ABSTRACT
The cytoskeleton provides structure and plays an important role in cellular function such as migration, resisting compression forces, and transport. The cytoskeleton also reacts to physical cues such as fluid shear stress or extracellular matrix remodeling by reorganizing filament associations, most commonly focal adhesions and cell-cell cadherin junctions. These mechanical stimuli can result in genome-level changes, and the physical connection of the cytoskeleton to the nucleus provides an optimal conduit for signal transduction by interfacing with nuclear envelope proteins, called nesprins, within the LINC (linker of the nucleus to the cytoskeleton) complex. Using single-molecule on single nuclei assays, we report that the interactions between the nucleus and the cytoskeleton, thought to be nesprin-cytoskeleton interactions, are highly sensitive to force magnitude and direction depending on whether cells are historically interfaced with the matrix or with cell aggregates. Application of ∼10–30 pN forces to these nesprin linkages yielded structural transitions, with a base transition size of 5–6 nm, which are speculated to be associated with partial unfoldings of the spectrin domains of the nesprins and/or structural changes of histones within the nucleus.
KEYWORDS: biomechanics, cytoskeleton, nesprin, nucleus, single molecule
Introduction
Cells are very sensitive to physical forces. A quintessential example of this is endothelial cells delineating between laminar and turbulent flow.1,2 Depending on the flow profile, endothelial cells are able to respond quickly and reprogram their entire protein expression profile.3,4 Mechanistic studies investigating which signaling cascades and mechanical connections are responsible for phenotype change were conducted by directly probing the cell surface and the associated cell-cell and cell-matrix interfaces.5,6 Because cytoskeletal elements conduct the majority of physical inputs on the cell, it is critical to study how these forces propagate within the cell, particularly as they propagate to a termination point such as the nucleus.
Much of the research in mechanotransduction has focused on the plasma membrane where cell-cell adhesion complexes and focal adhesions serve as mechanosensors.7 Similarly, proteins at each end of cytoskeletal filaments serve not only as anchors for filaments but also as mechanosensors.8 While interest in cytoskeletal anchoring at the nuclear envelope has developed over the past decade, it remains challenging to study because accessing the nuclear-cytoskeletal interface is difficult.
The first demonstration of a physical linkage between the nucleus and the plasma membrane represented a seminal shift in defining the mechanism by which the cell's external environment physically connects to organelles deep within the cytoplasm.9 In endothelial cells, shear stress applied to cells has been shown to displace cytoskeletal elements (e.g. actin, vimentin) and nodal structures (e.g., focal adhesions, cadherin junctions).10 It is logical that similar protein elements on the outer nuclear membrane could be just as mechanically rich. In fact, magnetic bead microrheology on lamin A-deficient fibroblasts allowed for greater bead displacement coupled with higher reported forces compared with wild type fibroblasts, thus further suggesting that subtle changes in nuclear envelope proteins can yield cellular phenotype changes.11 However, the microrheology experiment stopped short of single-molecule interrogations of the nuclear envelope.
The principal nuclear envelope proteins engaging the cytoskeleton are known as nesprins, of which there are many isoforms and splice variants, which are key components of the LINC (linker of nucleus to cytoskeleton) complex. The cytoskeleton is principally composed of 3 major protein filaments: actin, intermediate filaments, and microtubules (plus their associated molecules). Nesprins 1 and 2 primarily bind actin, nesprin 3 binds intermediate filaments via the actin-binding domain (ABD) of plectin, and nesprin 4 binds microtubules through kinesin intermediates,12-16 as seen in Fig. 1a. Disruption or inhibition of these proteins results in decreased cell stiffness and nucleus deformation.17 It should be noted that nesprins 1 and 2 have also been found to have some affinity for microtubule motor proteins,18,19 but this should not affect our results as the nuclei are isolated from cells in our assay, removing the cytoskeleton and any cytoskeleton associated machinery (Figures S1 and S2).
Figure 1.
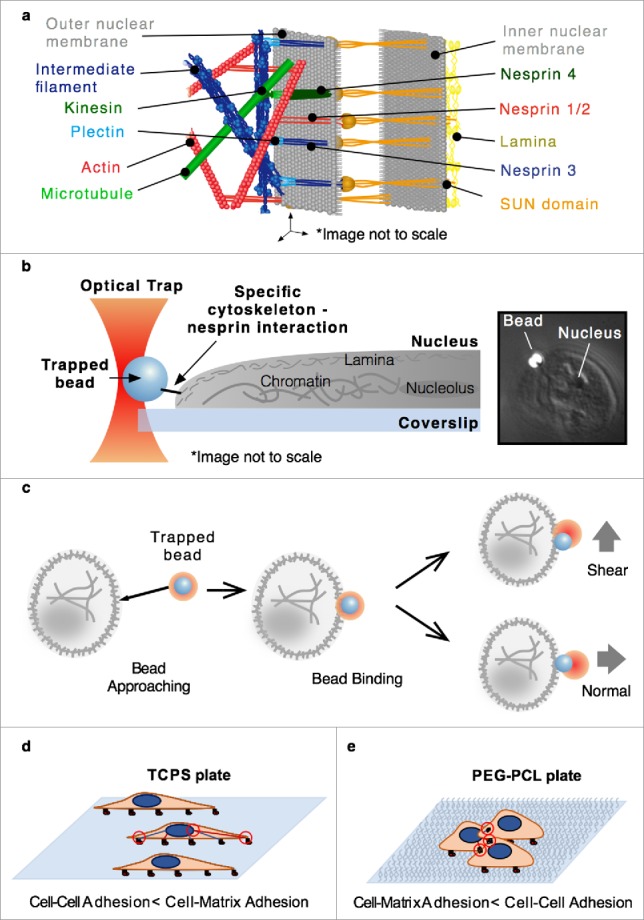
(a) The environment near the nuclear membrane is complex and includes the interactions of cytoskeletal elements with nesprins which are associated with the SUN protein in the periplasm. (b) A general representation of the optical tweezer based assay design for characterization of specific cytoskeleton-nesprin interactions. (c) Binding was achieved by bringing the bead close to the nucleus and waiting for binding, after which force was applied by moving the sample stage to apply either a shear or normal force. Cells were grown on either TCPS (d) or PEG-PCL (e) plates leading toward cell-matrix or cell-cell adhesion biases, respectively.
Methods to probe nesprin biology have been limited to basic in vitro and in vivo studies that manipulate gene expression or truncated forms of the nesprin protein itself.18,20,21 A direct connection to gene expression was recently seen in genetically engineered bacteria with fluorescent chromatin-binding proteins subjected to surface magnetic twisting cytometry. The study demonstrated that increasing force in specified loading directions allowed for greater chromatin unfolding and promoted transcription of sterically repressed genes.22 Another group also attempted to probe nuclear mechanical responses at the nesprin node using magnetic tweezers,23 but the study used antibodies rather than the native cytoskeletal ligands and the antibody epitopes do not allow for force to be applied in a manner similar to the native nesprin-cytoskeleton configuration.
