ABSTRACT
One of the major functions of DNA methylation is the repression of transposable elements, such as the long-interspersed nuclear element 1 (L1). The underlying mechanism(s), however, are unclear. Here, we addressed how retrotransposon activation and mobilization are regulated by methyl-cytosine modifying ten-eleven-translocation (Tet) proteins and how this is modulated by methyl-CpG binding domain (MBD) proteins. We show that Tet1 activates both, endogenous and engineered L1 retrotransposons. Furthermore, we found that Mecp2 and Mbd2 repress Tet1-mediated activation of L1 by preventing 5hmC formation at the L1 promoter. Finally, we demonstrate that the methyl-CpG binding domain, as well as the adjacent non-sequence specific DNA binding domain of Mecp2 are each sufficient to mediate repression of Tet1-induced L1 mobilization. Our study reveals a mechanism how L1 elements get activated in the absence of Mecp2 and suggests that Tet1 may contribute to Mecp2/Mbd2-deficiency phenotypes, such as the Rett syndrome. We propose that the balance between methylation “reader” and “eraser/writer” controls L1 retrotransposition.
KEYWORDS: DNA methylation, 5-hydroxymethylcytosine, genome stability, repetitive elements, Rett syndrome
Introduction
In humans, 17% of nuclear DNA consists of long interspersed nuclear element 1 (LINE1 or L1). The majority of L1s, however, are retrotransposition defective (RD-L1) due to 5′ truncations, internal rearrangements or mutations. Only 80–100 copies of the half a million human L1s are retrotransposition competent (RC-L1).1 Full length L1 has a total length of about 6 kilobase pairs and contains a 5′ untranslated region (5′UTR) with promoter activity in both, sense and antisense directions, 3 open reading frames (ORFs) and a 3′UTR that ends in an AATAAA polyadenylation signal. ORF1 encodes for a p40 protein with RNA-binding and chaperone activities,2 whereas ORF2 encodes for a protein of 150 kDa in size with endonuclease and reverse transcriptase activities.3 In contrast to ORF1 and ORF2, ORF0 is primate-specific and lies downstream the 5′UTR in antisense direction. It has 2 splice donor sites that can react with splice acceptors of downstream genomic sequences to generate fusion proteins.4 During the L1 retrotransposition procedure, ORF1 and ORF2 proteins bind to their own RNA in the cytosol to form a ribonucleoprotein particle (RNP), which facilitates the re-import of L1 RNA to the nucleus. The majority of genomic L1 integrations follow a mechanism termed target primed reverse transcription (TPRT),5 which involves endonuclease and reverse transcriptase activity of ORF2p. However, endonuclease independent L1 integration is also observed in non-homologous end joining (NHEJ) and p53 double deficient cells.6,7 Previous studies using engineered L1 elements showed that retrotransposition occurs not only in brain cells like neural progenitor cells of rat hippocampus8 and human fetal brain,9 but also in non-brain cells such as embryonic stem cells (ESC).10 Moreover, transgenic mice, harboring a fluorescently tagged human L1 under the control of its endogenous promoter showed only detectable L1 retrotransposition activity in mouse germ cells and brain. Furthermore, methylation level of L1 elements differed in brain and skin,8 indicating that L1s are differently regulated in tissues and cell types. Altogether, these studies demonstrate that retrotransposition of L1 elements can occur in embryonic and importantly, also in somatic cells and correlates with the L1 promoter methylation status.
Although host cells have multiple mechanisms to restrict L1 retrotransposition,11-14 sporadic insertions of a small number of RC-L1s accompanied by large chromosomal rearrangements can occur that lead to genomic instability.15 Insertions of L1 sequences into protein coding regions of the genome can decrease RNA levels by inhibiting transcriptional elongation.16 In addition to its ability to propagate itself, 3′ transductions of L1 occur in germ and cancer cells, whereby unique sequences downstream of L1 elements can also be retrotransposed, if transcription continues beyond the L1 sequence.17 Moreover, other studies showed that L1 expression leads to high levels of double strand breaks, as evidenced by the formation of γH2AX foci and the recruitment of repair proteins involved in the L1 retrotransposition process.18
One of the most important mechanisms repressing L1 retrotransposition depends on DNA methylation. Methylation of cytosines has been shown to recruit 5-methylcytosine (5mC) binding domain (MBD) proteins. Mecp2, the founding member of the MBD protein family, was subsequently shown to modulate L1 retrotransposition in a DNA methylation dependent manner.19,20 While the transcriptional repression domain (TRD) of Mecp2 was shown to be sufficient for repressing L1 retrotransposition in a reporter assay system,20 the mechanism(s) of L1 repression mediated by Mecp2 remain unknown. Mecp2 and Mbd2, another family member of the MBD protein family, specifically bind to methylated CpG dinucleotides in DNA.21 Hypomethylation of the L1 promoter is associated with overexpression of L1 transcripts,11 suggesting that a decrease in DNA methylation might play a role in L1 retrotransposition. Ten-eleven-translocation (Tet) proteins convert 5-methylcytosine to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) in an iterative iron- and oxoglutarate dependent oxidation reaction. These further modifications of methylated cytosines are proposed to be one of the long sought mechanisms leading to loss of DNA methylation.22 Tet1 and Tet2 depletion in ESCs has been shown to cause loss of 5hmC in the 5′ region of L1,23 but a connection to L1 regulation is lacking.
In this study, we investigate whether and how Mbd and Tet proteins affect L1 expression and mobilization in human cells. We detected increased transcription and transposition of human endogenous L1 in the presence of Tet1 proteins and showed that activation of L1 transposition depends on the catalytic activity of Tet1. By the use of an L1 retrotransposition reporter assay, we additionally showed that Tet1 proteins activate retrotransposition of engineered L1. Moreover, we found that Mbd2, Mecp2, as well as its methylcytosine binding and transcriptional repression subdomains counteract Tet1-mediated reactivation of L1 retrotransposons.
Results and discussion
Tet1 activates retrotransposition of endogenous L1
Three steps are involved in L1 retrotransposition, comprising loss of DNA methylation in the L1 5′UTR, L1 transcription and L1 transposition (Fig 1A). 5hmC, which is produced by Tet proteins, is thought to be an intermediate modification during loss of DNA methylation,24,25 as well as a stable epigenetic mark, which tunes a large number of CpG dinucleotides located at poised enhancers and actively transcribed regions.26,27 To test whether L1 retrotransposition is re-activated by Tet-mediated 5mC to 5hmC conversion, we transfected HEK-EBNA cells with the catalytically active domain of Tet1 fused to mcherry (mcherry-Tet1CD), which was previously shown to be sufficient to induce genome-wide hydroxymethylation in vivo.28-30 As control, mcherry-Tet1CDmut, which lacked catalytic activity due to 2 mutations in the Fe(II) binding sites (H1652Y, D1654A) was used.28 Since loss of DNA methylation in the L1 5′UTR is the first step that makes L1 retrotransposition possible, we performed GluMs-qPCR (DNA glucosylation, MspI digestion and quantitative PCR based 5hmC detection, described in the methods) to determine the methylation state of the L1 promoter. L1 has 20 mapped CpG sites within its 5′UTR. Methylation of only a subset of CpG dinucleotides, such as nucleotide position 482, were shown to correlate inversely with retrotransposition activity of L1 elements.9 Therefore, we chose CpG dinucleotide at position 482 to quantify the methylation status of the L1 promoter (Fig 1B) before and after transfection. In untransfected cells, we observed using GluMs-qPCR31 that around 60% of all CpG dinucleotides at position 482 were methylated upon Tet1CD expression, however, 5hmC levels at position 482 were increased compared with Tet1CDmut and mock transfected cells (Fig 1C), indicating that Tet1 oxidizes 5mC in the L1 promoter.
