Abstract
The molecular mechanisms involved in developmental biology and cellular differentiation have traditionally been considered to be primarily genetic. Environmental factors that influence early life critical windows of development generally do not have the capacity to modify genome sequence, nor promote permanent genetic modifications. Epigenetics provides a molecular mechanism for environment to influence development, program cellular differentiation, and alter the genetic regulation of development. The current review discusses how epigenetics can cooperate with genetics to regulate development and allow for greater plasticity in response to environmental influences. This impacts area such as cellular differentiation, tissue development, environmental induced disease etiology, epigenetic transgenerational inheritance, and the general systems biology of organisms and evolution.
Keywords: epigenetics, development, environment, transgenerational, systems biology, differentiation, disease etiology
Current Paradigm for The Molecular Control of Developmental Biology
Traditionally genetic mechanisms and processes have been thought to provide the primary control for cellular differentiation and developmental biology. This involves the regulation of genome activity through a series of genetic factors such as a cascade of critical transcription factors, regulation of gene expression to promote a programming of transcriptional events essential for cellular differentiation and eventual development of the tissue and organism. Abnormal development associated with events such as disease development have also been thought to be regulated primarily from genetic mutations. The role of genetics in the regulation of developmental biology is well established and many studies support the actions of numerous genes and genetic processes. As with any area of biology, genetics is a critical element of the normal and abnormal development of a cell, tissue or organism. However, there are several observations that suggest genetics alone is not sufficient to regulate the entire phenomena of development or systems biology.
Abnormal development and critical windows of exposure have demonstrated environmental factors can promote adult onset disease, which is associated with early alterations in normal development of a cell or tissue. These environmental factors include nutrition, environmental compounds, and stress. These factors generally do not have the ability to directly promote DNA sequence mutations nor permanently alter genetic processes. This fetal basis of adult onset disease clearly involves an abnormal developmental process, but is difficult to explain on the basis of genetic processes alone. Environmental factors can also promote alterations in normal development to induce developmental mutations and defects, but again most factors have not been shown to alter DNA sequence. On a population level, many evolutionary processes appear to be influenced by environmental factors to promote rapid alterations in development and adaptations, which are also difficult to explain with genetic mechanisms alone.
An additional molecular mechanism that can complement genetics to influence developmental biology is epigenetics. The ability of environment to influence development can also be facilitated by epigenetic processes. The paradigm that genetics is the primary factor to regulate developmental biology is limited and ignores the plasticity to respond rapidly to environment, nor does it explain abnormal development and disease etiology in the absence of genetic alterations. Epigenetics provides an additional molecular mechanism to complement genetics in the regulation of development. Therefore, the paradigm shift is that layers of molecular control and cascades of both epigenetic and genetic factors or processes are involved in regulating developmental biology, Figure 1. This provides a more robust and thorough control of development that integrates these two critical molecular processes.
Figure 1.
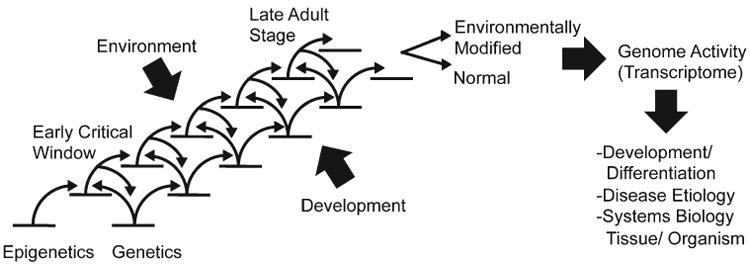
Epigenetic and genetic cascade of events involved in development.
Epigenetics
Epigenetics is defined as molecular factors and processes around DNA that regulate genome activity, independent of DNA sequence, and that are mitotically or meiotically stable (Skinner et al., 2010). The term heritable has been used, but is defined as generational inheritance by definition so does not include all elements of epigenetics. Therefore, mitotically stable is more precise and clarifies that as the cell undergoes mitosis the epigenome is replicated. An environmentally induced stable alteration in the epigenome can permanently alter the regulation of genome activity during cell differentiation and development processes.
