ABSTRACT
Among the many facets of DNA damage response (DDR), relocation of chromosome territories (CTs) is most intriguing. We have previously reported that cisplatin induced DDR in human dermal fibroblasts led to relocation of CTs 12, 15 from the nuclear periphery to its interior while CTs 19, 17 repositioned from the interior to its periphery. Studies of CT relocation remain nascent as we begin unraveling the role of key players in DDR to demonstrate its mechanistic basis. Consolidating our recent reports, we argue that γH2AX-signaling leads to enhanced recruitment of nuclear myosin 1 (NM1) to chromatin, which via its motor function, results in CT repositioning. Next, we invoke a novel systems-level theory that subsumed CTs as pairs, not solo entities, to present the physical basis for plasticity in interphase CT arrangement. Subsequently, we posited that our systems-level theory describes a unified physical basis for non-random positioning of CTs in interphase nuclei across disparate eukaryotes.
Keywords: 3D genome organization, cell cycle, chromosome territory, Cisplatin treatment, CT relocation, DDR, DNA damage, NM1 and Induced DNA damage, serum starvation, unified genome
Introduction
The eukaryotic DNA is packaged along with histone and non-histone proteins to form a 30 nm chromatin fiber,1 which in turn is non-randomly and hierarchically organized into chromosomes. In vertebrates, during interphase stage of the cell cycle, each chromosome is territorially confined and referred to as chromosome territory (CT)2 such that multiple chromosomes are non-randomly placed in a radial arrangement.2-5 However, in invertebrates, such as fungi,6 plants7 and insects,8 the interphase chromosomes remain tethered to the nuclear envelope in a polarized conformation, which is referred to as Rabl arrangement.
The DNA is subject to damage from endogenous as well as exogenous sources and such damages to the genetic material pose a threat in all organisms. As DNA damage can occur anywhere in the nucleus it is imminent for the DNA damage response (DDR) to occur spatially concordant with any damaged locus (or loci), anywhere in the nucleus. Therefore, DDR is best described as a systems-level response,9,10 (which for example involves over hundred genes in humans11,12), that is executed with high fidelity irrespective of the nature and extent of damage within a limit.13 Repair of linear collimated lesions (as that induced by radioactive α rays)14 as well as pan-nuclear damage (as that induced by mild dose of cisplatin)15 have been reported under in vitro conditions. Moreover, this evolutionarily conserved16 and precisely coordinated response is ubiquitous in both unicellular and multicellular eukaryotes, with many different cell-types.17,18 Here, in this Extra View article, we discuss results from 3 selected reports from our laboratory, in the context of some current findings and posit how disparate eukaryotic genomes adapt using remarkably similar strategies in large-scale CT repositioning, a unique facet seen during DDR.15 Our focus will incorporate instances of large-scale repositioning of specific CTs and a novel interpretation of the common underlying biology from an inter-disciplinary perspective.
CTs repositioning as a unique and specific DNA damage response
In vertebrates, the spatial positioning of CTs is cell-type specific19 and therefore the DDR stimuli thereof is also context driven. Large-scale CT repositioning has been reported during DDR in an ensemble of human dermal fibroblast (HDF) cells under controlled in vitro conditions. This unique facet of DDR involved the repositioning of 4 CTs, chromosome 12, 15, 17 and 19, from their respective native positions during a 4 hour treatment of 25 μM mild dose of cisplatin.15 The general methodology is schematically represented in Fig. 1, where HDFs were probed at 4 different time points during the entire 28 hour time window, which included 4 hours of cisplatin treatment. The time-series are so chosen that the window of observation encompasses robust DDR, with marginal, if at all, extent of cell death, where majority of damaged nuclei recover from damages due to DNA repair. The DDR time-series was monitored by quantifying the γH2AX response. Chromosomes 19 and 17, the 2 leading gene-rich CTs, relocated from their native positions in the interior of the nucleus to the periphery by the end of this treatment (after 4 hours). Additionally, chromosomes 12 and 15, also relatively gene-rich CTs, relocated from the nuclear periphery to the center during those 4 hours (Fig. 2).15 This response was very robust, with approximately 70–90% of cells exhibiting statistically significant relocations, from the native positions of the 4 CTs mentioned above. Among the remaining CTs, a few exhibited partial extents of spatial changes (chromosome 20) but most CTs did not show any perceptible relocation from the native non-random positions.15
Figure 1.
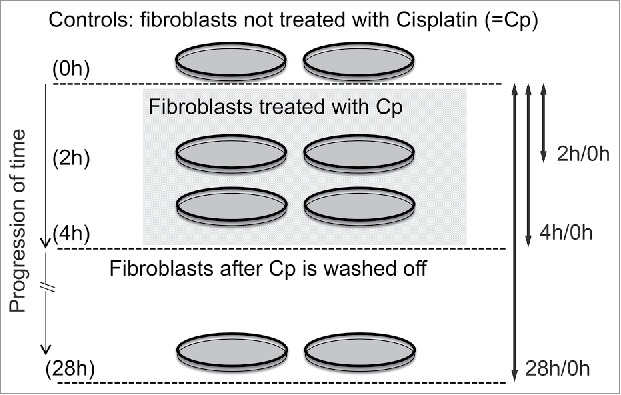
An overview of experimental assay and study design that demonstrated en masse CT repositioning during DNA damage response. This scheme is based on the experimental assay as reported by Mehta et al.15 Following controlled induction of DNA double strand breaks by cisplatin treatment of 4 h, it was established (using a clonal population of human dermal fibroblast cells) that CTs 12, 15, 17 and 19 relocated from their original position within the nucleus to newer ones. Upon washing off cisplatin (after 4 h), the original CT arrangement in the nucleus was restored in the population of treated fibroblast cells following DNA repair when probed after 24 h.
Figure 2.