Despite these insightful studies, little is known about the physical dynamics of the interaction of nesprins with their cytoskeletal partners that ultimately lead to potential changes in gene expression. Thus, there remains an unmet need for new assays that investigate the effects of force on LINC complex proteins, conformational changes and protein-protein/protein-chromatin interactions. Optical tweezer based single-molecule assays provide pick-and-place force application with high spatial (nm), temporal (ms), and force (pN) resolution, allowing for the elucidation of molecular mechanisms previously out of reach.8 Here, we isolated single nuclei and developed a semi-situ single-molecule on single nuclei assay to probe how force inputs are handled at the nuclear envelope at the single protein level via individual cytoskeletal elements (actin or plectin-ABD) bound to beads and actively coupled to the nuclear surface (Fig. 1b).
Results
We focused on the effects of three factors on the mechanotransduction of signals from the cytoskeleton to the nucleus: the specific interaction at the nuclear membrane (actin-nesprin 1/2 vs. plectin ABD-nesprin 3), the direction of force application (normal vs. shear) (Fig. 1c), and cell culture condition biases as dictated by pre-measurement growth conditions (tissue culture polystyrene (TCPS) vs. polyethylene glycol (PEG)-poly(ε-caprolactone) (PCL) block copolymer). TCPS permits uninhibited cell binding to the entire culture surface. PEG-PCL is a softer culture substrate than TCPS, and inclusion of PEG reduces binding to the substrate. Hence PEG incorporation forces increased cell-cell interactions to avoid anoikis (Fig. 1d–e). Human mesenchymal stem cells (hMSCs) were chosen due to their large nuclei and their known altered phenotype when cultured as single cells on TCPS versus as aggregates on PEG-PCL.24 The morphological changes that result from these growth conditions at the cellular level are mirrored in the nucleus and conserved throughout the 4 hour maximum experimental window in which isolated nuclei were used.
During purification, the cytoskeletal and cytoplasmic components of the cell are removed from the nucleus to yield isolated nuclei, as outlined in the methods. To ensure nucleus isolation was complete and residual cellular material was not interfering in our experiments, we fixed and stained isolated nuclei for calnexin (endoplasmic reticulum, ER, protein) and actin, as seen in Figures S1 and S2, respectively. Residual cellular material can sometimes be seen in DIC when a nucleus has not been fully isolated from the cell, but successfully isolated nuclei show no evidence of residual material. Additionally, we fixed and stained isolated nuclei from cells grown on both TCPS and PEG-PCL and confirmed the presence of nesprins 1–3 post-isolation (Fig. S3).
While it is possible F-actin could bind to proteins (other than nesprin 1/2) associated with the nucleus, such as FHOD1, as demonstrated in Kutscheidt et al.,25 it is unclear whether FHOD1 is present here as it is not a membrane-anchored protein. Additionally, if FHOD1 were present, it would be expected that F-actin would also be bound to nesprin 1/2 as FHOD1 crosslinks nesprin-2G and stabilizes its interactions with actin. This would result in off-path transitions if the interaction between actin and either nesprin or FHOD1 were interrupted and off-path results were eliminated from our analysis, as described later. Alternatively, plectin-ABD exclusively binds nesprin 3. Previous work by the Sonnenberg laboratory15,26 has shown that the recruitment of plectin-ABD to the nuclear envelope (NE) requires nesprin-3α and that even a single point mutation of nesprin-3α is sufficient to disrupt the recruitment of plectin-ABD to the NE. Taking the aforementioned studies and purity of our isolated nuclei into consideration, we are confident in our targeting of nesprin-cytoskeleton interactions.
Single molecule interactions were achieved through serial dilution of the cytoskeletal component during bead functionalization such that fewer than 50% of beads yielded productive binding. Control beads (non-functionalized streptavidin beads) showed minimal or no binding to the nuclear membrane (0–2 of 20 beads). Binding was achieved by holding a bead fixed in a trap on the nucleus surface. Force was then applied by moving the sample stage at 20 nm/s, resulting in a loading rate of 1.8 ± 1.2 (STD) pN/s, which varied with changes in trap stiffness and nuclear compliance. In the case of actin functionalized beads, only interactions forming free tethers were used for data collection ensuring that the beads are functionalized with F-actin (Figure S4 and videos S1 and S2). The tether length varies with the length of the specific actin filament and the location of binding, but it is estimated that tethers ranged from 300 nm – 1 μm in length.
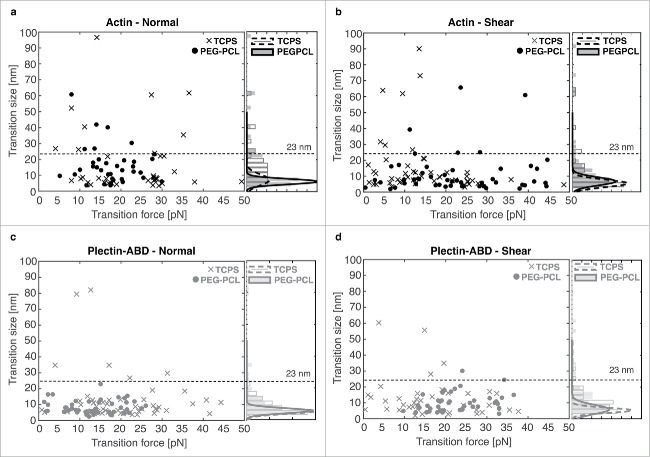
Abrupt changes in bead position (Fig. 2a), referred to as transitions, were observed for all conditions. A range of transition sizes were present and larger transitions (> 23 nm) were more prevalent in the actin data. Some transitions were clustered through quick succession of multiple smaller events. Each bead displacement was analyzed as a separate transition (see the Supplemental Note). Full ruptures, indicating the complete disruption of the nesprin-cytoskeleton interaction, were observed, but were rarer under PEG-PCL conditions (2.5% compared with 9.8% for TCPS nuclei) (Table S1).
Figure 2.
