ABSTRACT
Under waterlogged conditions, roots of gramineous plants form lysigenous aerenchyma (internal gas spaces) by inducing the death of cortical cells. Rice (Oryza sativa) roots induce aerenchyma formation through ethylene- and reactive oxygen species (ROS)-mediated signaling. Metallothionein (MT) is a small, cysteine-rich protein that acts as a ROS scavenger. In rice roots, the expression of MT1a, MT1b, MT1c and MT1Ld were higher than those of the other MT genes. In the root cortex, where aerenchyma forms exclusively, the expression of MT1a, MT1b and MT1Ld was reduced prior to aerenchyma formation. These findings suggest that ROS accumulation in the cortex, which is aided by downregulation of MT1 genes, is needed for aerenchyma formation in rice roots.
KEYWORDS: Laser microdissection, lysigenous aerenchyma, metallothionein (MT), reactive oxygen species (ROS), rice (Oryza sativa L.)
Lysigenous aerenchyma, which is created by cell death and lysis of cells in the root cortex, is essential for the internal transport of oxygen from shoots to roots of rice (Oryza sativa) and other gramineous plants under waterlogged conditions.1,2 In rice roots, lysigenous aerenchyma is constitutively formed under aerobic conditions (constitutive aerenchyma formation), and its formation is further induced under oxygen-deficient conditions (inducible aerenchyma formation).3
Programmed cell death (PCD), which is energy-dependent cell death, plays essential roles in development and in stress responses of multicellular organisms.4 Cell collapse during lysigenous aerenchyma formation is characterized as a type of PCD.5 The gaseous phytohormone ethylene stimulates PCD during inducible aerenchyma formation.6 Under waterlogged conditions, ethylene accumulates in roots due to its low diffusion rate to the rhizosphere.7 Moreover, expression of ethylene biosynthetic genes is induced, thereby increasing the activities of ethylene biosynthetic enzymes, during inducible aerenchyma formation.8–10
Reactive oxygen species (ROS), such as superoxide anion radical (O2·−) and hydrogen peroxide (H2O2), have a role in ethylene-dependent inducible aerenchyma formation in roots of gramineous plants.11,12 As further evidence of the importance of ROS in aerenchyma formation, the expression of respiratory burst oxidase homolog (RBOH), a plant enzyme that generates O2·−, increases in the cortex of rice roots under stagnant conditions that mimic waterlogged soil.13 Moreover, knockout of one RBOH isoform (RBOHH) in rice reduces H2O2 accumulation and aerenchyma formation under stagnant conditions.13
Metallothionein (MT) is a small, cysteine-rich protein that plays a role in metal homeostasis.14 Plant MTs were classified into four subfamilies (type/class 1, 2, 3 and 4) based on a study of MTs in Arabidopsis.15 Several lines of evidence show that plant MTs act as ROS scavenging enzymes. Indeed, rice MT2b17 and cotton (Gossypium hirsutum) MT3a18 have high antioxidative activities against O2·− and hydroxyl radicals in vitro. Moreover, H2O2 accumulation in leaves, as well as growth retardation of tobacco (Nicotiana tabaccum) plants in response to NaCl treatment is alleviated by overexpression of rice MT1a (OsMT1e).19 MTs have also been implicated in PCDs in plants. In rice, knockdown of MT2b expression was found to promote epidermal cell death in stems20 and to accelerate H2O2-mediated aerenchyma formation in the internodes.21 In maize, the expression level of MT1 in the root cortex was found to decrease during aerenchyma formation under waterlogged conditions.11 These findings suggest that MTs have a role in determining the fate of cells in roots during inducible aerenchyma formation.
Previous studies reported 9 MT genes17, 11 MT genes22 and 13 MT genes19 in the rice genome, for a total of 14 unique genes (Table 1). However, our search of the rice genome annotation databases revealed that rice genome has 15 MT genes, that is, we identified one more MT gene (MT1Ld) (Table 1). Our phylogenetic analysis of the predicted amino acid sequences of the 15 MT proteins (Fig. 1) classified them into four types (1–4) (Table 1). The four types were homologous to the four types described by Cobbett & Goldsbrough.15 One of the type 2 MTs (MT2La) lacks an N-terminal cysteine-rich motif, so that it might not have metal binding or ROS scavenging activities.
Table 1.
