ABSTRACT
The expression of cold-induced genes is critical for plants to survive under freezing stress. However, the underlying mechanisms for the decision of when, where, and which genes to express are unclear when a plant meets a sudden temperature drop. Previous studies have demonstrated epigenetics to play a central role in the regulation of gene expression in plant responses to environmental stress. DNA methylation and histone deacetylation are the two most important epigenetic modifications. This study was conducted to investigate the effects of inhibiting DNA methylation and histone deacetylation on gene expression, and to explore the potential role of epigenetics in plant responses to cold stress. The results revealed that histone deacetylase inhibitors (trichostatin A) and DNA methylation inhibitors (5-Aza-2′-deoxycytosine) treatment enhanced cold tolerance. DNA microarray analysis and the gene ontology method revealed 76 cold-induced differently expressed genes in Arabidopsis thaliana seedlings that were treated to 0°C for 24 h following Trichostatin A and 5-Aza-2′-Deoxycytidine. Furthermore, analyses of metabolic pathways and transcription factors of 3305 differentially expressed genes were performed. Each four metabolic pathways were significantly affected (p < 0.01) by Trichostatin A and 5-Aza-2′-Deoxycytidine. Finally, 10 genes were randomly selected and verified via qPCR analysis. Our study indicated that Trichostatin A and 5-Aza-2′-Deoxycytidine can improve the plant cold resistance and influence the expression of the cold-induced gene in A. thaliana. This result will advance our understanding of plant freezing responses and may provide a helpful strategy for cold tolerance improvement in crops.
KEYWORDS: Arabidopsis thaliana, cold-induced genes, cold tolerance, Trichostatin A, 5-Aza-2′-Deoxycytidine
Introduction
Fluctuations of atmospheric temperatures occur more frequently due to global warming, impacting all aspects of plant growth and development.1–4 Freezing damage in plants is a severe problem that can cause cellular dehydration due to extracellular formation of ice, consequently causing severe crop loss.5 Plants respond to environmental stresses by altering the expression of their genes. Freezing tolerance attributes to the expression of antifreeze genes.6,7 However, the regulatory mechanisms behind these gene expressions remain unclear.
Epigenetic modifications, such as DNA methylation and histone deacetylation provide stable gene silencing mechanisms that regulate gene expression by altering chromatin architecture, while regulating key cellular processes including transcription, DNA replication, and DNA repair. Epigenetic modifications have been studied as a regulatory mechanism associated with plant adaptation to environmental stresses.8–14 Histone deacetylation is a type of histone modification that may regulate key cellular processes, thus sensing changes of the external environment.15–18 When plant seedlings are exposed to environmental stress, a genome-wide methylation/demethylation occurs in different tissues.19, 20
Trichostatin A (TSA) has recently been applied in the treatment of serious diseases, e.g. latent HIV-1 provirus,21 Glioblastoma,22 and anti-tumor responses.23 However, the application of TSA penetrates into all aspects of plant science, e.g. the mediation of cellular dedifferentiation,24 the effect of pollen germination and pollen tube growth,25 the change of concentration of the superoxide anion within cells,26 and the inhibition of germination.27 Compared to Trichostatin A, the reports about 5-Aza-2′-Deoxycytidine (aza-dC) are few; however, it is also a very promising small molecular drug,28 and it has been used for studies of plant epigenetics. However, associations between both inhibitors and plant antifreeze are less well established.
The capability of plant cold tolerance attributes to rapid transcription of special genes29 and changes of gene expression before and after the freezing treatment in the whole genome transcriptome.30 The expressions of cold-induced genes are accurately regulated by epigenetic modification.31,32 Can the inhibitors of histone deacetylation and DNA methylation affect the expression of cold-induced genes? Is there any regularity? In this study, we investigated these questions.
