ABSTRACT
Metazoan nuclei are equipped with nuclear lamina - a thin layer of intermediate filaments (IFs) mostly built of nuclear lamins facing the inner nuclear membrane (INM). The nuclear lamina serves as an interaction hub for INM-proteins, soluble nuclear factors and DNA. It confers structural and mechanical stability to the nucleus, transduces mechanical forces and biochemical signals across the nuclear envelope (NE) and regulates the organization of chromatin. By using cryo-electron tomography (cryo-ET), we recently provided an unprecedented view into the 3D organization of lamin filaments within the lamina meshwork in mammalian somatic cells. Through implementation of averaging procedures, we resolved the rod and globular Ig-fold domains of lamin filaments. The density maps suggested that they assemble into 3.5 nm thick filaments. Our analysis revealed interesting structural differences between nucleoplasmic and cytoplasmic intermediate filaments, raising the question of which molecular cues define their assembly modes inside the cell.
Keywords: cryo-electron tomgraphy, intermediate filaments, lamin filaments, nuclear lamina
Introduction
Multicellular organisms have adapted for efficient response to mechanical strain by developing well-orchestrated molecular mechanisms that sense and transmit mechanical forces among the cells and their organelles. As a result, these signals are translated into biochemical and cellular alterations.1-3 To this effect, it becomes evident that metazoan cells have adjusted their overall architecture to resist these physically demanding conditions. One such mechanical feature is the presence of the so-called “nuclear lamina," which represents a dense fibrous protein meshwork at the periphery of the nucleus.4-6 It underpins the INM, supports the cross-talk between the cytoplasmic and nucleoplasmic compartment and attenuates the mechanical load to protect the genetic material.7-10 Besides its scaffolding and mechanosensory function, lamins also have essential roles in chromatin organization, DNA replication and repair, transcription regulation, resistance to oxidative stress, stem cell maintenance and differentiation, signaling and cell cycle progression.8,11-15
In mammalian cells, the lamin meshwork is mostly composed of 4 lamin isoforms: A, C, B1 and B2. Based on their sequence and structural properties, these type V intermediate filament (IF) proteins are sub-divided in 2 different classes, namely A-type and B-type. A-type lamins are represented by the 2 isoforms lamin A and C and are derived from a single gene by alternative splicing, whereas the B-type lamins, B1 and B2, are derived from 2 independent genes.16-19 While at least one B-type lamin is expressed at different stages of development, A-type lamins are mainly expressed in terminally differentiated cells.20
Like most cytoplasmic IF proteins, lamins are composed of a central α-helical “rod” domain, flanked by a non-α-helical N-terminal “head” and C-terminal “tail” domain. The head and tail domains of IF proteins are variable in size, whereas the ∼45 nm long central rod domains are composed of 4 conserved α-helices (1A, 1B, 2A, 2B) and 3 linker regions (L1, L12, L2). However, some characteristic features distinguish nuclear lamins from cytoplasmic IF proteins, namely 6 additional heptads (42 residues) within helix 1A of the rod domain, the presence of a nuclear localization signal (NLS), a globular immunoglobulin (Ig)-fold within the C terminus, and a CaaX motif at their C-terminal end.21-24