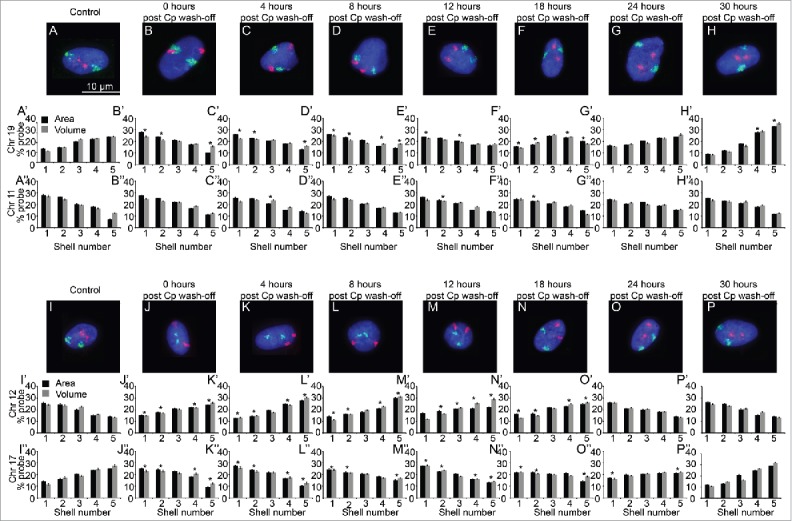
Restoration of native CT positions after washing off cisplatin. Normal human dermal fibroblasts (NHDFs) were treated with 25 µM cisplatin to induce DNA damage for 4 hours. The cisplatin (Cp) was then washed off (0 h post Cp wash-off) and the cells were then allowed to repair the damage by placing them in fresh medium and samples were collected at varying time-periods post cisplatin wash-off (as indicated in Fig. 1). 2D-FISH was then performed for a set of chromosomes and at least 100 images per sample were analyzed via IMACULAT.50 The software basically divides the nuclei of each cell in 5 shells of equal area (shell 1 being the most peripheral shell and shell 5 the innermost shell). The amount of FISH probe in each shell is then normalized with the amount of DNA and histograms are plotted. The positions of chromosomes 11 and 19 (panels A–H) and 12 and 17 (panels I–P) were determined at respective time points using 2D-FISH analyses. Cp = cisplatin. Adapted by permission from BioMed Central: reference 15, copyright 2013.
Spatial relocation of CTs induced by disparate exogenous stimuli is reversible
From the aforementioned experiment,15 it was most striking that after the DDR inducing agent was washed-off from the cells, the relocated CTs reverted back to their native positions.15 The repositioning of CTs is not merely an instance of a reversible physical change as they incurred biological “hysteresis” (CT relocation and reversal take different time durations). The reversal of CT positions back to their native positions needed at least 24 hours after the wash-off, which was significantly longer than the 4 hours duration for the forward relocation (Fig. 2).15 Absolute distance measurements revealed that relocated CTs (chromosomes 19, 17, 15 and 12) reverted to the native inter-CT distances vis-à-vis stationary CT landmarks following the completion of reversal. It is also interesting to note that this reversal of CTs to their native positions coincide with the loss of γH2AX foci (indicating completion of repair). Thus, these results suggest a possible crosstalk between DNA repair proteins and mechanisms that relocate chromosomes within a cell. Importantly, these results together reflect that CT reorganization is an integral part of cellular DDR. We speculate that CT repositioning is linked to “transcriptional rewiring” of the cell required during large perturbative changes such as DDR,15 serum starvation,20 etc. We surmised that the underlying molecular biology for CT changes associated with relocation and reversal might not be trivially similar which prompted us to consider other independent studies where such large-scale CT repositioning had been reported.
Next, we inferred some parallels from another in vitro experiment, which had resulted in CTs being relocated during serum starvation induced stress. That study used primary dermal fibroblast cells and reported that as normal fibroblasts exit the cell cycle, the rapid movement of 2 CTs, chromosomes 13 and 18 show rapid relocation within about 15 minutes in serum starved media.20 Here, the relocation of CTs from their quiescent positions to their proliferating location was observed after more than 24 hours from the point of serum re-addition (to offset the induced biological stress). In a different study, which only involved serum restimulated cells, more than 30 hours were required for CT repositioning.21 Interestingly, after their exit from mitosis, CT positions are set up in the G1 phase of the cell cycle.22,23 High chromatin mobility is observed in early G1 when the positions of those chromosomes is being set up, followed by little or no relocation throughout the cell cycle.22-24 However, the mechanistic details of regulation that connects CT relocation to cell cycle stages is currently far from clear and forms an exciting new area of research. Interestingly, much slower kinetics of CT reversals observed during both DDR and serum-starvation paradigms suggest that the CT dynamics is perhaps synchronous with the ongoing cell cycle changes. In consistent with this notion all these results indicate that the cells need to traverse through mitosis before CTs reverted during DDR (Figs. 7 and 8 in ref. 15). The two paradigms, cited so far, along with various other reports, suggest that genomes are dynamic both spatially and temporally at different length scales, which may vary from gene-level to pan-nuclear scales. Spatial dynamic changes are not limited to chromosomes alone but may even extend to other nuclear organelles beyond chromatin such as nuclear speckles and PML-bodies etc. during DDR (unpublished results). However, the underlying molecular mechanisms that initiate large-scale CT relocations during various nuclear responses are still unclear. Unlike the local dynamic changes associated with chromatin loops, those associated with large-scale CT relocations are expected to impact genome functions much more extensively. To understand the mechanistic basis of the same, we focused on the crosstalk between DDR signaling and the dynamics of CT relocation25 and addressed: (i) how is a systems-level biological response for chromosomal relocation, orchestrated? (ii) what are the motor proteins implicated in such “active” translations, if any? and (iii) whether motor proteins are impacted by any signaling events that are associated with DDR?
Figure 7.