List of genes encoding metallothionein in rice.
| Group |
Gene name |
Wong (2004)*1 |
Zhou (2006)*2 |
Kumar (2012)*3 |
RAP_Os IDs*4 |
MSU LOC_Os IDs*5 |
RAPDB_Description*6 |
| Type 1 | |||||||
| MT1a | OsMT1a | OsMT-I-1a | OsMT1e | Os11g0704500 | LOC_Os11g47809 | Metallothionein-like protein type 1. | |
| MT1b | OsMT1b | OsMT-I-4a | OsMT1a | Os12g0570700 | LOC_Os12g38270 | Similar to Metallothionein-like protein type 1. | |
| MT1c | OsMT1c | OsMT-I-4c | OsMT1d | Os12g0571100 | LOC_Os12g38300 | Similar to Metallothionein-like protein type 1. | |
| MT1La | OsMT-I-1b | OsMT1b | Os03g0288000 | LOC_Os03g17870 | Similar to Metallothionein. | ||
| MT1Lb | OsMT1f | Os12g0567800 | LOC_Os12g38010 | Plant metallothionein, family 15 protein. | |||
| MT1Lc | OsMT-I-4b | OsMT1c | Os12g0568200 | LOC_Os12g38051 | Metallothionein-like protein type 1. | ||
| MT1Ld | Os12g0568500 | LOC_Os12g38051 | Metallothionein-like protein type 1. | ||||
| MT1Le | OsMT1g | Os12g0571000 | LOC_Os12g38290 | Metallothionein-like protein type 1. | |||
| MT2a | OsMT2a | OsMT-I-2a | OsMT2a | Os01g0149800 | LOC_Os01g05650 | Metallothionein-like protein type 2. | |
| MT2b | OsMT2b | OsMT-I-2c | OsMT2b | Os05g0111300 | LOC_Os05g02070 | Similar to Metallothionein. | |
| MT2c | OsMT2c | OsMT-I-2b | OsMT2c | Os01g0974200 | LOC_Os01g74300 | Metallothionein-like protein. | |
| (MT2La)*7 | OsMT2d | Os01g0149200 | LOC_Os01g05585 | Hypothetical gene. | |||
| Type 3 | |||||||
| MT3a | OsMT3a | OsMT-I-3a | OsMT3a | Os01g0200700 | LOC_Os01g10400 | Similar to Metallothionein-like protein type 3. | |
| MT3b | OsMT3b | OsMT-I-3b | Os05g0202800 | LOC_Os05g11320 | Similar to Metallothionein-like protein 3B. | ||
| Type 4 | |||||||
| MT4 | OsMT4 | OsMT-II-1a | OsMT4 | Os10g0542100 | LOC_Os10g39610 | Plant EC metallothionein-like protein, family 15 protein. | |
Wong et al. Plant Physiol 2004; 135:1447-56.
Zhou et al. Biochem Mol Biol 2006; 39:595-606.
Kumar et al. BMC Plant Biol 2012; 12:107.
RAP Os IDs in Rice Annotation Project Database (RAP-DB; http://rapdb.dna.affrc.go.jp/).
MSU LOC_Os IDs in Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu/).
Descriptions in RAP-DB (IRGSP-1.0).
OsMT2La lacks the N-terminal cystein-rich motif.
Figure 1.