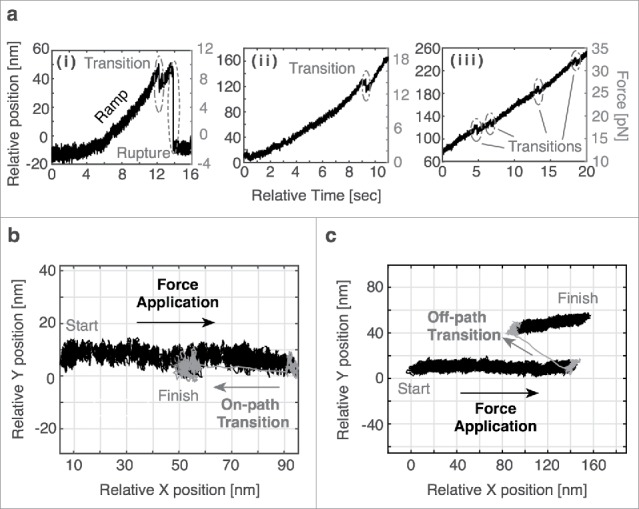
(a) Representative traces depicting transitions during force application. Full rupture (i) and small transitions (i-iii) are observed. In some cases, multiple transitions are present in the same trace. (b) A representative top-down plot depicting an on-path transition in which the transition follows the same path back toward the origin as during force application. (c) A representative top-down plot depicting an off-path transition in which the transition does not result in a position along the path of force application.
Top-down plots of these transitions were critical in distinguishing between on- and off-path transitions. On-path transitions were defined as transitions that followed the path of force application back toward the origin (Fig. 2b), while off-path transitions indicated a displacement that shifts to a point creating a new vector with respect to the direction of force application (Fig. 2c). Off-path transitions were twice as common in actin data (13.5%) as they were in plectin-ABD data (7.8%). Further transition analysis was only completed for on-path transitions. As nesprins 1/2 contain actin-binding domains, more than one nesprin may bind along an actin filament simultaneously.
A closer look at the transitions reveals shifts in transition forces given changes in experimental conditions, particularly with respect to force application direction and cell culture surface. For actin we see a shift to a higher transition force for TCPS nuclei, 22.8 ± 1.5 (SEM) pN, vs. PEG-PCL nuclei, 17.6 ± 0.9 (SEM) pN, when force is applied normal to the nuclei (Fig. 3a). This trend is reversed with a shear force, resulting in transition forces of 15.2 ± 1.6 (SEM) pN for TCPS nuclei and 23.1 ± 1.7 (SEM) pN for PEG-PCL nuclei (Fig. 3b). Additionally, the trends seen in plectin-ABD are consistent with those observed in actin. Again, we see a shift to higher transition forces for TCPS nuclei, 20.5 ± 1.3 (SEM) pN, vs. PEG-PCL nuclei, 13.4 ± 1.0 (SEM) pN, when force is applied normal to the nuclei (Fig. 3a). The trend is reversed with a shear force, resulting in transition forces of 18.6 ± 2.1 (SEM) pN for TCPS nuclei and 22.6 ± 1.1 (SEM) pN for PEG-PCL nuclei (Fig. 3b). Collectively, the above transition force results indicate that normal forces stabilize nuclear interactions for TCPS nuclei while shear forces stabilize interactions for PEG-PCL nuclei.
Figure 3.
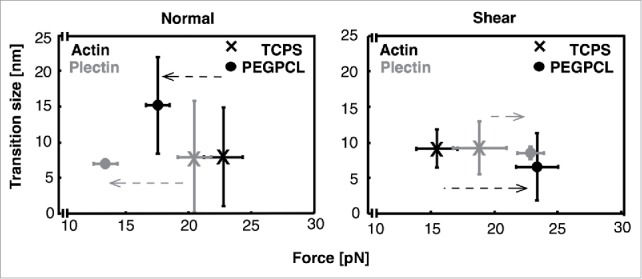
A comparison of median transition sizes and mean transition forces for actin and plectin-ABD interactions on nuclei from cells grown on both TCPS and PEG-PCL when force is applied (a) normal and (b) shear to the nuclei. The data indicate that the interactions of PEG-PCL nuclei are stronger when subjected to shear forces while TCPS nuclei are stronger when subjected to normal forces. All error bars are standard error on the mean (SEM). N incorporated into each point in the normal direction are as follows: Actin/TCPS = 42, Actin/PEG-PCL = 40, Plectin-ABD/TCPS = 59, Plectin-ABD/PEG-PCL = 48. N incorporated into each point in the shear direction are as follows: Actin/TCPS = 49, Actin/PEG-PCL = 54, Plectin-ABD/TCPS = 40, Plectin-ABD/PEG-PCL = 44.
Small transitions were conserved across all experimental conditions, yielding a consistent median transition size (Table S1). A closer look at transition sizes revealed a base transition size of 5–6 nm across all conditions (Fig. 4), with larger transition populations up to approximately 23 nm. A distribution of transition sizes pooled from all conditions (N = 276) exhibits structure fitting well to a multiple Gaussian (Fig. S6) with a strong peak representing the base transition size at 5.6 nm, a shoulder at 10.1 nm, and a weak tail centered near 19.2 nm. Transitions larger than approximately 23 nm were less abundant and no longer followed a specific trend.
Figure 4.
Scatter plots of transition size vs. transition force and a histogram of transition size for (a) actin interactions with normal force (b) actin interactions with shear force (c) plectin-ABD interactions with normal force, and (d) plectin-ABD interactions with shear force. The fits overlaying each histogram represent the base transition size of 5–6 nm present in each condition and the dashed line at 23 nm indicates the value at which trends in the transition sizes are no longer observed.
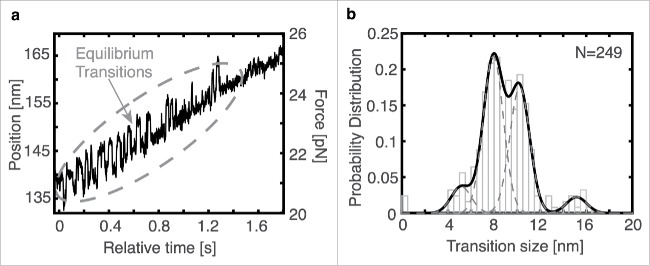
In a few cases, we observed reversible transition behavior within the trace. These transitions, occurring in quick succession, are characteristic of a structural change at equilibrium, and similar reversible behavior has been reported in other systems.27,28 The transitions, as observed in Fig. 5a, have a time constant, with respect to transition lifetime, of 0.011 s (Fig. S7). Interestingly, reversible transitions were observed over a range of forces including at 1.5–2.5 pN, 20–25 pN, and ∼32 pN. Similar to the transition sizes in the non-equilibrium data, multiple transition size peaks emerged (Fig. 5b) including a base transition size of 5.1 nm, consistent with the 5–6 nm base transition size seen earlier, two strong peaks at 7.9 and 10.2 nm, and a small additional peak centered near 15.2 nm (N = 249).
Figure 5.
Equilibrium transitions are present in some traces, usually during the force ramp, as seen in (a) and result in a distribution of transition sizes (b) with Gaussian means at 5.1, 7.9, 10.2, and 15.2 nm. The sum of the Gaussians is shown in black while the single Gaussian distributions are dashed gray lines.
