Abstract
Once obscure, the cilium has come into the spotlight during the past decade. It is now clear that aside from generating locomotion by motile cilia, both motile and immotile cilia serve as signaling platforms for the cell. Through both motility and sensory functions, cilia play critical roles in development, homeostasis, and disease. To date, the cilium proteome contains more than 1,000 different proteins, and human genetics is identifying new ciliopathy genes at an increasing pace. Although assigning a function to immotile cilia was a challenge not so long ago, the myriad of signaling pathways, proteins, and biological processes associated with the cilium have now created a new obstacle: how to distill all these interactions into specific themes and mechanisms that may explain how the organelle serves to maintain organism homeostasis. Here, we review the basics of cilia biology, novel functions associated with cilia, and recent advances in cilia genetics, and on the basis of this framework, we further discuss the meaning and significance of ciliary connections.
Keywords: left-right asymmetry, Joubert syndrome, intraflagellar transport, zebrafish, heterotaxy, polycystic kidney disease
INTRODUCTION: THE CILIUM TAKES CENTER STAGE
In recent years, the cilium has garnered immense attention because of the revelations of the indispensable role that the organelle plays in regulating vertebrate development and homeostasis. Although once considered vestigial, the cilium is now recognized to be a cell-surface signaling center that is well conserved from single-cell eukaryotes to humans. The organelle functions to sense extracellular factors and integrates these signals with a plethora of downstream functions, such as signal transduction, growth, and motility (55). As the cilium serves as the nexus of extracellular-to-intracellular signal integration, it is not surprising that ciliary defects have been linked to numerous disease etiologies, including polycystic kidney disease (PKD), obesity, and cancer (63).
The first evidence that linked the cilium to vertebrate disease came from studies of Kartagener syndrome (KS). The earliest such study, published in 1972, identified four male KS patients that exhibited immotile sperm, frequent bronchitis and sinusitis, situs inversus totalis, and the lack of mucociliary transport and ciliary motion due to the loss of flagellar dynein arms (2). Despite these findings, the crucial role of the cilium in disease and development remained poorly appreciated until proteins associated with cystic kidney disease were found on the cilium more than twenty years later. The first of such studies demonstrated the localization of polycystin 1 (PKD1), a protein implicated in autosomal dominant polycystic kidney disease (ADPKD), to the sensory cilia of Caenorhabditis elegans (11). However, the importance of this connection was not fully appreciated until it was found that the orpk mouse, a model for autosomal recessive polycystic kidney disease (ARPKD), contains a mutation in the ciliary gene Ift88/Polaris (115). This finding was further supported when polycystin 2 (PKD2), the second protein responsible for ADPKD, was found localized to renal cilia (116). In addition, genes responsible for nephronophthisis (NPHP), a juvenile cystic kidney disease, were also found to encode proteins that localize to the cilium (145). Finally, a genetic screen in zebrafish for mutations causing kidney cyst formation identified multiple cilia mutants, thus providing unbiased support for the importance of this organelle in PKD pathogenesis (138). The pleiotropic nature of cilia diseases was best emphasized by the revelation that the causative proteins behind Bardet-Biedl syndrome (BBS), a disorder that is characterized by the combination of polycystic kidney disease, obesity, diabetes, blindness, and polydactyly, localize as a large complex to the basal body and cilium (5, 104).
Over the past two decades, our knowledge of the cilium has largely been spearheaded by two core areas of investigation: (a) biochemical and cell biological studies using the ciliated green alga Chlamydomonas reinhardtii and (b) developmental and genetic studies in animal modules. The assembly and biogenesis of the cilium has been best characterized in the green alga, whereas the function of the cilium in signal transduction, development, and disease has been best demonstrated in humans, mice, zebrafish, and worms.
CILIA STRUCTURE AND CLASSIFICATION
The cilium is a microtubule-based structure that has been historically classified into two categories: motile and primary cilia. Motile cilia beat to generate fluid flow across an epithelial surface or to propel cell movement. Although mostly immotile, the primary cilium sits at the surface of most cells in the human body and senses physiological, chemical, and physical cues. Primary cilia are typically found one per cell (Figure 1g), such as in the luminal epithelium of the kidney duct. In some cases, clusters of primary cilia can be found on one cell, e.g., on the choroid plexus of the brain. Motile cilia are more frequently found in clusters on multiciliated cells (Figure 1h), such as tracheal cells and occasionally they are found individually on monociliated cells, such as the embryonic left-right organizer (LRO).
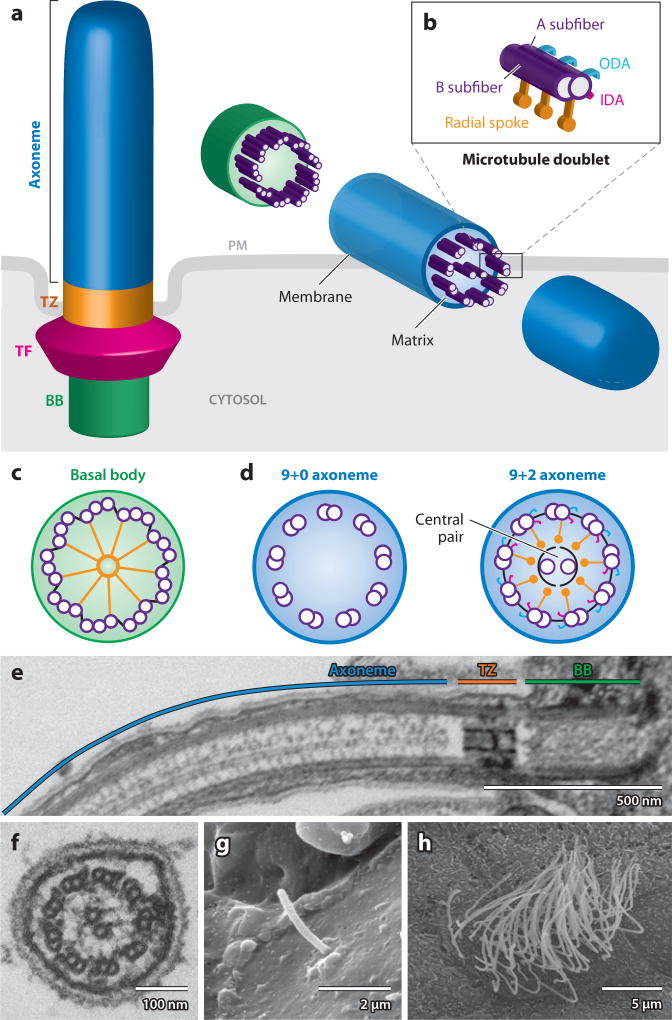
Figure 1.
Morphology and structure of the cilium. (a) Illustration depicting the cell surface location and three distinct structural segments of the cilium: axoneme (blue), transition zone (TZ) (orange), and basal body (BB) (green). The axoneme is composed of nine outer microtubule doublets surrounded by the ciliary membrane, which is contiguous with, but distinct from, the plasma membrane (PM) (gray). Transition fibers (TF) (magenta) found at the base of the cilium interconnect the microtubules and the plasma membrane. (b) Detailed examination of a microtubule doublet, which consists of A and B subfibers (violet). In motile cilia, the A subfiber displays outer dynein arms (ODAs) (cyan), inner dynein arms (IDAs) (magenta), and radial spokes (orange). (c) Transverse section of the basal body, detailing nine microtubule triplets (violet) and cartwheel (orange). (d) Transverse section of the axoneme, which is found in two configurations: with a central pair (9+2) and without the central pair (9+0). In the 9+2 configuration, nexins interconnect the nine microtubule doublets, and a sheath surrounds the central pair. (e) Longitudinal transmission electron microscopy (TEM) section of a 9+2 flagellum from a wild-type Chlamydomonas cell, with the axoneme, transition zone, and basal body highlighted. (f) Transverse TEM section of a 9+2 flagellar axoneme from a wild-type Chlamydomonas cell, with the microtubule doublets and central pair apparent. (g) Scanning electron microscopy (SEM) micrograph of an immotile cilium on the endoderm of a stage 17 Xenopus tropicalis embryo. (h) SEM micrograph of a motile multiciliated cell on the epidermis of a Xenopus tropicalis embryo. EM images courtesy of Mustafa Khokha, Dennis Diener, and Joel Rosenbaum (Yale University).
Both motile and primary cilia are comprised of a membrane-bound axoneme that consists of nine outer microtubule doublets (Figure 1a). Each doublet contains two microtubule subfibers, A and B (Figure 1b). Motile cilia contain two additional inner microtubules at the center of the axoneme, which compose the central pair and define the 9+2 axonemal configuration (Figure 1d, f) (117). Primary cilia lack these inner microtubules and are thus described as 9+0. Motile cilia also contain additional motors and components within the axoneme that assist with generating movement. Among the best studied of these proteins are the radial spokes, nexins, and outer (ODAs) and inner dynein arms (IDAs).
The proximal end of the axoneme is anchored to the cell body by the basal body, which is a modified centriole (Figure 1c). The ciliary membrane is a lipid bilayer that is an extension of the plasma membrane but with a distinct repertoire of membrane and channel proteins. The proximal region of the cilium between the axoneme and basal body is defined by the transition zone and transition fibers, which are believed to play a critical role in regulating traffic into and out of the cilium (Figure 1a,e). Intraflagellar transport (IFT) particles and cargo proteins have been shown to accumulate in this region as they enter and exit the cilium via IFT movement, suggesting that the transition fibers act as a filter that separates the ciliary compartment from the cytosol (39).
Recent progress has blurred the traditional distinction between motile and primary cilia. Exceptions exist to the 9+0 configuration of primary cilia and the 9+2 configuration of motile cilia. In the mouse node, a ciliated embryonic organ that establishes left-right (LR) asymmetrical body patterning, cilia of the 9+0 configuration have been proposed to be motile and beat in a circular motion to generate a directional flow (107). In the rabbit LRO, 9+4 motile cilia have been identified (48). Further, the choroid plexus epithelia in mice present 9+1 and 9+0 cilia, which may be partially motile (106). The inner ear of mammals contains hair cells with an immotile sensory kinocilium that is composed of a 9+2 axoneme (37). Curiously, in invertebrates, motile cilia of atypical axonemal configurations, such as 14+0 and 3+0 flagella on sperm of protura and parasitic protozoans, respectively, have been previously described (8, 122).