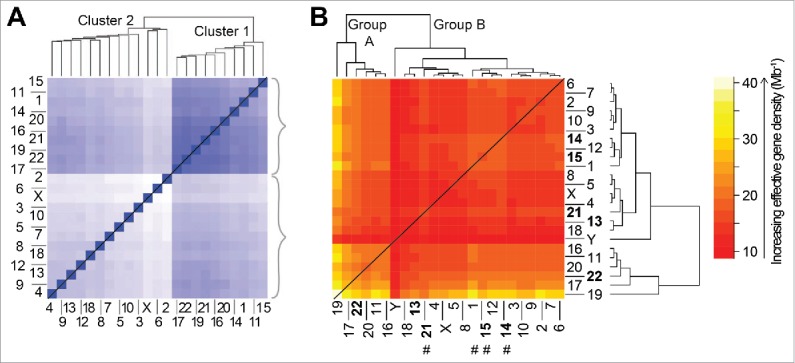
Comparison of the hierarchical organization for genome-level data generated independently from experiment (A) and theory (B). (A) Population-based analysis of Tethered Conformation Capture (TCC) Hi-C results,38 from the (46,X,X) diploid human nucleus, for chromosome territory localizations. Clustering of CTs was generated with respect to the average distance between the center of mass of a CT pair in the genome from the population of cells. The TCC Hi-C clustering dendrogram is shown on top and a matrix of the average distances between CT pairs is shown at the bottom. The darker blue shade signifies proximal genomic loci signifying closer contacts in space and therefore the intensity (of shade) changes inversely with the distance of separation between loci. Panel adapted by permission from Macmillan Publishers Ltd: reference 38, copyright 2012. (B) The hierarchical clustering of effective gene density elements from the effective gene density matrix that is generated for the diploid human male genome (46,X,Y). The 5 CTs associated with the Nucleolar Organizing Region are represented in bold. Four CTs that do not concur with “cluster 1” in panel A are highlighted using hash (#) in panel B.
Figure 8.
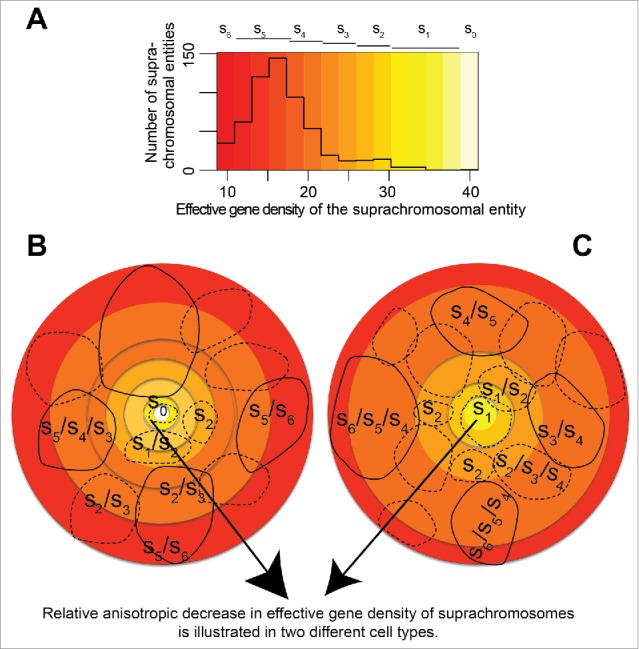
(A) schematic representing distinct CT positioning in the (46,X,Y) diploid human nuclei. (A) The effective gene density distribution from CT pairs is represented in a histogram using off-white color (s0 – highest effective gene density) to red color (s6 – lowest effective gene density). All pairs of chromosomal entities (suprachromosomal units), which were derived from the human genome represent the histogram in panel A. Lighter colored hues, from off-white to yellow, represent a shallow pool of degenerate suprachromosomal entities. However, deeper hues, from orange to red, represent progressively larger pools of degenerate suprachromosomal units with similar effective gene density. (B, C) Cartoons of 2 human nuclei, rendered in 2D, represent the plasticity of a hypothetical arrangement of those suprachromosomal entities. The cohorts of suprachromosomal entities s0 – s6 may be used to recreate different CT constellations in cell type-specific or a clonal population of human nuclei. As mandated by a hypothetical boundary condition, where suprachromosomal units are arranged with relatively high effective gene density suprachromosomal contributors (described by s0 – s2) toward the interior of the nuclei. This condition then sets the basis for a self-organized arrangement where the lower effective gene density contributors (described by s5 – s6) are toward the periphery (direction of arrowhead). Schematic suggests multiple ways (plasticity in spatial CT arrangement) in which suprachromosomal units described by s5 – s6 occupy the periphery of the 3D nucleus, those described by s0 – s2 occupy the interior and s3 – s4 are intermediate to the 2. Solid contours imply suprachromosomal entities in the foreground while those in the background are shown using dashed contours.
Outlined next are some of the highlights that have now begun to define the molecular basis for CT relocations in the context of induced stress during DDR.
Probing the mechanistic basis for CT relocation and reorganization during DDR
γH2AX signaling is essential for CT relocation during DDR
In the DDR paradigm, it was shown that CT repositioning required the activity of the 3 human apical kinases ATM/ATR along with DNA Protein Kinase C (DNA-PKcs)15 and that γH2AX signaling was also needed (Fig. 2 reference 25). To understand the role of γH2AX signaling, we overexpressed a non-phosphorylatable mutant of H2AX S139A. Cells expressing the mutant H2AX S139A showed fewer number of γH2AX foci upon cisplatin damage than those in either untransfected or H2AX wild type transfected control cells. We also confirmed that the decrease in γH2AX foci number led to a concomitant reduction of its downstream signaling in the mutant H2AX expressing cells following DNA damage. We observed that DNA damage induced relocation of chromosomes 19 and 15 was not observed in the cells expressing mutant H2AX S139A construct, while the same were observed as per expectations in untransfected or wild type H2AX transfected control cells (Fig. 3, reference 25). Thus γH2AX signaling at DNA double strand breaks (DSBs) by the phosphorylated histone variant (γH2AX) was a pre-requisite for damage induced CT relocation, as cells deficient in γH2AX signaling fail to exhibit such a response.25 Statistical analysis quantitated the experimentally derived frequency distributions of CT positions between the nuclear center and CT center in 3D confocal images (63x magnification) using Imaris software (N = 2, n = at least 30 nuclei) as represented in Fig. 3 (and Fig. 3 reference 25). Chromosomes 19 and 15 boxplots represented frequency distributions where the differences between treated and untreated samples were statistically significant (P < 0.05 and P < 0.01, from Fig. 3 reference 25). We reported CTs 19 and 15 as relocated, while CT 11 was used as control.25 Next, to check for the involvement of motor proteins, cells deficient and proficient (wild type) in a myosin isoform were sought.