Discussion
Using optical tweezers to actively present cytoskeletal ligands to nesprin proteins on single isolated nuclei, we observed a mechanically rich system, operating in the force range of 10s of piconewtons. The magnitude of these transition forces was highly dependent on the force application direction (normal or shear) and cell culture conditions, and defined the forces required for cytoskeletal machinery to interact with the nuclei as the cell adapts to external physical cues. The nature of the shift in transition force between cell growth conditions suggests that hMSCs (and perhaps many other cell types) have the ability to alter their force sensing mechanism, allowing for the cells to react to physical stimuli at a nuclear level in a function-specific way. This type of phenomenon has been shown for hMSC differentiation in tunable hydrogels where the mechanics of the initial gel impart a mechanical memory and influence cell differentiation lineage choice following an in situ gel stiffness switch.29
To start, it is important to consider why the observed transition forces are different between culture substrates. The answer may lie in the morphology of the nuclei that results from their cell culture conditions. hMSCs on TCPS exhibit a flatter, polarized morphology coupled with a cell-matrix interaction bias (Fig. 1d). Isolated TCPS nuclei adopt this morphology with dimensions approximated to be near 15–20 μm × 7–10 μm × 3–5 μm. The PEG-PCL environment yields cells, and nuclei, with a more spherical, morphology and a cell-cell interaction bias (Fig. 1e) with a nucleus diameter of ∼7–9 μm. While the volumes between these populations of nuclei should be similar, their surface areas are vastly different with TCPS nuclei easily having a surface area 25–30% greater than that of PEG-PCL nuclei. This means that variation in morphology requires differences in the internal architecture (including chromatin-histones and lamin mesh structure) as well as the spacing of nesprins on the surface of the nuclei, which could result in minute changes in the presentation of these heteropolymers. These unique geometries could then lead to differences in force requirements.30 In addition, growth conditions could affect several cellular and nuclear parameters not specifically tested here, including differential expression of LINC complex components, nuclear envelop protein organization, and chromatin structure/remodeling, all of which could contribute to the differences in behavior observed here.
Critical to the function of the LINC complex, the interaction of the SUN protein to the c-terminal region of a nesprin, the KASH domain, is a covalently linked disulfide bond. Previously published research used dominant negative KASH domains in fibroblasts to demonstrate that the lack of the disulfide bond decouples the physical link between the cytoskeleton to the nucleus,31 and it is unlikely any observed transitions in our data were caused by disruption of the SUN-KASH link. Therefore, transition forces observed in our data likely originate from either within the nucleus (LINC-chromatin interactions) or between the nesprin-cytoskeletal filaments.
Like focal adhesions, LINC complexes can be rather fluid within the nuclear membrane,17 allowing us to speculate that multiple LINC complexes may aggregate and resist higher forces.32,33 This focal adhesion-like networking has been found on the nuclear envelope. The transmembrane actin-dependent nuclear (TAN) line phenomena, which are generated by arrays of nesprin 2 and SUN2 proteins, shape the nucleus when cells migrate or reshape to their environment.34 In such cases the distribution of parallel LINC complexes provide anchor points to sustain higher force loads that manipulate nucleus shape. Networked LINC complexes could potentially subject the nucleus to much higher forces, mirroring focal adhesion networks, transmitting mechanical inputs directly to the nucleus and its contents.
In evaluating on- and off-path transitions, we observe that off-path and larger transitions are more common in actin interactions. The rod like structure of actin permits binding with nesprins at multiple points that can lead to large and/or off-path transitions if one interaction dissociates (Fig. S8a). Plectin is more accurately modeled as a single point (Fig. S8b) and is therefore expected to be on-path. Nesprins can also self-dimerize through interactions between spectrin repeats.35-37 Thus, loss of these dimers may also be responsible for off-path transitions.
Small on-path transitions are thought to represent structural unfoldings or conformational changes somewhere along the loading pathway into the nucleus. This is supported by the observed reversible equilibrium behavior described in the results (Fig. 5a). While it is impossible to pinpoint the location of structural changes given the complexity of the load pathway, possibilities include changes within the spectrin repeats of their respective nesprins, linker zones between spectrin repeats, sliding or rearrangement of spectrin repeat helicies, or chromatin rearrangement within the nucleus itself as chromatin is intricately linked to nuclear membrane proteins via the lamina layer.38
The roughly 100-residue long spectrin repeats consist of 3 helices in anti-parallel arrangement.39,40 Unfolding spectrin repeats with atomic force microscopy (AFM) revealed unfolding as an all-or-nothing event between 23 and 42 nm at forces ranging from 25–80 pN.41-44 The study by Lenne et al. using a tetramer R16 spectrin repeat, revealed a 15 nm partially unfolded state at forces of 60 pN.44 Their experiments, while at a much higher loading rate than our own (∼160 pN/s vs. 1.8 ± 1.2 (STD) pN/s), revealed that a 10-fold decrease in loading rate can result in as much as a 20% decrease in transition force.44 Based on this observation, the transition forces we observe are consistent. Our testing conditions also approach physiologic conditions as hMSCs can crawl 3 μm/hr over a stiffness gradient.45 Constant force measurements by Aubin-Tam et al. showed in a study using filamin substrate that their ∼98 aa extensions corresponded to 14–19 nm at forces of 7–14 pN.46 Our observed transitions near 15 nm are, therefore, consistent with full spectrin unfoldings while smaller transitions of 5–10 nm could be attributed to the unfolding of a single helix. It should also be noted that the studies of the folding of spectrins reveal a variety of stabilities and partially structured states that could parallel unfolding transitions with the application of force and may explain the observation of multiple transition sizes.47
Unlike the short polymers of previous studies,41,44 the native nesprins here contain high numbers of spectrin repeats with nesprin 1, 2, and 3 containing 74, 56, and 8 repeats, respectively.15,48 The greater diversity of spectrin sequence/strength in long spectrin repeats can lead to lower loading rates and partial unfolding states. Additionally, a study of the smaller, inner nuclear membrane protein, nesprin-1α, highlights the additional complexity of nesprins. The study found that the stability of the spectrin-like repeats (SLRs) of nesprin-1α is affected by a largely disordered adaptive domain (AD) between repeats 5 and 6.35 The giant nesprin 1/2 isoforms found at the outer nuclear membrane, which are the focus here, contain more of these ADs, making the biophysics of these proteins more complicated. Further investigation of the impact of ADs on SLRs in necessary to better understand nesprin structure and stability.
Nesprin size and complexity have meant that biophysical studies on full nesprin proteins have been rare, but it should be noted that Arsenovic et al.49 have previously observed mechanical force across nesprin-2G by incorporating a FRET biosensor into the protein. This work proves nesprins, and by extension the LINC complex, play a key role in the mechanotransduction pathway to the nucleus. The work also noted the sensitivity of nesprin-2G to cytoskeletal tension, cell shape, and nuclear stiffness (studied by lamin mutations). Our work also demonstrates the sensitivity of the nuclear mechanotransduction to force and cell shape (by way of growth conditions).