The functional differences between motile and primary cilia have also been questioned. Recent reports have demonstrated that in some cell types, motile cilia also serve sensory roles. Motile cilia of the mammalian airway epithelium, which beat to clear the lung of contaminants, display both mechanosensation and chemosensation functions (134). Interestingly, several classes of chemoreceptors are found on cilia of lung epithelial cells and addition of bitter compounds elevates intracellular calcium in these ciliated cells and stimulates cilia beating motility (134). In addition, mechanoreceptors have been shown to localize to the flagella of Chlamydomonas. Suppression of these mechanoreceptors by pharmacological treatment or genetic depletion results in aberrant swimming collision behavior in those cells (53). Similarly, mechanoreceptors that serve for collision avoidance behavior have also been found on motile cilia of Paramecium, a ciliated protozoan (72). As the number of reports that seemingly defy the canonical classification of distinct motile and primary cilia continue to grow, it becomes increasingly plausible that all types of cilia serve sensory roles and that motility is an additional function in a subset of cilia.
CILIA BIOGENESIS: INTRAFLAGELLAR TRANSPORT
The assembly of the cilium is believed to begin with the nucleation and growth of the microtubule axoneme onto the basal body, a modified centriole. Although the maturation, docking, and orientation of the basal body are not clearly understood, assembly of the cilium is highly influenced by these factors (71). Further, ciliogenesis is regulated by the cell cycle and is primarily induced during the G1 growth phase. Although initial growth of the axoneme is believed to involve the transition zone, continued elongation is restricted to the distal tip of the cilium and requires the transport of building proteins (71). Strikingly, the cilium does not contain its own translational machinery and instead utilizes proteins synthesized in the cell body to source the materials needed for construction and maintenance of the organelle (117). The transport of these proteins into the cilium depends on the bidirectional movement of IFT particles into and out of the cilium. IFT movement was first described in the green alga Chlamydomonas and remains the best-studied model of IFT-mediated ciliogenesis. IFT proteins are well conserved and required for ciliogenesis in nearly all ciliated organisms, from protozoans to vertebrates. When the final steady state of the cilium is reached, the IFT machinery remains active and continually turns over proteins, such as tubulin, at the distal tip (71, 117, 124).
Molecular motors power the movement of IFT particles and their cargo up and down the microtubule axoneme. The kinesin-2 family of motor complexes drives IFT particles toward the distal tip, which has been called anterograde movement. The first kinesin-2 complex to be associated with IFT was the canonical heterotrimeric kinesin-2, which was initially identified in sea urchin eggs (32). Heterotrimeric kinesin-2 is composed of two motor subunits, called kinesin superfamily protein 3A (KIF3A) and KIF3B, and one companion subunit, called kinesin-associated protein (KAP) (31). Genetic and biochemical studies in green alga, fruit fly, sea urchin, worm, and mouse have demonstrated that heterotrimeric kinesin-2 is essential for cilia assembly and biogenesis (131). A second complex, homodimeric kinesin-2, also contributes to ciliogenesis. Homodimeric kinesin-2 shares a redundant function with heterotrimeric kinesin-2 in worms and has been suggested to aid kinesin-2 in the assembly of distinct sensory cilia (131).
Retrograde movement is the return of IFT particles down the axoneme to the cell body and is driven by cytoplasmic dynein 2 (previously named cytoplasmic dynein 1b) (119). Cytoplasmic dynein 2 is a complex composed of four known subunits: a heavy chain (DYNC2H1), an intermediate chain (WD34), a light intermediate chain (DYNC2LI1), and a light chain (LC8). Genetic studies in green alga, worm, and mouse have demonstrated that depletion of cytoplasmic dynein 2 subunits results in disrupted retrograde IFT and stunted cilia with abnormal aggregations of IFT particles (130).
IFT particles were first purified from the flagellum of Chlamydomonas using sucrose gradient separation methods. Two distinct IFT protein complexes were identified and named IFT complex A and B, with each complex containing multiple proteins (33, 120). These studies and others have revealed 6 members of IFT complex A and 14 members of complex B. A large collection of studies from numerous ciliated organisms has revealed that IFT proteins are well conserved, and many are functionally required for cilia biogenesis (117). IFT complex B has been demonstrated to be important for anterograde movement, as disruptions in complex B proteins result in the failure to build cilia. Conversely, complex A proteins are not required for cilia assembly but seem to regulate retrograde movement (71, 117, 124). However, recent studies have revealed a more complicated picture for complex A components (90, 103). For example, IFT144 seems to be essential for cilia biogenesis and is required for anterograde transport of several membrane proteins (90).
THE EXPANDING FUNCTIONS OF CILIA
Many excellent reviews have been devoted to the well-established functions of cilia in development, disease, and intracellular signaling (4, 14,46,55,127, 132, 135). Here, we focus on novel and underappreciated functions for the organelle that have been recently revealed.
Neuronal Signaling and Development
The cilium plays a critical role in the patterning of the developing neural tube by regulating the Hedgehog (Hh) pathway (68). However, this may be the tip of the iceberg for the function of cilia in neural tissues. Cilia are detected throughout the developing and adult brain (15, 19,40,41, 60, 95). In addition, disrupting cilia biogenesis in a temporally and spatially controlled manner in the mouse, either by conditional knockout or targeted shRNA, is revealing an increasing list of ciliary involvement in the brain: telencephalic patterning (16), Hh-dependent expansion of the granule progenitor pool in the cerebellum (29), the migration and placement of interneurons in the developing cerebral cortex (62), hippocampal neurogenesis (22), the formation of adult neural stem cells (59), and glutamatergic synaptic integration of adult-born neurons (83). Consistently, multiple neuronal defects have been detected in ciliopathy patients. For example, a cerebellum malformation is the hallmark of the ciliopathy Joubert syndrome (JS). Axon guidance defects have also been detected in JS patients (136). Interestingly, there seems to be anecdotal evidence linking cilia and neuropsychiatric disorders. For example, a significant fraction of JS patients have been diagnosed with autistic disorder, and a common variant of AHI1, a JS gene, shows significant association with autism (3). Moreover, an RNAi screen revealed that out of 41 candidate genes associated with schizophrenia, bipolar disorder, autism, and intellectual disability, 20 affected ciliogenesis (96).
A possible connection between cilia and neurodegenerative diseases has also been hinted at by several studies. Keryer et al. (76) showed that Huntingtin (HTT), which has been implicated in Huntington’s disease (HD), regulates protein trafficking to the centrosome and controls ciliogenesis through a pathway consisting of HTT, huntingtin-associated protein 1 (HAP1), and pericentriolar material 1 protein (PCM1). Further, although deletion of Htt resulted in reduced ciliogenesis, polyQ expansion led to abnormally long cilia (76). However, whether ciliary defects contribute to the pathogenesis of HD remains unanswered. More recently, the Anderson group showed thatTTBK2, the causative gene of spinocerebellar ataxia type 11, is required for the initiation of axoneme assembly (56). Ttbk2 encodes a protein kinase that binds to microtubules and phosphorylates Tau, a microtubule-binding protein. Strikingly, Ttbk2 cells fail to make cilia, although the basal body appears to be normal. More importantly, expression of disease alleles of TTBK2 in wild-type cells inhibits ciliogenesis, thus providing a more direct correlation between cilia and this disease.
Both motile and primary cilia play a critical role in the development of the brain and neuronal tissues by regulating the cerebrospinal fluid (CSF). Primary cilia on radial glia and choroid plexus epithelial (CPE) cells coordinate with motile cilia on ependymal cells to transmit a host of signaling factors through the CSF to the developing and mature brain. A recent study demonstrated that cilia on CPE regulate the transcytosis of CSF into the ventricle by sensing levels of the neuropeptide FF, which is secreted by CPE cells themselves, thus suggesting a feedback mechanism in which CPE cilia act as chemosensors to control CSF production (106). Disruption of CPE cilia results in increased CSF production and hydrocephalus in mice (10).
Additionally, during late embryogenesis, radial glia progenitors differentiate into ventricular ependymal cells by establishing numerous motile cilia (99, 137). Sawamoto et al. (129) demonstrated that these ventricular cilia generate a fluid flow that establishes a Slit2 morphogen gradient in the CSF, which directs new neuron migration. Truncation or abnormal patterning of motile ependymal cilia results in motility defects, accumulation of CSF in the ventricles, and hydrocephalus (10). Interestingly, recent studies have demonstrated that planar cell polarity (PCP) regulates the proper establishment of CSF flow by coordinating the orientation of motile ependymal cilia in the ventricle (58).
DNA Damage Response
Emerging evidence has connected DNA damage response (DDR) and cilia. In an intriguing study, Chaki et al. (28) identified MRE11, ZNF423, and CEP164 as novel NPHP genes through the combination of homozygosity mapping and whole-exome resequencing. Interestingly, all three are involved in DDR. In addition, multiple proteins encoded by NPHP-related ciliopathy genes were found to colocalize with known DDR proteins to nuclear foci in response to DNA damage. Finally, knockdown of cep164 in zebrafish led to increased DDR (28). In a converse fashion, ATR, a protein previously known to be important for DDR, is found in the connecting cilium of photoreceptor cells, and partial reduction of its activity led to degeneration of photoreceptor cells in a fashion similar to retinitis pigmentosa (141). Together, these results suggest that multiple proteins are involved in both regulating DDR and maintaining a functional cilium. However, whether there is a causative relationship between the two is still unclear. Our recent results in zebrafish indicate that although DDR is increased in some cilia-associated mutants, there is no significant difference between several IFT mutants and their wild-type siblings, thus revealing that DDR is not intrinsic to ciliary defects (151). It is plausible that proteins involved in both DDR and ciliary function are involved in separate pathways, both ciliary and nonciliary.
