAT, Gal4paraxis, and ATF4. The total amount of plasmid DNA was equivalent in all trans-fections with the use of empty vector and transfected for 16 hr, after which the cells were rinsed in phosphate-buffered saline (PBS) to remove excess precipitate, and cells were maintained in growth medium. At 48 hr post-transfection, cells were harvested and lysates collected. The amount of protein in each sample was determined by protein assay (Bio-Rad, Hercules, CA), and chloramphenicol acetyltransferase (CAT) assays were performed with an equal amount of total protein.
Myogenic Conversion of C3H10T1/2 Cells
The procedure was completed as previously described (Wilson-Rawls et al., 1999b). Briefly, C3H10T1/2 cells were seeded onto gelatinized plates at a density of 6.0 × 104 cells/ml in growth medium (DMEM, 10% FBS) and transfected using Fugene6 (Roche BioSciences Co, Indianapolis, IN) with EMSV-MyoD, with and without CS2 +CREB1 or CS2 +ATF4, and CMV-EGFP as an internal transfection control. The amount of DNA transfected was maintained at a constant level with the addition of empty vector. After 24 hr, the efficiency of transfection was monitored by expression of GFP. The cells were rinsed in serum free medium and then cultured in differentiation medium (DMEM, 2% donor horse serum) and this was changed daily for 5 days. The transfected cells were fixed in cold methanol, and the expression of myosin heavy chain (MHC) was detected immunohistochemically using the MY-32 monoclonal antibody (Sigma Chemical Co.) and the histostainSP colorimetric detection kit (Zymed-Invitrogen, Carlsbad, CA). The number of MHC+ve multinucleated myotubes were counted in 10 random 10× fields per plate. The data are expressed as percent of MyoD induced myotubes, from three replicate experiments done in duplicate.
Electrophoretic Mobility Shift Assays
Coupled in vitro transcription–translation reactions were performed with 0.5 μg of each plasmid DNA and TNT reticulocyte lysates with T7 polymerase (Promega). The efficiency of translation was determined by performing duplicate translation reactions in the presence of Trans-[35S] (NEN-PerkinElmer, Wellesley, MA,). As a probe, double-stranded oligonucleotides corresponding to each gene-specific E-box site was used and end labeled with [γ-32P]ATP (NEN-PerkinElmer) and T4 polynucleotide kinase (New England Biolabs, Beverly, MA). For each DNA binding reaction 2 mg of the total translation products were added to a 20 μl total reaction mixture along with 40,000 cpm of probe in binding buffer (40 mM KC1; 15 mM HEPES, pH 7.9; 1 mM EDTA; 0.4 mM dithiothreitol, 50% [vol/vol] glycerol), and 2 μg of poly(dIdC) (Pharmacia Biotech, Piscataway, NJ). Binding reactions were carried out for 20 min at room temperature, and protein–DNA complexes were analyzed on 5% 0.5X TBE polyacrylamide gels. Protein concentrations were normalized to allow equal loading on the gels. Approximately equal ratios of different proteins were used when combined.
Immunoprecipitations
In vitro translations were performed with the coupled TNT T7 transcription and translation kit (Promega). Translation products were labeled with Trans-[35S] (NEN-PerkinElmer). For immunoprecipitations, 1 ml of ice-cold NTT lysis buffer (140 mM NaCl; 50 mM Tris, pH 7.5; 1 mM EDTA; 0.1% Triton X-100; 10% glycerol) containing the Complete protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, IN) was added to each reaction. Each immunoprecipitation received 5 μl of either anti-E12 or anti-ATF4 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and 25 μl of protein A/G-Plus agarose (Santa Cruz Biotechnology). Lysates were then incubated for 4 hr at 4°C with shaking, followed by centrifugation and then three washes in NTT buffer containing inhibitors. Proteins were analyzed on 12% SDS–PAGE with Kaleidoscope prestained molecular-weight markers (Bio-Rad). Aliquots of the translation products were analyzed electrophoretically on the SDS gels to correct for loading and levels of proteins used. Gels were dried and exposed to film.
RNA Preparation and Quantitative Polymerase Chain Reaction (PCR)
Cultured Sertoli cells were lysed using TRI reagent (Sigma Chemical Co.). The lysate was passed through a pasture pipette to form a homogenous lysate. The whole tissue samples were homogenized in a tissue homogenizer in the presence of TRI reagent. The homogenate was centrifuged at 12,000g for 10 min at 4°C. Total RNA was then isolated from the cell lysate and whole-tissue homogenate following the manufacturer’s protocol for RNA isolation using TRI reagent. The final RNA pellet was dissolved in distilled water. Total RNA from Sertoli cells, germ cells, and peritubular cells were isolated as described earlier using TRI reagent.
A two-step real-time PCR was carried out to analyze the expression pattern of ATF4. Reverse transcription (RT)-PCR was used to generate cDNA from either hormone treated cultured Sertoli cells, freshly isolated Sertoli cells or whole testis. Total RNA (2 μg) was reverse-transcribed into cDNA in a reaction primed by oligo(dT) using MMLV reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Reverse and forward oligonucleotide primers, specific to the chosen candidate genes, were designed using Primer Express 2.0 software (Applied Biosystems, Foster City, CA) as described by the manufacturer. The following primers were used: ATF4 5′-aag caa agc taa gcc tcc atc ttg tgc and 3′-aaa gga atg ctc tgg agt gga aga cag; and control ribosomal RNA (S2) 5′-ctg ctc ctg tgc cca aga ag and 3′-aag gtg gcc ttg gca aag tt. Real-time RT-PCR was performed in a 96-well plate using a 7000 ABI prism sequence detection system (Applied Biosystems). The previously synthesized cDNA was used as template. Samples from cultured Sertoli cells, freshly isolated Sertoli cells or testes were plated in triplicate PCR reactions. The PCR reaction contained 5–10 ng of cDNA, SYBR GREEN master mix (Applied Biosystems), and 100 nM of each reverse and forward primers of ATF4 in a final PCR reaction of 50 μl. Amplification parameters were: denaturation at 94°C for 10 min followed by 40 cycles of 94°C for 15 sec and 60°C for 60 sec. Samples were analyzed in triplicate and ribosomal RNA (S2) was used as an endogenous control and for calibration curves (Muir et al., 2005). A minimum of three different experiments, each done in triplicate, were performed.
Protein Structure Predictions
The previous crystal structures for bHLH (MyoD/E12; Ma et al., 1994) and CREB/ATF4 (Podust et al., 2001) proteins were used to build a model for bHLH/ATF4 heterodimer. Specifically, the PDB coordinates, 1MDY and 1CI6, corresponding to bHLH and CREB/ATF4, were used to predict the structure. The initial positions of two peptides were obtained through combination of superposition and the solid docking module on QUANTA (BioSYM/Micron Separations), which is based on conformational space, followed by a quick energy minimization by CNS v1.1 (Brunger et al., 1998) using potential function parameters of CHARMM19. The protein modeling was performed by the Washington State University Biomolecular X-ray Crystallography Center.
Whole Testis Microarray Analysis
Whole testis from mice of different development ages (E11.5 to P56) were collected as described (Shima et al., 2004; Small et al., 2005). Approximately 70 embryos were processed for a single 11.5 and 12.5 dpc time point while approximately 35 embryos were utilized for the remaining embryonic time points. Testes were placed in TRIZOL reagent (Invitrogen) and total RNA collected as per the manufacturers instructions. Once purified, RNA quality was determined by electrophoresis on a denaturing agarose gel and quantified by spectroscopy at 260 and 280 nm. Biotinylated complementary RNA (cRNA) was created from the total RNA. The biotinylated cRNA was generated using an oligo(dT) primer with a T7 promoter in a reverse transcription reaction. This was followed by in vitro transcription using the MEGAScript kit (Ambion, Austin, TX) with biotinylated cytosine and uridine triphosphate. The labeled cRNA was hybridized to Affymetrix arrays (Affymetrix, Santa Clara, CA), and stained in accordance with the manufacturers protocol. The same procedure was performed to create duplicate samples for each time point. The arrays were stained and washed utilizing the Affymetrix GeneChip Fluidics Station 400 and scanned using a GeneArray Scanner 2500A (Agilent, Palo Alto, CA). The resulting data were viewed and preliminary assessment was made using Microarray Suite 5.0 (Yang et al., 2004) software (Affymetrix). All probe sets from each microarray were scaled to a target signal of 125. All microarray hybridization and bioinformatics procedures were performed in the Genomics and Bioinformatics Cores in the Center for Reproductive Biology at Washington State University.
Knockout Mouse Strains
A targeted null ATF4 allele was generated by replacing the second exon of ATF4 with the neomycin-resistance gene (Tanaka et al., 1998). This mutation removes the basic leucine zipper domain that is essential for the function of the protein. Mice carrying the disrupted ATF4 gene were identified using a PCR genotyping strategy (Tanaka et al., 1998; Libby et al., 2002). A transgenic GFP reporter of scleraxis expression, ScxGFP, was generated by inserting the GFP gene at the ATG of the scleraxis gene in a 12KB genomic fragment from the scleraxis locus (Pryce et al., 2007). A null allele of the scleraxis gene was recently generated by deleting the 1st exon that contains most of the ORF of the scleraxis gene (Murchison et al., 2007). The experiments were carried out according to the principles and procedures outlined in the Guide for Care and Use of Laboratory Animals and were approved by the WSU Institutional Animal Care and Use Committee.