The unfolding of a single helix within a spectrin repeat may also explain the reversible transitions (hopping) we observed. Grum et al. noted that the α-helices between spectrin repeats can slide and undergo conformational rearrangement that shorten the end-to-end distance of the spectrin repeat and suggested this could be due to bending of the linker regions.50 Years later, Paramore and Voth found that once a linker region was disrupted, spectrin repeats became less stable, allowing lower forces to further unfold the repeat.51 Thus, our observed transitions may be the result of linker unfolding.
Another possible source of our transitions is chromatin-histone unbinding and reorganization. In single molecule studies, these have been shown to distort with 5–6 pN and can result in the dissociation of approximately 70 base pairs (∼23 nm).52,53 These forces are within the transition forces we observe and brings to light the possibility that gene rearrangement could result solely from mechanical manipulation. While these transitions are larger than what we typically observe here, the histone system is anchored to the lamina layer within the nucleus, which may reduce the observed transition magnitude due to its viscoelastic nature.38
There are many mechanisms used to transfer signals including diffusion, active transport, and mechanical coupling. As seen here and in previous studies,54,55 mechanical coupling is crucial to cellular function. The question remains: why has mechanotransduction evolved as a method of signal transfer? Diffusion can be severely limited within the cell due to the crowded intracellular environment56 and motor-based transport is limited to velocities of approximately 1 μm/s. However, physical coupling has been reported to allow signal transmission at up to 6 μm/s.57-59 From an energy perspective, mechanical transitions are similar to the chemical energy required for other biological processes. Using an average force of 20 pN and an unfolding distance of 5–23 nm, we estimate an energy requirement per transition per interaction of 24–112 kT at room temperature, which is equal to approximately 1–5 ATP hydrolysis events. Therefore, mechanotransduction allows for signal transduction that is not only fast and stable but also reconfigurable through efficient physical manipulation and structural variation.14,15,60
In conclusion, we have provided new insights into the behavior and response of specific cytoskeletal interactions with the nucleus, including the importance of the cell culture conditions and the resulting nuclear response to varying force conditions. Prior to the present study, the mechanical response of nuclei had been revealed through AFM and micropipette experiments. We supplement this through direct manipulation of single nuclei at physiologically relevant forces and loading rates. Despite this, there is still much to be learned about how the nucleus senses, receives, and translates mechanical signals. Further development of this system that includes fluorescence reporting of chromatin opening or rearrangement in real-time could directly connect mechanical input to chromatin alteration and transition states ripe for transcription or repression. Our work on single nuclei and single molecule experiments on nesprin domains and smaller spectrin homopolymers provide a bridge to live cell experiments that probe protein network function. Additionally, future technology that could produce and purify the 750–1000 kDa sized nesprin proteins may help identify specific zones of the protein that are susceptible to unfolding and further characterize the nature of nesprin conformational states. Moreover, CRISPR/Cas9 modulation of nesprin isoform expression can elucidate further mechanical and biochemical roles at the nuclear membrane.
Materials and methods
Polymer substrate preparation
5% PEG (MW = 2000 Da) -95% PCL (PEG-PCL) was synthesized using methods described previously.61 Spin-coated polymer films were prepared with a commercial spin-coater (Laurell Technologies, North Wales, PA, USA) on 10 cm Pyrex Petri dishes (Corning Inc.). Specifically, 1% weight/volume (w/v) solution of PEG-PCL in tetrahydrofuran (THF, Sigma Aldrich) was spun for 2 min at 1,500 RPM on Petri dishes (1 ml solution/sample). All samples were then exposed to constant cold-trap vacuum for ≥30 min to remove excess solvent. Dishes were UV sterilized for 60 min before use.
Cell culture
hMSCs were purchased from Lonza. hMSCs were maintained in complete media (CM) composed of α-minimum essential media with nucleosides (αMEM, Life Technologies) with 16.7% fetal bovine serum (Life Technologies), 1% penicillin/streptomycin (Life Technologies), and 4 μg/mL plasmocin (InvivoGen). Cells were kept in a humidified incubator at 37 °C and 5% CO2, and media was replaced twice each week. For all experiments, hMSCs (passage 5) were seeded at a density of 10,000 viable cells/cm2, as determined by exclusion of Trypan blue, and cultured for 3 to 4 d.
Nucleus enrichment
After hMSCs were cultured for 3 to 4 d on either tissue culture polystyrene (TCPS) or PEG-PCL, cells were washed with PBS-/-, trypsanized, diluted with culture media and centrifuged for 5 minutes at 400 g. The supernatant was removed and the pellet was resuspended in 15 mL of Buffer A (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT and 0.05% NP-40) with 1% v/v protease inhibitors. The suspension was incubated on ice for 20 minutes in a large dounce homogenizer and subsequently subjected to 15–20 dounce motions with plunger B. The dounced suspension was cold centrifuged (4 °C) for 10 minutes at 218 g. The supernatant was removed and the nucleus pellet was resuspended in 200 μL of nucleus wash buffer (0.2 M sucrose, 10 mM HEPES, and 1 mM MgCl2) with 1% v/v protease inhibitors.
Generation of actin-functionalized beads
Biotinylated actin was created such that actin and biotinylated actin were present in a 10:1 ratio. First, 5 μL of 10 mg/mL pure actin (Cytoskeleton - AKL99) in deionized water is mixed with 5 μL of 1 mg/mL biotinylated actin (Cytoskeleton - AB07). Then 100 μL of General Actin Buffer, GAB, (5 mM Tris-HCl, 0.2 mM CaCl2, 0.5 mM DTT, 0.2 mM ATP) was added and the mixture placed on ice for 1 hour. Actin was then polymerized by adding 11 μL of Actin Polymerization Buffer, APB, (50 mM Tris-HCl, 500 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 2 mM DTT, 5 mM ATP) and incubating on ice for 20 minutes. After polymerization, actin filaments were stabilized by adding 5 μL phalloidin (Life Technologies – A22282) and incubating on ice in the dark for 1 hour. Actin was then diluted as needed through dilution into a 10:1 GAB/APB buffer mixture. 8 μL of 1.09 μm streptavidin-coated polystyrene beads (Spherotech – SVP-10–5) were mixed with 5 μL biotinylated actin filaments (diluted 10,000 times from stock into 10:1 GAB/APB buffer), 0.6 μL ATP (100 mM), and 45 μL of nucleus wash buffer and allowed to mix on a rotator for 30 minutes at room temperature. The generation of F-actin before bead functionalization and its presence after bead functionalization is confirmed in Figure S5.