The interest in lamins and the structural organization of the nuclear lamina has increased because of the identification of more than a dozen distinct heritable diseases that are associated with mutations in the human lamin A gene. These diseases, collectively termed “laminopathies," affect adipose, bone, nerve and skin cells and comprise a variety of syndromes, such as muscular dystrophies (e.g. Emery-Dreifuss muscular dystrophy, EDMD), lipodystrophies (e.g., familial partial lipodystrophy, FPLD), neuropathies and premature aging (or progeroid) syndromes (e.g., Hutchinson Gilford progeria syndrome, HGPS; or restrictive dermopathy, RD).25-29
Visualization of the nuclear lamina by cryo-ET
The first view into the organization of the lamin meshwork was presented 30 y ago by visualizing chromatin-free and detergent-treated nuclear envelopes, isolated from nuclei of Xenopus laevis oocytes, with transmission electron microscopy (TEM).30 These images, represent the organization of lamin LIII, which is only expressed in the germline of fish, amphibians, reptiles and birds;31 however, the visualization of the lamin meshwork in somatic cells has proven more challenging. The main obstacle to the electron microscopic analysis of the nuclear lamina in situ is the thickness of the cellular sample, especially in nuclear regions. Although it was shown that the nuclear envelope of U2OS cells can be studied in toto,32 the crowded environment of the NE, particularly the bulk of chromatin that surrounds the filamentous structure, prohibits a detailed analysis of the nuclear lamina. Recent developments of imaging technologies such as focused ion beam scanning electron microscopy (cryo-FIB-SEM) and cryo-electron tomography (cryo-ET) have become very attractive tools to study supramolecular assemblies within the native environment of biological specimens.33,34 Two recent studies have implemented these approaches to investigate the nuclear periphery of nuclei in vitrified and milled C.elegans embryos and HeLa cells, respectively.33,35 The results suggested the presence of short filaments at the periphery of nuclei. However, experimental evidence that these filaments are indeed lamins was missing. The detection of only short filaments (most are ≤ 55 nm in HeLa cells, which is about the length of a lamin dimer) contradicts the presence of a fully assembled lamin meshwork. This might be explained by the dense environment within which the lamin filaments are embedded and the intimate interaction with heterochromatin, all of which reduce the contrast of lamins and make it challenging to track them carefully throughout their entire length.
By applying cryo-ET to purified ghost nuclei, i.e. nuclei devoid of chromatin, from vimentin knockout mouse embryonic fibroblasts (MEF), we acquired a first view into the molecular architecture of the mammalian nuclear lamina.36 For this, we subjected MEFs to a short exposure in mild detergent conditions and treatment with nuclease (Fig. 1a). Our cryo-ET analysis revealed that the nuclear lamina is composed of ∼3.5 nm thick globular-decorated filaments (Fig. 1b), assembled into a ∼14 nm thick meshwork that is localized adjacent to the INM, underneath nuclear pore complexes.37 Co-immunogold-labeling experiments showed that these filaments are composed of A- and B-type lamins. Interestingly, efforts in designing lamin A knockout cells in vimentin null background were unsuccessful, emphasizing the importance of IFs in single cells.