Histological Analysis
Whole testes were removed from ATF4 and scleraxis knockout and heterozygous mice, placed in 4% paraformaldehyde (PFA) for 24 hr then rinsed in 70% ethanol. Testis samples were submitted to the Washington State University histology core laboratory for sectioning at 4 μm and staining with hematoxylin and eosin. Sectioned testes were examined under light microscopy (Nikon, Melville, NY) for morphology. Whole testis from scleraxis GFP reporter transgenic mice were placed in 4% paraformaldehyde for 4 hr then transferred to 1% paraformaldehyde for 24 hr. One testis from each mouse was processed for cyrosectioning using sucrose as a cyroprotectant. Testis tissue was embedded in OTC (Miles Laboratories, Inc., Clifton, NJ) and sectioned frozen at 20 μm then analyzed using fluorescent confocal microscopy. The contralateral testis was processed for hematoxylin and eosin (H&E) staining by leaving the testis in 4% PFA for 24 hr then transferring to 70% ethanol. Testes were then submitted to the Washington State University Histology Core Laboratory for embedding and sectioning at 4 μm.
Testicular cells undergoing apoptosis were visualized by the TUNEL assay which labels fragmented DNA with digoxigenin-deoxy-UTP using terminal deoxynucleotidyl transferase, as detected by the anti-digoxigenin-conjugated reporter system. Briefly, serial sections were deparaffinized, rehydrated through a series of graded alcohols and PBS, digested with 20 μg/ml proteinase K for 15 min at 37°C, and washed with water. Slides were rinsed with PBS, covered with a coverslip and evaluated under fluorescence microscopy (Nikon). Visualization of apoptotic cells was performed using fluorescent microscopy with a Nikon Eclipse E800 microscope (Nikon). The number of apoptotic cells was normalized to total tubule number per testis. At least three knockout mice and heterozygous mice with a minimum of three cross-sections per testis were utilized for the assay. Germ cell nuclear antigen (GCNA) staining was performed on testis sections from ATF4 and scleraxis knockout and heterozygous mice as previously described (Cupp et al., 2002). Briefly, serial sections from testes were deparaffinized and rehydrated through a series of alcohol washes. Sections were boiled in sodium citrate for 10 min to quench endogenous peroxidase activity then blocked in 10% serum for 30 min at room temperature. Primary GCNA antibody was diluted 1:10 in 10% goat serum, added to slides, which were then placed in a humidified chamber, and incubated at 4°C for 16 hr. Slides were rinsed in PBS and biotinylated goat anti-mouse secondary antibody (1:200; Vector Laboratories, Burlington, CA). This was allowed to incubate at room temperature for 2 hr and secondary antibody was detected using the HistostainSp kit (Zymed Laboratories, San Francisco, CA). In each experiment, three serial sections of testis from at least three animals were analyzed.
Statistical Analysis
The values were expressed as the mean ± SEM of at least three representative cross-sections. Differences between means were determined using paired comparison Student’s t-test. Groups were considered significantly different at a P-value of ≤0.05.
RESULTS
bHLH and CREB Family Member Interactions
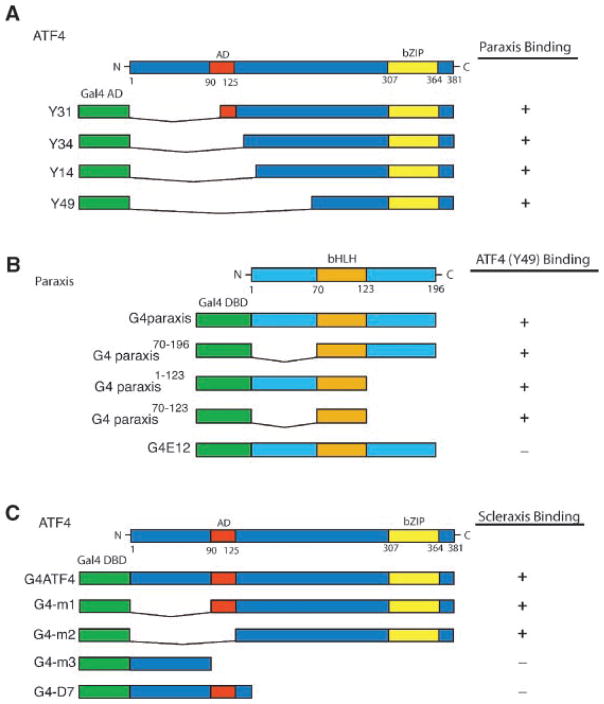
A yeast-2-hybrid screen was performed using a testicular Sertoli cell library and scleraxis as bait. Thirteen independent clones of ATF4 of varying lengths were identified from the screen. A yeast-2-hybrid screen was also performed using an E10.5 mouse cDNA library and paraxis as bait. Approximately 10% of the sequenced colonies were ATF4. Not all of the ATF4 clones identified in the embryo library were full-length, but all included the bZIP domain, Figure 1A. The shortest clone is restricted to the C-terminal region of the protein that encompasses the bZIP domain, suggesting that the interactions between paraxis and ATF4 are mediated by this domain. However, some C-terminal region outside the bZIP domain (200–300) was also present and needs to be considered in the interpretation of the interaction observed. Analysis of ATF4 mutants (Ord and Ord, 2003) in a yeast-2-hybrid assay also demonstrated that the C-terminal region is necessary for this protein to interact with scleraxis, Figure 1C. The C-terminal ATF4 clone containing the bZIP domain was used as a prey to study the region of paraxis that mediates the interaction. When challenged with paraxis deletion mutations, it was found that the bHLH domain was sufficient for the interaction with the C-terminal region of ATF4 (Fig. 1B). A similar experiment with scleraxis was performed and demonstrated the bHLH domain was involved in the interaction with ATF4 (data not shown). These results suggest binding of these proteins appears to be mediated by a novel bZIP/bHLH interaction, Figure 1.
Fig. 1.
Domain mapping of ATF4, paraxis, and scleraxis for sites of interaction. A yeast-2-hybrid screen with deletion mutant constructs and full-length activation domain partner listed in the right column, with (+) indicating positive yeast growth and β-galactosidase (control) activity and (−) indicating no growth or β-galactosidase. A: ATF4 truncations that were identified in the initial yeast two hybrid screens along with full-length ATF4. Y49 which contains the bZIP motif was used as a bait with paraxis mutants. B: Paraxis mutants containing the bHLH domain (G4 paraxis) interactions with the Y49 ATF4 construct were assessed. E12 was used as a control to verify that the interaction with ATF4 is specific for the bHLH domain of paraxis. C: ATF4 mutants (Ord and Ord, 2003) containing the bZip region (m1 and m2) lacking the bZip region (m3 and D7) and interactions with scleraxis were assessed. Data are from three separate experiments with three replicates per experiment.
To determine whether the interaction between ATF4 and scleraxis or paraxis represents a unique event or something shared between bZip and bHLH proteins, we used the yeast-2-hybrid system to test the ability of a broader set of bHLH proteins to bind to ATF4 and the highly related CREB1 protein, Table 1. In this yeast-2-hybrid screen both growth on amino acid selection plates and levels of β-galactosidase activity were examined. ATF4 was found to interact with scleraxis, MyoD, ITF2, and Id2, but not with E47, Table 1. Interestingly, while ATF4 interacts with a variety of bHLH proteins, CREB1 was found to interact with MyoD and ITF2, but not with scleraxis or Id2, Table 1. The individual bait and prey constructs did not provide false positive activation or binding when used alone (data not shown). Therefore, several bHLH and ATF factors can interact and the distinct binding specificity appears to exist between various CREB and bHLH proteins.
TABLE 1.
ATF4 Interaction With bHLH Transcription Factors
| Bait | Prey | -LTHA | β-galactosidase activity | |
|---|---|---|---|---|
| 1 | p53 | pSV40 | + | + |
| 2 | ATF4 | pSV40 | − | − |
| 3 | ATF4 | E47 | − | − |
| 4 | ATF4 | Scleraxis | + | +++ |
| 5 | ATF4 | ITF2 | + | + |
| 6 | ATF4 | MyoD | + | ++ |
| 7 | ATF4 | Id2 | + | +++ |
| 8 | CREB1 | Scleraxis | − | − |
| 9 | CREB1 | MyoD | + | ++ |
| 10 | CREB1 | ITF2 | + | + |
| 11 | CREB1 | Id2 | − | − |
| 12 | CREB1 | E47 | − | − |
| 13 | CREB1 | ATF4 | + | +++ |
A (+) in the -leu, -trp, -his, -ade (-LTHA) column represents growth of yeast on amino acid selection plates. A (+) in the β-galactosidase activity column represents a minimum of twofold increase in activity, (++) represents a minimum of fourfold increase in activity, and (+++) represents a minimum of sixfold increase in activity as compared to the negative control (ATF4 (bait) and pSV40 (prey)). The negative control is the baseline set at 1. Data are representative of a minimum of three different experiments.