Generation of plectin-ABD-functionalized beads
Plectin-ABD functionalized beads were created by binding purified plectin-ABD protein to streptavidin beads via a glutathione linker. Biotinylated glutathione was first created by mixing 4 μL of 5 mg/mL biotinylated maleimide (Sigma – B1267) in DMSO with 50 μL of 10 mg/mL glutathione (ThermoScientific - 78259) in deionized water and incubating at 4 °C on a rotator overnight to generate biotinylated glutathione. Streptavidin beads were then functionalized with glutathione by mixing 10 μL of 1.09 μm streptavidin beads pre-diluted 20 times in PBS (pH 7.4), 30 μL of the overnight biotinylated glutathione solution, 2 μL of biotinylated bovine serum albumin (b-BSA, 5 mg/mL), and 58 μL PBS. The solution was then incubated on a rotator at room temperature for 1 hour. Excess and unreacted reagents were removed from the bead solution and the buffer exchanged to nucleus wash buffer by centrifuging the sample 2 times for 3.5 minutes at 9250 rpm and resuspending the beads in nucleus wash buffer each time. After the second centrifuge step, the beads were resuspended in 200 μL of nucleus wash buffer, 2 times that in the original solution. Upon completion, the resulting bead solution was sonicated using a cup sonicator for 2 minutes at 20%. Plectin-ABD (Litjens et al.62) was then added to the bead solution at single molecule concentrations by mixing 50 μL glutathione bead solution, 50 μL plectin-ABD (diluted 1:1 million in nucleus wash buffer, 5 ng/mL), and 1 μL ATP (100 mM) and incubating the mixture on a rotator at room temperature for 2 hours.
Slide preparation
Slides were prepared by creating a 10–15 μL flow cell using potassium hydroxide (KOH) etched coverslips coated in poly-L-lysine, standard microscope slides, and double-sided tape. After etching, the coverslips were coated in poly-L-lysine (Sigma Aldrich – P8920) by applying a layer of 3% poly-L-lysine solution in ethanol to one side of a coverslip (∼200 μL). The coverslip was then placed in the oven at approximately 90 °C until the ethanol was fully evaporated. The flowcell was then created using double-sided tape, ensuring the poly-L-lysine coated side of the coverslip faces the slide. Upon formation of the flowcell, a 20 μL nuclei sample was loaded onto the slide and incubated at room temperature for 30 minutes to allow the nuclei to become immobilized on the surface. Passivation of the surface was then achieved by adding 20 μL of a 10 mg/mL solution of bovine serum albumin, BSA, (Sigma Aldrich – A7030) in nucleus wash buffer to the flowcell and allowing the slide to incubate at room temperature for 10 minutes. Finally, 20 μL of the appropriate bead solution was added to the flowcell. The flow cell was then sealed and the slide was placed on the microscope.
Data acquisition
A slide was loaded onto a custom built inverted microscope with an optical trap and an isolated nucleus found in the field of view. Then a functionalized bead was located, brought near a side of the nucleus that was oriented along an axis of the sample stage, and focused such that the edge of the nucleus was clear. The bead was then calibrated and the stiffness found using the variance. During calibration, the KhroneHite anti-alias filter is set to a cut-off frequency of 30 kHz to allow for complete sampling. Upon calibration, the bead was carefully brought into contact with the nucleus by manually moving the sample stage. This was repeated 3–4 times in an effort to initiate binding. If binding did not occur within a few tries, the bead was discarded and a new one found. If binding occurred, the bead became tethered to the nuclear membrane. The bead was then centered using a custom LabVIEW program and data acquisition initiated. Data was acquired at a sampling frequency of 3 kHz with an anti-alias cut-off of 1.5 kHz. Upon initiation, the sample stage was moved in the desired direction (normal or tangential to the nuclear membrane). Experiments used a slow force ramp with a velocity of 20 nm/s, resulting in a loading rate of 1.8 ± 1.2 (STD) pN/s). At the end of a ramp, the sample could be stepped back to the center (2 μm/s) and pulled again. This can be repeated several times. After force application, the user simply waits for a transition to occur. Upon observance of transition(s), force can be applied again in the same way.
Each bead and each nucleus was only used once, however, it is possible to obtain several transitions from a single bead-nucleus interaction.
Data analysis
All data analysis was completed using custom Matlab scripts. Data were collected at a 3 kHz sampling frequency and averaged using an exponentially weighted moving average over a 10 point time period during analysis. The force of each trace is found by removing any baseline in the position and the multiplying the nanometer position measurements by the trap stiffness. Transitions were then determined to be on- or off-path using a top-down view of the respective trace (Fig. 2). On-path transitions were kept while off-path transitions were discarded and omitted from further analysis. Distributions fit were found using the cftool Matlab tool and constraints were also set such that each Gaussian in a multiple Gaussian fit had equal variances and if more than 2 peaks were found, each peak was separated by the same distance. In the cases in which equilibrium-like transitions were observed, the transitions were analyzed in the same manner but kept as a separate data set.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research work was funded and supported by NSF DMR BMAT under 1506717 and NIH EB under 019509 (D.A.B. and H.J.S.). H.J.S. was also supported by the Faculty Research Assistance Program of Yonsei University College of Medicine for 2000 (6–2016–0031). U.H.K was supported by the National Research Foundation of Korea (NRF) under MEST-2015M3A9B3028216. This work was also supported, in part, by Singapore-MIT Alliance for Research and Technology – BioSym, NSF under 1330792, and GAANN under P200A090323 (S.K.B and M.J.L).