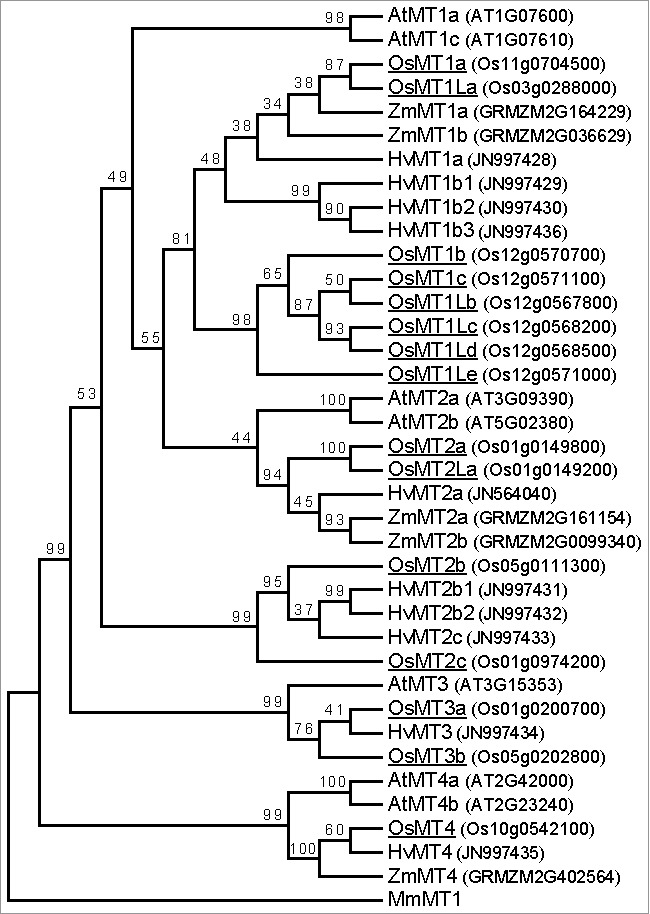
Phylogenetic analysis of MT proteins in plants. The amino acid sequences of the MT proteins were aligned by the ClustalW (MEGA6 package). MEGA6 (http://www.megasoftware.net) was used for construction of the neighbor-joining phylogenetic tree with bootstrap values calculated based on 1000 replicates. MT proteins from monocotyledonous plants [rice (Oryza sativa; Os), maize (Zea mays ssp. mays; Zm), and barley (Hordeum vulgare; Hv)], and dicotyledonous plants [Arabidopsis (Arabidopsis thaliana; At)] were used for the analysis. The rice OsMT proteins were indicated by underlines. MmMT1 from mouse (Mus musculus) was used as the out-group. Accession numbers or IDs of each protein were denoted in the parentheses. The amino acids sequences were obtained from each database as described by Yamauchi and colleagues.10
In rice (cv. Shiokari) roots, inducible aerenchyma formation starts at 24 to 36 h and peaks at 48 h after the transfer to stagnant conditions.13 To identify MT genes highly expressed in rice roots during inducible aerenchyma formation, absolute transcript levels of the 15 MT genes at 10 mm (±2 mm) from the tips of adventitious roots were investigated. Among these genes, four type 1 MT genes (MT1a, MT1b, MT1c and MT1Ld) had the highest transcript levels under aerated conditions (Fig. 2A). Moreover, the transcript levels of each of these genes except MT1c dramatically decreased under stagnant conditions (Fig. 2A). In a time-course analysis, the expression levels of MT1a and MT1b started to decrease at 12 h under stagnant conditions (Fig. 2B, C). The expression level of MT1c was comparable between aerated and stagnant conditions (Fig. 2D), whereas that of MT1Ld started to decrease at 24 h under stagnant conditions (Fig. 2E). These results indicate that expression of MT1a, MT1b and MT1Ld in rice roots strongly decreased prior to inducible aerenchyma formation.
Figure 2.
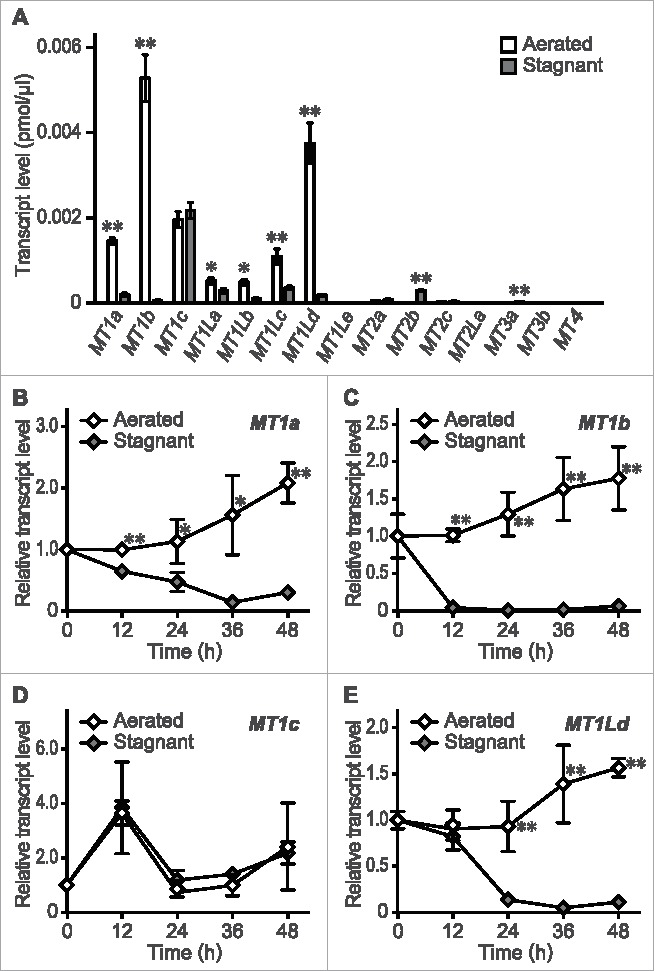
Expression of MT genes in rice roots during aerenchyma formation. (A) Absolute transcript levels of MT genes at 10 mm (± 2 mm) from the tips of adventitious roots of rice seedlings grown under aerated or stagnant conditions for 36 h. (B-E) Time-course relative transcript levels of MT1a (B), MT1b (C), MT1c (D), and MT1Ld (E) at 10 mm (± 2 mm) from the root tips under aerated or stagnant conditions. The 20 to 40 mm roots of the 10-d-old aerobically grown rice seedlings were subjected to the treatments. The gene encoding transcription initiation factor IIE, TFIIE, was used as a control. Values are means ± SD (n = 3). Significant differences between aerated and stagnant conditions at P < 0.01 and P < 0.05 (two sample t test) are denoted by ** and *, respectively. The methods are described in more detail by Yamauchi and colleagues.13