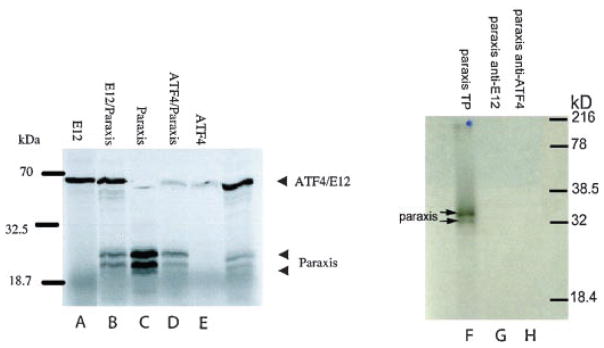
Previous studies demonstrated that interactions between CREB and bHLH factors were mediated by adaptor proteins such as CBP/p300 (Chaudhary and Skinner, 1999, 2001), yet the yeast-2-hybrid data suggests the potential ability of CREB and the bHLH proteins to directly interact. In order to confirm a direct interaction between ATF4 and paraxis the proteins were expressed using coupled transcription/translation reactions in reticulocyte lysates in the presence of Trans-[35S] and then immunoprecipitated with antibodies that recognize either E12 or ATF4. By using coupled in vitro transcription/translation to express the proteins, other adapter proteins and co-factors are not likely present and therefore cannot be involved in complex formation. There is a possibility that a nonspecific adaptor protein from the reticulocyte lysate may be present, but no other 35S-labeled protein was present. In Figure 2C, paraxis is expressed as two bands (lane marked paraxis), this lane is total translation products. Neither paraxis nor scleraxis form homodimeric structures that can bind DNA. Paraxis is known to form a complex with E12 (Wilson-Rawls et al., 2004), and when co-translated with E12 and immunoprecipitated using an anti-E12 antibody both proteins were detected (Fig. 2B, compare E12 and paraxis with E12/paraxis). Similarly, when paraxis and ATF4 were expressed in reticulocyte lysates they also co-immunoprecipitated with an anti-ATF4 antibody, indicating that these proteins form a protein complex (Fig. 2D, compare ATF4 with ATF4/paraxis). Similar observations were made with scleraxis and the scleraxis/ATF4 co-immunoprecipitation (data not shown). To control for equal levels of translation and loading, samples of the total translation products were taken before immunoprecipitation and demonstrated to contain approximately equal amounts of ATF4, E12, scleraxis, and paraxis in the lysates (data not shown). In addition, no nonspecific binding was detected in the immunoprecipitates, which had no translation product or lack of primary antibody (data not shown). Paraxis (Fig. 2F) could not be directly precipitated with the antibody to E12 or ATF4 (Fig. 2F–H). Taken together these data indicate that the CREB and bHLH proteins appear to form a heterodimer in the absence of adapter proteins.
Fig. 2.
Co-immunoprecipitation of ATF4 and paraxis. Proteins were translated in TNT reticulocyte lysates in the presence of Trans-35S and the proteins were immunoprecipitated and analyzed on 12% SDS–PAGE. Using an anti-E12 antibody, a protein corresponding to E12 (lane A, E12), or a complex containing E12 and Paraxis (lane B, E12/Paraxis) were immunoprecipitated. Similarly ATF4 could be immuno-precipitated with an anti-ATF4 antibody (lane E, ATF4), and paraxis was found in a complex with ATF4 when these proteins were co-translated (lane D, ATF4/paraxis). The lane C labeled paraxis is 1/10th of total translation, no immunoprecipitation. A control was run with paraxis translation product (lane F) with primary antibody to E12 (lane G) or ATF4 (lane H). Total translation products were run for comparison with equal amounts of protein (data not shown). Data are representative of three different experiments.
Structure Considerations for the bHLH/CREB Heterodimer
Based on the experimental data, dimerization occurs specifically through the bHLH and bZIP domains of the two proteins (Fig. 1). The presence of basic domains in both the bHLH and ATF4 suggests the heterodimer may have the capacity to bind DNA. Previous crystal structures for ATF4 (Podust et al., 2001) and bHLH proteins (Ma et al., 1994) were used to model the predicted structure of the heterodimer, Figure 3. The backbones of two modeled α-helices did not change significantly and clearly showed properly oriented DNA binding arms. All residues in the interface are within potentially interacting distances to each other (Fig. 3D) and the DNA oligomer can fit into the basic domains. The predicted structure does have an arrangement of the basic domains, which suggests a potential active DNA binding capacity. The proposed heterodimeric structure now needs to be confirmed with other approaches, such as X-ray crystallography or NMR.
Fig. 3.
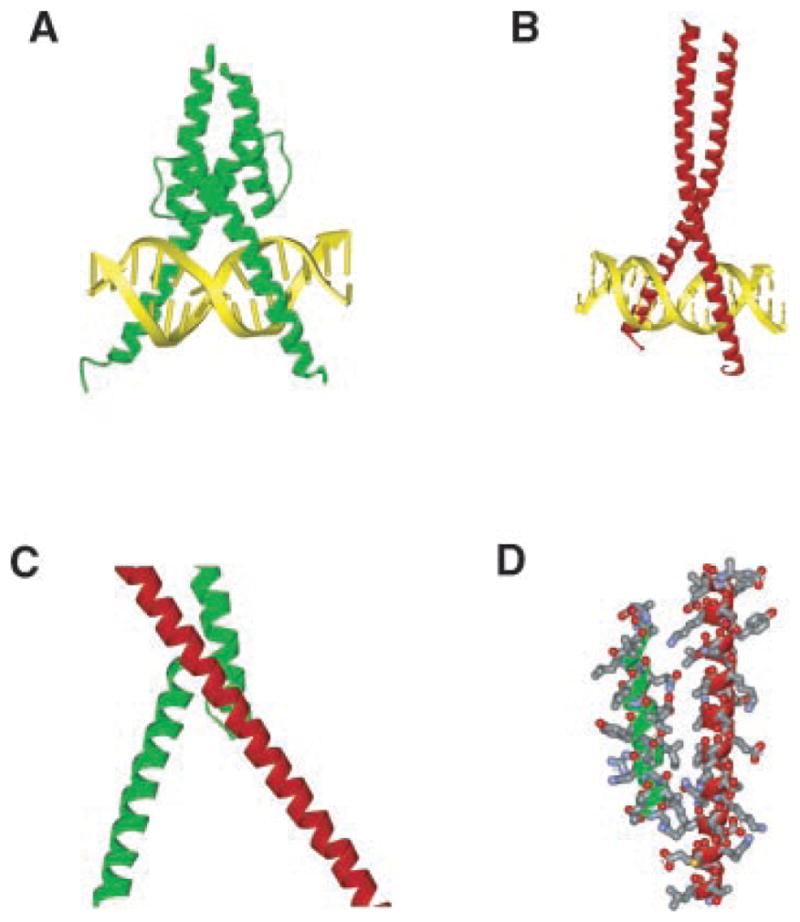
Predicted structure of a bHLH and CREB/ATF4 heterodimer. The bHLH (MyoD/E12) dimeric (1MDY, green) (A), CREB1/ATF4 (bZip) dimeric (1CI6, red) (B), and heterodimeric bHLH (scleraxis)-ATF4 heterodimer (C) is shown. The predicted basic domain interactions with DNA (yellow) are indicated (A,B). The amino acid interacting domains of scleraxis and ATF4 are shown (D).
Functional Role of bHLH and CREB Family Member Interactions
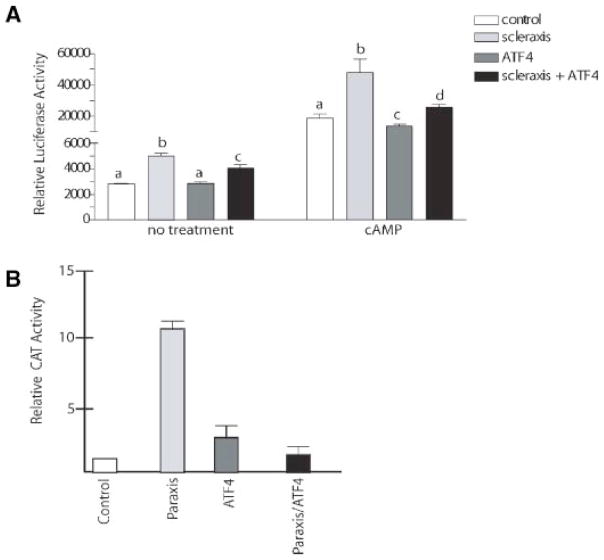
The functional significance of the bHLH and CREB2/ATF4 heterodimers was examined in testicular Sertoli cells isolated from 20-day-old rats and cultured. We have previously demonstrated that transferrin transcription during Sertoli cell differentiation is responsive to cAMP signaling through a CRE element and scleraxis (Chaudhary and Skinner, 1999, 2001). A reporter construct containing the transferrin promoter upstream of luciferase was transfected into Sertoli cells and tested for scleraxis and cAMP specific transcription. The basal level of transferrin promoter activity can be stimulated by cAMP treatment, Figure 4A. Scleraxis was found to increase both basal and cAMP stimulated luciferase activity through this promoter. Expression of ATF4 alone reduced the cAMP stimulated levels of the reporter, Figure 4A. Interestingly, when ATF4 was co-expressed with scleraxis both the basal and cAMP stimulatory actions of scleraxis were significantly inhibited. Dose curves with increasing or decreasing concentrations of the ATF4 expression construct were used to identify the optimal concentrations used in the current study and revealed approximately equal ratios of expression products were required (data not shown). Similar results were obtain when examining transcription regulation of paraxis. A paraxis/Gal4 DNA binding domain fusion protein can induce transcription from the Gal4-CAT reporter in C3H10T1/2 cells with an 11.5-fold stimulation over background, Figure 4B. Co-transfection of ATF4 with the paraxis fusion protein significantly inhibited CAT transcription. These data predict a role for ATF4 in inhibiting transcriptional activation by scleraxis and paraxis, Figure 4.
