ABSTRACT
In vitro studies of mitochondrial transcription often use linear templates that fail to replicate key features of transcription on a circular genome. We developed a plasmid-based system for the analysis of heavy-strand promoters that recapitulates key features of native mtDNA to study topological and protein requirements of promoter activation. The heavy-strand promoters (HSP1 and HSP2) are simultaneously active on a circular template. HSP2 requires supercoiling for maximal activation. Increasing TFAM concentrations suppress HSP2 at levels that result in HSP1 stimulation. This study shows distinct modes of promoter activation, providing opportunities for the regulation of mitochondrial gene expression by promoter selection.
KEYWORDS: mitochondrial DNA, TFAM, TFB2M, transcriptional regulation
Introduction
The mitochondrial genome is strikingly efficient in its organization, encoding both mitochondrial mRNAs and the tRNAs and rRNAs used in their translation. Transcription is bidirectional with most of the mRNAs and all of the rRNAs transcribed from the heavy-strand.1 Metabolic labeling studies of human mitochondrial RNA were initially used to identify two overlapping promoters on the heavy-strand.2 The proximal promoter (HSP1) is responsible for the synthesis of the rRNAs and the distal promoter is drives transcription of heavy-strand mRNAs (HSP2). Our group and others have previously shown that the isolation of HSP2 from HSP1 is required to examine the specific requirements for HSP2 activity in vitro.3,4 The failure to utilize HSP2 in the presence of HSP1 is striking, as both promoters must be active for the expression of all genes.
mtDNA exists in many distinct physical structures within the cell and these structural differences may determine whether mtDNA is participating in transcription, replication or is quiescent.5 The activities of mitochondrial-resident topoisomerases may alter the transcription and replication of mtDNA, and a recent study showed that mitochondrial topoisomerase I (TOP1MT) acts as a negative regulator for mRNA-productive mitochondrial transcription.6 In this study, we have examined the role DNA topology in the regulation of the mitochondrial promoters using a novel template system.
Results
Mitochondrial transcription on circular templates
Prior studies of mitochondrial heavy-strand transcription used linear templates for convenient monitoring of run-off transcription. The use of a circular template overcomes two disadvantages of linear systems. First, the mitochondrial RNA polymerase (POLRMT) initiates transcription from the ends of linearized DNA non-specifically, obscuring promoter-specific interactions. Second, linear templates cannot be used to study topological requirements.
The challenge with the use of circular templates is the monitoring of transcription. To solve this, we inserted the complement of the conserved sequence box 2 (CSB2) DNA sequence downstream from the heavy-strand promoters (Fig. 1A). CSB2 terminates transcription from the light-strand promoter (LSP) in a factor-independent but orientation-specific fashion by forming a G-quadruplex.7 We reasoned that cloning the complement of this sequence following the heavy-strand promoter would allow us to observe products with sizes specific to HSP1 and HSP2 activation.
Figure 1.
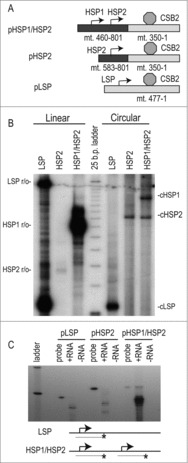
Circular templates allow activation of both heavy-strand promoters. (A) Circular heavy-strand promoter constructs were created by insertion of the CSB2 sequence in its active orientation following the heavy-strand promoters. A schematic of the six templates used in the study is shown. (B) Templates (25 nM) were transcribed with 50 nM POLRMT, TFB2M and POLRMT. The radiolabeled transcription products were isolated and resolved in the presence of a 25-base pair ladder. The expected lengths of the linear run-off products (r/o) are indicated at left and the expected lengths of the CSB-terminated products are indicated at right. (C) S1 analysis was performed on the product of unlabeled transcription reactions from the circular templates using single strand 5′ labeled DNA probes to identify the 5′ ends of RNA. Untreated full-length probe was loaded as control. +RNA lanes were treated with S1 following hybridization. –RNA lanes are a control for complete S1 digestion of unhybridized probe. For the pHSP1/HSP2 S1 reaction an HSP1-specific probe was used.