The central cylinder (CC), cortex (Co), and outer part of the roots (OPR) were isolated from sections at 10 mm (±2 mm) from the tips of adventitious roots by laser microdissection at 36 h under aerated or stagnant conditions. The expression levels of MT1a, MT1c and MT1Ld were highest at the OPR (Fig. 3A, C, D), and those of MT1b were highest at the Co (Fig. 3B). MT1a, MT1b and MT1Ld expression was significantly reduced in all the tissues examined under stagnant conditions (Fig. 3A, B, D), whereas MT1c expression was comparable between aerated and stagnant conditions (Fig. 3C). Under stagnant conditions, expression of MT1a and MT1Ld were higher in the OPR than in the CC and Co (Fig. 3A, D). Moreover, MT1c expression was specific to the OPR (Fig. 3C). These results suggest that ROS scavenging by MT1 proteins can still occur in the OPR, but that it is suppressed in the Co under stagnant conditions.
Figure 3.
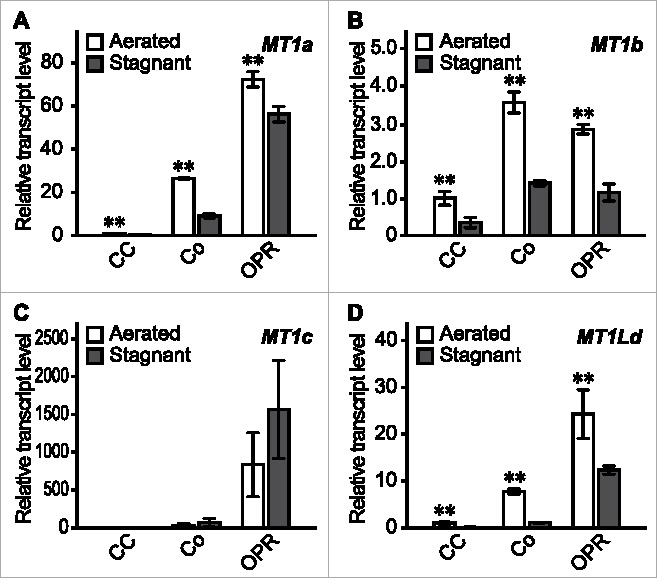
Tissue-specific expression of MT1 genes in rice roots during aerenchyma formation. (A-D) Relative transcript levels of MT1a (A), MT1b (B), MT1c (C), and MT1Ld (D) in the central cylinder (CC), cortex (Co), and outer part of the root (OPR) at 10 mm (± 2 mm) from the root tips under aerated or stagnant conditions for 36 h. The 30 to 50 mm roots of the 20-d-old aerobically grown rice seedlings were subjected to the treatments. The gene encoding transcription initiation factor IIE, TFIIE, was used as a control. Values are means ± SD (n = 3). Significant differences between aerated and stagnant conditions at P < 0.01 and P < 0.05 (two sample t test) are denoted by ** and *, respectively. The methods are described in more detail by Yamauchi and colleagues.13
Expression of RBOHH, whose product converts O2 to O2·−,23 is induced in the Co and OPR under stagnant conditions, although its transcript levels are higher in the Co than in the OPR.13 In the Co, expression of MT1 genes (e.g., MT1a, MT1b and MT1Ld) is suppressed under stagnant conditions (Fig. 3A, B, D), and thus high levels of ROS generated by RBOHH may be conserved, thereby inducing the PCD (i.e., aerenchyma formation) in the cortical cells. By contrast, ROS generated by RBOHH in the OPR may be reduced by MT proteins. Previously, we found that RBOHH expression was induced in all cell types of maize primary roots, but the expression of a gene encoding an MT1 (ZmMT1a in Fig. 1), which is a close homologue of rice MT1a, was specifically reduced in the Co during inducible aerenchyma formation under waterlogged conditions.11 These findings suggest that MT1-mediated ROS scavenging commonly underlies ROS-mediated inducible aerenchyma formation in gramineous plants.