Figure 1.
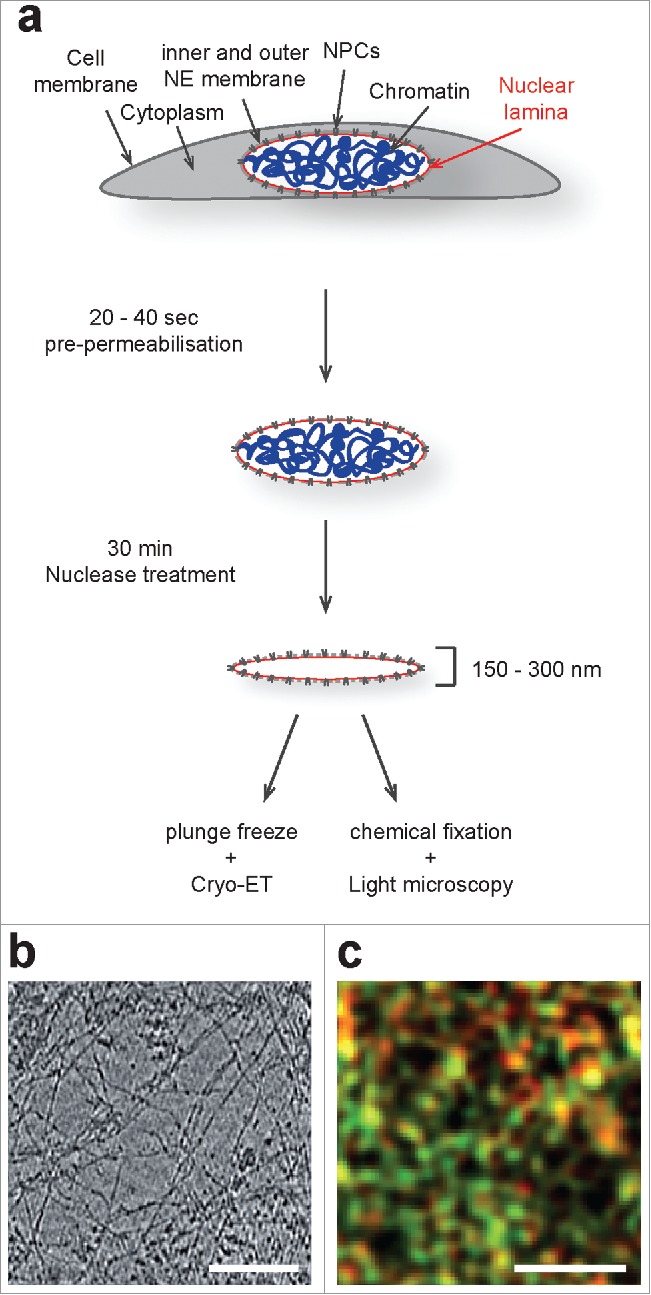
Sample preparation procedure and microscopic analysis of vimentin deficient MEFs by Cryo-ET and 3D-SIM. (a) Schematic illustration of the sample preparation procedure used to analyze lamins and the lamina by cryo-ET. This procedure does not alter the lamina organization as judged by 3D-SIM (see ref. 36). (b) A 4 nm thick xy-slice through a representative tomogram of the mammalian nuclear lamina. Scale bar, 100 nm. (c) A 3D-SIM image of pre-permeabilised, nuclease-treated and immuno-labeled MEFs shows localization and expression of lamin A (red) and lamin B1 (green). Scale bar, 1 µm.
Earlier studies on the organization of A- and B-type lamins within the nuclear lamina of MEF nuclei used 3D structured illumination microscopy (3D-SIM) and direct stochastic optical reconstruction microscopy (dSTORM) to visualize immuno-labeled lamins. The results of this super-resolution fluorescent microscopy approach showed a clear separation of the differently labeled antibodies that were used in the co-immuno-labeling experiments. This suggested that the different lamin isoforms assemble into separate meshworks, together forming a functional lamina.38,39 To deduce lamin filaments from the related label, it was supposed that the more closely the individual lamin labels are adjoined, the higher would be the probability that they are attached to the same filament. Based on this assumption they applied nearest-neighbor algorithms to connect the individual label and to create filaments. The modeled lamin A and lamin B1 filaments showed no obvious overlap and the composite meshworks displayed variations in the number, geometry and size of spaces that are surrounded by filaments. To compare the co-immuno-labeling results of our cryo-ET analysis with previously published super-resolution fluorescent microscopy images, we assembled a collage of images from our cryo-tomograms, containing the different sized lamin A and lamin B specific gold labels. Next, the coordinates of the 2 different sized gold labels were extracted and displayed in 2 different colors at a resolution of 120 nm, which is comparable to the resolution achieved with 3D-SIM. The resulting image showed similar numbers and distributions of the lamin A and lamin B1 label as compared with the results from the 3D-SIM analysis. This confirms that our sample preparation procedure did not induce major changes in the general organization of A- and B-type lamins within the nuclear lamina (Fig. 1c). However, visual inspection of our cryo-tomograms also revealed that the large amount of lamin filaments and the frequency of potential epitopes is much higher compared with the number of labeled antibodies that is detected in our and the reported co-immuno-labeling studies. This may reflect an insufficient decoration of lamin filaments with labeled antibody within the nuclear lamina, based on the compact organization of the lamin filaments and the presence of lamin binding proteins that may mask some of the epitopes.