Figure 3.
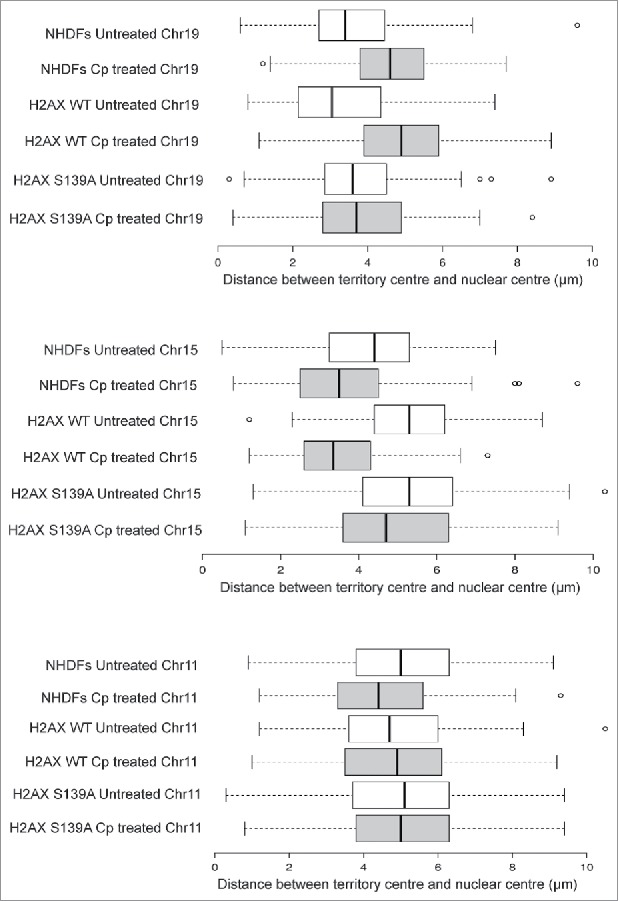
Chromosome territory repositioning requires γH2AX signaling. The box plots were generated from experimentally derived frequency distributions of CT positions between the nuclear center and CT center in 3D confocal images (63x magnification), quantitated using Imaris software (N = 2, n = at least 30 nuclei). Adapted by permission from Oxford University Press: reference 25, copyright 2016. Only top 2-sets among CT19 and CT15 boxplots represented frequency distributions where the differences between treated and untreated samples were statistically significant (P < 0.05 and P < 0.01 as shown in Fig. 3, reference 25).
NM1 is essential for DNA damage induced CT relocation
Addition of ATPase and/or GTPase Inhibitors to proliferating cell cultures depleted in serum impaired large-scale genome organization and no CT relocation was observed.20 Therefore, an active mechanism (of transport) was implicated early on in these studies, which possibly involved motor proteins such as actin and myosin. This was subsequently confirmed using cells that had impaired myosin function.
The human NM1 is the predominant nuclear isoform encoded by MYO1C gene.26 To check if this nuclear motor is essential for CT relocation during DDR, we used the siRNA mediated knockdown of MYO1C. As assessed by Western blotting analyses, this siRNA knockdown resulted in nearly 60–70% reduction in NM1 levels. By using chromosome-specific 2D-FISH analysis, we assessed the positions of chromosome 19, 15 and 11 in cells transfected with control and MYO1C siRNA. Therefore, we corroborated the results that control NHDFs (treated with the control siRNA) exhibited the expected relocation of chromosome 19 from the nuclear interior to the periphery upon cisplatin damage and vice versa for chromosome 15. However, under the same experimental conditions, MYO1C knockdown cells failed to show any damage induced relocation of chromosome 19 and 15 (Fig. 4). We also noted that the negative control chromosome 11 did not show any spatial relocation following DNA damage in cells treated with either siRNA. Additionally, we tested whether ATPase function was relevant for DNA damage induced CT relocation. To understand how the motor activity of NM1 influences its enhanced chromatin binding and CT relocation post DNA damage, cells over-expressing NM1 motor function defective mutant (G126S – that has defective ATPase function) or wild type NM1 were analyzed. As expected, in the control cells expressing wild type NM1, damage induced increase in chromatin recruitment of nuclear myosin protein, encoded by NM1, and CT relocation was observed. In contrast, the NM1-G126S mutant expressing cells failed to show such DNA damage induced changes (Fig. 5), indicating that the motor function of protein encoded by nuclear myosin gene NM1 was important for its binding to chromatin following damage and for the subsequent CT relocation.
Figure 4.
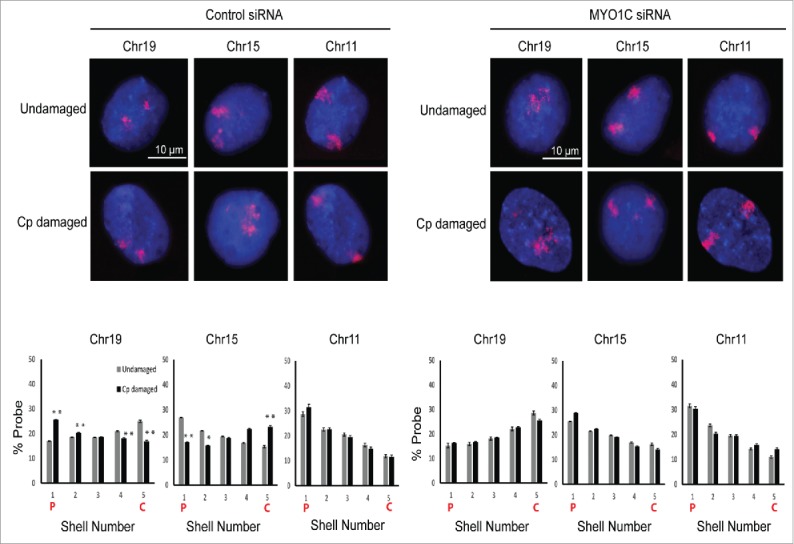
Nuclear myosin1 (NM1) is required for the relocation of chromosome territories. Positions of chromosomes 19, 15 and 11 were assessed by 2D-FISH in control siRNA and MYO1C siRNA treated cells. 2D-FISH images were analyzed and quantitated by using the computational script IMACULAT.50 X-axes of the graphs depict the nuclear shell number, with shell 1 being as the outermost and shell 5 being the innermost. Y-axes of the graphs show the percentage of probe normalized to DAPI. N = 2, n > 100 nuclei. Error bars represent the s.e.m. P-values calculated using Student's t-test. “*” and “**” indicate P < 0.05 and P < 0.01, respectively; n.s. = not significant. On graphs C = Center of nucleus, P = Periphery of nucleus. Adapted by permission from Oxford University Press: reference 25, copyright 2016.