The Arabidopsis and rice genomes have 7 and 15 MT genes (Fig. 1; Table 1), respectively. So far, there is little information about the subcellular localizations of MTs in plants possibly due to the instability of MT proteins.15 O2·− in the apoplast is thought to be spontaneously or enzymatically converted to H2O2, which then diffuses into the cytosol.24 It is thus likely that MTs act as scavengers of ROS in the cytosol, and would stall the ROS-mediated signal transduction that triggers PCD in the OPR. Overexpression of MT1a/OsMT1e reduced H2O2 accumulation in leaves of tobacco plants under high salinity conditions, thereby improving their growth.19 Alternatively, rice MT1a/OsMT1a has been proposed to indirectly enhance the activities of catalase and peroxidase, which detoxify H2O2.25 These antioxidant enzymes may also be involved in regulating ROS-mediated aerenchyma formation. Further studies are needed to understand how MT1s scavenge O2·− and/or H2O2 during inducible aerenchyma formation in roots of gramineous plants.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Kiyoaki Kato and Itsuro Takamure for providing seeds of rice (cv. Shiokari). We also thank Timothy D. Colmer, Ole Pedersen, Al Imran Malik, Nobuhiro Tsutsumi, Naoko K. Nishizawa, Hirofumi Yoshioka, Miki Yoshioka, Katsuhiro Shiono, Hirokazu Takahashi, Kazuyuki Kuchitsu, Takamitsu Kurusu, Kenji Hashimoto, Atsushi Oyanagi, Kentaro Kawaguchi, Fumitaka Abe, Yoshiro Mano, Mitsuhiro Obara, Tomomi Abiko, Shunsaku Nishiuchi and Kohtaro Watanabe for stimulating discussion.
Funding
This work was partly supported by a grant from the Bio-oriented Technology Research Advancement Institution (Promotion of Basic Research Activities for Innovative Biosciences) to MN; TY was supported by a post-doctoral fellowship from the Japan Society for the Promotion of Science.
References
- 1.Armstrong W. Aeration in higher plants. Adv Bot Res. 1979;7:225–332. doi: 10.1016/S0065-2296(08)60089-0. [DOI] [Google Scholar]
- 2.Colmer TD. Long-distance transport of gases in plants: a perspective on internal aeration and radial oxygen loss from roots. Plant Cell Environ. 2003;26:17–36. doi: 10.1046/j.1365-3040.2003.00846.x. [DOI] [Google Scholar]
- 3.Colmer TD, Voesenek LACJ. Flooding tolerance: suites of plant traits in variable environments. Funct Plant Biol. 2009;36:665–81. doi: 10.1071/FP09144. [DOI] [PubMed] [Google Scholar]
- 4.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans DE. Aerenchyma formation. New Phytol. 2003;161:35–49. doi: 10.1046/j.1469-8137.2003.00907.x. [DOI] [Google Scholar]
- 6.Jackson MB, Armstrong W. Formation of aerenchyma and the processes of plant ventilation in relation to soil flooding and submergence. Plant Biol. 1999;1:274–87. doi: 10.1111/j.1438-8677.1999.tb00253.x. [DOI] [Google Scholar]
- 7.Sasidharan R, Voesenek LACJ. Ethylene-mediated acclimations to flooding stress. Plant Physiol. 2015;169:3–12. doi: 10.1104/pp.15.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He CJ, Finlayson SA, Drew MC, Jordan WR, Morgan PW. Ethylene biosynthesis during aerenchyma formation in roots of maize subjected to mechanical impedance and hypoxia. Plant Physiol. 1996;112:1679–85. doi: 10.1104/pp.112.4.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamauchi T, Shiono K, Nagano M, Fukazawa A, Ando M, Takamure I, Mori H, Nishizawa NK, Kawai-Yamada M, Tsutsumi N, et al.. Ethylene biosynthesis is promoted by very-long-chain fatty acids during lysigenous aerenchyma formation in rice roots. Plant Physiol. 2015;169:180–93. doi: 10.1104/pp.15.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamauchi T, Tanaka A, Mori H, Takamure I, Kato K, Nakazono M. Ethylene-dependent aerenchyma formation in adventitious roots is regulated differently in rice and maize. Plant Cell Environ. 2016;39:2145–57. doi: 10.1111/pce.12766. [DOI] [PubMed] [Google Scholar]