Structural analysis of lamin assemblies
Structural analysis of in vitro assembled IFs by X-ray crystallography and electron microscopy suggested that assembly of cytoplasmic IFs and lamin filaments follows hierarchical ordered polymerization pathways. Both assemblies are initiated by parallel and in-register association of 2 monomers through their α-helical domains to form coiled-coil dimers.17,40,41 Next, lamin dimers interact longitudinally to form polar head-to-tail polymers that assemble laterally in an anti-parallel and half staggered fashion to build an apolar protofilament.30,42-45 Higher order assembly of lamins is based on the lateral interaction of protofilaments to form 10 nm filaments, ultimately leading to the formation of paracrystalline arrays. In comparison to lamin filament assembly, cytoplasmic IF formation starts with the lateral association of 2 dimers into anti-parallel and half-staggered tetramers. The tetramers subsequently associate in a lateral fashion to form full width unit-length type filaments (ULFs), which is followed by their longitudinal interaction to form ∼10 nm thick fibers.46 Whether in vivo intermediate filament assembly uses similar pathways and results in the formation of comparable structures is subject of current debate. Although it has been demonstrated that endogenous cytoplasmic IFs form ∼10 nm thick filaments in vitro and in vivo, little is known about the assembly state of lamin filaments in their native environment. Cryo-ET analysis of ectopically expressed C. elegans lamin in Xenopus oocytes showed 4–6 nm thick filaments at the nuclear periphery, suggesting that lamins may assemble into thinner filaments than cytoplasmic IFs.47 Indeed, this is further supported by our cryo-ET analysis of the mammalian nuclear lamina that revealed a composition of even thinner (∼3.5 nm) lamin filaments, raising the question of which cellular cues prevent higher order assembly. The most obvious reason that could explain this phenomenon might be the interaction of lamins with nucleoplasmic proteins, chromatin and membrane-proteins at the INM. Another important factor might be the specific concentration of each lamin isoform, based on the observation that overexpression of lamin A in Spodoptera frugiperda (Sf9) insect cells yields paracrystalline fiber formation.48
Structural classification and averaging of the lamin filaments from our cryo-tomograms revealed a globular-decorated fiber appearance, which resembles the filaments as observed at early stages in in vitro assembly experiments using purified lamins. The averaged structures displayed a uniform filament thickness of 3.5 nm. The dense globular domains (putative Ig-fold domains) appeared pairwise and in 20 nm steps alongside the rod of the filament and served as fiduciary marks, suggesting the assembly into tetrameric lamin filaments (Fig. 2, top filament). However, some of the structural classes showed that the position of the Ig-fold domains can vary, indicating a lateral displacement of up to 10 nm (Fig. 2, bottom filament), which may be attributable to the flexible stretch of amino acids between the end of the rod and the beginning of the Ig-fold in lamin proteins (∼50 aa in lamin A, C, B1 and ∼70 aa in lamin B249).
Figure 2.
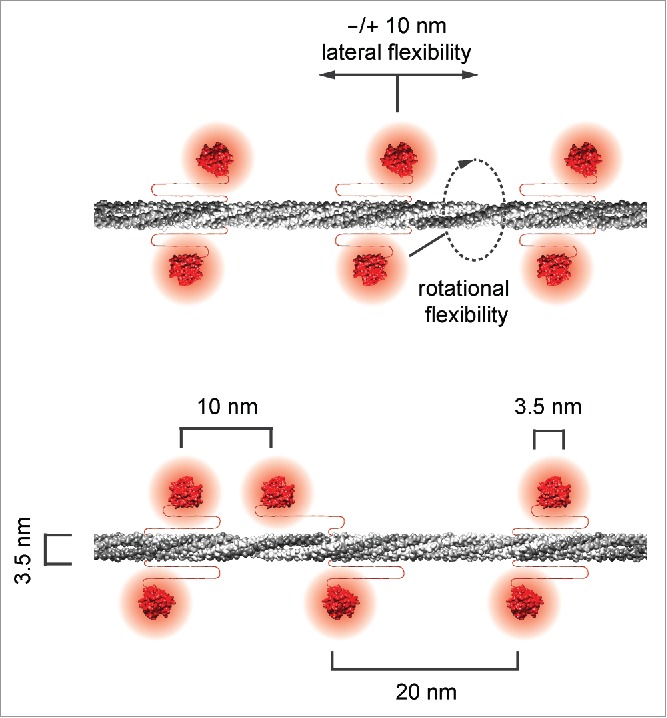
Lamin filaments exhibit a tetrameric structure in cross-section. The suggested models of lamin filaments illustrate the 3.5 nm uniform thickness of the rod domain (gray) and the positions of the Ig-fold domains (red) alongside the lamin filament, built from 2 head-to-tail polymers to form a tetrameric assembly, previously termed a protofilament. The Ig-fold domains repeat pairwise every 20 nm (top filament) and exhibit a high degree of spatial flexibility, most likely due to the linkers between the rod and the Ig-fold domain. In some cases, the Ig-fold domains along a filament are spaced as close as 10 nm.