Figure 1.
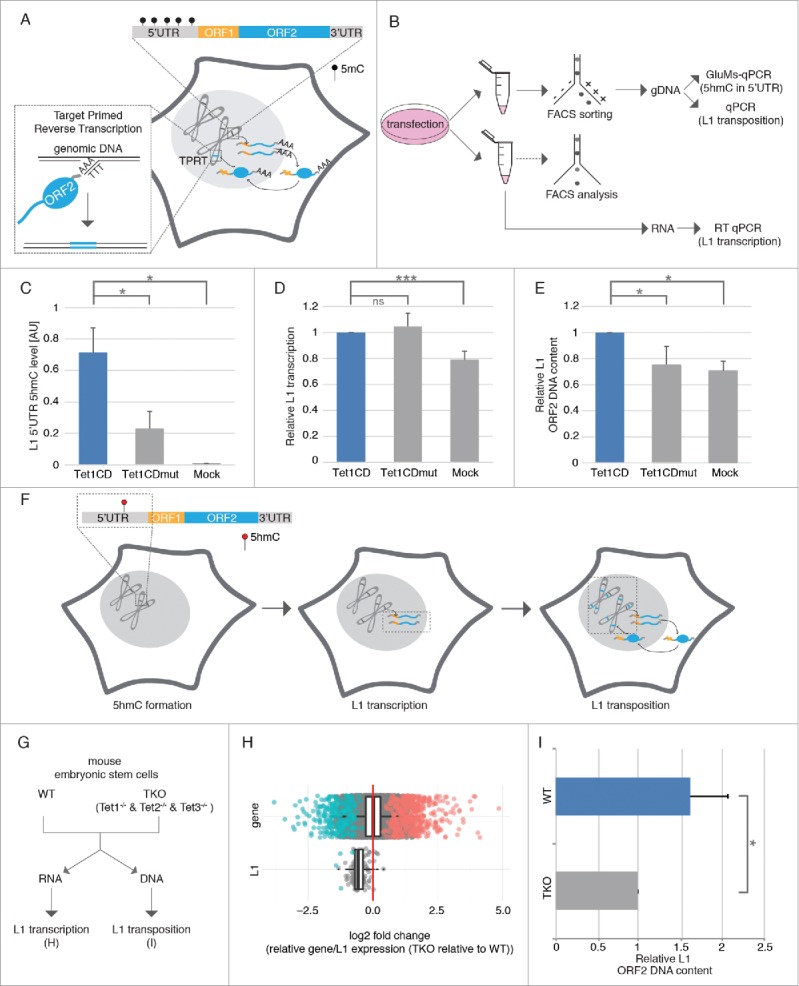
Tet1 reactivates retrotransposition of endogenous L1. (A) Schematic overview of L1 retrotransposition. (B) Experimental rationale of human L1 retrotransposition detection. (C) Relative 5hmC levels in L1 5′UTR (n = 3, *p < 0.05, independent 2-sample student's t-test), (D) relative L1 transcription levels (n ≥ 4; ns = non significant; ***p < 0.001, independent 2-sample student's t-test) and (E) relative L1 ORF2 DNA content was checked 48 hours after Tet1CD, Tet1CDmut and mock transfection (n = 4, *p < 0.05, independent 2-sample student's t-test). (F) Scheme summarizing the effect of Tet1 on the 3 steps of L1 retrotransposition. Black and red circles indicate 5mC and 5hmC nucleotides, respectively. Bars represent the mean + standard deviation (SD). (G) Experimental rationale of mouse L1 retrotransposition detection. (H) Boxplot of the log2-fold changes of the triple Tet-knockout mouse embryonic stem cells relative to wild-type (V6.5) for all genes and all L1 elements. Negative values indicate a downregulation in the knockout relative to the wild-type, positive values an upregulation. Significant elements are marked in color. The red line is at zero, i.e., the expected value if expression were identical in the wild type and mutant. (I) Relative mouse L1 ORF2 content. Bar represent mean+ SD. (n = 3, *p < 0.05, independent 2-sample student's t-test).
Consequently, we aimed to determine, whether Tet1-mediated 5hmC formation of the L1 promoter leads to reactivation of L1 transcription. Since RNA transcript levels might be affected by several exogenous factors during cell sorting,32 cells cotransfected with mcherry-Tet1CD and EGFP, as well as mcherry-Tet1CDmut and EGFP were analyzed by flow cytometry and only cell populations with comparable expression levels between samples were chosen for RNA preparation (Fig S1). Reverse transcription quantitative real-time PCR (RT qPCR) showed that besides Tet1CD, ectopic expression of Tet1CDmut leads to increased L1 transcription when compared with mock transfected cells (Fig 1D), indicating that L1 transcription does not depend on the catalytic activity of Tet1 proteins. Previous studies showed that besides DNA methylation, histone acetylation33 and chromatin structure34 affect L1 retrotransposition. In that regard, we found chromatin decondensation caused by Tet proteins in an oxygenase-independent manner (Fig S2), which might contribute to the reactivation of L1 transcription.
Using high content imaging, we next tested, whether the observed increase in L1 transcription results in elevated L1 ORF1p levels. Therefore, HEK cells ectopically expressing mcherry-Tet1CD, mcherry-Tet1CDmut or mcherry were fixed and immunostained for L1 ORF1p. While the catalytically active Tet1 variant increased mean protein levels of L1 ORF1 by ∼60%, ORF1p levels were elevated by ∼20% in Tet1CDmut expressing cells (Fig S3). Accordingly, L1 ORF1p formation requires, at least in part, the catalytic activity of Tet proteins.
Based on the knowledge that L1 elements mobilize via an RNA intermediate using a “copy and paste” mechanism and our observation that Tet1 proteins increase L1 transcription, we next analyzed L1 copy numbers in genomic DNA (gDNA) of Tet1CD and Tet1CDmut overexpressing cells, respectively. Since most of the newly inserted L1 elements are 5′ truncated, we used primers specific for the 5′UTR as a constant normalization control and ORF2 primers as an indication of newly inserted L1 elements to detect de novo transposition events. Compared to Tet1CDmut and mock transfected cells, genomic L1 ORF2 content was increased upon Tet1CD overexpression (Fig 1E), indicating that L1 transposition depends on the oxygenase activity of Tet1. As we observe that catalytically active and inactive Tet1 proteins generate similar L1 mRNA levels, but have different L1 transposition efficiencies, we suggest that the Tet 1 catalytic activity may enhance L1 transposition through a yet unknown mechanism. We found that the global Tet1-mediated 5hmC increase leads to accumulation of γH2AX (Fig S4). As L1 retrotransposition has been shown to be proportional to the number of γH2AX foci,35 we suggest that Tet1 induced formation of γH2AX might enhance L1 integration. Since, on the other hand, the catalytically inactive Tet1 does not produce 5hmC and, accordingly, γH2AX was not increased (Fig S4), L1 transposition was thus not elevated. In summary, our data demonstrate that Tet1 proteins induce loss of L1 promoter methylation and further activate L1 transcription and transposition in human HEK cells (Fig 1F).
As ESCs depleted of Tet1 and Tet2 showed loss of 5hmC in the 5′ region of L1,23 we further tested, whether loss of 5hmC affects L1 transcription and transposition. To this end, we performed RNA-seq analysis and determined L1 copy numbers using RNA and DNA from Tet1/Tet2/Tet3 triple knockout ESCs (Tet-TKO), as well as the corresponding wild type cells (v6.5 wt) (Fig 1G). Although global gene expression did not change, L1 expression was significantly reduced in Tet-TKO cells (Fig 1H), indicating that Tet proteins regulate L1 transcription. In agreement with this, L1 copy numbers were decreased in Tet-TKO cells, indicating that Tet proteins modulate L1 transposition.