References
- [1].Chiu JJ, Chien S. Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives. Physiol Rev 2011; 91(1):327-87; PMID:21248169; https://doi.org/ 10.1152/physrev.00047.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Davies PF, Remuzzi A, Gordon EJ, Dewey CF Jr, Gimbrone MA Jr. Turbulent fluid shear stress induces vascular endothelial cell turnover in vitro. Proc Natl Acad Sci U S A 1986; 83(7):2114-7; PMID:3457378; https://doi.org/ 10.1073/pnas.83.7.2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liu SQ, Yen M, Fung YC. On measuring the third dimension of cultured endothelial cells in shear flow. Proc Natl Acad Sci U S A 1994; 91(19):8782-6. ISSN 0027-8424 (Print); 0027-8424 (Linking); PMID:8090723; https://doi.org/ 10.1073/pnas.91.19.8782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chien S, Li S, Shyy YJ. Effects of mechanical forces on signal transduction and gene expression in endothelial cells. Hypertension 1998; 31(1 Pt 2):162-9. ISSN 0194-911X (Print); 0194-911X (Linking); PMID:9453297; https://doi.org/ 10.1161/01.HYP.31.1.162 [DOI] [PubMed] [Google Scholar]
- [5].Girard PR, Nerem RM. Endothelial cell signaling and cytoskeletal changes in response to shear stress. Front Med Biol Eng 1993; 5(1):31-6; PMID:8323880 [PubMed] [Google Scholar]
- [6].Tzima E. Role of small GTPases in endothelial cytoskeletal dynamics and the shear stress response. Circ Res 2006; 98(2):176-85; PMID:16456110; https://doi.org/ 10.1161/01.RES.0000200162.94463.d7 [DOI] [PubMed] [Google Scholar]
- [7].Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature 2011; 475(7356):316-23; PMID:21776077; https://doi.org/ 10.1038/nature10316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fedorchak GR, Kaminski A, Lammerding J. Cellular mechanosensing: getting to the nucleus of it all. Prog Biophys Mol Biol 2014; 115(2-3):76-92; PMID:25008017; https://doi.org/ 10.1016/j.pbiomolbio.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A 1997; 94(3):849-54; PMID:9023345; https://doi.org/ 10.1073/pnas.94.3.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Helmke BP, Rosen AB, Davies PF. Mapping mechanical strain of an endogenous cytoskeletal network in living endothelial cells. Biophys J 2003; 84(4):2691-9; PMID:12668477; https://doi.org/ 10.1016/S0006-3495(03)75074-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest 2004; 113(3):370-8; PMID:14755334; https://doi.org/ 10.1172/JCI200419670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, Karakesisoglou I, Korenbaum E. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp Cell Res 2004; 295(2):330-9; PMID:15093733; https://doi.org/ 10.1016/j.yexcr.2004.01.014 [DOI] [PubMed] [Google Scholar]
- [13].Rajgor D, Shanahan CM. Nesprins: from the nuclear envelope and beyond. Expert Rev Mol Med 2013; 15:e5; PMID:23830188; https://doi.org/ 10.1017/erm.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A 2009; 106(7):2194-9; PMID:19164528; https://doi.org/ 10.1073/pnas.0808602106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol 2005; 171(5):799-810; PMID:16330710; https://doi.org/ 10.1083/jcb.200506083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci 2002; 115(Pt 15):3207-22; PMID:12118075 [DOI] [PubMed] [Google Scholar]
- [17].Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res 2008; 314(8):1892-905; PMID:18396275; https://doi.org/ 10.1016/j.yexcr.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and syne/nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 2009; 64(2):173-87; PMID:19874786; https://doi.org/ 10.1016/j.neuron.2009.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wilson MH, Holzbaur ELF. Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development 2015; 142(1):218-28; PMID:25516977; https://doi.org/ 10.1242/dev.114769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wu J, Kent IA, Shekhar N, Chancellor TJ, Mendonca A, Dickinson RB, Lele TP. Actomyosin pulls to advance the nucleus in a migrating tissue cell. Biophys J 2014; 106(1):7-15; PMID:24411232; https://doi.org/ 10.1016/j.bpj.2013.11.4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Uzer G, Thompson WR, Sen B, Xie Z, Yen SS, Miller S, Bas G, Styner M, Rubin CT, Judex S, et al.. Cell mechanosensitivity to extremely low-magnitude signals is enabled by a linced nucleus. Stem Cells 2015; 33(6):2063-76; PMID:25787126; https://doi.org/ 10.1002/stem.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, Wang N. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater 2016; 15(12):1287-96; PMID:27548707; https://doi.org/ 10.1038/nmat4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, Burridge K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol 2014; 16(4):376-81; PMID:24609268; https://doi.org/ 10.1038/ncb2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Crowder SW, Balikov DA, Boire TC, McCormack D, Lee JB, Gupta MK, Skala MC, Sung HJ. Copolymer-mediated cell aggregation promotes a proangiogenic stem cell phenotype in vitro and in vivo. Adv Healthc Mater 2016; 5(22):2866-71. ISSN 2192-2659; https://doi.org/ 10.1002/adhm.201600819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kutscheidt S, Zhu R, Antoku S, Luxton GW, Stagljar I, Fackler OT, Gundersen GG. FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement. Nat Cell Biol 2014; 16(7):708-15; PMID:24880667; https://doi.org/ 10.1038/ncb2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Postel R, Ketema M, Kuikman I, de Pereda JM, Sonnenberg A. Nesprin-3 augments peripheral nuclear localization of intermediate filaments in zebrafish. J Cell Sci 2011; 124(Pt 5):755-64; PMID:21303928; https://doi.org/ 10.1242/jcs.081174 [DOI] [PubMed] [Google Scholar]
- [27].Fazal FM, Block SM. Optical tweezers study life under tension. Nat Photonics 2011; 5:318-21; PMID:22145010; https://doi.org/ 10.1038/nphoton.2011.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liphardt J, Onoa B, Smith SB, Tinoco J I, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science 2001; 292(5517):733-7; PMID:11326101; https://doi.org/ 10.1126/science.1058498 [DOI] [PubMed] [Google Scholar]
- [29].Yang C, Tibbitt MW, Basta L, Anseth KS. Mechanical memory and dosing influence stem cell fate. Nat Mater 2014; 13(6):645-52; PMID:24633344; https://doi.org/ 10.1038/nmat3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Makhija E, Jokhun DS, Shivashankar GV. Nuclear deformability and telomere dynamics are regulated by cell geometric constraints. Proc Natl Acad Sci 2016; 113(1):E32-40; PMID:26699462; https://doi.org/ 10.1073/pnas.1513189113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The interaction between nesprins and SUN proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem 2011; 286(30):26743-53; PMID:21652697; https://doi.org/ 10.1074/jbc.M111.233700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Schwarz US, Balaban NQ, Riveline D, Bershadsky A, Geiger B, Safran SA. Calculation of forces at focal adhesions from elastic substrate data: the effect of localized force and the need for regularization. Biophys J 2002; 83(3):1380-94; PMID:12202364; https://doi.org/ 10.1016/S0006-3495(02)73909-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, et al.. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol 2001; 3(5):466-72; PMID:11331874; https://doi.org/ 10.1038/35074532 [DOI] [PubMed] [Google Scholar]