Here, the effects of Trichostatin A and 5-Aza-2′-Deoxycytidine for plant cold response were investigated. Interestingly, the results revealed that Trichostatin A and 5-Aza-2′-Deoxycytidine treatment enhanced the cold tolerance, Trichostatin A in particular. Microarray data suggests that this disturbance of expression of several genes and several biological pathways may be the cause of this phenomenon. Moreover, the result revealed Trichostatin A and 5-Aza-2′-Deoxycytidine to be antagonists. Our study improves our understanding of the expression of plant cold-induced gene regulated by epigenetic modifications. With the appropriate dosage, these inhibitors of epigenetic modifications may help to improve crop traits.
Results
TSA and aza-dC improved cold resistance
To observe the effect of TSA and aza-dC for plant cold resistance, the lethal temperature was calculated via 50% electrolyte leakage. After repeated determination, the results unexpectedly revealed that TSA and aza-dC improved the cold resistance, which was particularly significant for TSA. As shown in Fig. 1, under the same experimental conditions, the half lethal temperature of the treatment group was lower than that of the control group. The difference between the TSA treatment (−2.24°C) and the control group (−1.63°C) was significant (p < 0.05). The half lethal temperature remained consistent with the control group when TSA and aza-dC were applied jointly (Fig. 1).
Figure 1.
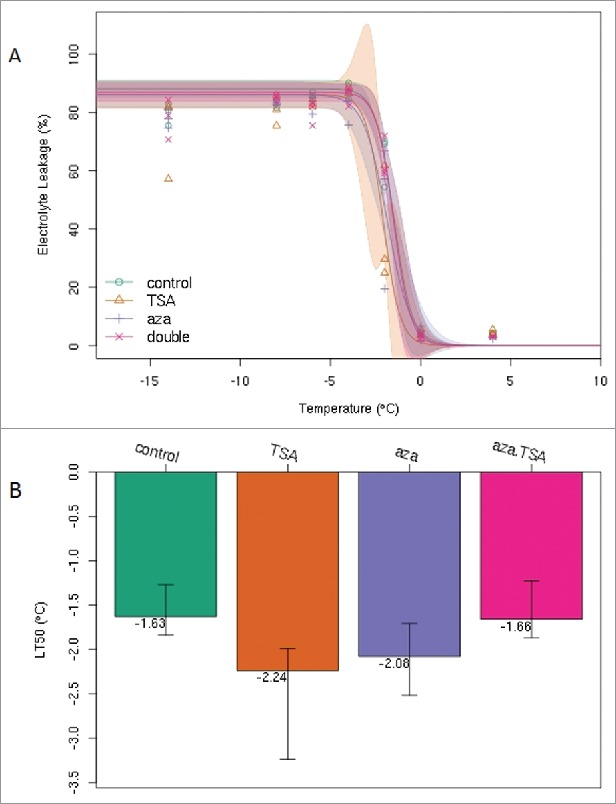
The half lethal temperature (LT50) was determined according to the electrical conductivity between control group and treatment groups. Error bars indicate standard errors (n = 12), and the experiment was repeated more than ten times. (A) Conductivity curve treated with TSA, aza-dC, and TSA + aza-dC at different temperatures (10°C – 20°C). (B) Semi-lethal temperature for different treatments.
Identification of differentially expressed genes, gene ontology (GO) analysis, and cold-induced gene digging
The data were analyzed using ANOVA to identify significant differential genes. A total of 3305 differentially expressed genes were identified (supplemental file 1). To excavate the functional groups associated with differentially expressed genes, gene ontology analysis was performed via the GSEABase package of BioConductor (http://www.bioconductor.org/).