The activity of L1 is tissue and cell type dependent.8 Low and high activity of L1 retrotransposition was observed in fibroblasts and neural stem cells, respectively, indicating that host cells adopt a protection mechanism to prevent L1 retrotransposition. To test whether Tet proteins could activate L1 in both cells types, we analyzed L1 copy number in cells expressing either mcherry-Tet1CD or mcherry. As shown in Figure S5, genomic L1 ORF2 content was increased in mouse neural stem cells, however, it was not changed in both mouse and human fibroblasts upon Tet1 expression. These results indicate that although Tet proteins can active L1 retrotransposition, cell type dependent protection mechanisms also play a role in preventing L1 activity.
Tet1 activates retrotransposition of engineered L1
To further validate Tet1 mediated L1 activation, we made use of a cell culture based retrotransposition assay, where the plasmid pLRE3-EGFP9 was episomally present in HEK-EBNA cells. The L1 cassette of the pLRE3-EGFP construct contains a full-length human L1 element, with a sense oriented γ-globin intron, which interrupts the antisense EGFP cassette in its 3′UTR region. Therefore, EGFP positive cells arise only when the EGFP is transcribed from the L1 promoter, spliced, reverse transcribed and integrated into the genome9 (Fig S6A). Self-replicating, viral-based vectors are capable of long-term episomal persistence in mammalian cells, in particular in HEK-EBNA cells. Two components are needed for episomal maintenance, the latent origin of replication (oriP) present in the vector and the transactivator protein EBNA-1 stably expressed in HEK-EBNA transgenic cells.36, 37 The episomal plasmid contains a puromycin resistance gene to facilitate selection of cells containing the episome. HEK-EBNA cells were transfected with the pLRE3-EGFP construct and treated with 2 µg/mL puromycin 2 d later. From 3 d after transfection, EGFP expression was observed in a small number of cells, indicating that HEK-EBNA cells are suitable to detect engineered L1 retrotransposition from the episome into the genome. Since we observed only a small number of EGFP expressing cells, i.e., where transposition took place, we wanted to test whether the EGFP negative cells still contained the episomal plasmid pLRE3-EGFP. To this end, PCR was performed on whole cell lysates of sorted, EGFP negative cells using primers for EGFP, which amplify both, spliced (genomically integrated copy) and unspliced (episomal copy) EGFP versions. As shown in Figure S7, only unspliced EGFP was amplified from EGFP negative cells, indicating that pLRE3-EGFP is present in these cells. A spliced, genomically integrated EGFP cassette (CMV promoter+EGFP), however, could not be detected indicating that no retrotransposition took place in these cells. After 15 d of antibiotic selection, puromycin-resistant colonies containing few EGFP positive cells formed (Fig S6B). Antibiotic resistance of a large number of EGFP negative cells, however, indicated that the pLRE3-EGFP was episomally present, but its ability to retrotranspose was most likely silenced by DNA methylation (Fig S6B).
To test whether Tet1 can activate L1 transposition from the silenced episome in these cells, we transfected the reporter cell line with mcherry-Tet1CD, mcherry-Tet1CDmut or mcherry, fixed them 24 and 48 hours post transfection and further quantified the EGFP positive cells by high content screening microscopy. The ratio of EGFP-positive cells in Tet1/mcherry-positive and -negative cells was used to show L1 retrotransposition events induced by Tet activity (Fig 2A). We observed an increase in the number of EGFP-positive cells upon overexpression of Tet1CD, compared with the overexpression of Tet1CDmut and mcherry alone at 48 hours (Fig 2C). 24 hours after transfection no increase of EGFP positive cells was detected in the presence of Tet1CD (Fig 2B). Consistent with the reporter assay using fixed cells, live-cell time lapse imaging analysis showed a large increase in the amount of EGFP positive cells after 40 hours with Tet1CD transfection, but not with Tet1CDmut and mcherry (Fig S8A, videos 1–3). This fits well the previously reported L1 de novo retrotransposition kinetics.38 To further validate the increase of L1 retrotransposition induced by Tet1CD, we used flow cytometry analysis. The number of mcherry positive (+) and negative (−), as well as EGFP positive (+) cells was counted by flow cytometry and the ratio between EGFP+/mcherry+ and EGFP+/mcherry- cells was calculated to show recent L1 integrations. The results showed an increase of EGFP positive cells in the presence of Tet1CD 48 hours after transfection (Fig S8B). These data indicate that the L1 promoter of the episomal plasmid was activated by Tet1CD ultimately leading to transposition into the host cell's genome and expression of the EGFP gene.
Figure 2.
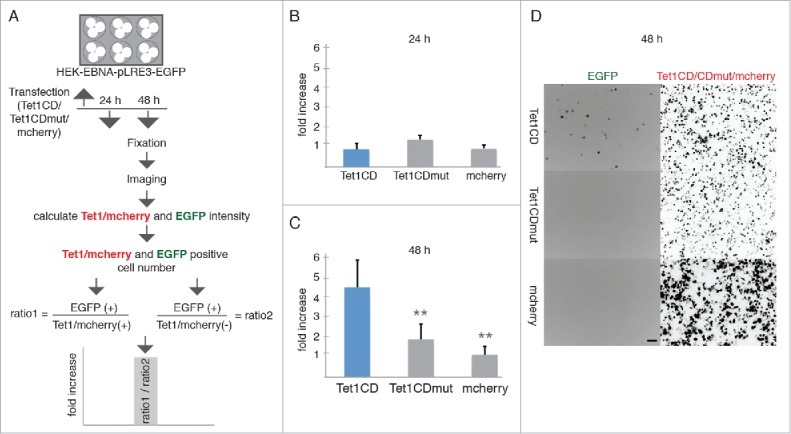
Tet1 reactivates retrotransposition of engineered L1. (A) Experimental rationale. The ratio of EGFP-positive cells in Tet1/mcherry-positive and negative cells, was quantified to detect recent L1 retrotransposition events. (B-C) Relative increases of retrotransposition events (B) 24 hours and (C) 48 hours after Tet1CD, Tet1CDmut and mcherry transfection. At least 3 independent experiments were performed and more than 250,000 cells for each group were analyzed and the bar represents the mean + SD. Independent 2-sample student's t-test was performed between Tet1CD and Tet1CDmut or Tet1CD and mcherry transfected cells. Only significant differences were indicated on the plots as 2 asterisks (p < 0.005). (D) Representative images of the pLRE3-EGFP reporter cell line 48 hours after mcherry-Tet1CD/CDmut and mcherry transfection, respectively. Scale bar: 100 µm.
To further verify that the activation of EGFP resulted from L1 retrotransposition and not from silenced genomically integrated copies, we cultured the reporter cell line in the absence of puromycin. Four and 11 d later, the cells were transfected with mcherry-Tet1CD plasmids and 2 d after transfection the EGFP positive cell numbers were counted using flow cytometry. As shown in Figure S9, EGFP positive cell numbers were decreased in the absence of puromycin. Previous studies showed that the episomal plasmids are easily lost during cell generations.39 In the absence of puromycin, the cells without pLRE3-EGFP plasmid are able to survive, but due to the lack of the episome no transposition can take place and, hence, those cells do not express EGFP. These results indicate that the activation of EGFP expression upon Tet expression results from L1 retrotransposition from the episome into the genome rather than from genomically integrated EGFP cassette, since the latter would be activated by Tet1CD in the absence or presence of puromycin.