To confirm the utility of this system we compared in vitro transcription using circular and linear templates containing the mitochondrial promoters. Linear LSP-containing templates (with sequences previously described in Ref. [3]) were incubated with equimolar POLRMT, TFB2M and TFAM to produce robust transcripts which can be visualized as radiolabeled products that either terminate at CSB2 or run off the linearized DNA (Fig. 1B). In agreement with our prior work,3 linear templates containing both HSP1 and HSP2 produce a run-off transcript initiating at HSP1, but no HSP2-initiated transcript. Only after the isolation of HSP2 is this activity observed and it is considerably less robust than transcription from HSP1 or LSP.
We first tested transcription from a circular template using a plasmid (pLSP) containing LSP followed by CSB2 in its native relationship. This drove the synthesis of a product corresponding to CSB2-dependent termination, identical in length to the product from the linear template as expected. S1 analysis of the RNA validated that LSP was the point of initiation ( Fig. 1C). A circular template containing HSP2 alone followed by CSB2 in the active orientation (pHSP2) produced a band corresponding to initiation from HSP2 with factor-independent termination at CSB2. The 5′ end of this product was confirmed to be the HSP2 initiation site by the use of S1 analysis. Remarkably, a template containing both the heavy-strand promoters (pHSP1/HSP2) was capable of producing products corresponding to HSP1 and HSP2-dependent transcription. Thus, the use of a circular template allows the recapitulation of mitochondrial transcriptional activity at both heavy-strand promoters simultaneously.
The effect of supercoiling of the mitochondrial promoters
We next used circular templates to examine the relationship between a promoter's activation and its topology. Plasmids prepared from Escherichia coli are negatively supercoiled, as is the major topoisoform of mtDNA.8 The linked heavy-strand promoters were used to simultaneously evaluate HSP1 and HSP2 (Fig. 2A – left). The supercoiled template produced both HSP1 and HSP2 products. Both promoters had improved activation in the presence of 50 nM TFAM, but were active in its absence. Next the template was relaxed with topoisomerase I, which was confirmed by monitoring the template's mobility through an agarose gel (Fig. 2B). Surprisingly, there was no visible transcription from HSP2 on a template that was relaxed by treatment with topoisomerase I. Relaxation of the template with topoisomerase also increased the sensitivity of HSP1 to TFAM, but HSP1 activity was robust under all conditions.
Figure 2.
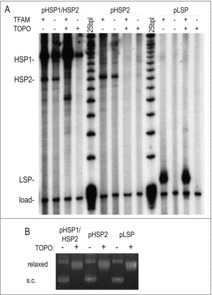
TFAM and DNA topology have distinct effects upon the different mitochondrial promoters. (A) Transcription was evaluated from circular templates with or without relaxation by topoisomerase I and in the presence or absence of TFAM. Added TFAM was equimolar (50 nM) to POLRMT and TFB2M. A loading reference oligonucleotide is shown at the bottom of the gel. (B) Topology of the plasmid templates after treatment with topoisomerase I or mock-treatment was confirmed by agarose gel electrophoresis.
We next evaluated the activity of HSP2 and LSP in isolation. We observed modest activation of isolated HSP2 by equimolar TFAM on the supercoiled template and again saw a requirement for supercoiled template (Fig. 3A – center). Transcription from LSP was entirely dependent upon the presence of TFAM and was unaffected by the topological state ( Fig. 3A – right side).
Figure 3.