- 11.Rajhi I, Yamauchi T, Takahashi H, Nishiuchi S, Shiono K, Watanabe R, Mliki A, Nagamura Y, Tsutsumi N, Nishizawa NK, et al.. Identification of genes expressed in maize root cortical cells during lysigenous aerenchyma formation using laser microdissection and microarray analyses. New Phytol. 2011;190:351–68. doi: 10.1111/j.1469-8137.2010.03535.x. [DOI] [PubMed] [Google Scholar]
- 12.Yamauchi T, Watanabe K, Fukazawa A, Mori H, Abe F, Kawaguchi K, Oyanagi A, Nakazono M. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J Exp Bot. 2014;65:261–73. doi: 10.1093/jxb/ert371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamauchi T, Yoshioka M, Fukazawa A, Mori H, Nishizawa NK, Tsutsumi N, Yoshioka H, Nakazono M. An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell. 2017;29:775–90. doi: 10.1105/tpc.16.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coyle P, Philcox JC, Carey LC, Rofe AM. Metallothionein: the multipurpose protein. Cell Mol Life Sci. 2002;59:627–47. doi: 10.1007/s00018-002-8454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobbett C, Goldsbrough P. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol. 2002;53:159–82. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- 16.Hassinen VH, Tervahauta AI, Schat H, Kärenlampi SO. Plant metallothioneins – metal chelators with ROS scavenging activity? Plant Biol. 2011;13:225–32. doi: 10.1111/j.1438-8677.2010.00398.x. [DOI] [PubMed] [Google Scholar]
- 17.Wong HL, Sakamoto T, Kawasaki T, Umemura K, Shimamoto K. Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 2004;135:1447–56. doi: 10.1104/pp.103.036384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue T, Li X, Zhu W, Wu C, Yang G, Zheng C. Cotton metallothionein GhMT3a, a reactive oxygen species scavenger, increased tolerance against abiotic stress in transgenic tobacco and yeast. J Exp Bot. 2009;60:339–49. doi: 10.1093/jxb/ern291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar G, Kushwaha HR, Panjabi-Sabharwal V, Kumari S, Joshi R, Karan R, Mittal S, Pareek SLS, Pareek A. Clustered metallothionein genes are co-regulated in rice and ectopic expression of OsMT1e-P confers multiple abiotic stress tolerance in tobacco via ROS scavenging. BMC Plant Biol. 2012;12:107. doi: 10.1186/1471-2229-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steffens B, Sauter M. Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell. 2009;21:184–96. doi: 10.1105/tpc.108.061887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steffens B, Geske T, Sauter M. Aerenchyma formation in the rice stem and its promotion by H2O2. New Phytol. 2011;190:369–78. doi: 10.1111/j.1469-8137.2010.03496.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhou G, Xu Y, Li J, Yang L, Liu J-Y. Molecular analyses of the metallothionein gene family in rice (Oryza sativa L.). Biochem Mol Biol. 2006;39:595–606. doi: 10.5483/BMBRep.2006.39.5.595. [DOI] [PubMed] [Google Scholar]
- 23.Torres MA, Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–67. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, Wu Y, Li Y, Ling H-Q, Chu C. OsMT1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice. Plant Mol Biol. 2009;70:219–29. doi: 10.1007/s11103-009-9466-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