HUMAN CILIOPATHIES
Although the remarkable role of the cilium in the etiology of human disorders was first hinted at nearly four decades ago (2), the genetic mechanisms underlying ciliopathies remained largely elusive until the past five years. With the recent advances in high-throughput genome sequencing technologies, numerous disease causing loci have been identified that have provided mechanistic insight into the pathogenesis of numerous motile and immotile ciliopathies.
Motile Ciliopathies
Among the first diseases to be linked to the cilium are diseases caused by defective motile cilia. Recent genetic studies of primary ciliary dyskinesia and heterotaxy have broadened our understanding of how defective motile cilia may contribute to disease states. Further, these findings support cilia-driven fluid flow as a crucial process during vertebrate development.
Primary ciliary dyskinesia
Primary ciliary dyskinesia (PCD) is a rare, autosomal recessive, genetically heterogeneous condition characterized by respiratory tract infections, chronic sinusitis, bronchiectasis, and infertility. When accompanied with heterotaxy (Htx), the disorder is known as KS, which accounts for more than 50% of PCD patients (49). The underlying causes are cilia motility defects, which affect multiple organs, notably the respiratory tract and reproductive system. The diagnosis of PCD has remained challenging because of the phenotypic, genetic, and cilia structural heterogeneity that is associated with the disorder. Historically, ciliary axonemal analysis by electron microscopy (EM) has been used for diagnosis, but approximately 28% of PCD patients do not display ciliary ultrastructural defects (73). Therefore, genetic testing has become increasingly important for diagnosis, but the current known mutations only account for approximately 50% of all PCD cases (87).
Recently, the application of next-generation genome sequencing has rapidly identified a large number of new disease-causing mutations for PCD. The majority of PCD-causing mutations have been characterized as defects in motile cilia components themselves, such as the ODAs, IDAs, and central pair apparatus. The first PCD-causative genes to be identified, DNAI1 and DNAH5, result in ODA defects and were isolated utilizing a candidate gene approach and homozygosity mapping (109, 118).
In addition, largely on the basis of studies performed in Chlamydomonas, it is understood that dynein arm subunits are preassembled in the cytosol, transported into flagella and docked onto the axoneme, although the mechanisms underlying these processes are poorly understood (52). Genetic studies in PCD patients have identified proteins that are critical for cytoplasmic dynein arm preassembly and axonemal trafficking, such as LRRC50, LRRC6, DNAAF3, and HEATR2, which are required for ciliary localization of both IDAs and ODAs (44, 65,79, 91, 100).
Studies in humans, dogs, mice, fish, and algae led to the identification of three uncharacterized genes encoding coiled-coil domain proteins (CCDC) as novel PCD candidate genes. Mutational analysis of PCD patients identified a number of families with mutations in CCDC39 and CCDC40 (6, 12, 20, 98). Immunofluorescence analysis of airway cilia samples from CCDC39 and 40 patients found a loss of GAS11, a component of the dynein regulatory complex (DRC), and DNALI1, a member of the IDA light chain family, from ciliary axonemes (12, 98). In Chlamydomonas, the CCDC39 ortholog, FAP59, was predicted to be critical for ciliary motility as orthologs do not exist in C. elegans, which only display immotile cilia. CCDC39 was found to localize to the entire ciliary axoneme in wild-type cilia but is lost in CCDC39 and CCDC40 patients, which suggests that these two proteins may physically interact with each other. Further, it is likely that these CCDC proteins interact with other axonemal components as new components of the DRC. This is supported by mutations in Chlamydomonas that affect known components of the DRC. For example, PF2 mutants exhibit a similar ultrastructural defect in flagella and failure of DRC and inner dynein arm assembly (121). Thus, like PF2, CCDC39 and 40 may encode DRC components that are required for the stability and function of the complex.
A separate study utilizing a similar approach identified ccdc103 from the zebrafish mutant smh, which displays paralyzed motile cilia and LR defects, as a critical modulator of ODA assembly (114). Genomic analysis of 146 PCD families identified 6 that displayed reduced ODAs and mutations in CCDC103. Immunofluorescence analysis of airway cilia from CCDC103 patients found partial loss of DNAH5, DNAH9, and DNAI2, all of which are ODA components, whereas IDA components were intact. The Chlamydomonas CCDC103 ortholog, PR46b, encodes a protein that binds to the flagellar axoneme, and dimerizes with LC2, an ODA component, in the cytoplasm. Together, these findings suggest that CCDC103 may regulate the docking of ODAs. Interestingly, two studies found PCD patients with mutations in CCDC114, which encodes a homolog of Chlamydomonas DCC2, an ODA microtubule docking-complex component (78, 110). This brings up the possibility that multiple CCDC proteins may form a complex that is essential for the docking of ODA components to the axoneme.
Heterotaxy
Congenital heart disease (CHD) encompasses a wide spectrum of structural cardiac defects that are present from birth and is the most common birth defect, affecting an estimated 1 in 130 live births. Although there is strong evidence to support a genetic contribution to CHD, the etiology of the disease has remained poorly understood. Htx is a rare congenital defect in which visceral organs, such as the heart, show discordant placement due to disruptions in normal LR patterning during embryogenesis. Htx is tightly correlated with CHD, as more than 90% of Htx patients have CHD and display relatively high mortality rates despite surgical intervention (30).
To elucidate the genetic network underlying human Htx, a recent study performed genome-wide analysis of copy number variations (CNVs) in 262 Htx subjects and identified 61 genes (47). To functionally verify the role that these genes play in LR development, they were investigated in a high-throughput manner in Xenopus. Among the 61 genes, 22 had orthologs in Xenopus that were heavily expressed in tissues relevant to LR development, such as the LRO and early heart. To functionally verify the role of these genes in LR development, loss-of-function analysis was performed by morpholino knockdown. This analysis demonstrated that five of these genes produced abnormal cardiac looping, gut looping, and Pitx2 expression, all of which are hallmarks of LR defects. Intriguingly, all five genes (GALNT11, NEK2, ROCK2, TGFBR2, and NUP188) were novel to LR development.
Patients with CHD and Htx display a high prevalence of respiratory disease, which has been assumed to be linked to the complications of surgeries that are commonly needed to correct structural heart defects (148). However, a recent genetic study of Htx patients has identified an enrichment of mutations in PCD genes, suggesting that defective cilia motility could be a common mechanism linking these two diseases (105). Among 43 CHD patients with Htx, 18 showed symptoms reminiscent of PCD and abnormal beating of cilia in the airway. Curiously, 11 of these patients were examined for ultrastructural defects in the ciliary axoneme and none were identified. However, sequencing of 14 PCD genes in 13 of these patients identified 3 patients with mutations in known PCD-causing genes, such as DNAI1, DNAH5, and DNAH11.
Immotile Ciliopathies
Although the connection between immotile cilia and disease states began with PKD, the breadth of this relationship has significantly broadened in recent years. The revelation that dysfunctional primary cilia are associated with numerous pleiotropic human disorders, many with overlapping abnormalities, began to suggest that ciliary signaling is a fundamental mechanism for maintaining a wide spectrum of cellular and developmental processes. Further, these findings revealed the utility of large-scale human genomic studies for identifying novel mechanisms of disease.
Renal and retinal-renal syndromes
ADPKD, which is characterized by the formation of bilateral renal cysts that commonly lead to end stage renal disease (ESRD), was among the first human disorders to be linked to defects affecting immotile cilia (as discussed above) (11, 115). The scope of renal cystic diseases caused by defective immotile cilia has expanded dramatically since the initial pioneering work. Among the most striking examples is NPHP, an autosomal recessive cystic kidney disease that is the most common cause of renal failure in children (64). NPHP has been linked to defects in 11 nephrocystins (NPHP1–11) that localize to the cilium, transition zone, and centrosome (64). Another ciliary protein, fibrocystin (PKHD1), is responsible for ARPKD, which is commonly characterized by renal cyst formation in utero (111, 142, 143).
NPHP patients may present extrarenal manifestations, such as retinal degeneration, laterality defects, and skeletal abnormalities. When in conjunction with retinal degeneration, the disorder is referred to as Senior-Løken syndrome and is commonly attributed to mutations in NPHP5, which encodes nephrocystin-5 (113). As photoreceptor cells contain modified cilia, it is not surprising that retinal and renal defects are commonly presented together in ciliopathies.
A recent study utilizing a novel candidate exome capture approach identified SDCCAG8, which encodes a centriolar protein, as a retinal-renal ciliopathy-causing gene (112). As exon capture with massively parallel sequencing often produces a large number of possible disease causing mutations for heterogeneous monogenetic disorders, Otto et al. (112) designed a combinatorial approach around homozygosity mapping in single families, followed by exon capture based around candidate ciliopathy genes and finally consecutive massively parallel sequencing. 12 SDCCAG8 mutations were identified as causing NPHP with retinal degeneration in 10 affected families. SDCCAG8 protein was found to localize to the basal body and transition zone of photoreceptor cells and to physically interact with OFD1, a ciliopathic protein. In addition, loss of sdccag8 in zebrafish resulted in kidney cysts and tail curvature, while loss of this gene in epithelial cells cultured led to luminal cell polarity defects.
Joubert syndrome and Meckel-Gruber syndrome
JS is a pleiotropic ciliopathy that is associated with NPHP and characterized by defects affecting the development of the cerebellum and brain stem, which results in a hallmark “molar-tooth sign” (54). JS patients often lack balance and coordination, which can manifest as ataxia, hypernea, and sleep apnea, and are commonly found to have mental retardation. In addition, a wide spectrum of secondary features is associated with JS, such as polydactyly, cleft lip, seizures, and retinal degeneration. A large number of disease-causing recessive mutations have been found through genomic analyses of JS patients, most notably ARL13B, INPP5E, TMEM216, MKS3, OFD1, and NPHP1 and 6 (42, 128). Many of these genes have been implicated in overlapping ciliopathies, such as NPHP and Meckel-Gruber syndrome (MGS), emphasizing the shared etiology of immotile ciliopathies. One of the most prominent examples is NPHP6/CEP290, which encodes a protein that localizes to the centrosome in a cell cycle–dependent manner and has been implicated in many ciliopathies, including NPHP, MGS, and BBS (63). Interestingly, the mutational load affecting NPHP6 directs the severity of the resulting phenotype. Relatively strong mutations in NPHP6, such as two truncating mutations, can result in a severe, early-onset disorder that affects several organ systems (as in MGS). In contrast, a relatively weak single missense mutation can result in a mild, late-onset disorder that affects one or two organ systems (as in NPHP) (63).