In summary, more genes were down-regulated than up-regulated following TSA treatment, indicating that TSA was a negtive regulator for gene expression (Fig. 2A). Three hundred forty six genes of stress response were differentially expressed with aza-dC treatment, and the number of up-regulated genes was greater than that of down-regulated genes in response stress genes, indicating that aza-dC played an activating role for most abiotic stress genes during freezing (Fig. 2B). The annotation of cellular components was enriched in membrane components and cell wall components, suggesting response regions within the genome regulated by TSA for abiotic stress in plants (Fig. 2A). Interestingly, with aza-dC treatment, many gene expressions were inhibited in the chloroplast and cytoplasm (Fig. 2B), indicating the aza-dC had a positive effect on the regulation of gene expression in the chloroplast and cytoplasm when the plants meet stress. This provides clues for the research on the plant cold tolerance mechanism. According to an enrichment analysis of molecular functions, the differentially expressed genes were primarily involved in iron ion binding factors, transferases and redoxase activities, suggesting TSA and aza-dC affected photosynthetic rates, plasma membrane electron transport, and a variety of metabolic pathways. The details of differentially expressed genes can be accessed via supplemental file 2 and 3.
Figure 2.

Enrichment analysis of differently expressed genes. The enrichment analysis of different expressional genes at 0°C for 24 h following TSA (A) or aza-dC (B) on the biological process, molecular function, and cellular components are displayed.
Following the results of GO ontology analysis, 76 cold related genes were selected for hierarchical cluster analysis (Fig. 3 and supplemental file 4). The results reveal that 33 cold stress response genes were up-regulated and 43 down-regulated following aza-dC treatment; 32 genes were up-regulated and 44 were down-regulated following TSA treatment; 44 genes were up-regulated and 32 down-regulated following both inhibitors (Fig. 3). For the membrane related protein At1g55960 for example, the expression was up regulated by TSA treatment, while two FAD-linked oxidoreductases (At1g30720 and At4g20830) were down regulated by TSA treatment, suggesting TSA involvement in membrane protection and a reduction of energy dissipation.
Figure 3.

Cytosine methylation and histone deacetylation modulate the expression of cold responsive genes. (A) Hierarchical cluster analysis of 76 cold related genes selected according to the results of gene ontology analysis. (B) A. thaliana seedlings treated with TSA, aza-dC, and TSA+aza-dC were cultured in petri-dishes for a 0°C cold stress of 24 h. (C) Venn diagrams of 76 cold-induced genes.
Real time quantitative PCR was performed to validate the microarray data (Fig. 4). These results support the validity of the GeneChip data analysis.
Figure 4.
Expression analysis of ten genes via qPCR. Expression levels relative to 22°C control seedlings are plotted and represent mean values of four independent PCRs ± SD.
Biological pathway enrichments affected by TSA and aza-dC under freezing
Differentially expressed genes were projected to the KEGG pathway database via GenMAPP v2.1. To find significantly different pathways, one-way ANOVA analysis was carried out via SPSS. For aza-dC and TSA treatments, 61 pathways (supplemental file 9) and 34 pathways were identified (supplemental file 10). Of these, four had p-values < 0.01 in both groups separately (Table 1).
Table 1.
Changes in biological pathways with the TSA and aza-dC treatment.
| Treatment | Term | Count | P-Value |
|---|---|---|---|
| TSA | Phenylpropanoid biosynthesis | 24 | 8.14E-09 |
| Methane metabolism | 14 | 6.31E-06 | |
| Phenylalanine metabolism | 14 | 7.27E-06 | |
| Metabolism of xenobiotics by cytochrome P450 | 5 | 0.005927 | |
| aza-dC | Photosynthesis | 30 | 1.50E-07 |
| Photosynthesis – antenna proteins | 13 | 2.07E-06 | |
| Carbon fixation in photosynthetic organism | 23 | 9.70E-04 | |
| Nitrogen metabolism | 15 | 0.003602 | |
| TSA+aza-dC | = = = = = = = = = = = = = | = = | = = = = = = |
These results revealed that multiple biological pathways are associated with these differentially expressed genes, suggesting that TSA and aza-dC affected plant metabolic activities. Using the nitrogen metabolic pathway as an example, clarifies this: the results of the analysis are presented in Table 2 and revealed that the expression of genes encoding glutamate dehydrogenase and nitrilase was up-regulated by aza-dC treatment. However, gene expressions encoding carbonic anhydrase, amino methyl transferase, phenylalanine ammonia-lyase, asparagine synthetase, glutamine synthetase, and formamidase synthetase were down-regulated. This reveals that aza-dC affected the activity of glutamine synthetase and glutamyl dehydrogenase in amino acid synthesis pathways.