The origin of the use of the term epigenetics was by Conrad Waddington in the 1940s while he was studying environment—gene interactions to promote phenotypes (Van Speybroeck, 2002; Waddington, 1940, 1956). The first molecular epigenetic factor identified was DNA methylation (Holliday and Pugh, 1975) in the 1970s which was subsequently shown to influence processes such as X chromosome inactivation and gene expression (Chen and Riggs, 2005). In the late 1980s imprinted genes were identified and shown to be regulated by DNA methylation as well (Chen and Riggs, 2005). In the 1990s histone modifications and chromatin structure were identified and shown to regulate promoter activity and gene expression (Turner, 1998). In 2000, small noncoding RNAs were found to have epigenetic activity and regulate gene expression (Berdasco and Esteller, 2010). Around 2005 the first genome wide mapping of the epigenome appeared (Pokholok et al., 2005). Therefore, the molecular characterization of epigenetics is relatively new and additional epigenetic marks are likely to be identified in the future (e.g., hydroxy-methylcytosine) (Kriaucionis and Heintz, 2009).
The functions of the various epigenetic marks and factors are distinct. The DNA methylation has a role in early development to help establish early cell lineages (e.g., stem cells) and can regulate the activity of promoters and general genome regions (e.g., repeat elements) (Kazazian, 2004). Many DNA methylation events are distal to promoters, but can influence gene expression (Illingworth and Bird, 2009). Histone modifications are primarily localized in the promoter and gene regions and fine tune the regulation of specific genes (Turner, 1998). These histone modifications are not generally involved in establishing the absence or presence of expression of a specific gene, but the subsequent expression and responsiveness to transcriptional control (Turner, 1998). The chromatin structure can modulate gene expression at a distance through looping, nuclear matrix association and nucleosome positioning (Biddie, 2011). The noncoding sRNAs can act at a distance to regulate gene expression through promoter modulation and can influence other epigenetic factors (Wan and Bartolomei, 2008). The combination of all these elements creates the epigenome and a complex regulation of genome-wide activity. All these factors are critical and play distinct roles in the process. In regards to development, the DNA methylation is thought to have the initial programming role followed by chromatin structure and histone modifications to fine tune the regulation of gene expression at the various stages of differentiation and development.
An important aspect of epigenetics, first clarified by Arthur Riggs (Russo et al., 1996), is that the epigenetic marks are mitotically and meiotically stable. The initial reference was to heritable, but the term heritable is most commonly defined as generational transmission (i.e., inheritance). Therefore, a more accurate term is “mitotically stable” and implies as a cell divides or proliferates the epigenetic marks constituting the epigenome are replicated. If an epigenetic mark or change was not mitotically stable, then the mark would only be relevant in the individual cell and would not be important outside that cell's function. When an epigenetic mark is mitotically stable, then all cells that come from that initial cell will have the same epigenome. Therefore, early in life an environmental signal could modify a cell's epigenetic marks that then would be mitotically stable and appear later in development in the tissue the cells reside. Therefore, the epigenome is programmed and maintained in a cell population as it further differentiates and is associated with the development of any tissue or organism. As the epigenome regulates gene expression, the environmental or developmental shifts in the epigenome can become a critical element affecting the developmental process. The process of mitotic stability for DNA methylation is understood, but how histone modifications and chromatin structure are replicated and transmitted through cellular division is not as well understood. For example, during DNA replication associated with mitosis, the parental DNA strand has a methylated nucleotide and as the new strand of DNA is synthesized the associated methyltrasferase methylates the hemimethylated DNA to replicate the original parental strand epigenetic mark. Further research is needed to elucidate how the complete epigenome is replicated and mitotically stable. This aspect of epigenetics is critical and allows for a dramatic influence on development and biology.