A recent study identified CEP41 as a causative gene for JS by combining patient sequencing with mechanistic analysis in model organisms (86). CEP41 was found to localize to the ciliary axoneme and centrosome and loss of CEP41 in mice and zebrafish produced multiple ciliary phenotypes, including cardiac LR defects and exencephaly. Strikingly, CEP41 complexes with and controls the ciliary trafficking of TTLL1, a conserved polyglutamylase enzyme that is required for glutamylation of microtubules. Microtubules are the critical structural backbone of the ciliary axoneme and acquire several posttranslational modifications, including acetylation, detyrosination, glycylation, and glutamylation. Depletion of cep41 in zebrafish did not disrupt normal cilia morphology yet resulted in defects in the A-tubules of the outer doublet microtubules and paralysis of motile cilia in the Kupffer’s vesicle (KV) and kidney. These results suggest that CEP41 functions in ciliary tubulin glutamylation by mediating transport of TTLL6 and links tubulin posttranslational modifications at the cilium to a ciliopathy.
MGS is a perinatally lethal ciliopathy that is characterized by renal cyst formation, posterior encephalocele, polydactyly, and situs inversus. Multiple causative genes for MGS are also shared by other ciliopathies, such as MKS1, TMEM216, CEP290, CC2D2A, NPHP3, and TMEM67 (63). Thus, as in JS, MGS arises from mutations affecting an overlapping pool of ciliopathy-causing genes, and the severity and combination of these mutations can dictate the severity and breadth of the manifested phenotype.
To define the molecular mechanisms underlying MKS-JBTS-NPHP and to test for a potential physical interaction among the disease proteins, the Jackson group performed high-throughput proteomics by combining a streamlined method for establishing stable cell lines expressing a double epitope tag fused to a single MKS-JBTS-NPHP protein with tandem affinity purification, followed by mass spectrometry to identify interacting proteins (126). In all, 850 interactors were copurified with 9 MKS-JS-NPHP bait proteins. These interactors were organized into a ciliary proteome network and clustered into three distinct functions: axonemal, centrosomal, and signaling-related. Strikingly, utilizing a candidate gene approach with 38 identified interactors, linkage and sequence analysis of 250 ciliopathic patients identified two novel genes associated with both NPHP and JS: ATXN10 and TCTN2. Thus, this work presents a new strategy for identifying ciliopathy-causing mutations: coupling human genetics with candidate genes that are verified by large-scale proteomic networks.
Bardet-Biedl syndrome
BBS is a genetically heterogeneous autosomal recessive disorder with a wide spectrum of phenotypes, including retinal degeneration, renal cysts, polydactyly, CHD, cognitive impairment, diabetes, and obesity (150). Approximately 30% of BBS patients display anosmia due to defective cilia on olfactory neurons (82). BBS genes were first linked to ciliopathies when it was shown that numerous BBS proteins localize to the ciliated sensory neurons of C. elegans (5). Mutations in 17 genes have been linked to BBS, with 7 encoding proteins that form a large protein complex (referred to as the BBSome) that is involved in ciliary membrane biogenesis and trafficking (5, 104). A second BBS complex consisting of three BBS proteins has been recently shown to be critical for regulating the assembly of the core BBSome complex (133).
FUNCTIONAL HUMAN GENETICS AND THE ROLE OF MODEL ORGANISMS
Next-generation sequencing technologies are revolutionizing human genetics and are revealing new genetic lesions at an unprecedented pace. With seven billion people on this planet, it is now feasible to systematically identify collections of mutations that lead to a specific symptom, in a manner analogous to traditional genetic screens performed in model organisms yet with the advantage of indisputable relevance to human biology and disease. Although this advantage seemingly disparages the relevance of model organisms, the advance of human genomic methodologies has heightened and emphasized the importance of model organisms for biomedical research. As human genetics moves into increasingly rare diseases and genetic modifications other than single gene lesions, such as CNVs, it will be difficult to determine the causative mutation(s) even by combining mapping and state-of-the-art sequencing technologies. For example, CNVs in a region at 16p11.2 have been associated with a range of neurocognitive defects (57, 144, 152). However, this region contains at least 29 different genes. In an interesting study, Golzio et al. (57) rapidly screened through the 29 candidates using overexpression and morpholino knockdown in zebrafish and identified KCTD13 as the major contributor of the phenotypes associated with CNVs in this region. Although this study was conducted for diseases with no clear connection to cilia, it illustrates the power of correlating human genetics with functional analysis in model organisms to pinpoint and validate disease-causing genes. Moreover, many genes or genetic loci identified by human genetic studies have no obvious known function and thus cannot immediately provide mechanistic insight into the disease. Only in model systems can we obtain designed genotypes and perform experiments that are not feasible in humans to dissect out the functions of these genes.
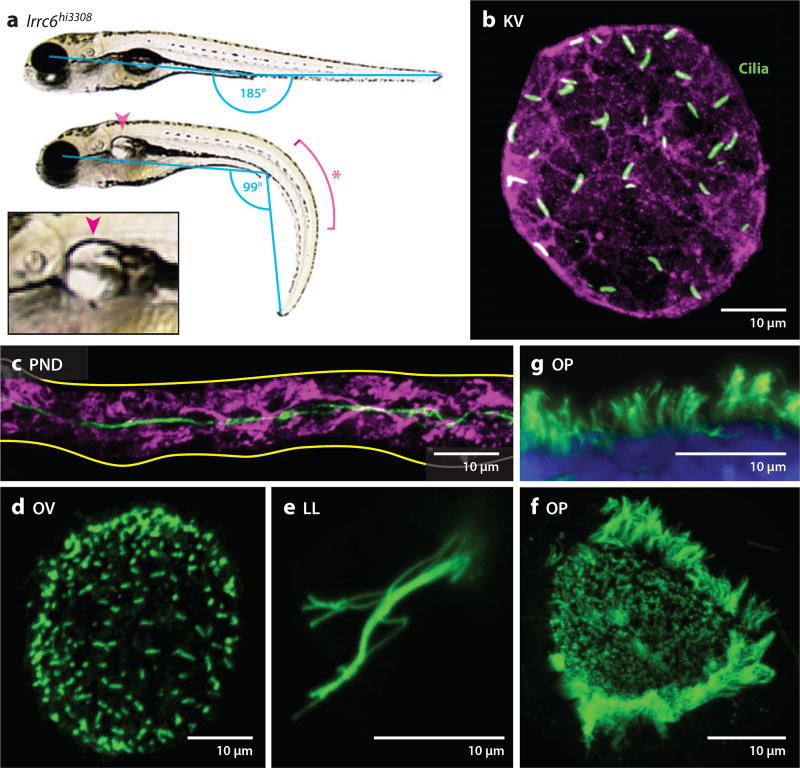
Many model organisms serve important roles in our quest to understand cilia and ciliopathies. Here, we discuss the use of zebrafish for the study of cilia and ciliopathies. One of the most attractive features of zebrafish is genetic tractability. Small in size, a large number of zebrafish can be housed in a laboratory setting and each pair of adult fish can produce hundreds of offspring at weekly intervals. In addition, the optical transparency and ex utero development of zebrafish embryos allows for monitoring of ciliary phenotypes in live and fixed embryos (Figure 2). Combined, these features make zebrafish accessible to large-scale genetic screens. In addition, zebrafish can be used to evaluate gene function in a relatively high-throughput fashion. Microinjection of morpholino oligos can be used to perform loss-of-function studies, while mRNA microinjection can be used to perform gain-of-function studies of different alleles both qualitatively and quantitatively. Furthermore, it is well established that many disease genes are functionally conserved in zebrafish. For example, vHNF1, a gene associated with human familial GCKD (glomerulocystic kidney disease), and PKD2, a gene associated with ADPKD, can cause kidney cysts when mutated in zebrafish (138, 139). Conversely, arl13b/scorpion, first identified as a novel ciliary and cystic kidney gene in a zebrafish mutagenesis screen (138), was later linked to JS (25). lrrc6/seahorse (Figure 2a), identified as another novel gene in the same genetic screen (138), was recently linked to PCD (79). Zebrafish are also uniquely situated for studying cilia-mediated signaling. Despite the remarkably high level of conservation in cilia structure and biogenesis in species ranging from the green alga Chlamydomonas to human, signaling mediated by cilia has diverged significantly, notably between vertebrate and invertebrate animals. For example, in contrast to the wide distribution of cilia on mammalian cells, most fly cells are devoid of cilia. Consistent with this distribution difference, the cilium provides a signaling platform for the Hh pathway in mammals, but it does not appear to be involved in this pathway in the fly. Similarly, in C. elegans, only some neuronal cells contain cilia (9, 70). Further, a functional Hh pathway seems to be absent in this organism. In contrast to worm and fly, cilia are widely distributed on zebrafish cells (Figure 2b–g). Moreover, as shown by the Hh pathway, cilia-mediated signaling is conserved in zebrafish. This is supported by defective Hh signaling in maternal-zygotic cilia mutants and the conserved cilia-targeting of Smo and Gli in zebrafish (35, 66, 77). Finally, zebrafish are highly amenable to pharmacological screens, albeit at a relatively moderate scale compared with cell models. Typically, thousands of compounds can be screened in a single effort by a typical laboratory utilizing zebrafish. However, the ability to evaluate the impact of chemical treatment in an intact vertebrate animal is attractive, as hits are more readily transferrable to mammals and screening can be performed in engineered disease models. For these reasons, chemical screens in zebrafish have already yielded novel insights and promising leads for several diseases, although with a very short history (45, 108, 149). In the ciliopathy field, histone deacetylase (HDAC) inhibitors were identified as suppressors of zebrafish pkd2 mutants in a pilot screen (27), and this result was further validated by mouse studies (27, 147). In the same screen, a number of chemicals with known biological activities were found to modify the phenotypes of IFT mutants (27). Some of the activities identified in the screen have been previously linked to cilia biogenesis and function, including TGF-β signaling (TGFβRI inhibitor), calcium signaling (K-252a and thapsigargin), cAMP signaling (IBMX and etazolate), and the microtubule cytoskeleton (nacodazole and paclitaxel). The connection between other candidates from this screen and cilia is less clear, which provides new avenues for future studies.