Fig. 4.
A: Constitutive expression of scleraxis and/or ATF4 in cultured Sertoli cells. Cultured 20-day-old rat Sertoli cells were co-transfected with the proximal mouse transferrin promoter-luciferase construct (mTf-Luc) in the presence of scleraxis expression vector, ATF4 expression vector or both expression vectors. A basal no-insert plasmid (control) was used as a control. Data are mean ± SEM from three separate experiments with three replicates per experiment. Different letter superscripts are statistically different from each other with ANOVA with P <0.05. B: Expression of paraxis and/or ATF4 in cultured muscle/neuronal progenitor cells. The effect of ATF4 on the ability of paraxis to transactivate a heterologous reporter was tested using transfected 10T1/2 cells and the GAL4E1b5CAT promoter. Data are mean ± SEM from three separate experiments with three replicates per experiment. A significant difference was observed between paraxis and ATF4, as well as paraxis and paraxis +ATF4 at P <0.01 by Student’s t-test.
Inhibition by ATF4 may occur by forming a protein complex that is unable to bind to E-boxes in the promoters. MyoD was found to directly interact with ATF4 and CREB1 (Table 1), the effects of ATF4 and CREB1 expression on MyoD induced myogenic conversion of C3H10T1/2 cells was examined (Wilson-Rawls et al., 1999b; Fig. 5). C3H10T1/2 cells can be induced to differentiate in the presence of MyoD, and transfection of these cells with MyoD effectively induced the formation of myotubes (Fig. 5). The expression of ATF4 or CREB1 alone did not induce myotube formation (Fig. 5D,E). Interestingly, the co-expression of CREB1 with MyoD was found to significantly inhibit myo-tube formation (Fig. 5). Although, ATF4 had a small inhibitory effect on MyoD actions, this was not statistically significant. Therefore, CREB1 inhibits the ability of MyoD to induce myotube formation in culture, which supports the above differentiation marker reporter gene data (Fig. 4).
Fig. 5.
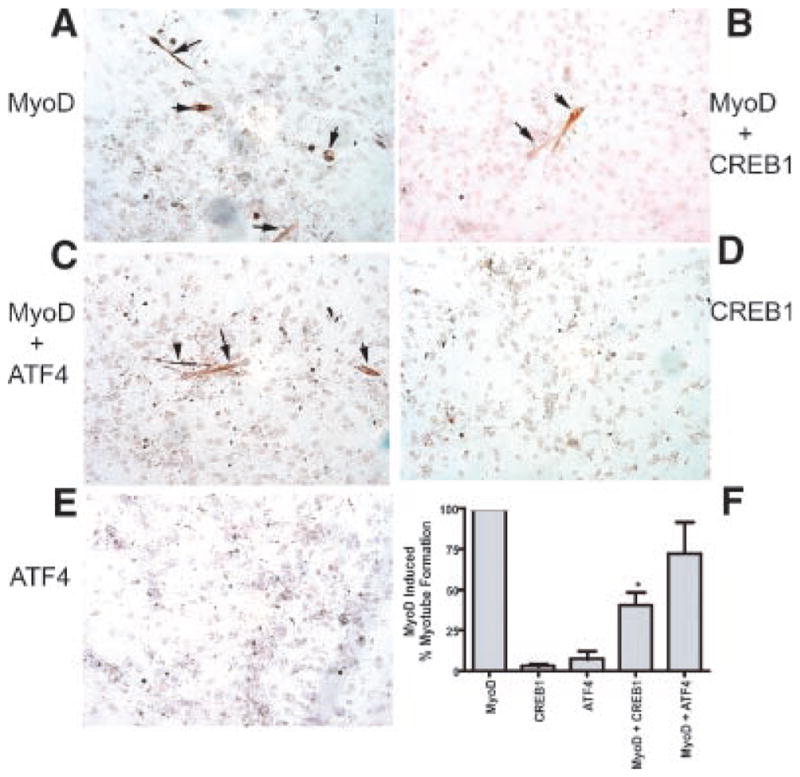
CREB1 inhibits MyoD induced myogenic conversion of 10T1/2 cells. 10T1/2 cells were transfected using Fugene6 with MyoD or MyoD +CREB1 or ATF4. At 24 hr post-transfection the growth medium was replaced with differentiation medium and the cells were maintained for 5 days with daily medium changes. Cells were fixed and IHC was done using MY-32 an anti-MHC antibody and the proteins visualized using DAB. MHC positive, multinucleated myotubes were counted and representative fields are presented. A: 10T1/2 cells in the presence of EMSV-MyoD. B: Expression of MyoD and CREB1. C: Expression of MyoD and ATF4. D,E: Expression of CREB1 or ATF4 constructs only. F: Quantitation of myotube formation in the presence of MyoD, and/or CREB1 or ATF4 expression constructs. The mean +SEM is presented for three experiments in duplicate, and (*) indicates a statistical difference from MyoD and MyoD +CREB1. Equivalent levels of expression constructs were used for each treatment.
Observations suggest CREB1 and ATF4 may block the ability of specific bHLH proteins to bind to E-boxes in the promoters. Therefore, electrophoretic mobility gel shift assays (EMSA) were done using in vitro translated proteins and an E-box probe to determine if paraxis or scleraxis can bind DNA in the presence of ATF4. Neither paraxis nor scleraxis will bind E-boxes as homodimers, they bind as heterodimers with the ubiquitous class A bHLH proteins such as E12. In contrast, E12 will bind E-boxes as a homodimer (Wilson-Rawls et al., 2004). The proteins were translated using reticulocyte lysates and added to EMSA reactions, equal amounts of protein were loaded in all lanes and gel shifts, with approximately equal ratios of different proteins present. Using an end-labeled INS-1 E-box that is bound by a paraxis/ E12 heterodimer (Wilson-Rawls et al., 2004) it was found that formation of the complex was blocked in the presence of ATF4 (Fig. 6A). Paraxis and E12 preferentially form a heterodimer when co-translated (Fig. 6A). However, when ATF4 was co-translated with paraxis and E12, an E12 homodimer complex was present in the EMSA, indicating that ATF4 had displaced E12 from a heterodimer with paraxis. Similarly, scleraxis/E12 heterodimers also were inhibited from binding the scleraxis E-box in this assay by ATF4 (Fig. 6B). Thus these data indicate that ATF4 displaced E12 and formed a complex with paraxis and scleraxis and this blocked binding of the E-box.
Fig. 6.
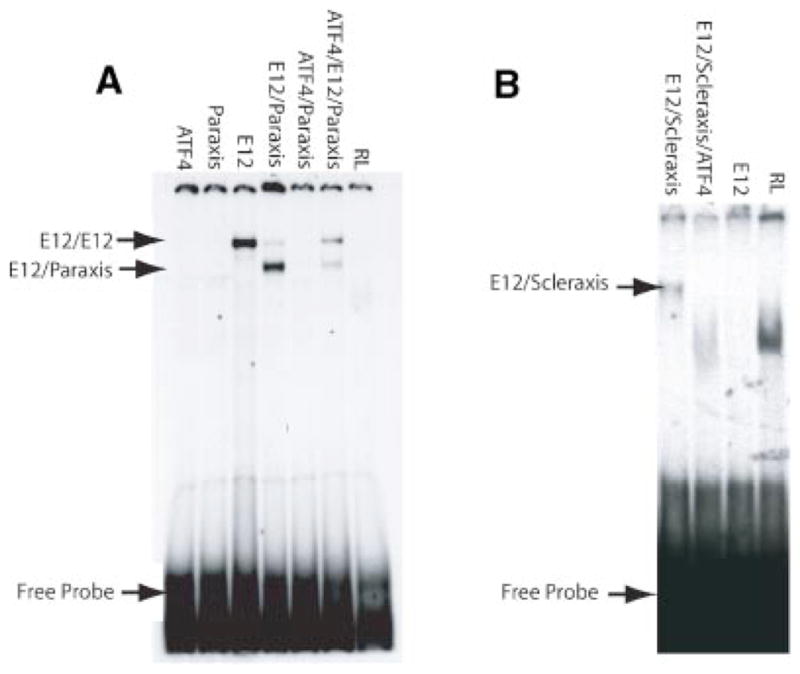
A: EMSA done with the INS-1 E-box in the absence or presence of ATF4. This E-box can be shifted by E12/E12 or E12/paraxis dimers. In the presence of ATF4, there is a decrease in the E12/paraxis complex, and a reappearance of the E12 homodimer complex. An ATF4/ paraxis complex does not appear. B: EMSA done with the scleraxis E-box. This E-box from the scleraxis promoter is bound by a scleraxis/ E12 heterodimer. E12 will not shift this E-box as a homodimer. E12/ scleraxis dimers shift this E-box and this was abolished in the presence of ATF4. RL is unprogrammed reticulocyte lysate. Equivalent amounts of each protein were used and ratios of two proteins were comparable. Data are representative of a minimum of three different experiments.