Conclusion
In Turgay et al., 2017,36 we report the molecular organization of lamin filaments at the NE in mammalian somatic cells. Our structural characterization of single lamin filaments revealed important insights into the lamina organization and lamin assembly in vivo. Further data acquisition and analysis may help improve the resolution of the lamin structures to <1nm, which is essential to delineate the precise organization of the nuclear lamins, both A- and B-type, and to pinpoint specific alterations on the single filament level. Moreover, this will help elucidate the molecular mechanisms that underlie laminopathies.
Two general models may explain these scenarios. First, the “structural hypothesis” proposes that mutations in lamin A/C render structural alterations of lamins, which produce more fragile nuclei, therefore causing cell death and eventually disease in mechanically stressed tissues. Second, the “gene regulation hypothesis” proposes that specific lamin mutations lead to distinct alterations in gene regulation, and this may be the underlying cause for the development of different disease phenotypes.50 Other models hint on defective nucleo-cytoskeletal coupling or imbalanced stem-cell and differentiation homeostasis.51-54 However, so far none of these models can convincingly explain the variety of diseases caused by lamin mutations. Hence, identifying the alterations of lamina structures and the mechanical properties due to specific mutations will be an instrumental step in our understanding of the disease mechanisms underlying laminopathies.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank R. Irobalieva for critical reading of the manuscript and the Center for Microscopy and Image Analysis at the University of Zurich (ZMB).
Funding
This work was funded by a Swiss National Science Foundation Grant (SNSF 31003A 159706/1), the Mäxi Foundation and GIF I-1289–412.13/2015 to O.M., and the Forschungskredit of the University of Zurich to Y.T.
References
- [1].Piccolo S. Embracing mechanical forces in cell biology. Differentiation 2013; 86:75-6; PMID:24054842; 20822892https://doi.org/10.1016/j.diff.2013.08.001 [DOI] [PubMed] [Google Scholar]
- [2].Medalia O, Geiger B. Frontiers of microscopy-based research into cell-matrix adhesions. Curr Opin Cell Biol 2010; 22:659-68; PMID:20822892; https://doi.org/ 10.1016/j.ceb.2010.08.006 [DOI] [PubMed] [Google Scholar]
- [3].Sorrentino S, Studt JD, Medalia O, Tanuj Sapra K. Roll, adhere, spread and contract: structural mechanics of platelet function. Eur J Cell Biol 2015; 94:129-38; PMID:25655000; https://doi.org/ 10.1016/j.ejcb.2015.01.001 [DOI] [PubMed] [Google Scholar]
- [4].Gruenbaum Y, Medalia O. Lamins: the structure and protein complexes. Curr Opin Cell Biol 2015; 32:7-12; PMID:25460776; https://doi.org/ 10.1016/j.ceb.2014.09.009 [DOI] [PubMed] [Google Scholar]
- [5].Irianto J, Pfeifer CR, Ivanovska IL, Swift J, Discher DE. Nuclear lamins in cancer. Cell Mol Bioeng 2016; 9:258-67; PMID:27570565; https://doi.org/ 10.1007/s12195-016-0437-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bell ES, Lammerding J. Causes and consequences of nuclear envelope alterations in tumour progression. Eur J Cell Biol 2016; 95:449-64; PMID:27397692; https://doi.org/ 10.1016/j.ejcb.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Burke B, Stewart CL. The nuclear lamins: Flexibility in function. Nat rev Mol Cell Biol 2013; 14:13-24; PMID:23212477; https://doi.org/ 10.1038/nrm3488 [DOI] [PubMed] [Google Scholar]