Table 2.
Differentially expressed genes in nitrogen metabolism pathway with the aza-dC treatment.
| Enzymology Code Description Gene ID and Name |
| Up-Regulation |
| EC3.5.5.1 nitrilase AT3G44300 (NIT2) |
| AT5G22300 (NIT4) |
| EC1.4.1.3 mitochondria glutamate dehydrogenase (NAD(P)+) AT5G07440 (GDH2) |
| AT5G18170 (GDH1) |
| cytosolic glutamine synthetase AT5G37600 (GSR1) |
| Down-Regulation |
| EC4.2.1.1 carbonic anhydrase AT1G70410 (BCA4) |
| AT3G01500 (CA1) |
| AT5G14740 (CA2) |
| EC2.1.2.10 aminomethyltransferase AT1G11860 |
| EC4.3.1.24 phenylalanine ammonialyase AT3G10340 (PAL4) |
| EC3.5.1.49 formamidase AT4G37560 |
| T4G37550 |
| EC6.3.5.4 asparagine synthase (glutamine-hydrolysing) AT5G65010 (ASN2) |
| EC1.4.7.1 glutamate synthase (ferredoxin) AT5G04140 (Fd-GOGAT) |
| EC6.3.1.2 chloroplastic glutamine synthetase AT5G35630 (GS2) |
Discussion
Interestingly, repeated experiments revealed that Trichostatin A and 5-Aza-2′-Deoxycytidine improves plant cold tolerance. TSA and aza-dC could have affected the expression of particular genes, and microarray data suggests their involvement in biological pathway adjustment. Moreover, the result revealed that Trichostatin A and 5-Aza-2′-Deoxycytidine are antagonists. This is consistent with previous research.33 Our study suggests that TSA and aza-dC may serve agricultural production as novel agents.
Our previous studies revealed an interaction between plant histone deacetylase and DNA methytransferase,34 and the interacting activities are involved in retaining genomic stability and stress responses in plants.18,35 In further analysis, the results from KEGG pathway analysis demonstrated that treatment of A. thaliana seedlings with 0°C and TSA significantly affected the biosynthetic pathway of phenylpropanes, the metabolic pathway of methane, the symbiotic metabolic pathway of phenylalanine, and the symbiotic metabolism-related pathway caused by cytochrome p450, indicating that histone deacetylation plays a critical role in plant freezing responses (Table 1). Plant cytochrome p450 is found in various subcellular locations in soluble and membrane bound forms and plays an important role in preventing injury of harmful substances via catalyzing in plants.36 In the metabolism of xenobiotics via the cytochrome P450 pathway (supplementary file 10), alcohol dehydrogenase family proteins and plant glutathione transferases were of particular interest, as these have been known to play a functional role in anti-exotoxicants.37,38 Treating A. thaliana seedlings with freezing and TSA may affect ADH1 and GSTU27 activity.