The hormonal regulation of scleraxis and ATF4 were compared to further investigate the interactions between bHLH proteins and ATF4 in Sertoli cells. A previous study demonstrated both FSH and cAMP induced a transient increase in scleraxis expression at 2–4 hr after treatment, then subsequently declined to basal levels (Muir et al., 2005). Therefore, scleraxis appears to be an early event gene in the actions of FSH on Sertoli cells (Muir et al., 2005). In contrast, analysis of ATF4 expression in Sertoli cells demonstrated FSH has no effect on ATF4 mRNA levels at any treatment duration for 24 hr, Figure 7. The cAMP stimulation was only observed after 24 hr of treatment in contrast to the regulation of scleraxis, Figure 7. These data regarding ATF4 regulation was extended with a micro-array analysis of Sertoli cells after 72 hr of treatment with FSH versus cAMP. Again FSH had no effect on ATF4 mRNA levels and cAMP increased ATF4 expression levels at the 72 hr treatment duration (data not shown). Therefore, scleraxis and ATF4 appear to have a distinct relationship in regards to the hormonal (i.e., cAMP) regulation of Sertoli cells, with scleraxis being an early event responsive gene and ATF4 being a later responding gene, Figure 7. The inhibitory actions of ATF4 may allow for a negative feedback on bHLH actions induced by hormones.
Fig. 7.
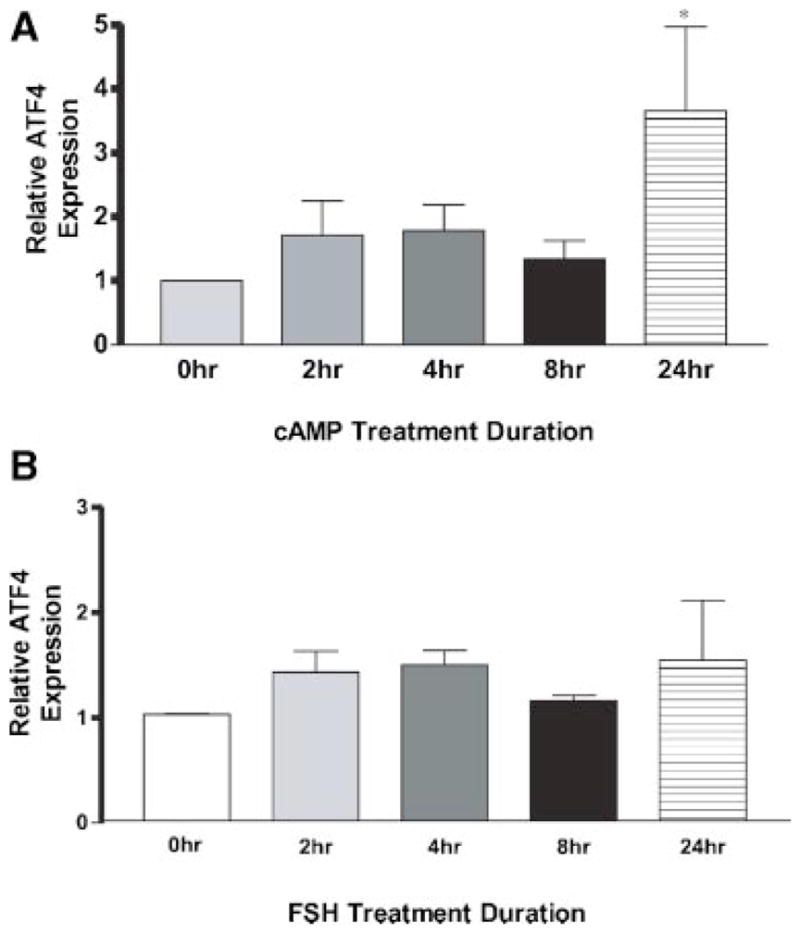
ATF4 mRNA expression in response to dibutryl (A) cAMP or (B) FSH. Analysis of ATF4 mRNA and ribosomal RNA (S2) levels in Sertoli cells treated with FSH or cAMP. Sertoli cells were cultured for 0, 2, 4, 8, and 24 hr in the presence of FSH or cAMP. ATF4 and ribosomal RNA (S2) levels were analyzed using quantitative real-time-PCR. All data were normalized to the 0 hr time point for each treatment group. Data are mean ± SEM from three separate experiments with three replicates per experiment. Asterisk indicates a significant difference between the 0 hr treatment of cultured Sertoli cells and the other treatment time points at P < 0.05 by Student’s t-test.
Developmental and Functional Role of ATF4 and Scleraxis in the Testis
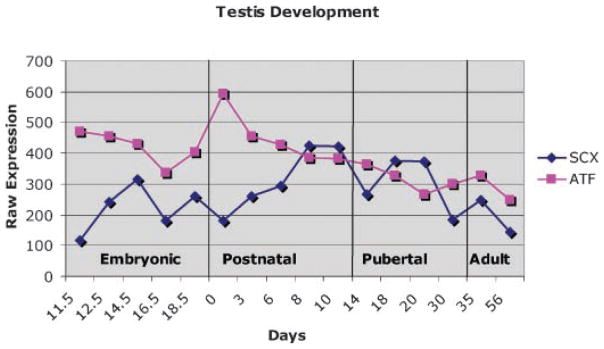
Since the data indicated a potential regulatory relationship between scleraxis and ATF4 during Sertoli cell differentiation, relative levels of gene expression were measured with a microarray analysis. Testis development microarray data were used to determine the relative expression of scleraxis from embryonic day 11.5 (E11.5) through postnatal day 56 (P56), Figure 8. During the embryonic stages of Sertoli cell development, expression of scleraxis increases from E11.5 to E14.5, correlating to the proliferation of Sertoli cells during this period. This trend of increased expression continues through the later postnatal period of Sertoli cell proliferation where expression of scleraxis increases approximately fourfold between postnatal days 0–10. ATF4 is expressed in all cell types of the testis and its relative expression, as seen by microarray analysis, is relatively constant during mouse testis development, Figure 8.
Fig. 8.
Expression of scleraxis and ATF4 in the mouse testis from embryonic day 11.5 through postnatal day 56 as depicted by microarray analysis. The raw signal expression level is presented for each developmental time point.
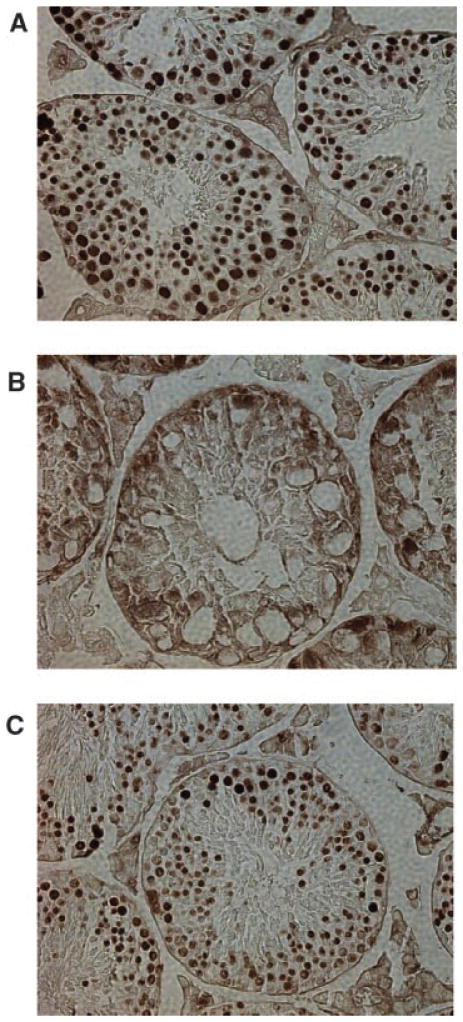
Scleraxis plays a known role in tendon differentiation in the developing fetus but until recently had not been identified in the adult testis. Northern blot analysis revealed that the expression of scleraxis in the testis is limited to Sertoli cells (Muir et al., 2006). A transgenic mouse model has been developed where GFP expression is driven by the scleraxis promoter (Pryce et al., 2007). Observations confirm the results of Muir et al. (2006) and show expression of scleraxis is limited to Sertoli cells in the testis, Figure 9. Since the Sertoli cells are critical for normal testicular development both pre- and post-natally, histological analysis of testis cross-sections from ATF4 and scleraxis knockout adult mice was performed. It has previously been determined that ATF4 knockout mice are infertile due to an obstruction of the vas deferens (Fischer et al., 2004). Sections of adult 65-day-old mouse testis from ATF+/− and ATF4−/− males were analyzed, Figure 10. ATF4+/− males are fertile and testis appear morphologically normal. Seminiferous tubules of ATF4−/− males contain vacuoles and the lumen appear to have fewer spermatozoa as compared to the ATF4+/− mice (Fig. 10). There was variation in the observed defects of the ATF4−/− mouse seminiferous tubules. Some mice appeared to have almost normal spermatogenesis, as was found by Fischer et al. (2004), while others were almost completely devoid of spermatogenesis with a dramatic increase in the number of vacuoles in the seminiferous tubules (Fig. 10). Sections of 65-day-old mouse testis from scleraxis−/− and scleraxis+/− males were analyzed. GFP expression driven by the scleraxis promoter demonstrated the localization of scleraxis in both the null and heterozygote testis in Sertoli cells, Figure 9A,B. Histological analysis demonstrated testis of scleraxis−/− mice appear to have reduced spermatogenesis, some tubules with no spermatogenic cells and some vacuolization occurring within the seminiferous tubules when compared to scleraxis+/− (Fig. 9C,D). Some tubules also had no lumen and the testis were also smaller in the scleraxis −/− mice. Similar results are seen with the ATF4−/− mouse testis (Fig. 10A–D).
