ABSTRACT
Brassinosteroids (BRs) are critical for the plant growth and development. BRs signal through the plasma membrane localized receptor-like kinases to downstream transcription factors BES1/BZR1 to regulate the expression of thousands of genes for various BR responses. In addition to the role in plant growth and development, BRs have been implicated in responses to environmental stresses such as drought. However, the mechanism through which BRs regulate drought have just begun emerging. We have recently found that a group of WRKY transcription factors, WRKY46, WRKY54, WRKY70, which are well known for the function in abiotic and biotic stress, cooperates with BES1 to mediate BR-regulated drought response. The wrky46 wrky54 wrky70 triple mutants showed growth defect, likely due to impaired BR signaling as well as some reduction of endogenous BR level. WRKY46/54/70 cooperates with BES1 to regulate the expression of BR target genes to promote growth. We also found that WRKY46/54/70 negatively modulates drought tolerance by globally repressing drought-inducible gene expression. Thus, our result uncovers a new role for WRKY transcription factors in BR signaling and provides the molecular mechanism for BR-regulated plant growth and drought stress through WRKY46/54/70 and BES1 transcription factors.
KEYWORDS: Brassinosteroids, BES1, BIN2, drought responses, plant growth, WRKY46, WRKY54, WRKY70
Brassinosteroids (BRs), a group of plant steroidal hormones, regulates a wide range of plant growth and development, such as cell elongation, cell division, xylem differentiation and stress responses.1,2 BR is perceived by receptors BRI1 and its co-receptor BAK1, which initiates a signaling cascade via several intermediates, such as BSK1, BSU1 and BIN2, to downstream transcription factors BES1/BZR1, regulating thousands of target genes for BR response.3,4 In addition to its critical role in plant growth, genetic studies have shown that BRs play a negative role in drought stress response. Loss-of-function BR mutants displayed increased drought tolerance.5–8 Recent studies started to shed light on the mechanism of the crosstalk between BR signaling and drought response. RD26, a stress-inducible NAC family transcription factor, antagonizes BES1 to mediate the crosstalk between BR-regulated growth and drought responses.7 Under drought and other stress conditions, BES1 is targeted for autophagy-mediated degradation through SINAT E3 ubiquitin ligase and ubiquitin receptor protein DSK2, thereby regulating the trade-off between plant growth and stress response.8,9 Here, we identified a group of WRKY transcription factors that cooperate with BES1 to mediate the crosstalk between BR-regulated plant growth and drought responses, in which, unlike RD26 and its homologs, WRKYs play a negative role in drought responses.
WRKY transcription factors, a family of plant-specific transcription factors, comprises more than 70 members in higher plants.10 WRKY is defined by its WRKY domain, a conserved DNA-binding domain containing 60 amino acid residues, which binds to W-box (TTGACC/T) at the target promoters.11 In addition, WRKY possesses zinc-finger motif at its C-terminus, either Cx4–5Cx22–23HxH (CCHH) or Cx7Cx23HxC (CCHC).12 WRKY family members are involved in various biologic processes, including trichome and seed development, senescence, biotic and abiotic stress response.13–18 In Arabidopsis, WRKY54 and WRKY70 negatively control osmotic stress tolerance by inhibiting stomatal closure.19 WRKY46 was also found to modulate plant responses to drought stress and stomatal aperture.18 Besides their negative role in drought stress, in this study, we presented multiple lines of evidence that WRKY46, WRKY54 and WRKY70 function as positive regulators in BR-regulated plant growth.20
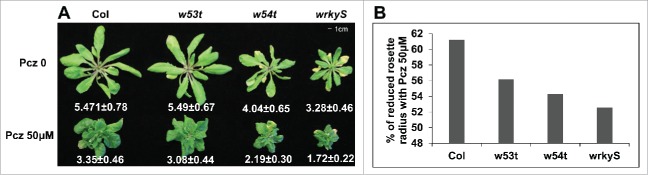
The knockout triple mutant wrky46 wrky54 wrky70 (w54t) displayed growth defects with shorter blade lengths, blade width and petiole length, suggesting that they play positive roles in plant growth.20 The w54t mutant has a slightly reduced accumulation of endogenous BR content and disrupted BR signaling.20 The w54t mutant has compromised BR response in hypocotyl elongation.20 To assess if w54t have compromised BR response at adult stage, we treated the WT and mutant plants with propiconazole (Pcz) that reduces endogenous BR level.21 The w54t mutant plants are more sensitive to Pcz in adult stage (Fig. 1A). Under Pcz treatment, the maximum radius of 4-week-old adult WT decreased to 61% compared with mock conditions, whereas w54t mutant deceased to 54% (Fig. 1B). The wrky30 wrky41 wrky53 (w53t) showed a reduction of radius to 56%, which was further decreased in wrky30 wrky41 wrky53 wrky46 wrky54 wrky70 (wrkyS) to 52% compared with mock treatment (Fig. 1B). Furthermore, our transient expression showed that WRKY46/54/70 cooperates with BES1 to regulate the expression of BR target genes through direct physical interaction.20 Taken together, all the evidence suggested that WRKY46/54/70 play positive roles in BR-regulated plant growth.