Treatment with aza-dC significantly impacted the photosynthesis pathway, the pathway of carbon fixation in photosynthetic organisms, and the nitrogen metabolic pathway. Several subtle links between these were found, with nitrogen metabolic pathway as an example; nitrate reduction is closely related to carbon metabolism, and the energy of photosynthesis 25% was for nitrate reduction.39 Photosynthetic carbon metabolism and NO2− assimilation occur in chloroplasts, and both consume organic carbon and energy from carbon assimilation, photosynthesis, and electron transport chain. The expression of glutamate dehydrogenase (GDH) encoding genes was up-regulated (Table 2). GDH plays a unique role in the synthesis of glutamic acid and the metabolism of NH4+. It plays an initial role in the synthesis and re-synthesis of NH4+, and is a supplementary in the glutamic acid synthesis cycle; however, its physiological role is still controversial in higher plants.40 GDH has two forms, one is dependent on NADH and is found in the mitochondria, another is dependent on NADPH and can be found in the chloroplasts. The former was regulated by DNA methylation, while the encoding glutamate synthase (Fd-GOGAT) was down-regulated. Furthermore, GOGAT have two forms, one depends on NADH as the electron donor, the other uses ferredoxin as donor and us called Fd-GOGAT.41 Our results revealed that the latter (Fd-GOGAT) was regulated by DNA methylation and involved in photorespiration and nitrogen assimilation. More interestingly, we found that two kinds of isoenzymes of glutamine synthetase (GS) have different patterns of aza-dC regulation. Our results revealed glutamine synthetase in the cytosol to be up-regulated by aza-dC; however, glutamine synthetase in the chloroplast was down-regulated. GS1 and GS2 localize in both cytoplasm and chloroplast, and they perform different physiological functions. GS1 is mainly involved in the synthesis of nitrogen in the roots, and the function of GS2 is the synthesis of glutamine (Gln) from photorespiration.42 Our results illustrate that the status of GS in plant N metabolism is complex, and the expression of genes encoding different GS isozyme is regulated by aza-dC (supplemental file 9). Nitrogen assimilation and metabolism is not only one of the basic processes of plant physiology, but also one of the important parts of the global chemical cycle. However, the role of epigenetic regulation in the nitrogen metabolism still remains unclear. From this study, some clues about TSA affected plant secondary metabolism, and aza-dC affected the photosynthetic pathway as well as the nitrogen metabolism. Therefore, TSA and aza-dC may provide a novel chemical treatment for improvement and breeding in plants.
Furthermore, 76 cold-induced genes were identified from GO physiological processes analyses (Fig. 3 and supplemental file 4). Some of these were typical antifreeze genes, e.g., transcriptional activator CBF1, cold regulated protein COR15b, RNA helicase, and DRE&CRT-binding protein DREB1C. The expression of these genes was increasing with TSA treatment, and increased or slightly decreased with aza-dC treatment. One report is consistent with our result that Trichostatin A can suppress the cold-induced DREB1 gene in Maize.43
In addition, TSA and aza-dC were antagonistics for combined treatment. However, the mechanism for this antagonistic behavior of TSA and aza-dC remains unclear.
Currently, weather extremes occur frequently. Cold and frost are harm plant growth and development; even seriously threating the survival of several species. Therefore, it is a meaningful goal to improve cold resistance in plants. This study found that TSA and aza-dC can improve cold tolerance in plants, and identified differentially expressed genes associated with TSA and aza-dC. Future studies will further reveal the mechanisms of TSA and aza-dC to enable agricultural application.
Materials and methods
Plant materials, growth conditions, chemical and cold treatments
Arabidopsis thaliana seeds after vernalization were sown on half MS germination medium,44 pH 5.7. Seedlings were cultivated in Petri dishes for 12 days, then transferred to new Petri dishes containing semisolid half-strength MS medium. 5-aza-2′-deoxycytidine (aza-dC, Sigma) and/or trichostatinA (TSA, Sigma) were added to the medium. The final concentration was 7 g/ml aza-dC in water and 0.5 M TSA in methanol.33 The seedlings in the control group grew in half-strength MS medium without chemical treatment. All seedlings were kept in the same growth chamber at 25°C (16 h light and 8 h darkness) for three days, then the cold treatment was started at 12 PM at 0°C under light and was continued for 24 h. Liquid nitrogen cryopreservation was ready for RNA extraction and microarray analysis.