Figure 5.
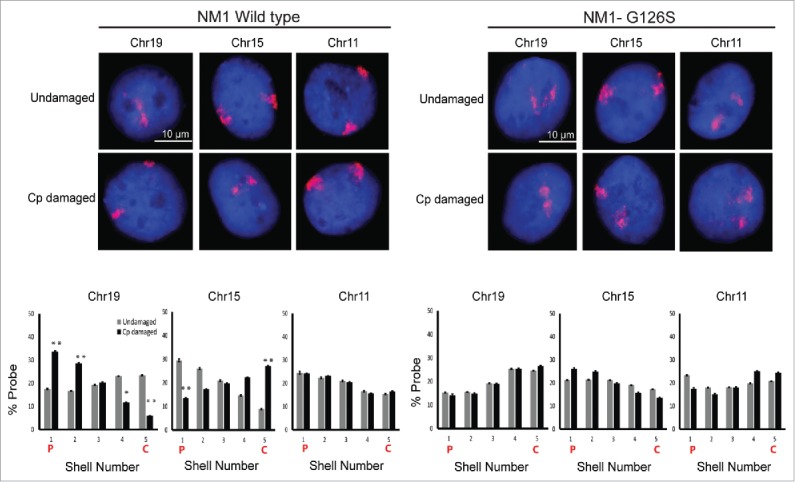
Motor function of nuclear myosin NM1 is required for CT relocation due to damage induced DDR. FISH was used to analyze the positions of CTs 19, 15 and 11 in the NM1 wild type or motor defective over-expressing cells. X-axes of the graphs depict the nuclear shell number, with shell 1 being the outermost and shell 5 being the innermost. Y-axes of the graphs show the percentage of probe normalized to DAPI. N = 2, n = at least 100 nuclei. On graphs C = Center of nucleus, P = Periphery of. Adapted by permission from Oxford University Press: reference 25, copyright 2016.
A hypothetical molecular basis for CT mobility and active transport
Based on our results described here as well as published earlier, we speculate that a co-ordination between early DDR sensing by apical kinases ATM, DNA-PK followed by γH2AX signaling; subsequent chromatin remodeling and NM1 recruitment to the chromatin is required for DNA damage induced CT relocation, which is an integral part of DDR. CT relocation perhaps is a means of orchestrating large-scale remodeling of the nuclear milieu, thereby rewiring the transcriptional program for efficient DDR and maintenance of genomic stability (Fig. 6). One of the intriguing aspects of this model is that NM1 puncta on chromatin do not seem to colocalize with γH2AX repair foci in the nuclei during DDR. However our biochemical assays suggest that γH2AX signaling is functionally coupled to NM1 recruitment, both of which are mandatory for mediating CT relocation changes. The nature of functional coupling between these 2 spatially separate signaling centers is unclear. We are currently probing DDR specific NM1 interactome with the hope that candidate(s) from the same might mediate a cross talk with γH2AX foci. We believe that even though this model captures the essential mechanistic steps underlying CT relocations, a precise nature of molecular cross talk that may finally culminate in systemic changes of CT dynamics is far from clear which forms an active area of future research.
Figure 6.
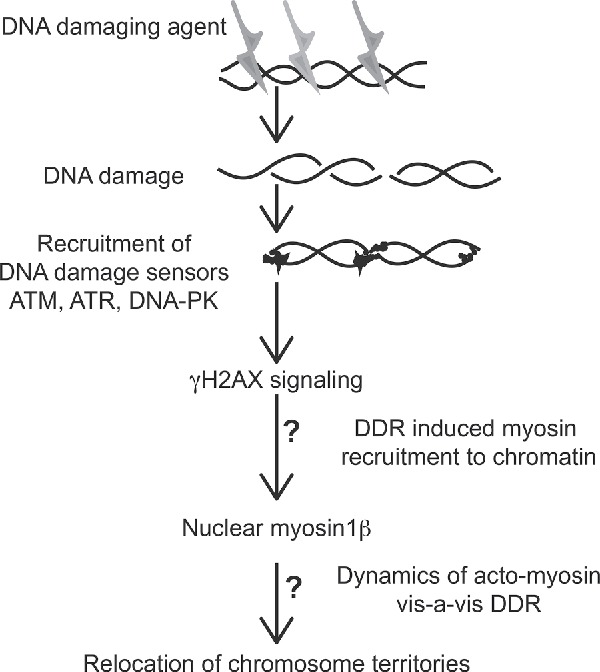
Data derived schematic describing our current understanding of the molecular basis for CT repositioning. The roles of γH2AX-signaling axis of DNA damage response leads to enhanced recruitment of nuclear myosin protein, encoded by NM1 gene, to chromatin. “?” Indicates significant steps of functional coupling whose molecular basis remain unclear.
In addition to the molecular insights on CT dynamics described above, we also sought to learn more about CT repositioning at a more coarse-grained level – at a level that subsumed individual CTs into pairs of CTs. As such studies were unconventional, we used a first principles approach from physics to investigate how CT positioning and its subsequent repositioning was architecturally feasible in the crowded milieu of the human nucleus.
Organization of CTs from kilobase scales to beyond solitary CTs
For nearly 2 decades, long before the noncoding part of the human genome was described and annotated, 2 empirical rules outlined the basis for spatial positioning of chromosomes in interphase nuclei. The first rule contended that gene rich CTs were localized to the nuclear interior,27,28 while in the second one was that small-sized CTs were localized to the nuclear interior.29 However, ambiguities in chromosomal arrangement could not be explained. Moreover, new Hi-C technology provided deep insight in to a fractal-like pattern of chromosomal arrangement,30 which melded well with a crumpled globule model of the DNA that had been proposed using polymer physics approaches.31 Therefore, we asked what information could be obtained from diverse disciplines to gain an insight into the physical basis for CT repositioning?