Figure 1.
w54t is more sensitive to propiconazole (Pcz) treatment. (A) The growth phenotype of 4-week-old Col, wrky30 wrky41 wrky53 (w53t), wrky46 wrky54 wrky70 (w54t) and wrky30 wrky41 wrky53 wrky46 wrky54 wrky70 (wrkyS) watered with Pcz 50μM or mock. The number below the plants are the average maximum radius of rosette leaves and standard deviation. N = 12. (B) The bar graph of percentage of reduced rosette radius with Pcz treatment. Percentage is average maximum radius of rosette leaves with Pcz 0 divided by that with Pcz 50μM and then multiplying by 100.
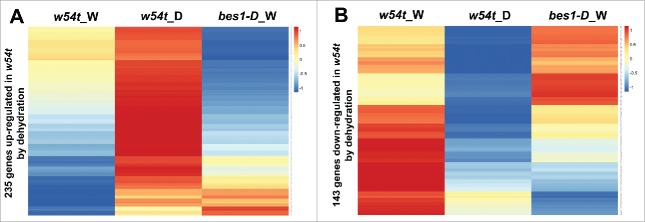
Consistent with the previous studies of the phenotype of WRKY46/54/70 in response to osmotic stress, the w54t mutant showed enhanced tolerance to drought with a higher survival rate.18–20 Our global gene expression analysis revealed that 53% of dehydration upregulated genes are constitutively upregulated in w54t mutant, which is further induced upon dehydration treatment in the mutant. Similarly, 64% of dehydration downregulated genes are constitutively off in w54t mutant, further decreased upon dehydration..20 Furthermore, large portions of genes differentially expressed in w54t by dehydration are also regulated in bes1-D, a gain-of-function mutant of BES1 gene.20 More specifically, most dehydration upregulated genes in w54t are downregulated in bes1-D, and most dehydration downregulated genes in w54t are upregulated in bes1-D (Fig. 2A, B). WRKY54 and BES1 cooperate to inhibit the transcription of GLYI7, a drought-inducible gene, by binding to the W-box and G-box regions, respectively.20 Thus, our result suggested that WRKY46/54/70 are negative regulators of BR-regulated drought response.
Figure 2.
Clustering analysis of genes upregulated (A) and downregulated (B) in w54t mutant by dehydration treatment in bes1-D, a gain-of-function mutant. Values indicate normalized expression levels.
BIN2, a glycogen synthase kinase (GSK)-3 like kinase, is a central negative regulator in BR signaling.22 BIN2 substrates share a conserved phosphorylation motif S/TXXXS/T (S is serine, T is threonine and X is any amino acid). WRKY54 has 29 putative phosphorylation sites and is phosphorylated by BIN2 via direct interaction.20 Similar to BIN2 phosphorylation of BES1, phosphorylation of WRKY54 by BIN2 also leads to the protein destabilization.20,23 It is plausible that BIN2 phosphorylation and destabilization of WRKY54 release the inhibitory effect of WRKY54 on the drought-inducible gene expression during drought. Interestingly, we found that the protein level of WRKY54 and BES1 decreases under the drought stress, which is likely caused by BIN2 phosphorylation.20 Abscisic acid (ABA), a stress-inducible phytohormone, inhibits BR signaling by regulating BIN2.24 Therefore, it is most likely that drought stress leads to the elevated BIN2 level causing the protein destabilization of WRKY54 and BES1, resulting in increased expression of drought-inducible genes to combat drought for survival (Fig. 3). On the other hand, under the normal condition, BR inhibits BIN2 activity, causing increased protein level of WRKY54 and BES1, which then activate the growth-related gene expression to favor growth (Fig. 3).
Figure 3.