Figure 2.
Zebrafish as a vertebrate model for cilia analysis. (a) Brightfield image of a wild-type and lrrc6hi3308 zebrafish embryo at five days postfertilization (dpf). lrrc6hi3308 mutants display bilateral renal cysts (arrowhead and inset image) and a ventral body curvature (asterisk) due to ciliary defects. Body curvature is a hallmark of ciliary zebrafish mutants and the severity of the defect can be quantified by measuring the angle between the eye, yolk extension, and tail tip (cyan). (b–g) Fluorescent immunostaining of numerous ciliated tissues in wild-type zebrafish embryos, with cilia labeled by an antibody against acetylated-α-tubulin (green). (b) Kupffer’s vesicle (KV) at the 8-somite stage. The apical membrane is labeled with an antibody against atypical protein kinase C (PKC) (violet). (c) Pronephric duct (PND) at 2 dpf. The basolateral membrane is labeled with an antibody against Cdh17 (violet). (d) Otic vesicle (OV) at 20 h postfertilization (hpf). (e) Lateral line (LL) at 36 hpf. (f) Olfactory placode (OP) at 36 hpf. (g) Magnification of a peripheral edge of the OP. Nuclei are labeled with TOTO3 (blue).
As in human genetics, new technologies are transforming zebrafish genetics. Transposon-based vectors have made transgenesis in zebrafish routine (1, 74, 85, 140). Transgenic lines with fluorescent cilia combined with the optical transparency of zebrafish allow live imaging of cilia, both motile and immotile, in intact embryos (21, 94). Next-generation sequencing technologies greatly facilitate the cloning of mutations in zebrafish, previously a major bottleneck in large-scale mutagenesis screens in zebrafish. Another exciting development is the advance of new genome-editing technologies. In the past, TILLING (targeting induced local lesions in genomics), which identifies mutations in specific genes from a randomly mutagenized pool, was used to isolate mutations in a target gene (101). This technique is very labor intensive and therefore cannot be readily applied to groups of genes by individual laboratories. Alternatively, the custom-designed zinc-finger nuclease (ZFN) emerged as a truly targeted strategy. ZFNs are fusion proteins of C2H2 zinc fingers, which recognize and bind to 3–4 base pairs of DNA, and the cleavage domain of the endonuclease FokI. ZFNs can be engineered to contain several fingers to mediate sequence-specific cleavage in genomic DNA. Consequently, mutations at the target site are introduced via nonhomologous end-joining (NHEJ) repair (43, 92, 97). However, the sequence specificity of individual fingers can be influenced by its context, hampering its rapid and wide use in zebrafish. On the basis of the same principle, fusions of endonucleases with transcription activator-like (TAL) effectors have been developed to induce mutations in target genes. Because of the truly modular specificity of TAL effectors, this technique was embraced by the zebrafish community in a short time (13, 24, 38, 102). Recently, the clustered regularly interspaced short palindromic repeat (CRISPR) system, a bacteria defense system that uses short RNA to direct the degradation of foreign DNA, has been used to edit vertebrate genomes (34, 93). Because the only target-specific element in this system is a short RNA, this technique is even more straightforward to use. Excitingly, the CRISPR system seems to work efficiently in zebrafish as well, although germ-line transmission of induced mutations remains to be demonstrated (69). The concern of potential off-target effects is less an issue in zebrafish, whose 25 pairs of chromosomes make it feasible to breed out unintended mutations, and the specificity of mutant phenotypes can be validated by rescue experiments.
Owing to the development of such technologies, we now have a toolbox that allows us to perform large-scale genetic screens, generate human disease-specific alleles, and analyze cilia morphology, motility, and function in live embryos in depth. The eventual identification of all the Mendelian disease genes in the near future will generate a flood of information that will fuel the formulation of new concepts and hypotheses, all of which could be tested in model systems. In addition, model organisms and cultured cells also provide an opportunity to screen for therapeutic agents for ciliopathies, for which there is currently no directed therapy.
HORIZONS BEYOND CILIA: DIFFERENT DEGREES OF CILIARY CONNECTION
With more than 2,500 entries in the cilia database and more than 1,000 proteins in identified cilia proteomes (http://v3.ciliaproteome.org/cgi-bin/index.php), it is time to ask what constitutes a cilia connection and what this connection means. The most classic and perhaps most specific ciliary proteins are IFT proteins. First identified in Chlamydomonas (80), IFT proteins were thought to be functionally specific to cilia, even though their localization is not exclusive to the cilium, because multiple Chlamydomonas IFT mutants divide normally despite impaired ciliogenesis. In addition, elegant comparative genetic analyses have shown that IFT proteins are only conserved in ciliated organisms, suggesting that these proteins are specialized for cilia (7, 89). However, some evidence suggests that IFT proteins may function outside of the cilium in addition to their role in the cilium. For example, partial knockdown of IFT27, a complex B gene, in Chlamydomonas led to cytokinesis defects, whereas knockout resulted in lethality (123). This hints that IFT27 may serve additional functions, as many other Chlamydomonas IFT mutants do not display these phenotypes. It is worth noting that although conserved in Chlamydomonas and vertebrates, IFT27 homologs are not found in either Drosophila or C. elegans (75), whereas most other IFT proteins are conserved from the green alga to human, indicating that IFT27 may be unique among IFT components.
In vertebrates, there are rare but intriguing reports of nonciliary functions of IFT genes as well. The Baldari group showed that multiple IFT proteins, including IFT20, IFT88, and IFT57, are expressed in T cells and form a complex, even though these cells do not contain cilia (50). Further, by knockdown experiments, it was shown that IFT20 is involved in recycling signaling molecules to the immune synapse, which forms at the contact site when T cells are challenged by antigen-presenting cells (APCs) and serves as a signaling platform (50). Although this role of IFT20 in the immune synapse is clearly nonciliary, it is very similar on the molecular level to its role in IFT movement in ciliated cells, where it is predominantly localized in the golgi complex and is thought to be involved in delivering membrane proteins destined for the cilium to the ciliary base (51). There are additional similarities between the two processes. For example, both the immune synapse and the cilium are specialized signaling platforms with a high concentration of signaling molecules. During the formation of the immune synapse in T cells, the microtubule organizing center and golgi translocate toward the contact area to facilitate the delivery of signaling molecules to the immune synapse. This feature is reminiscent of cilia biogenesis and suggests that microtubule-mediated targeted delivery seems to be a common theme in these two processes. Teasing out nonciliary functions in nonhematopoietic cells proves to be more challenging because most of these cells contain cilia at many stages, making the exclusion of ciliary involvement beyond a reasonable doubt technically difficult.
In zebrafish, IFT mutants show characteristic ventral body curvature and kidney cyst phenotypes (Figure 2a), although cilia are able to form during early embryonic development because of the maternal contribution of the gene products (26, 81, 138). Nonetheless the essential role of IFT genes in cilia biogenesis is conserved in this model organism, as shown by the complete abolishment of cilia formation in maternal-zygotic ift88 mutants (66). Interestingly, mz-ift88 shows very similar, albeit slightly more severe phenotypes as the zygotic mutant (66). Multiple IFT mutants establish a constellation of phenotypes, including LR asymmetry defects, body curvature, bilateral kidney cysts, and retinal degeneration, as cilia-associated phenotypes in zebrafish. Intriguingly, overexpression of a truncated Ift172, a B-complex component, leads to an early morphogenesis defect not seen in the mzift88 mutants, whereas similarly overexpressed full-length proteins incur no deleterious effect (26). Whether this indicates the existence of additional IFT functions that are masked by redundancy or nonspecific dominant negative effects through unknown mechanisms is currently unclear. A possible scenario is that the truncated or tagged IFT proteins disrupt the functions of their partners more profoundly than the complete absence of the proteins. Because these phenotypes are not seen in mutants without cilia, whether these effects of mutant IFT proteins should be considered as cilia-associated or nonciliary is open to debate.
KIF3A and KIF3B can be considered as special IFT proteins. They are components of kinesin-2, an anterograde motor of IFT, and are essential for cilia biogenesis. Kif3a and Kif3b mutants have been frequently used to show a ciliary involvement in observed phenotypes. In many cases, Kif3a or Kif3b mutants do display similar phenotypes as IFT mutants. However, kinesin-2, as a microtubule motor, is also involved in cytosolic processes, such as melanosome transport and mitotic spindle formation (23, 61, 84, 88). Consistently, it was shown that KIF3A’s role in the canonical Wnt pathway is through both ciliary and nonciliary mechanisms (36). Combined, these results suggest that IFT proteins could be involved in nonciliary processes, although these processes may be closely related to IFT, and that individual IFTs may have unique functions.
Aside from IFT components, numerous proteins have been found on the cilium, basal body, and centrosome. Although a ciliary localization strongly suggests a role at the cilium, it is unclear how significant this connection is, especially when many of these proteins are found elsewhere. In the case of the Hh pathway components, some of which localize to the cilium, the connection is clearly very significant. The critical role of the cilium in this important signaling cascade is well established by many studies and has been reviewed extensively (4, 14, 46, 55, 67, 70, 125, 127, 132, 135, 146). Nevertheless, it is noteworthy that the pathway can still function in the absence of a cilium. An extreme example is Drosophila, where most of the cells are devoid of cilia and yet a large part of the Hh pathway is preserved and performs essential functions. In fact, the core components of this pathway were originally identified in Drosophila. Even in vertebrates, it has been demonstrated that Hh can function through cilia-independent mechanisms. For example, multiple cell lines that lack cilia still respond to acute treatment of the ligand, suggesting that the pathway still functions in these cells (18). In addition, in the noncanonical Hh pathway, Hh functions as a chemoattractant to guide axons in a Smoothened-dependent and Gli-independent manner even in the absence of the cilium (17). Taken together, existing evidence suggests that although the cilium is a critical site for Hh function, it is not the only compartment in which Hh signaling is critical.