- [8].Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol 2010; 2:a000547; PMID:20826548; https://doi.org/ 10.1101/cshperspect.a000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fawcett DW. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat 1966; 119:129-45; PMID:6007824; https://doi.org/ 10.1002/aja.1001190108 [DOI] [PubMed] [Google Scholar]
- [10].Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, et al.. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev 2008; 22:3409-21; https://doi.org/ 10.1101/gad.1735208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Camozzi D, Capanni C, Cenni V, Mattioli E, Columbaro M, Squarzoni S, Lattanzi G. Diverse lamin-dependent mechanisms interact to control chromatin dynamics. Focus on laminopathies. Nucleus 2014; 5:427-40; PMID:25482195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Meister P, Mango SE, Gasser SM. Locking the genome: nuclear organization and cell fate. Curr Opin Genet Dev 2011; 21:167-74; https://doi.org/ 10.1016/j.gde.2011.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Naetar N, Korbei B, Kozlov S, Kerenyi MA, Dorner D, Kral R, Gotic I, Fuchs P, Cohen TV, Bittner R, et al.. Loss of nucleoplasmic LAP2alpha-lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nat Cell Biol 2008; 10:1341-8; PMID:18849980; https://doi.org/ 10.1038/ncb1793 [DOI] [PubMed] [Google Scholar]
- [14].Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol 2008; 10:452-9; PMID:18311132; https://doi.org/ 10.1038/ncb1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shumaker DK, Kuczmarski ER, Goldman RD. The nucleoskeleton: lamins and actin are major players in essential nuclear functions. Curr Opin Cell Biol 2003; 15:358-66; PMID:12787780; https://doi.org/ 10.1016/S0955-0674(03)00050-4 [DOI] [PubMed] [Google Scholar]
- [16].Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem 1993; 268:16321-6; PMID:8344919 [PubMed] [Google Scholar]
- [17].Parry DA, Conway JF, Steinert PM. Structural studies on lamin. Similarities and differences between lamin and intermediate-filament proteins. Biochem J 1986; 238:305-8; PMID:3800939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Peter M, Kitten GT, Lehner CF, Vorburger K, Bailer SM, Maridor G, Nigg EA. Cloning and sequencing of cDNA clones encoding chicken lamins A and B1 and comparison of the primary structures of vertebrate A- and B-type lamins. J Mol Biol 1989; 208:393-404; PMID:2795656; https://doi.org/ 10.1016/0022-2836(89)90504-4 [DOI] [PubMed] [Google Scholar]
- [19].Vorburger K, Lehner CF, Kitten GT, Eppenberger HM, Nigg EA. A second higher vertebrate B-type lamin. cDNA sequence determination and in vitro processing of chicken lamin B2. J Mol Biol 1989; 208:405-15; PMID:2477550 [DOI] [PubMed] [Google Scholar]
- [20].Worman HJ, Lazaridis I, Georgatos SD. Nuclear lamina heterogeneity in mammalian cells. Differential expression of the major lamins and variations in lamin B phosphorylation. J Biol Chem 1988; 263:12135-41; PMID:3403563 [PubMed] [Google Scholar]
- [21].Dhe-Paganon S, Werner ED, Chi YI, Shoelson SE. Structure of the globular tail of nuclear lamin. J Biol Chem 2002; 277:17381-4; PMID:11901143; https://doi.org/ 10.1074/jbc.C200038200 [DOI] [PubMed] [Google Scholar]
- [22].Krimm I, Ostlund C, Gilquin B, Couprie J, Hossenlopp P, Mornon JP, Bonne G, Courvalin JC, Worman HJ, Zinn-Justin S. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure 2002; 10:811-23; PMID:12057196; https://doi.org/ 10.1016/S0969-2126(02)00777-3 [DOI] [PubMed] [Google Scholar]