Fig. 9.
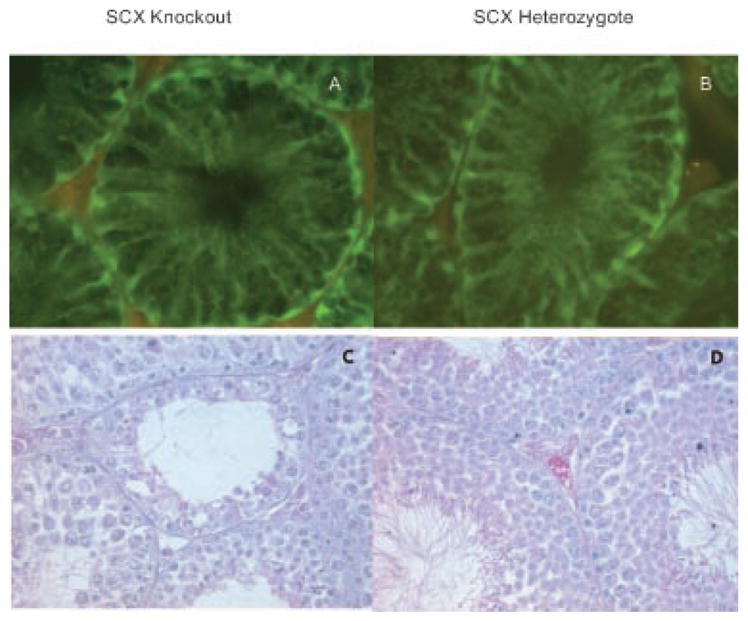
The expression of GFP in both the scleraxis −/− (A) and scleraxis +/− (B) testis as driven by the scleraxis promoter at 400× magnification. Fluorescent GFP staining restricted to Sertoli cells and data representative of two different animals and testis with replicate sections. Histological analysis of scleraxis −/− (C) and scleraxis +/− (D) testis at 400× magnification. Vacuoles and reduced spermatogenesis are indicated with arrows. Observations are representative of a minimum of three different animals and testes.
Fig. 10.
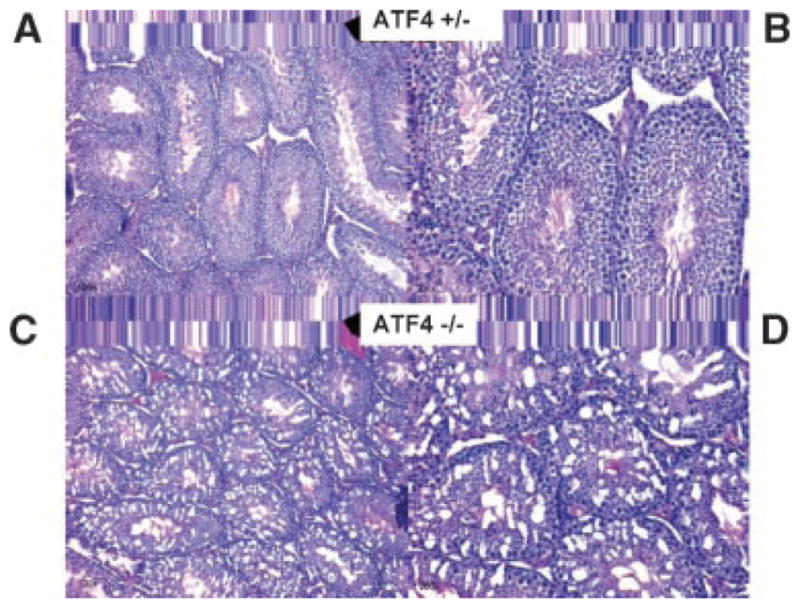
Histology of representative testis cross-sections from ATF4+/− (A,B) and ATF4−/− (C,D) mouse testis. The left panels (A,C) are 100× and the right (B,D) are 200×. Data are from three different animals and testis with replicate sections for ATF4 (−/−).
Histological analysis of ATF4−/− mice indicates a range of defects from seminiferous tubules containing large vacuoles and almost being devoid of spermatogenesis to seminiferous tubules that appear almost histologically normal (Fig. 11). An apoptosis (i.e., TUNEL) assay was performed on the testis cross-sections from 65-day-old ATF4+/− and ATF4−/− testis to determine if there was a difference in the number of cells with DNA fragmentation. There was no difference in the number of apoptotic cells between ATF4−/− and ATF4+/− testis (data not shown). Since histological analysis of ATF4−/− mice indicated a range of phenotypic variation, an immunocytochemical analysis for the germ cell marker GCNA was performed to assess total germ cell numbers between ATF4−/− and ATF4+/− testis. Observations indicate a similar number of germ cells between ATF4+/− mice and some of the ATF4−/− mice, while the ATF4−/− mice with vacuoles throughout the seminiferous tubules have many fewer germ cells that stain positive for GCNA, Figure 11. Therefore, the germ cell apoptosis required to create the vacuoles likely occurred prior to the adult stage testis analysis. The limited availability of scleraxis adult knockout homozygous animals and sections prevented a similar analysis of apoptosis and GCNA staining.
Fig. 11.
Testis germ cell GCNA staining of ATF4+/− (A), ATF4−/− (B) showing many vacuoles and ATF4−/− (C) showing an almost normal phenotype. The dramatic difference in phenotype is displayed between B and C, with B having no germ cells in the seminiferous tubule. All pictures taken at 400× magnification and replicate of a minimum of three experiments.
DISCUSSION
The ability of bHLH and CREB/ATF proteins to interact was initially observed through promoter analysis when E-box and CRE elements were found to be important for gene expression (Winter et al., 1993; Chaudhary and Skinner, 1999, 2001). These interactions were indirectly mediated by adaptor/linker proteins such as CREB binding protein (CBP/P300) that has the ability to bind both bHLH and bZip domain proteins. These protein complexes allowed the bHLH and CREB proteins to indirectly interact, but did not integrate the transcriptional pathways. These observations were extended through the demonstration of specific protein complexes involving bHLH dimers with bZip protein dimers (Kuras et al., 1997; Kurschner and Morgan, 1997). A tetrameric complex of a bHLH dimer and bZip dimer was observed (Kurschner and Morgan, 1997), as well as complexes containing additional proteins that influenced binding specificity to E-box elements (Kuras et al., 1997). More recently direct interactions using recombinant proteins and co-immunoprecipitations, as used in the current study, have demonstrated direct interactions of CREB1 and MyoD (Kim et al., 2005). The current study extends this observation to different members of the bHLH and CREB families. As shown with the EMSA, neither paraxis nor scleraxis can homodimerize. Therefore, a tetrameric complex of the bHLH and CREB would not be possible. The possibility of a dimeric ATF4 and monomeric bHLH complex could not be excluded, but the structural analysis data presented did not support this as the optimal conformation. In contrast, the structure analysis suggests the heterodimeric structure involving the bHLH and bZip domains is optimal, Figure 3. The gel shift data also demonstrate the ability of ATF4 to disrupt the bHLH-E12 heterodimer to apparently form an ATF4-bHLH heterodimer. Observations suggest a heterodimeric structure involving bHLH and CREB family members is possible and the structure of the heterodimer has a potential DNA binding capacity.