Increase of L1 retrotransposition and the conversion of 5mC to 5hmC induced by Tet1 prompted us to test, whether the observed L1 reactivation arose from DNA methylation changes, so we made use of the cytidine analog 5-azacytidine (5-aza-C) to induce DNA hypomethylation.40 The stable cell line was treated with either 5 µM or 50 µM of 5-aza-C and 48 hours later the number of EGFP positive cells was quantified by flow cytometry analysis. Compared to untreated cells, the 5-aza-C treatment increased the number of EGFP positive cells in a dose dependent manner (Fig S8C), indicating that decreased DNA methylation is involved in the activation of L1. This effect is in line with results from non-transformed cells that were treated with 5-aza-C.41 Since 5mC oxidation products such as 5hmC are considered to be intermediates of DNA demethylation, we suggest that the activation of L1 by Tet1 is mainly dependent on its DNA demethylation activity potentially involving subsequent repair processes.
Mecp2 and Mbd2 repress endogenous human L1 retrotransposition
As Mecp2 has been shown to regulate L1 retrotransposition in a methylation dependent manner,19,20 we wanted to test whether Tet1 mediated activation of L1 can be counteracted by MBD proteins. To this end, we analyzed the effects of co-expressed Tet1CD, Tet1CDmut, Mecp2 and Tet1CD+Mecp2 on transcription of a luciferase reporter plasmid driven by the internal L1.3 promoter (nucleotides 1–909) in response to its methylation by HpaII methyltransferase20,42 (Fig S10A and S10B). While in the absence of any effector protein (Tet1CD/Tet1CDmut), methylation of the L1.3 promoter led to a weak transcriptional decrease, overexpression of Tet1CD/Tet1CDmut resulted in similar transcription rates for the methylated and unmethylated promoter (Fig S10C). Ectopic expression of Mecp2, in contrast, reduced transcription from the methylated promoter in the absence and presence of Tet1CD (Fig S10C), indicating that Mecp2 represses Tet1 mediated activation of L1 transcription.
Next, we co-expressed Tet1 with Mbd2, Mecp2, or its subdomains MBD and IDTRD, respectively in HEK-EBNA cells and analyzed 5hmC levels at the endogenous L1 promoter by GluMS-qPCR. The results showed decreased 5hmC levels in the 5′UTR of L1 upon co-overexpression of MBDs together with Tet1CD (Fig 3A), indicating that MBDs block Tet mediated 5mC to 5hmC conversion in the 5′UTR of L1. The failure of Tet binding to DNA in the presence of Mbd2, Mecp2,43 as well as its subdomains MBD and IDTRD30 might thus be causative for the observed decrease of 5hmC at the L1 5′UTR. Previous studies showed activation of L1 retrotransposition in the absence of Mecp2,19 and our data indicate that this activation might be due to Tet induced 5hmC formation. In accordance with the decreased 5hmC levels, L1 mRNA levels and copy numbers were lower in cells co-expressing Tet1CD and MBDs (Fig 3B and Fig 3C), indicating that MBDs can prevent L1 transcription and transposition. In summary, the activation of L1 by Tet1 can be prevented by the action of MBD proteins.
Figure 3.
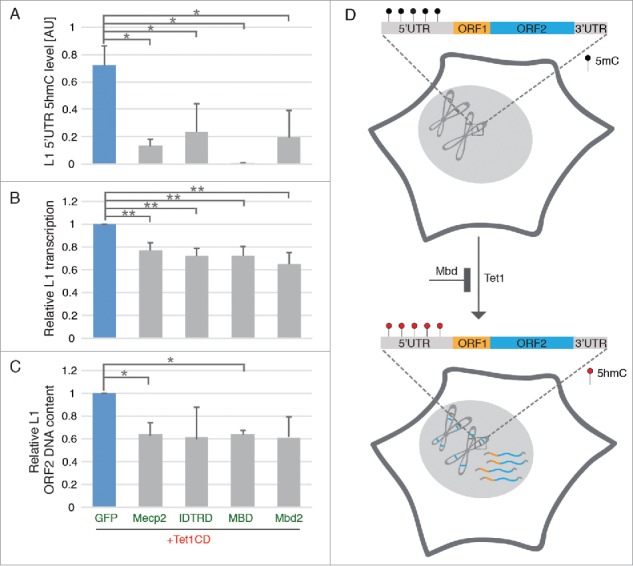
MBD proteins prevent Tet-mediated reactivation of endogenous L1. (A) Relative 5hmC in L1 5′UTR (n = 3, *p < 0.05, independent 2-sample student's t-test), (B) relative L1 transcription levels (n = 4; **p < 0.01, independent 2-sample student's t-test) and (C) relative L1 ORF2 DNA content were checked 48 hours after cotransfection with plasmids coding for Tet1CD- and MBD proteins (n = 3, *p < 0.05, independent 2-sample student's t-test). All of the 3 independent experiments for IDTRD and Mbd2 showed decreased, but variable L1 copy number when compared with Tet1CD, giving rise to an apparent non-significant difference. (D) Scheme illustrating the effect of Tet1- and MBD proteins on L1 retrotransposition. Black and red circles indicate 5mC and 5hmC nucleotides, respectively. Bars represent the mean + SD.
Here, we show that Tet1 mediated formation of 5hmC activates the L1 retrotransposition. Previous studies have shown that the activation of endogenous L1 transcription does not depend on Tet3 in mouse zygotes,44 but the potential effect of Tet3 on L1 transposition is still unknown. To further check the effect of Tets on L1 transposition, we took advantage of Tet1, Tet2 and Tet3 triple knockout ESCs, which do not express any Tets but their corresponding wild type cells express high levels of Tets, especially Tet1 and Tet2. We showed that the transcription and transposition of mouse L1 is significantly decreased in Tet1, Tet2 and Tet3 triple knockout mouse ESCs as compared with the corresponding wild type cells, indicating that Tets are involved in L1 retrotransposition activation. Since 5hmC is not only involved In loss of DNA methylation, but also a stable epigenetic mark in mouse embryonic stem cells, we propose that both, loss of DNA methylation and the 5hmC mark itself are involved in L1 retrotransposition activation.
Rett syndrome (RTT), a postnatal occurring neurologic disorder with an incidence of ∼1 in 10,000 female births,45 is mostly caused by mutations in the MBD and IDTRD of Mecp2.46,47 The increased L1 retrotransposition events in RTT patients suggest that L1 activity is facilitated upon loss of Mecp2 function in human cells,19 but the mechanism is still unknown. Here, we show that L1 retrotransposition is activated by Tet1 and repressed by MBD proteins. In the absence of Mecp2, Tet proteins oxidize methylated DNA, which is usually bound by Mecp2, leading to L1 element activation. Although previous studies showed that Mecp2 binds to 5hmC enriched within active genes,48 we found, on the other hand, that the MBD of Mecp2 does not bind to 5hmC.49 Therefore, our data are not consistent with sustained inhibition of L1 activation through binding of Mecp2 to 5hmC. However, Mecp2 binds with high affinity to DNA and this could explain the results of Mellen et al.48
Previous studies showed that transgenic mice harboring either DNA binding-incompetent MBD or TRD of Mecp2 is sufficient to cause RTT.47 Here, we show that MBD as well as the IDTRD of Mecp2 are also each sufficient to mediate repression of L1 mobilization. In addition to Mecp2, we show that Mbd2 can also repress human L1 retrotransposition, indicating that the repression of L1 is not Mecp2 specific. Besides 5mC specific binding domains, such as the MBD of Mecp2, the non sequence specific DNA binding domain IDTRD of Mecp2 can also repress the L1 retrotransposition. When compared with in vitro methylated L1 5′UTR using HpaII methyltransferase,20 human endogenous L1 5′UTR41 might provide more binding sites to Mbd2. Thus, 4 methylated CpG sites seem to be not enough for the repression of L1 retrotransposition by Mbd2, but it is enough for Mecp2 because of its sequence unspecific strong DNA binding ability.50
In summary, we show that human L1 can be re-activated by Tet1 proteins and this might lead to decreased genome stability15 and activation of proto-oncogenes in cancer.51 Finally, our data indicates that the Tet1 mediated activation of L1 can be repressed by Mecp2 and Mbd2 revealing a role of Mecp2 and Mbd2 as guardians of genome stability by preventing retrotransposition.