A physical organization of CT arrangement that subsumes individual CTs
It has been long realized that CTs intermingle with neighboring CTs and other nuclear structures, such as nuclear lamina, within the inter-chromosomal space.32 The spatial proximity among CTs may in turn manifest as translocations among two or more chromosomes at high frequency—beyond random occurrences,19,33 and furthermore it has also been independently reported that intermingling volumes between specific CTs correlate with the frequency of chromosome translocations.32 Although, at chromosomal length scales, these CTs remain relatively stationary during interphase, it has been long reported that chromatin strands (at resolutions finer than nucleosomes) experience constrained diffusion-like dynamics.34 Following that, it was suggested that CTs were not distinct entities that were separated by spaces depleted in chromatin35 and then subsequently confirmed by a series of relatively recent high throughput chromosome conformation capture (Hi-C) experiments from disparate mammals.30,36-41 Evidence for chromosomal-level positional ambiguity (degeneracy) among intermingling CTs has also been strengthened by the results from various ensemble-based Hi-C experiments.30,36-41 Those experiments have generated unflinching support for the possible physical contacts among genomic loci across long-ranges spanning mega bases and across CTs, which were generated from unbiased all-to-all genome-spanning contact maps (heat map as shown in Fig. 7A) at inter-CT length scales, across species. Again, from unbiased Hi-C data, in the context of genome activity it has also been reported that active chromatin marks (such as H3K36 trimethylation sites) correlate with long-range genomic loci, including inter-CT loci.30,42 Additionally, single-cell Hi-C study has revealed statistically significant cell-to-cell variability in inter-CT arrangement, substantiating the organizational plasticity in the murine genome.43 Despite such organizational plasticity, the inter-CT contact interfaces seem to be patterned such that active gene domains are positioned where the CTs contact or intermingle.43 Therefore, we speculate that the 3D organizational plasticity is linked with the genome activity as represented by H3 lysine 4 tri-methylation sites from the single-cell Hi-C study.43
Clearly solitary CTs failed to capture the overall inter-CT dynamics that concerned us, and so we sought a physical scale that was beyond solitary CTs. In an alternate approach, we proposed a physics-based theory for the first time, using linear vector spaces (matrix algebra), whereby it was possible for us to assess the physical basis of such positional ambiguity.44,45 In our novel theory, which we referred to as the paired chromosome gene count (PCGC) theory, we considered pairs of chromosomes as minimal physically interacting units across the genome.45 These abstract pairs provided us with a theoretical suprachromosomal basis to rationalize the ambiguities seen during CT positioning in a radial format, and during CT relocations. Moreover, if the eukaryotic nucleus is perceived as a complex physical system with multiple chromosomes, it is but natural that a solitary chromosome cannot be considered as an independent player that shapes the architecture of the eukaryotic 3D genome. Therefore, we proposed to investigate the complexities of the genome at a higher, but abstract physical dimension, which we termed as the suprachromosomal scale. Our formalism involved solving the mathematics using linear vector algebra (matrices).
A higher order organization in the eukaryotic genome takes the form of chromosomes, each of which encodes several hundred genes. One may mathematically characterize the total number of genes per chromosome (say on the chromosome labeled ) by the product of the chromosomal length and a coarse-grained average gene density . In this formalism, we propose an alternate higher order organization – at a suprachromosomal scale. Analogous to a solitary CT, here we have quantitatively described all possible pairs of nearest-neighbor CT (a pair from CTs labeled and ), which we termed as a suprachromosomal pair, using its effective total gene count ().45 Then, analogous to an isolated chromosomal entity, where the total number of genes is the product of an average coarse-grained gene density and the chromosomal length, we hypothesized that an effective total gene count is a product of the effective gene density () and an effective length of the suprachromosomal pair:
| (1) |
This formalism has, for the first time, delineated a new way of describing the “suprachromosomal” organization comprising of inter-CT couplings. Since the biologic framework of the same is unknown currently, we resorted to a computational description of the same, which enabled us uncover the basic rules of CT hierarchy in the mammalian nuclei.45 Next, we derived this effective gene density term as a sum of the original gene density term representing a solitary CT (intrinsic parameter) plus a correction term (extrinsic parameter), which lends it a semblance of a systems-level parameter that is incumbent on its neighborhood, subsuming a solitary CT. This effective gene density is:
| (2) |
For a diploid male nucleus with 46 CTs, chromosomes 1–22, X and Y, the effective gene density may be mathematically represented as a matrix as shown in Fig. 7B. An unbiased hierarchical clustering generates two primary subgroups of CTs, which we refer to a Group A and Group B.45 We note a remarkable similarity in the contents of Group A and Group B (Fig. 7B)45 to the Clusters 1 and 2 respectively represented in Fig. 7A.38 The subgrouping of all CTs corroborate in the 2 instances except for 4 CTs, 3 of which (chromosomes 14, 15 and 21) are primarily associated with the Nucleolar Organizer Region (NOR). The multiple nucleoli within the human nucleus may therefore represent an assortment, which is specific to the NOR, and involves all 10 homologous acrocentric CTs, 2 each of: chr13–15, chr21 and chr22. The biological basis of NOR heterogeneity, if any, with respect to the assortment of NOR-CT clustering in mammalian cells is an interesting unsolved problem in cell biology. Here, it is important to highlight that our theoretical method does not mandate any spatial information with regard to the positioning of CTs in the interphase nucleus.45 However, on mandating (as boundary condition imposed on a physical system) that the most gene-rich CT (chromosome 19) occupy the interior of the nucleus (which is an experimentally known fact), we can justify the physical basis of a self-organized constellation of all the remaining CTs in the interphase human nucleus (Fig. 8), irrespective of cell-type specificity.