The CREB and bHLH heterodimer can integrate these two transcriptional control pathways. Members of the bHLH family and CREB/ATF family differentially interact with each other to form specific heterodimers. The bHLH proteins scleraxis and paraxis are important for somite development and proper segmentation of somite derived tissues, as well as chondrocyte and tendon development (Burgess et al., 1996; Wilson-Rawls et al., 1999a). Paraxis is restricted to muscle and heart in the adult (Wilson-Rawls et al., 2004). Recently, scleraxis has been shown to be present in a variety of adult tissues, including the testicular Sertoli cell (Muir et al., 2005). Both scleraxis and paraxis were found to interact with ATF4. Co-immunoprecipitation of recombinant proteins, the yeast-2-hyrbid interaction assays, protein structural analysis and gel shift analysis all suggested scleraxis or paraxis and ATF4 directly interact. Several of the procedures utilized do have limitations. The rabbit reticulocyte lysate used to generate the recombinant proteins for co-immunoprecipitation could contain proteins that could foster protein–protein interactions. In addition, the yeast cells used in the yeast-2-hybrid assays could potentially contain proteins that may bridge protein–protein interactions. Although these limitations need to be considered in any data interpretations, the combined data from the different procedures, including the structural analysis, strongly indicate a direct heterodimerization of the bHLH and bZip proteins. ATF4 also interacted with ITF2, MyoD, and Id2, but not E47 or E12. Interestingly, CREB1 and MyoD or ITF2 also form complexes, but none of the other bHLH proteins tested interacted with the ATF/CREB proteins. Therefore, specific complexes between bHLH and CREB family members appear to form, suggesting potential unique functions and integration of the two transcriptional control pathways. Future studies will require the functional analysis of various potential heterodimers.
Analysis of the hormonal regulation of scleraxis and ATF4 expression in Sertoli cells demonstrated different responses between the two. As hormones and cAMP pathways promoted an early event response in scleraxis expression, a delayed late response is seen for ATF4. One possible interpretation is that hormones such as FSH act through a cAMP pathway to initiate a scleraxis/ bHLH mediated transcriptional event as an early response (e.g., 2–4 hr) to initiate cellular differentiation. This is then followed by a delayed response in ATF4 expression that inhibits the scleraxis/bHLH E-box mediated transcription and promotes a downstream response with the scleraxis/ATF4 heterodimer. This distinct hormonal response between bHLH and CREB factors could provide a novel mechanism for negative feedback events and/or provide a link for a cascade of transcriptional events leading to cellular differentiation. Further elucidation of the interactions between bHLH and CREB factors through heterodimerization will provide insights into the molecular endocrinology of cellular differentiation.
The functional data presented suggests that the bHLH–ATF4 interaction inhibits binding to the E-box and suppresses the activity of bHLH proteins. The reporter systems used in the current study were inhibited by ATF4, but the bHLH–ATF4 complex may activate an alternative response element that remains to be identified. CREB1 can inhibit MyoD directed differentiation of 10T1/2 cells in culture. Interestingly, ATF4 and MyoD interacted in the yeast-2-hybrid assay, but in the functional assay ATF did not significantly inhibit MyoD. This may reflect a difference in affinity of the various heterodimers or functional differences. Previous observations have shown that CREB family members can form heterodimers with other classes of bZIP containing transcription factors such as fos and jun (Hai and Curran, 1991; Hai and Hartman, 2001), and the CNC-bZIP protein Nrf2, and these heterodimers bind unique response elements (Hai and Curran, 1991; He et al., 2001). The myogenic bHLH factors have also been shown to form complexes with MEF2 proteins and with GATA factors and are able to activate promoters through either E-boxes or MEF2 (Molkentin et al., 1995) or GATA sites (Kim et al., 2005). Therefore, the bHLH–ATF4 heterodimer provides a possible alternate transcriptional control mechanism for these two transcription factor family members. Although the potential unique response element remains to be identified, the observations made in the current study demonstrates these two transcription factor families can regulate each other.
The inhibitory actions of ATF4 on scleraxis and paraxis is consistent with observations that cAMP-dependent PKA blocks myogenic differentiation induced by myf5, MyoD, and myogenin (Li et al., 1992; Winter et al., 1993). These previous studies noted that PKA did not directly affect the bHLH proteins and proposed a mechanism of repression in which activation of CREB was indirectly targeting the E-box (Li et al., 1992; Winter et al., 1993). The formation of a bHLH-ATF/ CREB heterodimer could provide a mechanism for this inhibitory action of cAMP-dependent PKA on bHLH induced cellular differentiation. The specific mechanistic interactions between the CREB and bHLH pathways, the role of phosphorylation, actions of specific heterodimers, and function of potentially novel response elements remains to be elucidated. However, the ability of the bHLH and CREB families of transcription factors to directly interact adds a novel integration not previously considered.
The current study demonstrates the capacity of CREB and bHLH family members to directly interact and influence cellular differentiation. As with many developmentally important pathways and molecules, expression constructs are required to establish an in vitro model to study the system. This experimental limitation needs to be taken into account in extrapolating the current in vitro observations to in vivo physiological considerations. To address this issue an in vivo analysis of ATF4 and scleraxis null-mutant knockout testis was performed. Initially a microarray analysis of whole mouse testis was assessed to investigate the expression pattern of scleraxis and ATF4 from E11.5 through postnatal day (P) 56. Scleraxis expression in the testis is known to be specific to the Sertoli cell (Muir et al., 2006; Fig. 9) while the expression of ATF4 is ubiquitous (unpublished observation). During testis development, scleraxis expression increased approximately fourfold during the time of Sertoli cell proliferation and declined when germ cell number increased. Therefore, the decrease in expression of scleraxis in whole adult testis is due to the increased number of germ cells. ATF4 expression during this same time stayed relatively consistent.
Morphological analysis of ATF4 and scleraxis null-mutant knockout mouse testis revealed a range of defects associated with a lack of either gene. ATF4−/− testis are infertile (Fischer et al., 2004) and histology revealed the process of spermatogenesis is disrupted. The vacuoles that are present in the seminiferous tubules of the ATF4−/− male mice suggest a decrease in germ cell development and number. The presence of vacuoles in the seminiferous tubules is due to a spermatogenic cell defect and germ cell loss. No difference in the number of apoptotic cells was found between the ATF4−/− and ATF4+/− testis. Since there is not a decrease in the number of apoptotic germ cells in the testis, the number of germ cells was assessed. GCNA analysis was performed on testis cross-sections from ATF4−/− and ATF4+/− mouse testis. The difference in the number of germ cells between the ATF4−/− mice with the phenotype containing seminiferous tubules full of vacuoles is readily apparent. There are very few germ cells being produced in these mice and many tubules appear to be completely devoid of spermatogenesis (Fig. 11). In contrast, the ATF4−/− mice with the mild phenotype have small pockets within the seminiferous tubules that are devoid of spermatogenesis. The difference in the phenotypes of ATF4−/− mice indicates a difference in the stage of germ cell loss and suggests a defect in the Sertoli cell rather than a direct germ cell defect. Therefore there is either a loss of germ cells prior to the analysis or the rate of spermatogenesis is decreased resulting in fewer total cells. The scleraxis−/− testis phenotype data was similar to the ATF4−/− with an increase in seminiferous tubule vacuoles and a spermatogenic cell defect. More extensive analysis of the scleraxis−/− testis was not preformed. Observations suggest the ATF4−/− and scleraxis−/− testis phenotypes are similar.
The observations presented that specific bHLH and CREB family members appear to heterodimerize to potentially regulate gene expression adds a unique complexity and specificity to the transcriptional regulation of cellular function and differentiation. The current study suggests these heterodimers may have a critical role in the integration of bHLH and CREB transcriptional regulation pathways. For example, there are both CRE and E-box elements in the transferrin and follistatin promoters (Chaudhary and Skinner, 1999; Iezzi et al., 2004). Both of these promoters are up-regulated in an additive fashion by bHLH and CREB factors. The presence of the bHLH antagonist Id1 suppresses both E-box and CRE mediated transcription, suggesting promoter activation is dependent on potential interactions between bHLH and CREB activators (Iezzi et al., 2004; Chaudhary et al., 2005). These observations are consistent with the heterodimerization observations presented in the current study. Future studies will need to investigate the functional role of specific heterodimers between different bHLH and CREB family members. Analysis of testis development indicates a role for both CREB2/ATF4 and scleraxis in Sertoli cells and in the maintenance of normal spermatogenic cell capacity. The observations presented in the current study demonstrate the ability of these two different transcription factor families to potentially heterodimerize and integrate the transcriptional signaling pathways.
Acknowledgments
We acknowledge the technical contributions of Dr. Ingrid Sadler-Riggleman, Dr. Eric Nilsson, Dr. Jianrong Liu, Mr. Shane Rekow, Mr. Nathan Meyer, and Mr. John Hughs. We thank the WSU Biomolecular X-ray Crystallography Center for assistance with the protein modeling. This research was supported in part by grants from the USA National Institutes of Health (NIH to MKS), and the USA National Science Foundation (NSF to JWR and AR).
Grant sponsor: USA National Institutes of Health (NIH); Grant sponsor: USA National Science Foundation (NSF).