Materials and methods
Plasmids, cell culture and transfection
Plasmids coding EGFP tagged MBDs21,52-54 (Mecp2: pc1121, MBD: pc0841, IDTRD: pc1852, Mbd2: pc2399) and mcherry-tagged catalytic active30 (mcherry-Tet1CD: aa 1367–2007, pc2547) and inactive30 (mcherry-Tet1CDmut: aa 1367–2007, H1652Y, D1654A, pc2815) domain of mouse Tet1 were described in previous publications.
The reporter plasmid pGL3-L1.3-Luc20 (pc3342) coding for a firefly luciferase under the control of the L1.3 promoter was a generous gift of G. Schumann (Paul Ehrlich Institute, Langen, Germany).
HEK-EBNA cell line was purchased from Invitrogen (catalog #R620–07). Cells were cultured and transfected as described previously.46 For genomic DNA (gDNA) extraction, HEK-EBNA cells were transfected with mcherry-Tet1CD/CDmut and EGFP-MBDs and flow cytometry sorted (Biorad S3 sorter, Bio-Rad Laboratories) according to EGFP and mcherry expression levels. 488 and 561 nm excitation lasers and 525 ± 30 and 586 ± 25 nm emission filters are used for EGFP and cherry detection. For RNA preparation, HEK-EBNA cells were transfected with mcherry-Tet1CD/CDmut and EGFP-MBDs encoding plasmids, and the expression of mcherry and EGFP was analyzed by flow cytometry. Cells with similar mcherry-Tet1CD/CDmut expression were used for RNA preparation.
C2C12 mouse myoblast cell line55 were cultured using the conditions described previously.56
C2C12 cells were grown to 70% confluency on glass coverslips and transfected with EGFP-Tet1CD/CDmut and EGFP expression constructs 24 hours post seeding using Lipofectamine (Life Technologies) according to the manufacturer's instructions.
Human AG01522D foreskin fibroblasts cells (obtained from Coriell Cell Repository) were cultured in DMEM medium supplemented with 15% FCS. The cells were transfected by electroporation as described previously.57
Mouse tail fibroblasts (MTF) cells were a gift from A. Bird (Wellcome Trust Center for Cell Biology, Edinburgh, UK) and were cultured using the conditions as described previously.58
Mouse neural stem cells were a gift from B. Hendrich (Cambridge Stem Cell Institute, Cambridge, UK) and cultured using the conditions as described previously.59
Human fibroblast cells (BJ-hTERT; ATCC CRL-4001)60 were cultured in DMEM medium supplemented with 15% FCS. The cells were transfected by electroporation as described previously.57
V6.5 wt and Tet-TKO mouse embryonic stem cells61 were a gift from R. Jaenisch (Whitehead Institute for Biomedical Research, Cambridge, USA) and were maintained under serum-free and feeder-free conditions on Geltrex-coated flasks in N2B27 (50% neurobasal medium (Life Technologies) and 50% DMEM/F12 (Life Technologies) containing 2 mM L-glutamine (Life Technologies), 0.1 mM β-mercaptoethanol, N2 supplement (Life Technologies), B27 serum-free supplement (Life Technologies), 100 U/ml Penicillin-Streptomycin, 1000 U/ml LIF and 2i (1 μM PD032591 and 3 μM CHIR99021 (Axon Medchem).
L1 retrotransposition reporter assay
HEK-EBNA cells were transfected with the pLRE3-EGFP plasmid9 (gift from J. V. Moran, U. Michigan Medical School, USA, pc3341). Two days after transfection, cells were cultured in DMEM medium containing 2 µg/mL puromycin (Invitrogen) for resistance selection. 18 d later, single colonies were picked and passaged in DMEM medium containing 2 µg/mL puromycin. For retrotransposition reporter assays, cells were cultured in DMEM medium without puromycin selection.
For fixed-cell analysis, the reporter cell line was grown on glass coverslips and transfected with mcherry-Tet1CD/CDmut and only mcherry using PEI. 24 hours and 48 hours after transfection, cells were fixed with 3.7% formaldehyde (Carl Roth GmbH), DNA was counterstained with DAPI (Invitrogen) and cells were mounted in Vectashield antifade medium (Vector Laboratories, Burlingame, CA, USA). DAPI, EGFP and mcherry were imaged by high content microscopy with 20x long/0.45 NA objective (Operetta), a xenon fiber optic as light source, 360–400, 460–490 and 560–580 nm excitation- and 410–480, 500–550 and 590–640 emission filters, respectively. And the intensities were analyzed using the Harmony software (PerkinElmer, UK).
For flow cytometry analysis of the aforementioned reporter cell line, cells either ectopically expressing mcherry-Tet1CD/CDmut or mcherry or treated with 5-azacytidine (5-aza-C, Sigma-Aldrich) were grown in 6-well plates. 48 hours later, cells were harvested and EGFP positive cells were counted by flow cytometry.
RNA-Seq library preparation and data analysis
Total RNA was isolated from mouse embryonic stem cells (V6.5) in biological quadruplicates using the nucleospin triprep kit from Macherey-Nagel. 50 ng RNA was reverse transcribed. cDNA was pre-amplified as described elsewhere.62 1 ng of cDNA was used as input for tagmentation by the Nextera XT Sample Preparation Kit (Illumina), where a second amplification round was performed for 12 cycles. For each sample, 2.5 ng of final library was pooled. The library pool was sequenced 1 × 100 bases on a Illumina HiSeq1500. The average sequencing depth was 1.2 million reads per replicate.
Sequencing reads were demultiplexed from the Nextera (i5 and i7) indices. Demultiplexed reads were mapped to the Mouse genome build mm10 using STAR version STAR 2.5.163 with the specific settings: –outFilterMultimapNmax 100 –outFilterMismatchNmax 4 –winAnchorMultimapNmax → 100. The junction annotation was taken from ensembl GRCm38.75 and the index was created as recommended using the option –sjdbOverhang → 99. The resulting bam-files were then processed using TEtranscript64 to obtain read count tables for transcripts and transposons, using the TE annotation as provided by the authors of TEtranscript (http://labshare.cshl.edu/shares/mhammelllab/www-data/TEToolkit/TE_GTF/mm10_rmsk_TE.gtf.gz). Normalization and differential expression analysis was done using DESeq2.65
Quantitative PCR (q-PCR)
q-PCR was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems) with Platinum SYBR Green qPCR SuperMix-UDG w/ROX (Invitrogen) according to the manufacturer's instruction. The program used for the amplification of all fragments consisted of 1) inactivation of UDG for 2 min at 50°C, 2) denaturation of DNA for 10 min at 98°C, 3) 40 cycles of PCR (98°C for 15 sec, 60°C for 1 min), followed by 4) dissociation (melting) curve analysis to confirm the specificity of the amplicon.