We had presented this systems-level perspective of CTs as an important step forward in our understanding of eukaryotic genome organization because we could corroborate the hierarchy tree from independent ensemble based Hi-C experiments.30,38 Furthermore, in the same report we have reasoned why spatial ambiguity of CT positioning emerges as a natural consequence of this hierarchical tree and corroborated our findings in disparate vertebrate species.45 By applying our theory to other vertebrates with known radial CT arrangement, we correctly predicted their inner core CTs.45 When we sought the human chromosomes with shared conserved synteny to the predicted inner CTs from disparate vertebrates, we discovered that chromosomes 19 and 17 featured prominently.45 Interestingly, our theory does not include any DNA sequence information, but our theoretical results underscore a highly conserved nature of inner CTs across disparate vertebrates in the proposed abstract effective gene density space as well as in the realm of sequence-based information theory.
There is conclusive evidence that spatial organization of chromosomes is dynamic,34 which is further enhanced during DNA damage;13,46-48 but the question persists why? During the discussion of single-cell Hi-C results we highlighted that the crosstalk of genome activity and genome plasticity was revealed from the murine genome. Here, we hypothesize that the same crosstalk may be constrained during evolution, which in turn may suggest that genome plasticity could have evolved across eukaryotes, and not abruptly appear in the murine lineage. It has been proposed that chromatin compartments influence DDR and sub-nuclear compartments, like the nuclear envelope and the nucleolus affect DSB repair,13,46-48 but the component of spatial ambiguity during CT repositioning (or its lack of) has been unexplained. Using this physics-based method, we hypothesize that CT positioning may fundamentally manifest the spatial degrees of freedom that are mandated during DDR in the 3D space. Therefore, hypothetically speaking and irrespective of species, such conformational plasticity may merely be a consequence of genome-level constraints that remain conserved during the course of evolution – from unicellular species with Rabl CT arrangement to multicellular species having radial CT organization. The 3D spatial plasticity of CT arrangement is much restricted for Rabl arrangement because the CTs remain tethered during interphase. However, we hypothesize that in the radial format of CT arrangement, 3D spatial plasticity evolved along with the genome activity and its expansion, both features enabling CT relocation during stress – such as induced serum starvation or controlled cisplatin treatment. Radial format of CT arrangement provides the necessary plasticity in suprachromosomal organization in higher eukaryotes where cell-type specific epigenome function is a critical cellular requirement. Therefore, we believe that this systems-level theoretical approach (PCGC formalism) can capture the genome-centric hierarchy changes during Rabl to radial transition.49 Moreover, PCGC formalism provides a unified and genome-level approach that integrates multiple suprachromosomal organizational states ranging from Rabl to radial formats, perhaps also encompassing intermediate states.49 Therefore, it is of great importance to theoretically predict Rabl and radial arrangements using such physics-based approaches to complement experiments.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Felsenfeld G, McGhee JD. Structure of the 30 nm chromatin fiber. Cell 1986; 44:375-7; PMID:3510744; https://doi.org/ 10.1016/0092-8674(86)90456-3 [DOI] [PubMed] [Google Scholar]
- [2].Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2001; 2:292-301; PMID:11283701; https://doi.org/ 10.1038/35066075 [DOI] [PubMed] [Google Scholar]
- [3].Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell 2013; 152:1270-84; PMID:23498936; https://doi.org/ 10.1016/j.cell.2013.02.001 [DOI] [PubMed] [Google Scholar]
- [4].Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet 2013; 14:390-403; PMID:23657480; https://doi.org/ 10.1038/nrg3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 2015; 16:245-57; PMID:25757416; https://doi.org/ 10.1038/nrm3965 [DOI] [PubMed] [Google Scholar]
- [6].Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J Cell Biol 2005; 168:375-87; PMID:15684028; https://doi.org/ 10.1083/jcb.200409091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Berr A, Schubert I. Interphase chromosome arrangement in Arabidopsis thaliana is similar in differentiated and meristematic tissues and shows a transient mirror symmetry after nuclear division. Genetics 2007; 176:853-63; PMID:17409060; https://doi.org/ 10.1534/genetics.107.073270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol Biol Cell 1996; 7:825-42; PMID:8744953; https://doi.org/ 10.1091/mbc.7.5.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al.. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316:1160-6; PMID:17525332; https://doi.org/ 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- [10].Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T. A systems approach to mapping DNA damage response pathways. Science 2006; 312:1054-9; PMID:16709784; https://doi.org/ 10.1126/science.1122088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutation Res 2005; 577:275-83; https://doi.org/ 10.1016/j.mrfmmm.2005.03.007 [DOI] [PubMed] [Google Scholar]
- [12].Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science 2001; 291:1284-9; PMID:11181991; https://doi.org/ 10.1126/science.1056154 [DOI] [PubMed] [Google Scholar]
- [13].Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 2009; 10:243-54; PMID:19277046; https://doi.org/ 10.1038/nrm2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 2004; 303:92-5; PMID:14704429; https://doi.org/ 10.1126/science.1088845 [DOI] [PubMed] [Google Scholar]
- [15].Mehta IS, Kulashreshtha M, Chakraborty S, Kolthur-Seetharam U, Rao BJ. Chromosome territories reposition during DNA damage-repair response. Genome Biol 2013; 14:R135; PMID:24330859; https://doi.org/ 10.1186/gb-2013-14-12-r135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res 1999; 27:1223-42; PMID:9973609; https://doi.org/ 10.1093/nar/27.5.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461:1071-8; PMID:19847258; https://doi.org/ 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 2011; 25:409-33; https://doi.org/ 10.1101/gad.2021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol 2004; 5:R44; PMID:15239829; https://doi.org/ 10.1186/gb-2004-5-7-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mehta IS, Amira M, Harvey AJ, Bridger JM. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol 2010; 11:R5; PMID:20070886; https://doi.org/ 10.1186/gb-2010-11-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bridger JM, Boyle S, Kill IR, Bickmore WA. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol 2000; 10:149-52; PMID:10679329; https://doi.org/ 10.1016/S0960-9822(00)00312-2 [DOI] [PubMed] [Google Scholar]