(A)model for WRKY in BR signaling and drought response. See text for details.
In conclusion, our study revealed a novel role for WRKY transcription factors in the crosstalk between BR-regulated growth and drought stress by cooperating with BES1, which is regulated by BIN2 kinase. In the future, identification of WRKY46/54/70 partners and target genes will further advance our understanding of the crosstalk between BR-regulated growth and drought responses.
Acknowledgments
The work was supported by grants from the National Institutes of Health (1R01GM120316–01A1), and by the Plant Sciences Institute at Iowa State University. We thank Data2Bio (Ames, IA) for performing RNA-seq analysis. J. Chen was partially supported by fellowship from China Scholarship Council.
References
- 1.Fujioka S, Yokota T. Biosynthesis and metabolism of brassinosteroids. Ann Rev Plant Biol. 2003;54:137–64. doi: 10.1146/annurev.arplant.54.031902.134921. [DOI] [PubMed] [Google Scholar]
- 2.Choe S. Brassinosteroid biosynthesis and inactivation. Physiol Plant. 2006;126:539–48. doi: 10.1111/j.1399-3054.2006.00681.x. [DOI] [Google Scholar]
- 3.Clouse SD. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23:1219–30. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo H, Li L, Aluru M, Aluru S, Yin Y. Mechanisms and networks for brassinosteroid regulated gene expression. Curr Opin Plant Biol. 2013;16:545–53. doi: 10.1016/j.pbi.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Beste L, Nahar N, Dalman K, Fujioka S, Jonsson L, Dutta PC, Sitbon F. Synthesis of hydroxylated sterols in transgenic Arabidopsis plants alters growth and steroid metabolism. Plant Physiol. 2011;157:426–40. doi: 10.1104/pp.110.171199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Northey JG, Liang S, Jamshed M, Deb S, Foo E, Reid JB, McCourt P, Samuel MA. Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nat Plants. 2016;2:16114. doi: 10.1038/nplants.2016.114. [DOI] [PubMed] [Google Scholar]
- 7.Ye H, Liu S, Tang B, Chen J, Xie Z, Nolan TM, Jiang H, Guo H, Lin HY, Li L, et al.. RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat Commun. 2017;8:14573. doi: 10.1038/ncomms14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nolan TM, Brennan B, Yang M, Chen J, Zhang M, Li Z, Wang X, Bassham DC, Walley J, Yin Y. et al.. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev Cell. 2017;41:33–46. doi: 10.1016/j.devcel.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang M, Li C, Cai Z, Hu Y, Nolan T, Yu F, Yin Y, Xie Q, Tang G, Wang X. et al.. SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev Cell. 2017;41:47–58. doi: 10.1016/j.devcel.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 2004;7:491-. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 11.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 12.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–58. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Johnson C.S. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14:1359–75. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao YL, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol. 2004;55:853–67. doi: 10.1007/s11103-005-2142-1. [DOI] [PubMed] [Google Scholar]
- 15.Luo MD, Dennis ES, Berger F, Peacoc WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:17531–6. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, Dong Q, Yu D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012;185–186:288–97. doi: 10.1016/j.plantsci.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Ding ZJ, Yan JY, Li GX, Wu ZC, Zhang SQ, Zheng SJ. WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 2014;79:810–23. doi: 10.1111/tpj.12597. [DOI] [PubMed] [Google Scholar]
- 18.Ding ZJ, Yan JY, Xu XY, Yu DQ, Li GX, Zhang SQ, Zheng SJ. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 2014;79:13–27. doi: 10.1111/tpj.12538. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Besseau S, Törönen P, Sipari N, Kollist H, Holm L, Palva ET. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013;200:457–72. doi: 10.1111/nph.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Nolan TM, Ye H, Zhang M, Tong H, Xin P, Chu J, Chu C, Li Z, Yin Y. Arabidopsis WRKY46, WRKY54 and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought response. Plant Cell. 2017;29:1425–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartwig T, Corvalan C, Best NB, Budka JS, Zhu JY, Choe S, Schulz B. Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS ONE. 2012;7:e36625. doi: 10.1371/journal.pone.0036625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Youn JH, Kim TW. Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol Plant. 2015;8:552–65. doi: 10.1016/j.molp.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–91. doi: 10.1016/S0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhang SC, Cai Z, Wang X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106:4543–8. doi: 10.1073/pnas.0900349106. [DOI] [PMC free article] [PubMed] [Google Scholar]