DNA glucosylation, MspI digestion and quantitative PCR based 5hmC and 5mC detection (GluMs-qPCR)
To detect 5hmC after Tet1 transfection, gDNA was extracted from HEK-EBNA cells as described previously.66 Concentration and purity of DNA was measured on a TECAN infinite M200 plate reader (Tecan Group Ltd.) by the absorbance at 260 nm and 280 nm. 1 µg of gDNA was treated with or without 0.18 µM of T4 phage β-glucosyltransferase (T4-BGT)67 in a final volume of 50 µl supplemented with 1x NEB cut smart buffer (NEB) and 1 mM of UDP-Glucose (Sigma-Aldrich) for 18 hours at 37°C. Then 0.5 µg of glucosylated or mock treated DNA was used for digestion with 100 units of MspI (NEB) at 37°C for 18 hours in a final volume of 20 µL, which was followed by treatment with 20 µg of proteinase K (PK, Carl Roth GmbH) for 30 min at 50°C. Following proteolysis, PK enzymatic activity was inactivated for 10 min at 98°C. The MspI-resistant fraction was amplified using qPCR with primers flanking the MspI site (F: 5'- ATCCCACACCTGGCTCAGAGGG -3′ and R: 5′- GTCAGGGGTCAGGGACCCACTT -3′). After qPCR, the relative amounts of 5hmC were analyzed as described previously.31 To detect 5mC at position 482 in L1 5′UTR before Tet1 transfection, the gDNA was treated with or without T4-BGT as described above. Then, the gDNA was further treated with MspI, HpaII (50 units, NEB) or mock for 18 hours at 37°C. The qPCR and data analysis were performed as above.
cDNA preparation and reverse transcription quantitative real-time PCR (RT qPCR)
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instruction. To remove traces of genomic DNA, RNA was treated with RNase-free recombinant DNaseI (Macherey Nagel) for 30 min at 37°C and further purified with the Qiagen RNeasy Mini Kit. To assess the concentration and purity of RNA, the ratio of absorbance at 260 nm and 280 nm was measured on a TECAN infinite M200 plate reader. 500 ng of total RNA were used for cDNA synthesis using 200 units M-MuLV reverse transcriptase (NEB), 0.01 OD units random primer from the Prime-It II Random Primer Labeling Kit (Stratagene), 0.5 mM dNTPs (Carl Roth GmbH) and 40 units recombinant ribonuclease inhibitor RNaseOUT (Invitrogen) in a total reaction volume of 20 µL. Cycles were set to 5 min at 25°C, 90 min at 50°C and 15 min at 70°C. For qPCR, 0.5 ng of cDNA were used for each reaction. Primers for quantitative real-time PCR contained the following sequences: Gapdh F: 5′- CAT GAG AAG TAT GAC AAC AGC CT-3′, Gapdh R: 5′-AGT CCT TCC ACG ATA CCA AAG T-3′,68 hL1 5′ UTR F: 5′-GAA TGA TTT TGA CGA GCT GAG AGA A-3′, hL1 5′ UTR R: 5′-GTC CTC CCG TAG CTC AGA GTA ATT -3′.9 5′ UTR expression level was normalized to Gapdh and calculated using the comparative CT method (ΔΔCT method).69
L1 copy number analysis
To detect newly integrated L1 ORF2 sequences in human embryonic kidney or human fibroblast cells, qPCR was performed as described above. For each reaction, 80 pg of gDNA were used. Primers contained the following sequences: hL1 ORF2 F: 5′- CAAACACCGCATATTCTCACTCA-3′, hL1 ORF2 R: 5′-CTTCCTGTGTCCATGTGATCTCA-3′.9 Computational estimates using the UCSC genome browser in silico PCR function (genome: human, assembly: December 2013 (GRCh38/hg38), target: genome assembly) indicated that at least 2734 endogenous human L1 elements could be detected using the hL1 ORF2 primer set.
As control we used primers for hL1 5′ UTR. This sequence is usually truncated during the course of retrotransposition. Accordingly, the number of hL1 5′ UTR sequences remains constant. Primers contained following sequences: hL1 5′ UTR F: 5′-ACAGCTTTGAAGAGAGCAGTGGTT-3′, hL1 5′ UTR R: 5′-AGTCTGCCCGTTCTCAGATCT-3′.9
To determine the content of L1 ORF2 sequences in mouse embryonic stem cells (wild type and Tet1/Tet2/Tet3 TKO) and mouse fibroblast cells ectopically expressing mcherry or mcherry-Tet1CD, gDNA was isolated and qPCR was performed as described above. For each reaction, 80 pg of gDNA were used. Primers contained the following sequences: mL1 ORF2 F: 5′-CTGGCGAGGATGTGGAGAA-3′, mL1 ORF2 R: 5′-CCTGCAATCCCACCAACAAT-3′.19 Computational estimates using the UCSC genome browser in silico PCR function (genome: mouse, assembly: December 2011 (GRCm38/mm10), target: genome assembly) indicated that at least 1308 endogenous mouse L1 elements could be detected using the mL1 ORF2 primer set.
As control we used non-mobile genomic repetitive sequences: 5SRNA F: 5′-ACGGCCATACCACCCTGAA-3′; 5SRNA R: 5′-GGTCTCCCATCCAAGTACTAACC-3′.19
To calculate the relative copy number of genomic L1 ORF2, the comparative CT method (ΔΔCT method) was used. For HEK cells, as well as for mouse and human fibroblast cells, relative ORF2 content was further normalized Tet1CD expressing cells. For mouse ESCs, relative ORF2 content of wild type cells was further normalized to the mean relative ORF2 content of Tet-TKO cells.
Chromatin decondensation analysis
3D structured illumination microscopy images were acquired as described previously.70 To quantify the grade of chromatin decondensation, binary nuclear masks were generated. Therefore, images were processed using a 3D median filter. Filtered images of the DAPI channel were then thresholded using the basic algorithm. Next, all DAPI pixels below the threshold were set to 0 and all pixels above the threshold were set to 1. For further improvement of the nuclear masks, binary images were additionally processed using the “fill holes” and “watershed” algorithms. Finally, the standard deviation of all DAPI histograms was calculated automatically. To automate this procedure, a routine was written in the programming language python.
Immunofluorescence staining
Human AG522D fibroblasts cells were fixed for 10 minutes in 4% formaldehyde in PBS and permeabilized for 20 minutes with 0.5% Triton X-100. For detection of genomic 5hmC and γH2AX, the cells were further fixed with ice-cold methanol for 5 minutes. After RNaseA treatment (10 µg/mL) for 30 minutes at 37°C, cells were washed and blocked for 60 minutes in 0.2% fish skin gelatin (Sigma Aldrich) at 37°C. Then 5hmC and γH2AX were detected using a rabbit anti-5hmC (1:250, catalog number: 39769, Active Motif) and a mouse Anti-phospho-Histone H2A.X (1:400, catalog number: 05–636, Merck), antibody in conjunction with 25 U/mL DNaseI (Sigma Aldrich) for 70 minutes at 37°C. To stop DNaseI digestion, cells were washed 3 time with PBS containing 1mM EDTA and 0.01% Tween. Following incubation with secondary antibody mixture of AMCA donkey anti-rabbit IgG (1:100, catalog number: 715–155–151, The Jackson Laboratory) and Alexa Fluor 488 goat anti-mouse IgG (1:250, catalog number: A11029, Invitrogen) for 50 minutes at RT. After 3 time washing with PBS containing 0.01% Tween the cells were mounted in Vectashield Medium (Vector Labs) and imaged with high content screening microscopy with 20x long/0.45 NA objective (Operetta). The intensities were further calculated using the Harmony software (Operetta) and blotted by RStudio (https://www.rstudio.com).
L1 ORF1p proteins were detected in HEK using the polyclonal rabbit anti-L1 ORF1p antibody71 as described before.72 As secondary antibody the AMCA donkey anti-rabbit IgG was used (1:100, catalog number: 715–155–151, The Jackson Laboratory).