- [23].Loewinger L, McKeon F. Mutations in the nuclear lamin proteins resulting in their aberrant assembly in the cytoplasm. EMBO J 1988; 7:2301-9; PMID:3056713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shumaker DK, Lopez-Soler RI, Adam SA, Herrmann H, Moir RD, Spann TP, Goldman RD. Functions and dysfunctions of the nuclear lamin Ig-fold domain in nuclear assembly, growth, and Emery-Dreifuss muscular dystrophy. Proc Natl Acad Sci U S A 2005; 102:15494-9; PMID:16227433; https://doi.org/ 10.1073/pnas.0507612102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: Laminopathies and their role in premature ageing. Physiol Rev 2006; 86:967-1008; PMID:16816143; https://doi.org/ 10.1152/physrev.00047.2005 [DOI] [PubMed] [Google Scholar]
- [26].Worman HJ, Courvalin JC. Nuclear envelope, nuclear lamina, and inherited disease. Int Rev Cytol 2005; 246:231-79; PMID:16164970 [DOI] [PubMed] [Google Scholar]
- [27].Worman HJ, Fong LG, Muchir A, Young SG. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest 2009; 119:1825-36; PMID:19587457; https://doi.org/ 10.1172/JCI37679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Worman HJ, Ostlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol 2010; 2:a000760; PMID:20182615; https://doi.org/ 10.1101/cshperspect.a000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zuela N, Zwerger M, Levin T, Medalia O, Gruenbaum Y. Impaired mechanical response of an EDMD mutation leads to motility phenotypes that are repaired by loss of prenylation. J Cell Sci 2016; 129:1781-91; PMID:27034135; https://doi.org/ 10.1242/jcs.184309 [DOI] [PubMed] [Google Scholar]
- [30].Aebi U, Cohn J, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature 1986; 323:560-4; PMID:3762708; https://doi.org/ 10.1038/323560a0 [DOI] [PubMed] [Google Scholar]
- [31].Hofemeister H, Kuhn C, Franke WW, Weber K, Stick R. Conservation of the gene structure and membrane-targeting signals of germ cell-specific lamin LIII in amphibians and fish. Eur J Cell Biol 2002; 81:51-60; PMID:11893082; https://doi.org/ 10.1078/0171-9335-00229 [DOI] [PubMed] [Google Scholar]
- [32].Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure 2012; 20:998-1006; PMID:22632834; https://doi.org/ 10.1016/j.str.2012.03.025 [DOI] [PubMed] [Google Scholar]
- [33].Harapin J, Bormel M, Sapra KT, Brunner D, Kaech A, Medalia O. Structural analysis of multicellular organisms with cryo-electron tomography. Nat Methods 2015; 12:634-6; PMID:25961413; https://doi.org/ 10.1038/nmeth.3401 [DOI] [PubMed] [Google Scholar]
- [34].Harapin J, Eibauer M, Medalia O. Structural analysis of supramolecular assemblies by cryo-electron tomography. Structure 2013; 21:1522-30; PMID:24010711; https://doi.org/ 10.1016/j.str.2013.08.003 [DOI] [PubMed] [Google Scholar]
- [35].Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, Baumeister W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 2016; 351:969-72; PMID:26917770; https://doi.org/ 10.1126/science.aad8857 [DOI] [PubMed] [Google Scholar]
- [36].Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD, Medalia O. The molecular architecture of lamins in somatic cells. Nature 2017; 543:261-4; PMID:28241138; https://doi.org/ 10.1038/nature21382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Eibauer M, Pellanda M, Turgay Y, Dubrovsky A, Wild A, Medalia O. Structure and gating of the nuclear pore complex. Nat Commun 2015; 6:7532; PMID:26112706; https://doi.org/ 10.1038/ncomms8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Xie W, Chojnowski A, Boudier T, Lim JS, Ahmed S, Ser Z, Stewart C, Burke B. A-type lamins form distinct filamentous networks with differential nuclear pore complex associations. Curr Biol 2016; 26:2651-8; PMID:27641764; https://doi.org/ 10.1016/j.cub.2016.07.049 [DOI] [PubMed] [Google Scholar]