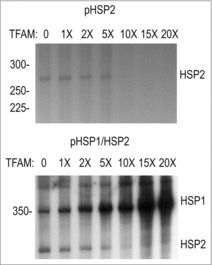
TFAM dosage restricts HSP2 activity while activating HSP1. Transcription reactions were performed using supercoiled pHSP1 (A) or pHSP1/HSP2 (B). POLRMT, TFB2M and template concentrations were held constant with increasing ratios of TFAM (as compared to POLRMT) added to the reaction.
TFAM dosage balances heavy-strand promoter selection
Previous studies by our group and others have shown that elevated ratios of TFAM to POLRMT reduce the activity of HSP2.3,4 We were interested in repeating this study using a circular template where both HSP1 and HSP were active. We first confirmed that isolated HSP2 was inhibited by the presence of high stoichiometric multiples of TFAM compared to POLRMT and TFB2M using transcription from a supercoiled, circular template ( Fig. 3). When the experiment was repeated with pHSP1/HSP2, we observed the progressive inactivation of HSP2 at higher molar ratios of TFAM. By contrast, HSP1 was activated by higher ratios. This suggests that the suppression of HSP2 by TFAM is independent of the presence or activity of HSP1 when the two promoters are in their native relationship. One important possibility is that nonspecific interactions of TFAM with the template repress HSP2 by preventing POLRMT interaction, while the specific interaction with HSP1 is enhanced at higher concentrations.
Discussion
In this study, we introduce the use of circular templates for simplified analysis of in vitro mitochondrial transcription. The templates are easy to produce, can be readily modified by site-directed mutagenesis to study point mutations and are simple to monitor. Nonspecific, competitive initiation from blunt or overhanging ends of template is eliminated. In this study, we have observed that the templates recapitulate the circular nature of mtDNA to allow the simultaneous evaluation of HSP1 and HSP2 activity that had previously been impossible using linearized templates.
We found that TFAM was not absolutely required for the transcription of either HSP1 or HSP2 on circular templates in vitro, but that both promoters were sensitive to TFAM dosage. For HSP1, activation by TFAM was apparent whether the template was supercoiled or relaxed. TFAM was not required for HSP2 activation and high levels of TFAM inhibit HSP2 activity.
In vitro systems may make it difficult to establish consensus about the activities of mitochondrial transcription factors. Conflicting conclusions have been reached about the role of TFAM. There is a good agreement on a requirement for TFAM for effective transcription from LSP.9-11 The role of TFAM at the heavy-strand promoters is more complex. One group showed TFAM-independent HSP1 transcription using a linear template with HSP1 and LSP in their native orientation with modulation by different TFAM/POLRMT ratios.10 Other studies have not identified HSP1 activation in the absence of TFAM.11 Studies of HSP2 have both shown that TFAM is not required for activation and represses HSP2 at high concentrations.3,4 It is likely that the loss of HSP2 activity with higher concentrations of TFAM is due to nonspecific interactions of TFAM with the template,12 which render it inaccessible to POLRMT.
We have also identified a novel HSP2-specific requirement with DNA supercoiling. After relaxation of the template by topoisomerase I, transcription failed to initiate. The requirement for supercoiling at HSP2 existed regardless of the presence of the HSP1 promoter. One plausible explanation for these observations is that while HSP2 lacks high-affinity binding sites for TFAM that are used to alter LSP and HSP1, POLRMT and TFB2M may still require local modification of DNA in order to initiate transcription. At HSP2, this role may be served by changes in DNA topology, rather than the TFAM-mediated DNA bending seen at LSP.13,14 The differential regulation of very nearby promoters by is unusual, although there are precedents seen in the use of ATP-dependent supercoiling of DNA as a signal for increasing transcription by prokaryotic RNA polymerases.15 Supercoiling has previously been shown to produce promoter-independent activation of transcription by POLRMT,8 but our S1 analysis of transcription demonstrates that transcription initiated at the mitochondrial promoters.