- [34].Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 2010; 329(5994):956-9; PMID:20724637; https://doi.org/ 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhong Z, Chang SA, Kalinowski A, Wilson KL, Dahl KN. Stabilization of the spectrin-like domains of nesprin-1α by the evolutionarily conserved “adaptive” domain. Cell Mol Bioeng 2010; 3(2):139-50; PMID:20563238; https://doi.org/ 10.1007/s12195-010-0121-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mislow JM, Holaska JM, Kim MS, Lee KK, Segura-Totten M, Wilson KL, McNally EM. Nesprin-1α self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett 2002; 525(1-3):135-40; PMID:12163176; https://doi.org/ 10.1016/S0014-5793(02)03105-8 [DOI] [PubMed] [Google Scholar]
- [37].Ketema M, Wilhelmsen K, Kuikman I, Janssen H, Hodzic D, Sonnenberg A. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J Cell Sci 2007; 120(Pt 19):3384-94; PMID:17881500; https://doi.org/ 10.1242/jcs.014191 [DOI] [PubMed] [Google Scholar]
- [38].Dahl KN, Kalinowski A. Nucleoskeleton mechanics at a glance. J Cell Sci 2011; 124(Pt 5):675-8; PMID:21321324; https://doi.org/ 10.1242/jcs.069096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Parry DA, Dixon TW, Cohen C. Analysis of the three-alpha-helix motif in the spectrin superfamily of proteins. Biophys J 1992; 61(4):858-67; PMID:1581500; https://doi.org/ 10.1016/S0006-3495(92)81893-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Speicher DW, Marchesi VT. Erythrocyte spectrin is comprised of many homologous triple helical segments. Nature 1984; 311(5982):177-80; PMID:6472478; https://doi.org/ 10.1038/311177a0 [DOI] [PubMed] [Google Scholar]
- [41].Law R, Carl P, Harper S, Dalhaimer P, Speicher DW, Discher DE. Cooperativity in forced unfolding of tandem spectrin repeats. Biophys J 2003; 84(1):533-44; PMID:12524305; https://doi.org/ 10.1016/S0006-3495(03)74872-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Randles LG, Rounsevell RW, Clarke J. Spectrin domains lose cooperativity in forced unfolding. Biophys J 2007; 92(2):571-7; PMID:17085494; https://doi.org/ 10.1529/biophysj.106.093690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rief M, Pascual J, Saraste M, Gaub HE. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J Mol Biol 1999; 286(2):553-61; PMID:9973570; https://doi.org/ 10.1006/jmbi.1998.2466 [DOI] [PubMed] [Google Scholar]
- [44].Lenne PF, Raae AJ, Altmann SM, Saraste M, Hrber JKH. States and transitions during forced unfolding of a single spectrin repeat. FEBS Lett 2000; 476(3):124-8; PMID:10913598; https://doi.org/ 10.1016/S0014-5793(00)01704-X [DOI] [PubMed] [Google Scholar]
- [45].Vincent LG, Choi YS, Alonso-Latorre B, del Alamo JC, Engler AJ. Mesenchymal stem cell durotaxis depends on substrate stiffness gradient strength. Biotechnol J 2013; 8(4):472-84; PMID:23390141; https://doi.org/ 10.1002/biot.201200205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell 2011; 145(2):257-67; PMID:21496645; https://doi.org/ 10.1016/j.cell.2011.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nilsson OB, Nickson AA, Hollins JJ, Wickles S, Steward A, Beckmann R, von Heijne G, Clarke J. Cotranslational folding of spectrin domains via partially structured states. Nat Struct Mol Biol 2017; 24(3):221-5; PMID:28112730; https://doi.org/ 10.1038/nsmb.3355 [DOI] [PubMed] [Google Scholar]
- [48].Autore F, Pfuhl M, Quan X, Williams A, Roberts RG, Shanahan CM, Fraternali F. Large-scale modelling of the divergent spectrin repeats in nesprins: giant modular proteins. PLoS One 2013; 8(5):e63633; PMID:23671687; https://doi.org/ 10.1371/journal.pone.0063633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Arsenovic PT, Ramachandran I, Bathula K, Zhu R, Narang JD, Noll NA, Lemmon CA, Gundersen GG, Conway DE. Nesprin-2 g, a component of the nuclear linc complex, is subject to myosin-dependent tension. Biophys J 2016; 110(1):34-43; PMID:26745407; https://doi.org/ 10.1016/j.bpj.2015.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Grum VL, Li D, MacDonald RI, Mondragon A. Structures of two repeats of spectrin suggest models of flexibility. Cell 1999; 98(4):523-35; PMID:10481916; https://doi.org/ 10.1016/S0092-8674(00)81980-7 [DOI] [PubMed] [Google Scholar]
- [51].Paramore S, Voth GA. Examining the influence of linkers and tertiary structure in the forced unfolding of multiple-repeat spectrin molecules. Biophys J 2006; 91(9):3436-45; PMID:16891371; https://doi.org/ 10.1529/biophysj.106.091108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cui Y, Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc Natl Acad Sci U S A 2000; 97(1):127-32; PMID:10618382; https://doi.org/ 10.1073/pnas.97.1.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Xiao B, Freedman BS, Miller KE, Heald R, Marko JF. Histone H1 compacts DNA under force and during chromatin assembly. Mol Biol Cell 2012; 23(24):4864-71; PMID:23097493; https://doi.org/ 10.1091/mbc.E12-07-0518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Khatau SB, Bloom RJ, Bajpai S, Razafsky D, Zang S, Giri A, Wu PH, Marchand J, Celedon A, Hale CM, et al.. The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci Rep 2012; 2:488; PMID:22761994; https://doi.org/ 10.1038/srep00488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science 2014; 345(6200):1062-5; PMID:25170155; https://doi.org/ 10.1126/science.1256965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wiegert JS, Bengtson CP, Bading H. Diffusion and not active transport underlies and limits ERK1/2 synapse-to-nucleus signaling in hippocampal neurons. J Biol Chem 2007; 282(40):29621-33; PMID:17675293; https://doi.org/ 10.1074/jbc.M701448200 [DOI] [PubMed] [Google Scholar]
- [57].Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol 2009; 10(1):75-82; PMID:19197334; https://doi.org/ 10.1038/nrm2594 [DOI] [PubMed] [Google Scholar]
- [58].Na S, Collin O, Chowdhury F, Tay B, Ouyang M, Wang Y, Wang N. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc Natl Acad Sci U S A 2008; 105(18):6626-31; PMID:18456839; https://doi.org/ 10.1073/pnas.0711704105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hu S, Wang N. Control of stress propagation in the cytoplasm by prestress and loading frequency. Mol Cell Biomech 2006; 3(2):49-60; PMID:16903256 [PubMed] [Google Scholar]
- [60].Isermann P, Lammerding J. Nuclear mechanics and mechanotransduction in health and disease. Curr Biol 2013; 23(24):R1113-21; PMID:24355792; https://doi.org/ 10.1016/j.cub.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Crowder SW, Gupta MK, Hofmeister LH, Zachman AL, Sung HJ. Modular polymer design to regulate phenotype and oxidative response of human coronary artery cells for potential stent coating applications. Acta Biomaterialia 2012; 8(2):559-69; PMID:22019760; https://doi.org/ 10.1016/j.actbio.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Litjens SH, Koster J, Kuikman I, van Wilpe S, de Pereda JM, Sonnenberg A. Specificity of binding of the plectin actin-binding domain to β4 integrin. Mol Biol Cell 2003; 14(10):4039-50; PMID:14517317; https://doi.org/ 10.1091/mbc.E03-05-0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.