- [39].Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell 2015; 26:4075-86; PMID:26310440; https://doi.org/ 10.1091/mbc.E15-07-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat rev Mol Cell Biol 2007; 8:562-73; PMID:17551517; https://doi.org/ 10.1038/nrm2197 [DOI] [PubMed] [Google Scholar]
- [41].Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol 1998; 122:42-66; PMID:9724605; https://doi.org/ 10.1006/jsbi.1998.3987 [DOI] [PubMed] [Google Scholar]
- [42].Ben-Harush K, Wiesel N, Frenkiel-Krispin D, Moeller D, Soreq E, Aebi U, Herrmann H, Gruenbaum Y, Medalia O. The supramolecular organization of the C. elegans nuclear lamin filament. J Mol Biol 2009; 386:1392-402 [DOI] [PubMed] [Google Scholar]
- [43].Goldberg MW, Huttenlauch I, Hutchison CJ, Stick R. Filaments made from A- and B-type lamins differ in structure and organization. J Cell Sci 2008; 121:215-25; PMID:18187453; https://doi.org/ 10.1242/jcs.022020 [DOI] [PubMed] [Google Scholar]
- [44].Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Ann Rev Biochem 2004; 73:749-89; PMID:15189158; https://doi.org/ 10.1146/annurev.biochem.73.011303.073823 [DOI] [PubMed] [Google Scholar]
- [45].Stick R, Goldberg MW. Oocytes as an experimental system to analyze the ultrastructure of endogenous and ectopically expressed nuclear envelope components by field-emission scanning electron microscopy. Methods 2010; 51:170-6; PMID:20085817; https://doi.org/ 10.1016/j.ymeth.2010.01.015 [DOI] [PubMed] [Google Scholar]
- [46].Winheim S, Hieb AR, Silbermann M, Surmann EM, Wedig T, Herrmann H, Langowski J, Mücke N. Deconstructing the late phase of vimentin assembly by total internal reflection fluorescence microscopy (TIRFM). PloS one 2011; 6:e19202; PMID:21544245; https://doi.org/ 10.1371/journal.pone.0019202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Grossman E, Dahan I, Stick R, Goldberg MW, Gruenbaum Y, Medalia O. Filaments assembly of ectopically expressed Caenorhabditis elegans lamin within Xenopus oocytes. J Struct Biol 2012; 177:113-8; PMID:22085746; https://doi.org/ 10.1016/j.jsb.2011.11.002 [DOI] [PubMed] [Google Scholar]
- [48].Klapper M, Exner K, Kempf A, Gehrig C, Stuurman N, Fisher PA, Krohne G. Assembly of A- and B-type lamins studied in vivo with the baculovirus system. J Cell Sci 1997; 110(Pt 20):2519-32; PMID:9372441 [DOI] [PubMed] [Google Scholar]
- [49].Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev 2008; 22:832-53; https://doi.org/ 10.1101/gad.1652708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hutchison CJ, Worman HJ. A-type lamins: guardians of the soma? Nat Cell Biol 2004; 6:1062-7; PMID:15517000; https://doi.org/ 10.1038/ncb1104-1062 [DOI] [PubMed] [Google Scholar]
- [51].Chen CY, Chi YH, Mutalif RA, Starost MF, Myers TG, Anderson SA, Stewart CL, Jeang KT. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell 2012; 149:565-77; PMID:22541428; https://doi.org/ 10.1016/j.cell.2012.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science 2014; 344:527-32; PMID:24786082; https://doi.org/ 10.1126/science.1252651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li P, Meinke P, Huong le TT, Wehnert M, Noegel AA. Contribution of SUN1 mutations to the pathomechanism in muscular dystrophies. Human mutation 2014; 35:452-61; PMID:24375709; https://doi.org/ 10.1002/humu.22504 [DOI] [PubMed] [Google Scholar]
- [54].Schwartz C, Fischer M, Mamchaoui K, Bigot A, Lok T, Verdier C, Duperray A, Michel R, Holt I, Voit T, et al.. Lamins and nesprin-1 mediate inside-out mechanical coupling in muscle cell precursors through FHOD1. Sci Rep 2017; 7:1253; PMID:28455503; https://doi.org/ 10.1038/s41598-017-01324-z [DOI] [PMC free article] [PubMed] [Google Scholar]