Luciferase reporter assay
Methylated pGL3-L1.3-Luc reporter plasmids were obtained by incubation with M.HpaII and controlled by digestion with HpaII and MspI. Cells were seeded in 6-well dishes at 7 × 105 cells/well. Three hours post seeding, cells were cotransfected with the unmethylated or methylated reporter plasmid pGL3-L1.3-Luc and effector constructs coding for Tet1CD/Tet1CDmut/ Mecp2/Tet1CD+Mecp2, respectively. Luciferase activity was determined using the “Luciferase Assay System” (Promega) as described by the manufacturer on a TECAN infinite M200 plate reader (Tecan Group Ltd.). To control for consistent transfection of methylated and unmethylated reporter plasmids, the fluorescent signal emanating from the effector proteins was quantified in parallel (GFP: excitation 475 nm, emission 520 nm; RFP: excitation 585 nm, emission 630 nm) and was used for normalization of the luciferase signal. In addition, fluorescent signals were used to control for homogeneous expression of Tet1CD in single (Tet1CD) and double (Tet1CD + Mecp2) transfected cells.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the all above-mentioned scientists for the generous gift of biologic reagents. We are grateful to Corella S. Casas-Delucchi for advice in image analysis and Andreas Maiser for structured illumination microscopy.
Funding
PZ received a fellowship of the China Scholarship Council. This work was supported in part by grants of the Deutsche Forschungsgemeinschaft (DFG CA 198/7 and DFG CA 198/10) to MCC.
Author contributions
PZ, AKL, FDH, CR, MS and AL performed experiments. PZ, AKL, FDH, CR and IH analyzed the data. PZ, AKL, HL and MCC conceived and designed the experiment and PZ, AKL and MCC wrote the manuscript.
References
- [1].Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al.. Initial sequencing and analysis of the human genome. Nature 2001; 409:860-921; PMID:11237011; https://doi.org/ 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- [2].Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol 2001; 21:467-75; PMID:11134335; https://doi.org/ 10.1128/MCB.21.2.467-475.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kazazian HH Jr., Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet 1998; 19:19-24; PMID:9590283; https://doi.org/ 10.1038/ng0598-19 [DOI] [PubMed] [Google Scholar]
- [4].Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MC, Diedrich JK, Aslanian A, Ma J, Moresco JJ, et al.. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell 2015; 163:583-93; PMID:26496605; https://doi.org/ 10.1016/j.cell.2015.09.025 [DOI] [PubMed] [Google Scholar]
- [5].Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J 2002; 21:5899-910; PMID:12411507; https://doi.org/ 10.1093/emboj/cdf592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet 2002; 31:159-65; PMID:12006980; https://doi.org/ 10.1038/ng898 [DOI] [PubMed] [Google Scholar]
- [7].Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MC, Muotri AR, Mu Y, Carson CT, Macia A, Moran JV, Gage FH. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc Nat Acad Sci U S A 2011; 108:20382-7; PMID:22159035; https://doi.org/ 10.1073/pnas.1100273108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 2005; 435:903-10; PMID:15959507; https://doi.org/ 10.1038/nature03663 [DOI] [PubMed] [Google Scholar]
- [9].Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature 2009; 460:1127-31; PMID:19657334; https://doi.org/ 10.1038/nature08248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, Moran JV. LINE-1 retrotransposition in human embryonic stem cells. Hum mol Genet 2007; 16:1569-77; PMID:17468180; https://doi.org/ 10.1093/hmg/ddm105 [DOI] [PubMed] [Google Scholar]
- [11].Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, Yang AS, Jones PA, Liang G. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet 2010; 6:e1000917; PMID:20421991; https://doi.org/ 10.1371/journal.pgen.1000917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Horn AV, Klawitter S, Held U, Berger A, Vasudevan AA, Bock A, Hofmann H, Hanschmann KM, Trösemeier JH, Flory E, et al.. Human LINE-1 restriction by APOBEC3C is deaminase independent and mediated by an ORF1p interaction that affects LINE reverse transcriptase activity. Nucleic Acids Res 2014; 42:396-416; PMID:24101588; https://doi.org/ 10.1093/nar/gkt898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 2013; 14:447-59; PMID:23732335; https://doi.org/ 10.1038/nrg3462 [DOI] [PubMed] [Google Scholar]
- [14].Cook PR, Jones CE, Furano AV. Phosphorylation of ORF1p is required for L1 retrotransposition. Proc Nat Acad Sci U S A 2015; 112:4298-303; PMID:25831499; https://doi.org/ 10.1073/pnas.1416869112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 2002; 110:327-38; PMID:12176320; https://doi.org/ 10.1016/S0092-8674(02)00839-5 [DOI] [PubMed] [Google Scholar]
- [16].Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 2004; 429:268-74; PMID:15152245; https://doi.org/ 10.1038/nature02536 [DOI] [PubMed] [Google Scholar]
- [17].Tubio JM, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, Gundem G, Pipinikas CP, Zamora J, Raine K, et al.. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 2014; 345:1251343; https://doi.org/ 10.1126/science.1251343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol 2006; 357:1383-93; PMID:16490214; https://doi.org/ 10.1016/j.jmb.2006.01.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature 2010; 468:443-6; PMID:21085180; https://doi.org/ 10.1038/nature09544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yu F, Zingler N, Schumann G, Stratling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res 2001; 29:4493-501; PMID:11691937; https://doi.org/ 10.1093/nar/29.21.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol 1998; 18:6538-47; PMID:9774669; https://doi.org/ 10.1128/MCB.18.11.6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013; 502:472-9; PMID:24153300; https://doi.org/ 10.1038/nature12750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 2011; 473:398-402; PMID:21460836; https://doi.org/ 10.1038/nature10008 [DOI] [PubMed] [Google Scholar]
- [24].Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009; 324:929-30; PMID:19372393; https://doi.org/ 10.1126/science.1169786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al.. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324:930-5; PMID:19372391; https://doi.org/ 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, Wang Y, Xie J, Zhang Y, Song C, et al.. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol 2014; 15:R49; PMID:24594098; https://doi.org/ 10.1186/gb-2014-15-3-r49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al.. Global epigenomic reconfiguration during mammalian brain development. Science 2013; 341:1237905; PMID:23828890; https://doi.org/ 10.1126/science.1237905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Muller U, Bauer C, Siegl M, Rottach A, Leonhardt H. TET-mediated oxidation of methylcytosine causes TDG or NEIL glycosylase dependent gene reactivation. Nucleic Acids Res 2014; 42:8592-604; PMID:24948610; https://doi.org/ 10.1093/nar/gku552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen H, Kazemier HG, de Groote ML, Ruiters MH, Xu GL, Rots MG. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res 2014; 42:1563-74; PMID:24194590; https://doi.org/ 10.1093/nar/gkt1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ludwig AK, Zhang P, Hastert FD, Meyer S, Rausch C, Herce HD, Müller U, Lehmkuhl A, Hellmann I, Trummer C, et al.. Binding of MBD proteins to DNA blocks Tet1 function thereby modulating transcriptional noise. Nucleic Acids Res 2017; 45:2438-57; PMID:27923996; https://doi.org/ 10.1093/nar/gkw1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Davis T, Vaisvila R. High sensitivity 5-hydroxymethylcytosine detection in Balb/C brain tissue. J Vis Exp 2011; https://doi.org/ 10.3791/2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nishimoto KP, Newkirk D, Hou S, Fruehauf J, Nelson EL. Fluorescence activated cell sorting (FACS) using RNAlater to minimize RNA degradation and perturbation of mRNA expression from cells involved in initial host microbe interactions. Journal of microbiological methods 2007; 70:205-8; PMID:17512621; https://doi.org/ 10.1016/j.mimet.2007.03.022 [DOI] [PubMed] [Google Scholar]