The impact of mtDNA topology on transcription was considered in a study that looked at the effects of depletion or forced expression of the mitochondria-resident class IB topoisomerase Top1mt.6 Here, the overexpression of TOP1MT (which should relax mitochondrial DNA) inhibited mitochondrial gene expression. Although the authors found that Top1mt generally antagonized mitochondrial transcription, the effect appeared less pronounced on mRNA synthesized from the light strand (ND6) when compared to mRNA synthesized from heavy-strand transcription. This finding is consistent with our observation that LSP activation in vitro is independent of supercoiling and suggests that topology may be important for regulating the expression of mitochondrial genes in the cell.
We conclude from our data that the fundamental requirements of the mitochondrial promoters are different. At LSP, TFAM bends the mtDNA and its presence is mandatory for initiation. At HSP1, TFAM is strongly stimulatory under all conditions. At HSP2, TFAM is largely dispensable and inhibitory at high concentration and DNA topology is a major activating feature. Thus, the promoters may use different means to allow partially unwind the promoter sites and allow access to the transcriptional machinery.
Studies of in vitro transcriptional system are limited by their inability to observe the full complement of proteins that interact with POLRMT. Although the circular system better approximates the physical state of mtDNA, it still may have limitations because it fails to recapitulate the complex interactions between potentially modified bases in mtDNA or very local changes in DNA topology. A further understanding of the balancing of mitochondrial transcription may show how mitochondrial gene expression is integrated with nuclear expression to maintain mitochondrial function.
Materials and methods
In vitro transcription
Linear templates for mitochondrial transcription have been previously described.3 Circular templates were generated in pCR2.1 (LifeTech). For the light-strand promoter, the CSB2 sequence was amplified in its natural relationship with the promoter. For heavy-strand templates, a fragment containing the CSB2 site was amplified as a NotI-ApaI fragment and inserted downstream of mt.460–801 (for pHSP1/HSP2) or mt.583–801 (for pHSP2). Recombinant proteins were produced as described previously described.16 Plasmids were relaxed by treatment with 10U topoisomerase I (NEB-0301) according to the manufacturer's instructions or were mock treated. Topological changes were confirmed by agarose gel electrophoresis with post-run staining by ethidium bromide.
Except where noted, transcription was performed using 25 nM circular or linear template, 50 nM recombinant proteins, 400 µM GTP, CTP and ATP, 40 µM UTP, 1µCi (α-32P) UTP, 10 mM HEPES (pH 7.4), 1 mM DTT, 10 mM MgCl2, 100 μg/mL BSA, 40 mM NaCl, 40 mM KCl as previously described.3 As a control for equal loading, a 120-nt fragment was end labeled and added during sample processing.
S1 analysis of transcription
S1 analysis of HSP2-dependent transcription was performed as described previously using products generated from unlabeled transcription reactions.3 The probe for HSP1-dependent transcription was complimentary to mt.554–607. The LSP probe was mt.364–411.
Abbreviations
- HSP
heavy-strand promoter
- LSP
light-strand promoter
- mtDNA
mitochondrial DNA
- POLRMT
mitochondrial RNA polymerase
- TFAM
mitochondrial transcription factor A
- TFB2M
mitochondrial transcription factor B2
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by a Grant from the National Institutes of Health (HD058022) to NS.