For other ciliary proteins and signaling cascades, the relationship between location and function is less clear because of the lack of detailed and in-depth information. Yet, we can envision different types of ciliary connections.
Essential. In this case, the cilium is the primary functional site for a protein. The protein is localized on the cilium and disruption of ciliary structure would have major impact on the function of this protein, or disruptions of the protein would affect ciliary structure.
Regulatory. A protein interacts with a cilia-centric protein (or proteins) and regulates ciliary function or is regulated by cilia. Because many cilia-centric proteins shuttle between the cilium and other compartments of the cell, a ciliary localization is not obligatory for these proteins.
Guilt by association. A protein has multiple functions, both ciliary and nonciliary, and the only connection between the two is that the same protein is involved. A ciliary localization in this case is not sufficient evidence for assigning all functions of the protein to the cilium.
Although varying in the degree of their association with the cilium, all of the above are relevant to our understanding of cilia and ciliopathies in an integrated fashion, particularly in a complex and in an in vivo context. As in any maturing field, new findings in the cilia and ciliopathy field are providing a more in-depth and complicated picture. At this stage, finding a superficial ciliary connection for an uncharacterized protein would not be very satisfactory or illuminating. Fortunately, the exciting new developments in both human genetics and model organisms make it feasible to dissect the molecular and cellular functions of each unique cilia-associated protein. More specifically, as a crucial step in understanding the function of ciliary proteins, it has become increasingly important to have a clear grasp of the dynamics of the protein’s subcellular localization and its impact on the structure and function of the cilium as well as how the status of the cilium may influence the localization and function of the protein. In addition, considering that the cilium is now well established as an important organelle for the cell, it is safe to postulate that there are novel reciprocal interactions between the cilium and many cellular pathways or basic cellular processes that are yet to be discovered. Therefore, the role of the cilium should be taken into consideration when exploring cell signaling pathways and developmental processes, particularly in the context of disease phenotypes. This is truly an exciting and interesting time for studying cilia and ciliopathies.
SUMMARY POINTS.
Although the structure and biogenesis of the cilium have been extensively studied, many questions still exist pertaining to the regulated trafficking of proteins into the cilium and the many signaling pathways and biological processes regulated by this organelle.
Although cilia function has been historically associated with the homeostasis of the kidney and the lung, as well as the reproductive system, evidence points to broader functions.
Many ciliary proteins, including IFT components, are not exclusive to the cilium and have functions outside of the organelle.
Next-generation sequencing technologies have identified numerous disease-causing mutations in human ciliopathies. This has provided novel insights into the genetic mechanisms of numerous ciliopathic disorders.
Correlating gene discovery in humans with target validation and functional analysis in model organisms has begun to enable detailed mechanistic dissection of ciliopathic disease processes.
FUTURE ISSUES.
As the cilium is found on nearly all cell types in humans, the function of the organelle is likely to be broad. However, our knowledge of the versatility of ciliary function is only now beginning to expand. Future studies are needed to characterize the role of the cilium in tissues that have high relevance for development and homeostasis, particularly regenerative and metabolic tissues.
Only a limited number of studies have coupled human gene discovery with in-depth mechanistic analysis in model organisms for ciliopathies. Further studies are needed to extend our knowledge of the function of disease genes in the etiology of ciliopathies.
Not all the cilia-related genes are the same, particularly in regard to location and function. Although some cilia-related proteins function exclusively at or depend on the cilium, others do not, and these may serve nonciliary functions. In-depth analysis on each individual gene is essential to form a comprehensive understanding of cilia and ciliopathies in vivo.
Our understanding of the function of IFT has been hampered by the lack of cargos that directly interact and transport with the IFT complex. Live imaging approaches may aid in the identification of such cargo.
A limited number of studies illustrate the feasibility of identifying new therapeutic strategies for ciliopathies via model organisms. Continued work in the area of drug discovery and gene therapy in model organisms as the point of entry is necessary for the development of effective therapies.
Acknowledgments
We kindly thank M. Khokha, D. Diener, and J. Rosenbaum for contributing EM images. We apologize to our colleagues for citing reviews rather than original articles in some instances due to space limitation. S.Y. was supported by NIH training grant 5T32HD00709436. Research in the Sun laboratory is supported by NIDDK (R01 DK092808 and core A, 1P30DK090744) and American Cancer Society (RSG-10-247-01-DDC).
Glossary
- Bardet-Biedl syndrome (BBS)
a pleiotropic ciliopathy that affects many organ systems, including the eyes, nose, heart, kidney, and limbs
- Left-right organizer (LRO)
an embryonic and transient organ in vertebrates where bilateral symmetry is initially broken via cilia-driven directional fluid flow
- Intraflagellar transport (IFT)
a transportation process that moves cargo up and down the microtubule axoneme of the cilium
- Left-right (LR) asymmetry
although vertebrates may appear bilaterally symmetric when viewed externally, many internal organs are asymmetrically positioned across the left-right axis
- Primary ciliary dyskinesia (PCD)
an autosomal recessive disorder characterized by recurrent airway infections and sterility due to impaired ciliary motility
- Heterotaxy (Htx)
a human disorder characterized by discordant positioning of numeral visceral organs across the left-right axis
- Congenital heart disease (CHD)
a disorder of structural heart defects present at birth
- Kupffer’s vesicle (KV)
the ciliated left-right organizer in zebrafish
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abe G, Suster ML, Kawakami K. Tol2-mediated transgenesis, gene trapping, enhancer trapping, and the Gal4-UAS system. Methods Cell Biol. 2011;104:23–49. doi: 10.1016/B978-0-12-374814-0.00002-1. [DOI] [PubMed] [Google Scholar]
- 2.Afzelius BA. A human syndrome caused by immotile cilia. Science. 1976;193:317–19. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez Retuerto AI, Cantor RM, Gleeson JG, Ustaszewska A, Schackwitz WS, et al. Association of common variants in the Joubert syndrome gene (AHI1) with autism. Hum. Mol. Genet. 2008;17:3887–96. doi: 10.1093/hmg/ddn291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson KV. Cilia and Hedgehog signaling in the mouse embryo. Harvey Lect. 2006;102:103–15. doi: 10.1002/9780470593042.ch5. [DOI] [PubMed] [Google Scholar]
- 5.Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–33. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 6.Antony D, Becker-Heck A, Zariwala MA, Schmidts M, Onoufriadis A, et al. Mutations inCCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum. Mutat. 2013;34:462–72. doi: 10.1002/humu.22261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, et al. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–39. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- 8.Baccetti B, Dallai R, Giusti F. The spermatozoon of Arthropoda. VI. Ephemeroptera. J. Ultrastruct. Res. 1969;29:343–49. doi: 10.1016/s0022-5320(69)90112-9. [DOI] [PubMed] [Google Scholar]
- 9.Bae YK, Barr MM. Sensory roles of neuronal cilia: cilia development, morphogenesis, and function in C. elegans. Front. Biosci. 2008;13:5959–74. doi: 10.2741/3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banizs B, Pike MM, Millican CL, Ferguson WB, Komlosi P, et al. Dysfunctional cilia lead to altered ependyma and choroid plexus function, and result in the formation of hydrocephalus. Development. 2005;132:5329–39. doi: 10.1242/dev.02153. [DOI] [PubMed] [Google Scholar]
- 11.Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–89. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- 12.Becker-Heck A, Zohn IE, Okabe N, Pollock A, Lenhart KB, et al. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat. Genet. 2011;43:79–84. doi: 10.1038/ng.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–18. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berbari N, O’Connor A, Haycraft C, Yoder B. The primary cilium as a complex signaling center. Curr. Biol. 2009;19:R526–35. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berbari NF, Bishop GA, Askwith CC, Lewis JS, Mykytyn K. Hippocampal neurons possess primary cilia in culture. J. Neurosci. Res. 2007;85:1095–100. doi: 10.1002/jnr.21209. [DOI] [PubMed] [Google Scholar]
- 16.Besse L, Neti M, Anselme I, Gerhardt C, Ruther U, et al. Primary cilia control telencephalic patterning and morphogenesis via Gli3 proteolytic processing. Development. 2011;138:2079–88. doi: 10.1242/dev.059808. [DOI] [PubMed] [Google Scholar]
- 17.Bijlsma MF, Damhofer H, Roelink H. Hedgehog-stimulated chemotaxis is mediated by Smoothened located outside the primary cilium. Sci. Signal. 2012;5:ra60. doi: 10.1126/scisignal.2002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop CL, Bergin AM, Fessart D, Borgdorff V, Hatzimasoura E, et al. Primary cilium-dependent and -independent Hedgehog signaling inhibits p16(INK4A) Mol. Cell. 2010;40:533–47. doi: 10.1016/j.molcel.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 19.Bishop GA, Berbari NF, Lewis J, Mykytyn K. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J. Comp. Neurol. 2007;505:562–71. doi: 10.1002/cne.21510. [DOI] [PubMed] [Google Scholar]
- 20.Blanchon S, Legendre M, Copin B, Duquesnoy P, Montantin G, et al. Delineation of CCDC39/CCDC40 mutation spectrum and associated phenotypes in primary ciliary dyskinesia. J. Med. Genet. 2012;49:410–16. doi: 10.1136/jmedgenet-2012-100867. [DOI] [PubMed] [Google Scholar]
- 21.Borovina A, Superina S, Voskas D, Ciruna B. Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat. Cell Biol. 2010;12:407–12. doi: 10.1038/ncb2042. [DOI] [PubMed] [Google Scholar]