- [33].Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, Hammer GD, Collins KL, O'Shea KS, Menendez P, et al.. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 2010; 466:769-73; PMID:20686575; https://doi.org/ 10.1038/nature09209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun 2014; 5:5011; PMID:25247314; https://doi.org/ 10.1038/ncomms6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Farkash EA, Kao GD, Horman SR, Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res 2006; 34:1196-204; PMID:16507671; https://doi.org/ 10.1093/nar/gkj522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Black J, Vos JM. Establishment of an oriP/EBNA1-based episomal vector transcribing human genomic beta-globin in cultured murine fibroblasts. Gene Ther 2002; 9:1447-54; PMID:12378407; https://doi.org/ 10.1038/sj.gt.3301808 [DOI] [PubMed] [Google Scholar]
- [37].Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res 2002; 30:E9; PMID:11788735; https://doi.org/ 10.1093/nar/30.2.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH Jr.. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res 2000; 28:1418-23; PMID:10684937; https://doi.org/ 10.1093/nar/28.6.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nanbo A, Sugden A, Sugden B. The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. The EMBO journal 2007; 26:4252-62; PMID:17853891; https://doi.org/ 10.1038/sj.emboj.7601853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Schermelleh L, Spada F, Easwaran HP, Zolghadr K, Margot JB, Cardoso MC, Leonhardt H. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat Methods 2005; 2:751-6; PMID:16179921; https://doi.org/ 10.1038/nmeth794 [DOI] [PubMed] [Google Scholar]
- [41].Woodcock DM, Lawler CB, Linsenmeyer ME, Doherty JP, Warren WD. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J Biol Chem 1997; 272:7810-6; PMID:9065445; https://doi.org/ 10.1074/jbc.272.12.7810 [DOI] [PubMed] [Google Scholar]
- [42].Thayer RE, Singer MF, Fanning TG. Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1-encoded protein. Gene 1993; 133:273-7; PMID:7693554; https://doi.org/ 10.1016/0378-1119(93)90651-I [DOI] [PubMed] [Google Scholar]
- [43].Zhang P, Rausch C, Hastert FD, Boneva B, Filatova A, Patil SJ, Nuber UA, Gao Y, Zhao X, Cardoso MC Methyl-CpG binding domain protein 1 regulates localization and activity of Tet1 in a CXXC3 domain-dependent manner. Nucleic Acids Res 2017; PMID:28449087; https://doi.org/ 10.1093/nar/gkx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Inoue A, Matoba S, Zhang Y. Transcriptional activation of transposable elements in mouse zygotes is independent of Tet3-mediated 5-methylcytosine oxidation. Cell Res 2012; 22:1640-9; PMID:23184059; https://doi.org/ 10.1038/cr.2012.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 1999; 23:185-8; PMID:10508514; https://doi.org/ 10.1038/13810 [DOI] [PubMed] [Google Scholar]
- [46].Agarwal N, Hardt T, Brero A, Nowak D, Rothbauer U, Becker A, Leonhardt H, Cardoso MC. MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res 2007; 35:5402-8; PMID:17698499; https://doi.org/ 10.1093/nar/gkm599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Brown K, Selfridge J, Lagger S, Connelly J, De Sousa D, Kerr A, Webb S, Guy J, Merusi C, Koerner MV, et al.. The molecular basis of variable phenotypic severity among common missense mutations causing Rett syndrome. Hum Mol Genet 2015; 25(3):558-70; https://doi.org/ 10.1093/hmg/ddv496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 2012; 151:1417-30; PMID:23260135; https://doi.org/ 10.1016/j.cell.2012.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, Leonhardt H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PloS One 2011; 6:e21306; PMID:; PMID:21731699; https://doi.org/ 10.1371/journal.pone.0021306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hansen JC, Ghosh RP, Woodcock CL. Binding of the Rett syndrome protein, MeCP2, to methylated and unmethylated DNA and chromatin. IUBMB life 2010; 62:732-8; PMID:21031501; https://doi.org/ 10.1002/iub.386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, Boland CR, Goel A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2014; 63:635-46; PMID:23704319; https://doi.org/ 10.1136/gutjnl-2012-304219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Becker A, Allmann L, Hofstatter M, Casa V, Weber P, Lehmkuhl A, Herce HD, Cardoso MC. Direct homo- and hetero-interactions of MeCP2 and MBD2. PloS One 2013; 8:e53730; PMID:23335972; https://doi.org/ 10.1371/journal.pone.0053730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Brero A, Easwaran HP, Nowak D, Grunewald I, Cremer T, Leonhardt H, Cardoso MC. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J Cell Biol 2005; 169:733-43; PMID:15939760; https://doi.org/ 10.1083/jcb.200502062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jost KL, Rottach A, Milden M, Bertulat B, Becker A, Wolf P, Sandoval J, Petazzi P, Huertas D, Esteller M, et al.. Generation and characterization of rat and mouse monoclonal antibodies specific for MeCP2 and their use in X-inactivation studies. PloS one 2011; 6:e26499; PMID:22140431; https://doi.org/ 10.1371/journal.pone.0026499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 1977; 270:725-7; PMID:563524; https://doi.org/ 10.1038/270725a0 [DOI] [PubMed] [Google Scholar]
- [56].Cardoso MC, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell 1993; 74:979-92; PMID:8402887; https://doi.org/ 10.1016/0092-8674(93)90721-2 [DOI] [PubMed] [Google Scholar]
- [57].Becker A, Zhang P, Allmann L, Meilinger D, Bertulat B, Eck D, Hofstaetter M, Bartolomei G, Hottiger MO, Schreiber V, et al.. Poly(ADP-ribosyl)ation of Methyl CpG Binding Domain Protein 2 Regulates Chromatin Structure. J Biol Chem 2016; 291:4873-81; PMID:26772194; https://doi.org/ 10.1074/jbc.A115.698357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet 2001; 27:322-6; PMID:11242117; https://doi.org/ 10.1038/85899 [DOI] [PubMed] [Google Scholar]
- [59].Martin Caballero I, Hansen J, Leaford D, Pollard S, Hendrich BD. The methyl-CpG binding proteins Mecp2, Mbd2 and Kaiso are dispensable for mouse embryogenesis, but play a redundant function in neural differentiation. PloS One 2009; 4:e4315; PMID:19177165; https://doi.org/ 10.1371/journal.pone.0004315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science 1998; 279:349-52; PMID:9454332; https://doi.org/ 10.1126/science.279.5349.349 [DOI] [PubMed] [Google Scholar]
- [61].Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F, et al.. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell 2014; 29:102-11; PMID:24735881; https://doi.org/ 10.1016/j.devcel.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 2013; 10:1096-8; PMID:24056875; https://doi.org/ 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- [63].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29:15-21; PMID:23104886; https://doi.org/ 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Jin Y, Tam OH, Paniagua E, Hammell M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 2015; 31:3593-9; PMID:26206304; https://doi.org/ 10.1093/bioinformatics/btv422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550; PMID:25516281; https://doi.org/ 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res 1997; 25:4692-3; PMID:9358185; https://doi.org/ 10.1093/nar/25.22.4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res 2010; 38:e181; PMID:20685817; https://doi.org/ 10.1093/nar/gkq684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Clarke LA, Sousa L, Barreto C, Amaral MD. Changes in transcriptome of native nasal epithelium expressing F508del-CFTR and intersecting data from comparable studies. Respir Res 2013; 14:38; PMID:23537407; https://doi.org/ 10.1186/1465-9921-14-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8; https://doi.org/ 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- [70].Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, et al.. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 2008; 320:1332-6; PMID:18535242; https://doi.org/ 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, Löwer J, Strätling WH, Löwer R, Schumann GG. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res 2012; 40:1666-83; PMID:22053090; https://doi.org/ 10.1093/nar/gkr863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Klawitter S, Fuchs NV, Upton KR, Munoz-Lopez M, Shukla R, Wang J, Garcia-Cañadas M, Lopez-Ruiz C, Gerhardt DJ, Sebe A, et al.. Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nat Commun 2016; 7:10286; PMID:26743714; https://doi.org/ 10.1038/ncomms10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