References
- [1].Battey J, Clayton DA. The transcription map of human mitochondrial DNA implicates transfer RNA excision as a major processing event. J Biol Chem 1980; 255:11599-11606; PMID:6254975 [PubMed] [Google Scholar]
- [2].Montoya J, Gaines GL, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell 1983; 34:151-159; PMID:6883508; https://doi.org/ 10.1016/0092-8674(83)90145-9 [DOI] [PubMed] [Google Scholar]
- [3].Zollo O, Tiranti V, Sondheimer N. Transcriptional requirements of the distal heavy-strand promoter of mtDNA. Proc Natl Acad Sci USA 2012; 109:6508-6512; PMID:22454497; https://doi.org/ 10.1073/pnas.1118594109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lodeiro MF, Uchida A, Bestwick M, Moustafa IM, Arnold JJ, Shadel GS, Cameron CE. Transcription from the second heavy-strand promoter of human mtDNA is repressed by transcription factor A in vitro. Proc Natl Acad Sci USA 2012; 109:6513-6518; PMID:22493245; https://doi.org/ 10.1073/pnas.1118710109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kolesar JE, Wang CY, Taguchi YV, Chou SH, Kaufman BA. Two-dimensional intact mitochondrial DNA agarose electrophoresis reveals the structural complexity of the mammalian mitochondrial genome. Nucleic Acids Res 2013; 41:e58; PMID:23275548; https://doi.org/ 10.1093/nar/gks1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sobek S, Dalla Rosa I, Pommier Y, Bornholz B, Kalfalah F, Zhang H, Wiesner RJ, von Kleist-Retzow JC, Hillebrand F, Schaal H et al.. Negative regulation of mitochondrial transcription by mitochondrial topoisomerase I. Nucleic Acids Res 2013; 41:9848-9857; PMID:23982517; https://doi.org/ 10.1093/nar/gkt768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wanrooij PH, Uhler JP, Simonsson T, Falkenberg M, Gustafsson CM. G-quadruplex structures in RNA stimulate mitochondrial transcription termination and primer formation. Proc Natl Acad Sci USA 2010; 107:16072-16077; PMID:20798345; https://doi.org/ 10.1073/pnas.1006026107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fukuoh A, Ohgaki K, Hatae H, Kuraoka I, Aoki Y, Uchiumi T, Jacobs HT, Kang D. DNA conformation-dependent activities of human mitochondrial RNA polymerase. Genes Cells 2009; 14:1029-1042; PMID:19624753; https://doi.org/ 10.1111/j.1365-2443.2009.01328.x [DOI] [PubMed] [Google Scholar]
- [9].Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet 2002; 31:289-294; PMID:12068295; https://doi.org/ 10.1038/ng909 [DOI] [PubMed] [Google Scholar]
- [10].Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc Natl Acad Sci USA 2010; 107:12133-12138; PMID:20562347; https://doi.org/ 10.1073/pnas.0910581107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shi Y, Dierckx A, Wanrooij PH, Wanrooij S, Larsson NG, WIlhelmsson LM, Falkenberg M, Gustafsson CM. Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. Proc Natl Acad Sci USA 2012; 109:16510-16515; PMID:23012404; https://doi.org/ 10.1073/pnas.1119738109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell 2007; 18:3225-3236; PMID:17581862; https://doi.org/ 10.1091/mbc.E07-05-0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rubio-Cosials A, Sidow JF, Jiménez-Menéndez N, Fernández-Millán P, Montoya J, Jacobs HT, Coll M, Bernadó P, Solà M. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat Struct Mol Biol 2011; 18:1281-1289; PMID:22037172; https://doi.org/ 10.1038/nsmb.2160 [DOI] [PubMed] [Google Scholar]
- [14].Ngo HB, Kaiser JT, Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat Struct Mol Biol 2011; 18:1290-1296; PMID:22037171; https://doi.org/ 10.1038/nsmb.2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hatfield GW, Benham CJ. DNA topology-mediated control of global gene expression in Escherichia coli. Annu Rev Genet 2002; 36:175-203; PMID:12429691; https://doi.org/ 10.1146/annurev.genet.36.032902.111815 [DOI] [PubMed] [Google Scholar]
- [16].Lodeiro MF, Uchida AU, Arnold JJ, Reynolds SL, Moustafa IM, Cameron CE. Identification of multiple rate-limiting steps during the human mitochondrial transcription cycle in vitro. J Biol Chem 2010; 285:16387-16402; PMID:20351113; https://doi.org/ 10.1074/jbc.M109.092676 [DOI] [PMC free article] [PubMed] [Google Scholar]