- [22].Thomson I, Gilchrist S, Bickmore WA, Chubb JR. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol 2004; 14:166-72; PMID:14738741; https://doi.org/ 10.1016/j.cub.2003.12.024 [DOI] [PubMed] [Google Scholar]
- [23].Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol 2003; 160:685-97; PMID:12604593; https://doi.org/ 10.1083/jcb.200211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Essers J, van Cappellen WA, Theil AF, van Drunen E, Jaspers NG, Hoeijmakers JH, Wyman C, Vermeulen W, Kanaar R. Dynamics of relative chromosome position during the cell cycle. Mol Biol Cell 2005; 16:769-75; PMID:15574874; https://doi.org/ 10.1091/mbc.E04-10-0876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kulashreshtha M, Mehta IS, Kumar P, Rao BJ. Chromosome territory relocation during DNA repair requires nuclear myosin 1 recruitment to chromatin mediated by Upsilon-H2AX signaling. Nucleic Acids Res 2016; 44(17):8272-91; PMID:27365048; https://doi.org/ 10.1093/nar/gkw573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dzijak R, Yildirim S, Kahle M, Novak P, Hnilicova J, Venit T, Hozák P. Specific nuclear localizing sequence directs two myosin isoforms to the cell nucleus in calmodulin-sensitive manner. PLoS One 2012; 7:e30529; PMID:22295092; https://doi.org/ 10.1371/journal.pone.0030529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet 2001; 10:211-9; PMID:11159939; https://doi.org/ 10.1093/hmg/10.3.211 [DOI] [PubMed] [Google Scholar]
- [28].Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 1999; 145:1119-31; PMID:10366586; https://doi.org/ 10.1083/jcb.145.6.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sun HB, Shen J, Yokota H. Size-dependent positioning of human chromosomes in interphase nuclei. Biophys J 2000; 79:184-90; PMID:10866946; https://doi.org/ 10.1016/S0006-3495(00)76282-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al.. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326:289-93; PMID:19815776; https://doi.org/ 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Grosberg AY, Nechaev SK, Shakhnovich EI. The role of topological constraints in the kinetics of collapse of macromolecules. J Phys France (Journal de physique) 1988; 49(12):2095–2100; https://doi.org/ 10.1051/jphys:0198800490120209500 [DOI] [Google Scholar]
- [32].Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol 2006; 4:e138; PMID:16623600; https://doi.org/ 10.1371/journal.pbio.0040138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bickmore WA, Teague P. Influences of chromosome size, gene density and nuclear position on the frequency of constitutional translocations in the human population. Chromosome Res 2002; 10:707-15; PMID:12575798; https://doi.org/ 10.1023/A:1021589031769 [DOI] [PubMed] [Google Scholar]
- [34].Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol 1997; 7:930-9; PMID:9382846; https://doi.org/ 10.1016/S0960-9822(06)00412-X [DOI] [PubMed] [Google Scholar]
- [35].Fedorova E, Zink D. Nuclear architecture and gene regulation. Biochim Biophys Acta 2008; 1783:2174-84; PMID:18718493; https://doi.org/ 10.1016/j.bbamcr.2008.07.018 [DOI] [PubMed] [Google Scholar]
- [36].Ay F, Bunnik EM, Varoquaux N, Bol SM, Prudhomme J, Vert JP, Noble WS, Le Roch KG. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res 2014; 24:974-88; PMID:24671853; https://doi.org/ 10.1101/gr.169417.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012; 485:376-80; PMID:22495300; https://doi.org/ 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol 2012; 30:90-8; https://doi.org/ 10.1038/nbt.2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell United States: Elsevier Inc 2012; 148:458-72; PMID:22265598; https://doi.org/ 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- [40].Varoquaux N, Liachko I, Ay F, Burton JN, Shendure J, Dunham MJ, Vert J-P, Noble WS. Accurate identification of centromere locations in yeast genomes using Hi-C. Nucleic Acids Res 2015; 43:5331-9; PMID:25940625; https://doi.org/ 10.1093/nar/gkv424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res 2015; 25:246-56; PMID:25367294; https://doi.org/ 10.1101/gr.170332.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yaffe E, Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet 2011; 43:1059-65; PMID:22001755; https://doi.org/ 10.1038/ng.947 [DOI] [PubMed] [Google Scholar]
- [43].Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013; 502:59-64; PMID:24067610; https://doi.org/ 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fatakia SN, Mehta IS, Rao BJ. Unified theory of human genome reveals a constrained spatial chromosomal arrangement in interphase nuclei. arXiv:150908074 [q-bioGN]: arXiv.org 2015; http://adsabs.harvard.edu/cgi-bin/bib_query?arXiv:1509.08074 [Google Scholar]
- [45].Fatakia SN, Mehta IS, Rao BJ. Systems-level chromosomal parameters represent a suprachromosomal basis for the non-random chromosomal arrangement in human interphase nuclei. Scientific Reports 2016; 6:36819; PMID:27845379; https://doi.org/ 10.1038/srep36819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Adam S, Dabin J, Polo SE. Chromatin plasticity in response to DNA damage: The shape of things to come. DNA Repair 2015; 32:120-6; PMID:25957486; https://doi.org/ 10.1016/j.dnarep.2015.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Polo SE. Reshaping chromatin after DNA damage: the choreography of histone proteins. J Mol Biol 2015; 427:626-36; PMID:24887097; https://doi.org/ 10.1016/j.jmb.2014.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Polo SE, Almouzni G. Chromatin dynamics after DNA damage: The legacy of the access-repair-restore model. DNA Repair 2015; 36:114-21; PMID:26429064; https://doi.org/ 10.1016/j.dnarep.2015.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fatakia SN, Rao BJ. Genome-level parameters describe the pan-nuclear fractal nature of eukaryotic interphase chromosomal arrangement. bioRxiv/2016/12/09/083121: BIORXIV 2016; https://doi.org/ 10.1101/083121 [DOI] [Google Scholar]
- [50].Mehta I, Chakraborty S, Rao BJ. IMACULAT - an open access package for the quantitative analysis of chromosome localization in the nucleus. PLoS One 2013; 8:e61386; PMID:23577217; https://doi.org/ 10.1371/journal.pone.0061386 [DOI] [PMC free article] [PubMed] [Google Scholar]