- 22.Breunig JJ, Sarkisian MR, Arellano JI, Morozov YM, Ayoub AE, et al. Primary cilia regulate hippocampal neurogenesis by mediating sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA. 2008;105:13127–32. doi: 10.1073/pnas.0804558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruno L, Salierno M, Wetzler DE, Desposito MA, Levi V. Mechanical properties of organelles driven by microtubule-dependent molecular motors in living cells. PLoS ONE. 2011;6:e18332. doi: 10.1371/journal.pone.0018332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–10. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, et al. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am. J. Hum. Genet. 2008;83:170–79. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y, Park A, Sun Z. Intraflagellar transport proteins are essential for cilia formation and for planar cell polarity. J. Am. Soc. Nephrol. 2010;21:1326–33. doi: 10.1681/ASN.2009091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Y, Semanchik N, Lee SH, Somlo S, Barbano PE, et al. Chemical modifier screen identifies HDAC inhibitors as suppressors of PKD models. Proc. Natl. Acad. Sci. USA. 2009;106:21819–24. doi: 10.1073/pnas.0911987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaki M, Airik R, Ghosh AK, Giles RH, Chen R, et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012;150:533–48. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chizhikov VV, Davenport J, Zhang Q, Shih EK, Cabello OA, et al. Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool. J. Neurosci. 2007;27:9780–89. doi: 10.1523/JNEUROSCI.5586-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen MS, Anderson RH, Cohen MI, Atz AM, Fogel M, et al. Controversies, genetics, diagnostic assessment, and outcomes relating to the heterotaxy syndrome. Cardiol. Young. 2007;17(Suppl. 2):29–43. doi: 10.1017/S104795110700114X. [DOI] [PubMed] [Google Scholar]
- 31.Cole DG. Intraflagellar transport: keeping the motors coordinated. Curr. Biol. 2005;15:R798–801. doi: 10.1016/j.cub.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 32.Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–70. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- 33.Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cong L, Ran FA, Cox D, Lin S, Barretto R, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–21. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 36.Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, et al. Kif3a constrains β-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat. Cell Biol. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- 37.Dabdoub A, Kelley MW. Planar cell polarity and a potential role for a Wnt morphogen gradient in stereociliary bundle orientation in the mammalian inner ear. J. Neurobiol. 2005;64:446–57. doi: 10.1002/neu.20171. [DOI] [PubMed] [Google Scholar]
- 38.Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, et al. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deane JA, Cole DG, Seeley ES, Diener DR, Rosenbaum JL. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 2001;11:1586–90. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- 40.Del Cerro MP, Snider RS. The Purkinje cell cilium. Anat. Rec. 1969;165:127–30. doi: 10.1002/ar.1091650202. [DOI] [PubMed] [Google Scholar]
- 41.Del Cerro MP, Snider RS. Cilia in axolotl neurons (Siredon mexicanum) Experientia. 1970;26:774–76. doi: 10.1007/BF02232544. [DOI] [PubMed] [Google Scholar]
- 42.Doherty D. Joubert syndrome: insights into brain development, cilium biology, and complex disease. Semin. Pediatr. Neurol. 2009;16:143–54. doi: 10.1016/j.spen.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008;26:702–8. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duquesnoy P, Escudier E, Vincensini L, Freshour J, Bridoux AM, et al. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2009;85:890–96. doi: 10.1016/j.ajhg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Durand EM, Zon LI. Newly emerging roles for prostaglandin E2 regulation of hematopoiesis and hematopoietic stem cell engraftment. Curr. Opin. Hematol. 2010;17:308–12. doi: 10.1097/MOH.0b013e32833a888c. [DOI] [PubMed] [Google Scholar]
- 46.Eggenschwiler J, Anderson K. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 2007;23:345–73. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fakhro KA, Choi M, Ware S, Belmont J, Towbin J, et al. Rare copy number variants in congenital heart disease patients identify genes in left-right patterning. Proc. Natl. Acad. Sci. USA. 2011;108:2915–20. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feistel K, Blum M. Three types of cilia including a novel 9+4 axoneme on the notochordal plate of the rabbit embryo. Dev. Dyn. 2006;235:3348–58. doi: 10.1002/dvdy.20986. [DOI] [PubMed] [Google Scholar]
- 49.Ferkol TW, Leigh MW. Ciliopathies: the central role of cilia in a spectrum of pediatric disorders. J. Pediatr. 2012;160:366–71. doi: 10.1016/j.jpeds.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finetti F, Paccani SR, Riparbelli MG, Giacomello E, Perinetti G, et al. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat. Cell Biol. 2009;11:1332–39. doi: 10.1038/ncb1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the golgi complex and is required for cilia assembly. Mol. Biol. Cell. 2006;17:3781–92. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fowkes ME, Mitchell DR. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol. Biol. Cell. 1998;9:2337–47. doi: 10.1091/mbc.9.9.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujiu K, Nakayama Y, Iida H, Sokabe M, Yoshimura K. Mechanoreception in motile flagella of Chlamydomonas. Nat. Cell Biol. 2011;13:630–32. doi: 10.1038/ncb2214. [DOI] [PubMed] [Google Scholar]
- 54.Gleeson JG, Keeler LC, Parisi MA, Marsh SE, Chance PF, et al. Molar tooth sign of the midbrain-hindbrain junction: occurrence in multiple distinct syndromes. Am. J. Med. Genet. A. 2004;125A:125–34. doi: 10.1002/ajmg.a.20437. discuss. 17. [DOI] [PubMed] [Google Scholar]
- 55.Goetz S, Anderson K. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 2010;11:331–44. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goetz SC, Liem KF, Jr, Anderson KV. The spinocerebellar ataxia-associated gene tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell. 2012;151:847–58. doi: 10.1016/j.cell.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–67. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guirao B, Meunier A, Mortaud S, Aguilar A, Corsi JM, et al. Coupling between hydrodynamic forces and planar cell polarity orients mammalian motile cilia. Nat. Cell Biol. 2010;12:341–50. doi: 10.1038/ncb2040. [DOI] [PubMed] [Google Scholar]
- 59.Han YG, Spassky N, Romaguera-Ros M, Garcia-Verdugo JM, Aguilar A, et al. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat. Neurosci. 2008;11(3):277–84. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- 60.Handel M, Schulz S, Stanarius A, Schreff M, Erdtmann-Vourliotis M, et al. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience. 1999;89:909–26. doi: 10.1016/s0306-4522(98)00354-6. [DOI] [PubMed] [Google Scholar]
- 61.Haraguchi K, Hayashi T, Jimbo T, Yamamoto T, Akiyama T. Role of the kinesin-2 family protein, KIF3, during mitosis. J. Biol. Chem. 2006;281:4094–99. doi: 10.1074/jbc.M507028200. [DOI] [PubMed] [Google Scholar]
- 62.Higginbotham H, Eom TY, Mariani LE, Bachleda A, Hirt J, et al. Arl13b in primary cilia regulates the migration and placement of interneurons in the developing cerebral cortex. Dev. Cell. 2012;23:925–38. doi: 10.1016/j.devcel.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N. Engl. J. Med. 2011;364:1533–43. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hildebrandt F, Zhou W. Nephronophthisis-associated ciliopathies. J. Am. Soc. Nephrol. 2007;18:1855–71. doi: 10.1681/ASN.2006121344. [DOI] [PubMed] [Google Scholar]
- 65.Horani A, Druley TE, Zariwala MA, Patel AC, Levinson BT, et al. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2012;91:685–93. doi: 10.1016/j.ajhg.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang P, Schier AF. Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development. 2009;136:3089–98. doi: 10.1242/dev.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 68.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 69.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–29. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 2011;12:393–406. [Google Scholar]
- 71.Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 2011;12:222–34. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 72.Jennings HS. Behavior of the Lower Organisms. New York: Columbia Univ. Press; 1906. [Google Scholar]
- 73.Jorissen M, Willems T, Van der Schueren B, Verbeken E, De Boeck K. Ultrastructural expression of primary ciliary dyskinesia after ciliogenesis in culture. Acta Oto-Rhino-Laryngol. Belg. 2000;54:343–56. [PubMed] [Google Scholar]
- 74.Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–22. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- 75.Keady BT, Samtani R, Tobita K, Tsuchya M, San Agustin JT, et al. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev. Cell. 2012;22:940–51. doi: 10.1016/j.devcel.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keryer G, Pineda JR, Liot G, Kim J, Dietrich P, et al. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J. Clin. Investig. 2011;121:4372–82. doi: 10.1172/JCI57552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim HR, Richardson J, van Eeden F, Ingham PW. Gli2a protein localization reveals a role for Iguana/DZIP1 in primary ciliogenesis and a dependence of Hedgehog signal transduction on primary cilia in the zebrafish. BMC Biol. 2010;8:65. doi: 10.1186/1741-7007-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Knowles MR, Leigh MW, Ostrowski LE, Huang L, Carson JL, et al. Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;92:99–106. doi: 10.1016/j.ajhg.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kott E, Duquesnoy P, Copin B, Legendre M, Dastot-Le Moal F, et al. Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2012;91:958–64. doi: 10.1016/j.ajhg.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 1993;90:5519–23. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kramer-Zucker AG, Olale F, Haycraft CJ, Yoder BK, Schier AF, Drummond IA. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development. 2005;132:1907–21. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- 82.Kulaga HM, Leitch CC, Eichers ER, Badano JL, Lesemann A, et al. Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nat. Genet. 2004;36:994–98. doi: 10.1038/ng1418. [DOI] [PubMed] [Google Scholar]
- 83.Kumamoto N, Gu Y, Wang J, Janoschka S, Takemaru K-I, et al. A role for primary cilia in glutamatergic synaptic integration of adult-born neurons. Nat. Neurosci. 2012;15:399–405. doi: 10.1038/nn.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kural C, Serpinskaya AS, Chou YH, Goldman RD, Gelfand VI, Selvin PR. Tracking melanosomes inside a cell to study molecular motors and their interaction. Proc. Natl. Acad. Sci. USA. 2007;104:5378–82. doi: 10.1073/pnas.0700145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–99. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 86.Lee JE, Silhavy JL, Zaki MS, Schroth J, Bielas SL, et al. CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nat. Genet. 2012;44:193–99. doi: 10.1038/ng.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leigh MW, Pittman JE, Carson JL, Ferkol TW, Dell SD, et al. Clinical and genetic aspects of PCD/Kartagener syndrome. Genet. Med. 2009;11:473–87. doi: 10.1097/GIM.0b013e3181a53562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Levi V, Serpinskaya AS, Gratton E, Gelfand V. Organelle transport along microtubules in Xenopus melanophores: evidence for cooperation between multiple motors. Biophys. J. 2006;90:318–27. doi: 10.1529/biophysj.105.067843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–52. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 90.Liem KF, Jr, Ashe A, He M, Satir P, Moran J, et al. The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J. Cell Biol. 2012;197:789–800. doi: 10.1083/jcb.201110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Loges NT, Olbrich H, Becker-Heck A, Haffner K, Heer A, et al. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am. J. Hum. Genet. 2009;85:883–89. doi: 10.1016/j.ajhg.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mali P, Yang L, Esvelt KM, Aach J, Guell M, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–26. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Malicki J, Avanesov A, Li J, Yuan S, Sun Z. Analysis of cilia structure and function in zebrafish. Methods Cell Biol. 2011;101:39–74. doi: 10.1016/B978-0-12-387036-0.00003-7. [DOI] [PubMed] [Google Scholar]