References
- Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2007;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Ameri K, Lewis CE, Raida M, Sowter H, Hai T, Harris AL. Anoxic induction of ATF-4 through HIF-1-independent pathways of protein stabilization in human cancer cells. Blood. 2004;103:1876–1882. doi: 10.1182/blood-2003-06-1859. [DOI] [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24:3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Buschhausen-Denker G, Bober E, Tannich E, Arnold HH. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J. 1989;8:701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burgess R, Rawls A, Brown D, Bradley A, Olson EN. Requirement of the paraxis gene for somite formation and musculoskeletal patterning. Nature. 1996;384:570–573. doi: 10.1038/384570a0. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Skinner MK. E-box and cyclic adenosine mono-phosphate response elements are both required for follicle-stimulating hormone-induced transferrin promoter activation in Sertoli cells. Endocrinology. 1999;140:1262–1271. doi: 10.1210/endo.140.3.6597. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Skinner MK. Role of the transcriptional coactivator CBP/p300 in linking basic helix-loop-helix and CREB responses for follicle-stimulating hormone-mediated activation of the transferrin promoter in Sertoli cells. Biol Reprod. 2001;65:568–574. doi: 10.1095/biolreprod65.2.568. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Sadler-Riggleman I, Ague JM, Skinner MK. The helix-loop-helix inhibitor of differentiation (ID) proteins induce post-mitotic terminally differentiated Sertoli cells to re-enter the cell cycle and proliferate. Biol Reprod. 2005;72:1205–1217. doi: 10.1095/biolreprod.104.035717. [DOI] [PubMed] [Google Scholar]
- Cupp AS, Tessarollo L, Skinner MK. Testis developmental phenotypes in neurotropin receptor trkA and trkC null mutations: Role in formation of seminiferous cords and germ cell survival. Biol Reprod. 2002;66:1838–1845. doi: 10.1095/biolreprod66.6.1838. [DOI] [PubMed] [Google Scholar]
- Dai YS, Cserjesi P, Markham BE, Molkentin JD. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J Biol Chem. 2002;277:24390–24398. doi: 10.1074/jbc.M202490200. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Olson EN. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1989;3:628–640. doi: 10.1101/gad.3.5.628. [DOI] [PubMed] [Google Scholar]
- Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Hadzic DB, McDaid JR, Firulli AB. The basic helix-loop-helix transcription factors dHAND and eHAND exhibit dimerization characteristics that suggest complex regulation of function. J Biol Chem. 2000;275:33567–33573. doi: 10.1074/jbc.M005888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Krawchuk D, Centonze VE, Vargesson N, Virshup DM, Conway SJ, Cserjesi P, Laufer E, Firulli AB. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet. 2005;37:373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer C, Johnson J, Stillwell B, Conner J, Cerovac Z, Wilson-Rawls J, Rawls A. Activating transcription factor 4 is required for the differentiation of the lamina propria layer of the vas deferens. Biol Reprod. 2004;70:371–378. doi: 10.1095/biolreprod.103.021600. [DOI] [PubMed] [Google Scholar]
- Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: Activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- He CH, Gong P, Hu B, Stewart D, Choi ME, Choi AM, Alam J. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem. 2001;276:20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- Iezzi S, Di Padova M, Serra C, Caretti G, Simone C, Maklan E, Minetti G, Zhao P, Hoffman EP, Puri PL, Sartorelli V. Deacetylase inhibitors increase muscle cell size by promoting myoblast recruitment and fusion through induction of follistatin. Dev Cell. 2004;6:673–684. doi: 10.1016/s1534-5807(04)00107-8. [DOI] [PubMed] [Google Scholar]
- Kim CH, Xiong WC, Mei L. Inhibition of MuSK expression by CREB interacting with a CRE-like element and MyoD. Mol Cell Biol. 2005;25:5329–5338. doi: 10.1128/MCB.25.13.5329-5338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Barbey R, Thomas D. Assembly of a bZIP-bHLH transcription activation complex: Formation of the yeast Cbf1-Met4-Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J. 1997;16:2441–2451. doi: 10.1093/emboj/16.9.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurschner C, Morgan JI. USF2/FIP associates with the b-Zip transcription factor, c-Maf, via its bHLH domain and inhibits c-Maf DNA binding activity. Biochem Biophys Res Commun. 1997;231:333–339. doi: 10.1006/bbrc.1997.6097. [DOI] [PubMed] [Google Scholar]
- Li L, Heller-Harrison R, Czech M, Olson EN. Cyclic AMP-dependent protein kinase inhibits the activity of myogenic helix-loop-helix proteins. Mol Cell Biol. 1992;12:4478–4485. doi: 10.1128/mcb.12.10.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cserjesi P, Olson EN. Dermo-1: A novel twist-related bHLH protein expressed in the developing dermis. Dev Biol. 1995;172:280–292. doi: 10.1006/dbio.1995.0023. [DOI] [PubMed] [Google Scholar]
- Libby BJ, De La Fuente R, O’Brien MJ, Wigglesworth K, Cobb J, Inselman A, Eaker S, Handel MA, Eppig JJ, Schimenti JC. The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev Biol. 2002;242:174–187. doi: 10.1006/dbio.2001.0535. [DOI] [PubMed] [Google Scholar]
- Ma PC, Rould MA, Weintraub H, Pabo CO. Crystal structure of MyoD bHLH domain-DNA complex: Perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- Muir T, Sadler-Riggleman I, Skinner MK. Role of the basic helix-loop-helix transcription factor, scleraxis, in the regulation of Sertoli cell function and differentiation. Mol Endocrinol. 2005;19:2164–2174. doi: 10.1210/me.2004-0473. [DOI] [PubMed] [Google Scholar]
- Muir T, Sadler-Riggleman I, Stevens JD, Skinner MK. Role of the basic helix-loop-helix protein ITF2 in the hormonal regulation of Sertoli cell differentiation. Mol Reprod Dev. 2006;73:491–500. doi: 10.1002/mrd.20397. [DOI] [PubMed] [Google Scholar]
- Murchison ND, Price BA, Conner DA, Keene DR, Olson EN, Tabin CJ, Schweitzer R. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;134:2697–2708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- Olson EN, Brown D, Burgess R, Cserjesi P. A new subclass of helix-loop-helix transcription factors expressed in paraxial mesoderm and chondrogenic cell lineages. Ann NY Acad Sci. 1996;785:108–118. doi: 10.1111/j.1749-6632.1996.tb56248.x. [DOI] [PubMed] [Google Scholar]
- Ord D, Ord T. Mouse NIPK interacts with ATF4 and affects its transcriptional activity. Exp Cell Res. 2003;286:308–320. doi: 10.1016/s0014-4827(03)00070-3. [DOI] [PubMed] [Google Scholar]
- Podust LM, Krezel AM, Kim Y. Crystal structure of the CCAAT box/enhancer-binding protein beta activating transcription factor-4 basic leucine zipper heterodimer in the absence of DNA. J Biol Chem. 2001;276:505–513. doi: 10.1074/jbc.M005594200. [DOI] [PubMed] [Google Scholar]
- Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn. 2007;236:1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- Quertermous EE, Hidai H, Blanar MA, Quertermous T. Cloning and characterization of a basic helix-loop-helix protein expressed in early mesoderm and the developing somites. Proc Natl Acad Sci USA. 1994;91:7066–7070. doi: 10.1073/pnas.91.15.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: Characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod. 2005;72:492–501. doi: 10.1095/biolreprod.104.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Tsujimura T, Takeda K, Sugihara A, Maekawa A, Terada N, Yoshida N, Akira S. Targeted disruption of ATF4 discloses its essential role in the formation of eye lens fibres. Genes Cells. 1998;3:801–810. doi: 10.1046/j.1365-2443.1998.00230.x. [DOI] [PubMed] [Google Scholar]
- Wilson-Rawls J, Hurt CR, Parsons SM, Rawls A. Differential regulation of epaxial and hypaxial muscle development by paraxis. Development. 1999a;126:5217–5229. doi: 10.1242/dev.126.23.5217. [DOI] [PubMed] [Google Scholar]
- Wilson-Rawls J, Molkentin JD, Black BL, Olson EN. Activated notch inhibits myogenic activity of the MADS-Box transcription factor myocyte enhancer factor 2C. Mol Cell Biol. 1999b;19:2853–2862. doi: 10.1128/mcb.19.4.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Rawls J, Rhee JM, Rawls A. Paraxis is a basic helix-loop-helix protein that positively regulates transcription through binding to specific E-box elements. J Biol Chem. 2004;279:37685–37692. doi: 10.1074/jbc.M401319200. [DOI] [PubMed] [Google Scholar]
- Winter B, Braun T, Arnold HH. cAMP-dependent protein kinase represses myogenic differentiation and the activity of the muscle-specific helix-loop-helix transcription factors Myf-5 and MyoD. J Biol Chem. 1993;268:9869–9878. [PubMed] [Google Scholar]
- Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ohbo K, Takakura A, Takebayashi H, Okada T, Abe K, Nabeshima Y. Sgn1, a basic helix-loop-helix transcription factor delineates the salivary gland duct cell lineage in mice. Dev Biol. 2001;240:517–530. doi: 10.1006/dbio.2001.0473. [DOI] [PubMed] [Google Scholar]