As the epigenetic marks that make up the epigenome are mitotically stable, an alteration in the epigenome early during cell differentiation or development is transmitted through that cells lineage to later stages of development of the tissue or organism. Therefore, environmental factors that can alter the epigenome promotes an abnormal programming that permanently alters the cell, tissue or organism development and function. This environmental epigenetics will have a critical impact on the developmental process and functions of cells or tissues later in life after the environmental exposure is removed. Environmental factors can include nutritional factors, environmental compounds or stress (Jirtle and Skinner, 2007; Skinner et al., 2010). Any external factor that can modulate normal development and the epigenome can be considered an environmental insult that impacts genome activity without altering DNA sequence. The mitotic stability of the epigenome suggests an environmental insult, even after its removal, will have a lasting effect on cell differentiation and development to promote alterations in physiology later in life. This provides a molecular mechanism for the fetal basis of adult onset disease or the developmental basis of disease (Barker et al., 2009; Bruce and Hanson, 2010). In addition, this provides a mechanism for environmental toxicology not previously considered, and explains how an early life exposure can promote a later life physiological effect. Environmental epigenetics will be a critical concept to consider in developmental biology, as well as most areas of biology (Jirtle and Skinner, 2007; Skinner et al., 2010).
Integration of Epigenetics and Genetics in Developmental Biology
Genetics has a central role in biology and in the control of development. Observations for the past several decades have identified many specific genes and genetic processes involved in the development of most cells, tissues and organisms. Epigenetics is an additional molecular process that can influence development and provides a mechanism for the environment and early life events to regulate cell differentiation and development. Therefore, epigenetics and genetics compensate and cooperate to control and regulate most biological processes, including developmental biology. They should not be viewed as conflicting or opposing processes, but when integrated provide a more robust molecular control of developmental biology.
A cascade of genetic and transcriptional events allows a cell or tissue to develop from a stem cell or basal state to a more mature or adult stage of development. This requires many steps and each step needs to be carefully controlled to allow the next step to proceed, Figure 1. As there is a cascade of genetics steps, there is also a cascade of epigenetic steps to cooperate with genetics to promote the developmental pathway. The epigenetic processes can respond to environmental factors to then integrate with the genetic processes to lead to a developmental step. The cascade of epigenetic and genetic steps during development is critical to promote the normal development process, Figure 1. Early in development there are critical windows of development that are more responsive to environmental factors to modify development. Later in development, the adult or at a more mature stage, the developmental process has been programmed and is less responsive to environmental factors to alter development. In the event an early critical window was influenced through an alteration in the cascade of epigenetic events, the final stage of development can be modified from the normal developmental state, Figure 1. This is generally reflected in the genome activity or transcriptome of the differentiated cells or tissue. The result of this cascade of epigenetic and genetic processes and the final influence on cell and tissue genome activity is the differentiation and development of the cell, tissue and organism. Abnormal development and alterations in the normal cascade of events through these mechanisms are likely to be a significant element of disease etiology. The overall process and integration of epigenetics and genetics leading to these developmental processes provides the systems biology of the tissue or organism, Figure 1.
Epigenetic Transgenerational Inheritance
The inheritance of environmentally induced phenotypes is the origin of the concept of epigenetics (Waddington, 1940; 1956). In the event these environmental factors modify the epigenome of the germ line and this becomes permanently programmed (imprinted) then the altered epigenome and phenotype become transgenerational and appear in subsequent progeny and generations in the absence of any further environmental exposures (Anway et al., 2005; Daxinger and Whitelaw, 2010; Jirtle and Skinner, 2007; Skinner et al., 2010). The mechanism involves the actions of an environmental factor at a critical time of gonadal sex determination in the mammalian fetus when the germ line cell fate is determined and the primordial germ cell differentiates into a male or female germ lineage (Skinner et al., 2010). During this critical period of development of the germ line an erasure of DNA methylation occurs and then upon gonadal sex determination the germ line DNA is remethylated in a male or female specific manner (Durcova-Hills et al., 2006). The actions of an environmental factor such as an endocrine disruptor (Anway et al., 2005; Anway et al., 2006b; Anway and Skinner, 2008) can modify this germ line methylation and promote a permanently altered epigenome (Anway et al., 2005; Guerrero-Bosagna C et al., 2010) in the