- 95.Mandl L, Megele R. Primary cilia in normal human neocortical neurons. Z. Mikrosk. Anat. Forsch. 1989;103:425–30. [PubMed] [Google Scholar]
- 96.Marley A, von Zastrow M. A simple cell-based assay reveals that diverse neuropsychiatric risk genes converge on primary cilia. PLoS ONE. 2012;7:e46647. doi: 10.1371/journal.pone.0046647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Merveille AC, Davis EE, Becker-Heck A, Legendre M, Amirav I, et al. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat. Genet. 2011;43:72–78. doi: 10.1038/ng.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mirzadeh Z, Han YG, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Cilia organize ependymal planar polarity. J. Neurosci. 2010;30:2600–10. doi: 10.1523/JNEUROSCI.3744-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mitchison HM, Schmidts M, Loges NT, Freshour J, Dritsoula A, et al. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat. Genet. 2012;44:381–89. S1–2. doi: 10.1038/ng.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moens CB, Donn TM, Wolf-Saxon ER, Ma TP. Reverse genetics in zebrafish by TILLING. Brief Funct. Genomic Proteomic. 2008;7:454–59. doi: 10.1093/bfgp/eln046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, et al. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS ONE. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, et al. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the sonic hedgehog pathway via cAMP signaling. Cell. 2013;152:210–23. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 104.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–13. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 105.Nakhleh N, Francis R, Giese RA, Tian X, Li Y, et al. High prevalence of respiratory ciliary dysfunction in congenital heart disease patients with heterotaxy. Circulation. 2012;125:2232–42. doi: 10.1161/CIRCULATIONAHA.111.079780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Narita K, Kawate T, Kakinuma N, Takeda S. Multiple primary cilia modulate the fluid transcytosis in choroid plexus epithelium. Traffic. 2010;11:287–301. doi: 10.1111/j.1600-0854.2009.01016.x. [DOI] [PubMed] [Google Scholar]
- 107.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, et al. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–37. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 108.North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–11. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Olbrich H, Haffner K, Kispert A, Volkel A, Volz A, et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat. Genet. 2002;30:143–44. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 110.Onoufriadis A, Paff T, Antony D, Shoemark A, Micha D, et al. Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;92:88–98. doi: 10.1016/j.ajhg.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Onuchic LF, Furu L, Nagasawa Y, Hou X, Eggermann T, et al. PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel β-helix 1 repeats. Am. J. Hum. Genet. 2002;70:1305–17. doi: 10.1086/340448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat. Genet. 2010;42:840–50. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Otto EA, Loeys B, Khanna H, Hellemans J, Sudbrak R, et al. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat. Genet. 2005;37:282–88. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 114.Panizzi JR, Becker-Heck A, Castleman VH, Al-Mutairi DA, Liu Y, et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat. Genet. 2012;44:714–19. doi: 10.1038/ng.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, et al. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 2000;151:709–18. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr. Biol. 2002;12:R378–80. doi: 10.1016/s0960-9822(02)00877-1. [DOI] [PubMed] [Google Scholar]
- 117.Pedersen L, Rosenbaum J. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr. Top. Dev. Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 118.Pennarun G, Escudier E, Chapelin C, Bridoux AM, Cacheux V, et al. Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am. J. Hum. Genet. 1999;65:1508–19. doi: 10.1086/302683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pfister KK, Fisher EM, Gibbons IR, Hays TS, Holzbaur EL, et al. Cytoplasmic dynein nomenclature. J. Cell Biol. 2005;171:411–13. doi: 10.1083/jcb.200508078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Piperno G, Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA. 1997;94:4457–62. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Piperno G, Mead K, Shestak W. The inner dynein arms I2 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J. Cell Biol. 1992;118:1455–63. doi: 10.1083/jcb.118.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Prensier G, Vivier E, Goldstein S, Schrevel J. Motile flagellum with a “3 + 0” ultrastructure. Science. 1980;207:1493–94. doi: 10.1126/science.7189065. [DOI] [PubMed] [Google Scholar]
- 123.Qin H, Wang Z, Diener D, Rosenbaum J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 2007;17:193–202. doi: 10.1016/j.cub.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rosenbaum J, Witman G. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–25. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 125.Roy S. Cilia and Hedgehog: When and how was their marriage solemnized? Differentiation. 2012;83:S43–48. doi: 10.1016/j.diff.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 126.Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–28. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 128.Sattar S, Gleeson JG. The ciliopathies in neuronal development: a clinical approach to investigation of Joubert syndrome and Joubert syndrome-related disorders. Dev. Med. Child Neurol. 2011;53:793–98. doi: 10.1111/j.1469-8749.2011.04021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sawamoto K, Wichterle H, Gonzalez-Perez O, Cholfin JA, Yamada M, et al. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science. 2006;311:629–32. doi: 10.1126/science.1119133. [DOI] [PubMed] [Google Scholar]
- 130.Scholey JM. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 2003;19:423–43. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- 131.Scholey JM. Intraflagellar transport motors in cilia: moving along the cell’s antenna. J. Cell Biol. 2008;180:23–29. doi: 10.1083/jcb.200709133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Scholey JM, Anderson KV. Intraflagellar transport and cilium-based signaling. Cell. 2006;125:439–42. doi: 10.1016/j.cell.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 133.Seo S, Baye LM, Schulz NP, Beck JS, Zhang Q, et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc. Natl. Acad. Sci. USA. 2010;107(4):1488–93. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ. Motile cilia of human airway epithelia are chemosensory. Science. 2009;325:1131–34. doi: 10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Singla V, Reiter JF. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science. 2006;313:629–33. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 136.Spampinato MV, Kraas J, Maria BL, Walton ZJ, Rumboldt Z. Absence of decussation of the superior cerebellar peduncles in patients with Joubert syndrome. Am. J. Med. Genet. A. 2008;146A:1389–94. doi: 10.1002/ajmg.a.32282. [DOI] [PubMed] [Google Scholar]
- 137.Spassky N, Merkle FT, Flames N, Tramontin AD, Garcia-Verdugo JM, Alvarez-Buylla A. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci. 2005;25:10–18. doi: 10.1523/JNEUROSCI.1108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–93. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- 139.Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–29. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Urasaki A, Kawakami K. Analysis of genes and genome by the tol2-mediated gene and enhancer trap methods. Methods Mol. Biol. 2009;546:85–102. doi: 10.1007/978-1-60327-977-2_6. [DOI] [PubMed] [Google Scholar]
- 141.Valdes-Sanchez L, De la Cerda B, Diaz-Corrales FJ, Massalini S, Chakarova CF, et al. ATR localizes to the photoreceptor connecting cilium and deficiency leads to severe photoreceptor degeneration in mice. Hum. Mol. Genet. 2013;22:1507–15. doi: 10.1093/hmg/dds563. [DOI] [PubMed] [Google Scholar]
- 142.Ward CJ, Hogan MC, Rossetti S, Walker D, Sneddon T, et al. The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat. Genet. 2002;30:259–69. doi: 10.1038/ng833. [DOI] [PubMed] [Google Scholar]
- 143.Ward CJ, Yuan D, Masyuk TV, Wang X, Punyashthiti R, et al. Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum. Mol. Genet. 2003;12:2703–10. doi: 10.1093/hmg/ddg274. [DOI] [PubMed] [Google Scholar]
- 144.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 145.Wolf MT, Hildebrandt F. Nephronophthisis. Pediatr. Nephrol. 2011;26:181–94. doi: 10.1007/s00467-010-1585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wong SY, Reiter JF. The primary cilium at the crossroads of mammalian hedgehog signaling. Curr. Top. Dev. Biol. 2008;85:225–60. doi: 10.1016/S0070-2153(08)00809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xia S, Li X, Johnson T, Seidel C, Wallace DP, Li R. Polycystin-dependent fluid flow sensing targets histone deacetylase 5 to prevent the development of renal cysts. Development. 2010;137:1075–84. doi: 10.1242/dev.049437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yildirim SV, Tokel K, Varan B, Aslamaci S, Ekici E. Clinical investigations over 13 years to establish the nature of the cardiac defects in patients having abnormalities of lateralization. Cardiol. Young. 2007;17:275–82. doi: 10.1017/s1047951107000479. [DOI] [PubMed] [Google Scholar]
- 149.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J. Clin. Investig. 2009;119:428–37. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhao L, Yuan S, Cao Y, Kallakuri S, Li Y, et al. Reptin/Ruvbl2 is a Lrrc6/Seahorse interactor essential for cilia motility. Proc. Natl. Acad. Sci. USA. 2013;110(31):12697–702. doi: 10.1073/pnas.1300968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zufferey F, Sherr EH, Beckmann ND, Hanson E, Maillard AM, et al. A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J. Med. Genet. 2012;49:660–68. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]