germ line (e.g., sperm) that gets transmitted to subsequent generations transgenerationally (Anway et al., 2005; Anway et al., 2006a,b; Anway and Skinner, 2008; Guerrero-Bosagna C et al., 2010). Therefore, the basic mechanism of epigenetic trans-generational inheritance involves the actions of an environmental factor (e.g., chemical or nutrition) during germ line remethylation at gonadal sex determination to permanently alter the germ line epigenome (Anway et al., 2005; Guerrero-Bosagna C et al., 2010; Skinner et al., 2010) to then transmit this altered germ line epigenome to subsequent generations (Anway et al., 2005; Anway et al., 2006a,b; Anway and Skinner, 2008; Guerrero-Bosagna C et al., 2010; Skinner et al., 2010). As the embryonic stem cell epigenome is altered due to this germ line transmission, all cell populations and tissues will have an altered epigenome and corresponding transcriptome (Anway et al., 2008; Skinner et al., 2010). The germ line generated by the next generation will also have this altered epigenome and transmit it to the subsequent generation (Guerrero-Bosagna C et al., 2010; Skinner et al., 2010). Exposure to the endocrine disruptors at other times of development do not appear to have the capacity to permanently alter the germ line epigenome (Anway et al., 2005; Anway and Skinner, 2008; Skinner et al., 2010). Of course, the vast majority of exposures will alter the somatic cells at critical periods of development to modify later cellular development and potential adult onset disease, Figure 1, but this does not have the capacity to become transgenerational as the germ line is not involved (Skinner et al., 2010). Epigenetic transgenerational inheritance through a permanently altered epigenome of the germ line has the capacity to have a dramatic influence on developmental biology, as well as other areas of biology such as evolution.
In the event the base-line epigenome is altered, then the cascade of epigenetic and genetic steps during development will be altered and a modified differentiated or developmental state achieved, Figure 1. Therefore, epigenetic transgenerational inheritance has a dramatic effect on the developmental biology of all cells and tissues derived from the germ line transmitting this modified baseline epigenome. Although not all cell types or tissues will develop a disease state, those tissues that have a sufficiently altered transcriptome will have a greater susceptibility to develop disease (Skinner et al., 2010). As all development and differentiation processes involve a cascade of epigenetic and genetic steps, alteration of the baseline epigenome, similar to alteration in the genetic baseline, will have the capacity to promote abnormal development which may lead to disease later in life. For this reason, environmentally induced epigenetic transgenerational inheritance through the germ line will have a significant impact on developmental biology. This mechanism and consideration of the cascade of integrated epigenetic and genetic events during development, Figure 1, will be an important factor in disease etiology not previously considered (Skinner et al., 2010).
In addition to the effects of epigenetic transgenerational inheritance on developmental biology and disease etiology, this phenomena will impact nearly all areas of biology, including evolutionary biology (Crews et al., 2007). If the base-line epigenome is modified, similar to the base-line effects of genetics, then the biological system will respond by altering the phenotype, physiology, and general biology of the organism. In considering the molecular mechanisms that promote an adaptation event and natural selection to allow an evolutionary event, an integration of epigenetics and genetics will be equally as important. The current paradigm of DNA mutational events promoting evolution is accurate, but the inclusion of epigenetics allows for a much higher degree of variability in the biological system to facilitate an adaptation event. In addition, the inclusion of epigenetics allows for a mechanism to have environment influence evolutionary processes. Therefore, epigenetic transgenerational inheritance is a novel process to consider in evolutionary biology not previously considered. This does not alter the basic Darwinian evolutionary paradigm, but simply provides a neo-Lamarckian component and more diverse molecular mechanism to be involved. Future research into the integration of epigenetics and genetics will likely reveal more powerful mechanistic considerations to be applied to all areas of biology, including evolutionary biology (Skinner et al., 2010).
Conclusions
Epigenetics will have a critical role in developmental biology and differentiation due to its function in regulating genome activity and the mitotic stability of the epigenetic marks to be replicated as cells proliferate. Therefore, epigenetic marks and alterations in the epigenome have the capacity to be maintained within a cell type or population for the life span of the organism. As an integration and cascade of epigenetic and genetic processes are required throughout development, Figure 1, modification of an epigenetic state has the capacity to create a new phenotype, and if abnormal a disease state. In contrast to genetics, epigenetics can be modified readily by environmental factors, such that epigenetics provides a molecular mechanism for the environment to alter genome activity and developmental biology. Future considerations of the molecular processes involved in developmental biology, as well as other areas of biology, requires a consideration of epigenetic processes.
The germ line creates through developmental biology nearly all species. Genetic modifications in DNA sequence have the capacity to dramatically alter the development of a species. As epigenetics is intimately integrated with genetics, alterations in the germ line epigenome also has the capacity to dramatically alter the development of a species. Therefore, epigenetic transgenerational inheritance induced by environmental factors will have a critical role in the biology, disease etiology and general development of most, if not all, species. A change in the base-line epigenome in the germ line through its integration with genetic processes and the cascade of events required in a developmental system, Figure 1, will have the capacity to alter phenotype (Waddington, 1940, 1956) and biological processes such as evolution (Crews et al., 2007). The complementary and integrated roles of epigenetics and genetics is what allows these new developmental events to occur and create new phenotypes.
References
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006a;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Memon MA, Uzumcu M, Skinner MK. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J Androl. 2006b;27:868–879. doi: 10.2164/jandrol.106.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Rekow SS, Skinner MK. Transgenerational epigenetic programming of the embryonic testis transcriptome. Genomics. 2008;91:30–40. doi: 10.1016/j.ygeno.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Skinner MK. Transgenerational effects of the endocrine disruptor vinclozolin on the prostate transcriptome and adult onset disease. Prostate. 2008;68:517–529. doi: 10.1002/pros.20724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Osmond C, Kajantie E, Eriksson JG. Growth and chronic disease: findings in the Helsinki Birth Cohort. Ann Hum Biol. 2009;36:445–458. doi: 10.1080/03014460902980295. [DOI] [PubMed] [Google Scholar]
- Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Biddie SC. Chromatin architecture and the regulation of nuclear receptor inducible transcription. J Neuroendocrinol. 2011;23:94–106. doi: 10.1111/j.1365-2826.2010.02079.x. [DOI] [PubMed] [Google Scholar]
- Bruce KD, Hanson MA. The developmental origins, mechanisms, and implications of metabolic syndrome. J Nutr. 2010;140:648–652. doi: 10.3945/jn.109.111179. [DOI] [PubMed] [Google Scholar]
- Chen ZX, Riggs AD. Maintenance and regulation of DNA methylation patterns in mammals. Biochem Cell Biol. 2005;83:438–448. doi: 10.1139/o05-138. [DOI] [PubMed] [Google Scholar]
- Crews D, Gore AC, Hsu TS, Dangleben NL, Spinetta M, Schallert T, Anway MD, Skinner MK. Transgenerational epigenetic imprints on mate preference. Proc Natl Acad Sci USA. 2007;104:5942–5946. doi: 10.1073/pnas.0610410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Trans-generational epigenetic inheritance: more questions than answers. Genome Res. 2010;20:1623–1628. doi: 10.1101/gr.106138.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcova-Hills G, Hajkova P, Sullivan S, Barton S, Surani MA, McLaren A. Influence of sex chromosome constitution on the genomic imprinting of germ cells. Proc Natl Acad Sci USA. 2006;103:11184–11188. doi: 10.1073/pnas.0602621103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Settles M, Lucker BJ, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS ONE. 2010;5:e13100. doi: 10.1371/journal.pone.0013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. CpG islands—“a rough guide. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethyl-cytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Russo VEA, Martienssen RA, Riggs AD. Epigenetic mechanisms of gene regulation. Woodbury: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21:214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Speybroeck L. From epigenesis to epigenetics: the case of C. H. Waddington. Ann NY Acad Sci. 2002;981:61–81. [PubMed] [Google Scholar]
- Waddington CH. Organisers and genes. Cambridge: Cambridge University Press; 1940. [Google Scholar]
- Waddington CH. Principles of embryology. London: George Allen & Unwin Ltd; 1956. [Google Scholar]
- Wan LB, Bartolomei MS. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]