Electrolyte leakage tests and lethal temperature calculation
Electrolyte leakage tests were performed and details of the handling as previously described.45,46 The leaves of 18-day old plants were added to 15 ml tubes and placed in the freezer (XT5201-D31-R40C, XuTemp, China). The plants were exposed to freezing temperatures ranging from 0 to −20°C (0, −4, −6, −8, −10, −12, −14, −16, −18, and −20°C). Leaves without damage were then immersed in 10 ml ultrapure water (Milli-Q Advantage) and placed on a shaker at 4°C for 2 h. Electrolyte leakage was calculated as the ratio of before and after leaves boiling via a conductivity meter (DDSJ-308, Leici, China). TEL50 was calculated using t-tests by SPSS software 18.0 version (IBM SPSS software, USA).
Microarray data analysis
20 μg of total RNA from the seedlings at 0°C under light for 24 h with or without chemical treatments were extracted using a RNA purification kit (Norgen, Canada) following the manufacturer's manual. Subsequently, the product was sent to Génome Québec Innovation Centre (McGill University, Canada) for the production of biotin-labeled cRNA targets. The Affymetrix Arabidopsis ATH1 genome array GeneChip was used for this study. The raw data was pre- and normalized processing using BioConductor (http://www.bioconductor.org/). Probe sets were used for genes and gene functional annotations.
The raw data of the transcript profiles can be found in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo/) with the accession number GSE63131.
Acquisition of differentially expressed genes
For acquisition of differentially expressed genes, the data was analyzed with R version 2.15.2 (R Core Team, 2012), utilizing the limma package47 to statistically analyze differentially expressed genes. The chip data were transformed with log2, as the input of the algorithm of hierarchical clustering, and the distance and linkage were retained via Euclidean distance and Average, respectively.
Biological pathway analysis of the differentially expressed genes
All metabolic and non-metabolic pathways were acquired from the KEGG pathways database (http://www.kegg.jp/kegg/pathway.html),48 and the KEGG pathways clustering analysis of differentially expressed genes was achieved via expression analysis systematic explorer (EASE).49
Gene otology (GO) analysis of the differentially expressed genes
The GO accessions were mapped to GO terms according to molecular function, biological process, and cellular component ontologies (http://www.geneontology.org/),50 and were performed with the GSEABase package of BioConductor (http://www.bioconductor.org/). GO enrichment was statistically tested using the hypergeometric test and the differentially expressed genes from biological processes were clustered. The Bonferroni correction method was used for multiple test correction.
Real-time Quantitative RT-PCR
Total RNA from the seedlings was extracted using the RNA purification kit (Norgen, Canada) following the manufacturer's instructions. One microgram of total RNA was used to produce the first strand of cDNA via reverse transcriptase (Fermentas). cDNA was transcribed reversely via 200U M-MLV reverse transcriptase (Promega, USA) and analyzed with Platinum SYBR green qPCR supermix-UDG reagents (Invitrogen). The ACTIN2 gene (AT3G18780.2) was amplified as a reference gene, as preliminary experiments had shown its expression to be unaffected by freezing treatment and chemical inhibitors. PCR conditions were 3 min at 95°C followed by 45 cycles of the following: 15 s at 95°C, 15 s at 60°C, and 15 s at 72°C. The primer pairs used were at1g14360 (5′-ctccggcgagttctgttatt-3′, 5′-gctcgaatctcttcccatctt-3′), at1g72430 (5′-caaagttgggaagctgacaaag-3′, 5′-ctgagagttcgtcggaaacag-3′), at4g19420 (5′-gagtgcagtgataggtttaggg-3′, 5′-cttgccagaaccatgaagtttg-3′), at4g39950 (5′-ctctgtcgatttccggttcat-3′, 5′-cgtcaggtgcagtgttctta-3′), at1g55960 (5′- tcttcttcttcctctccctctac-3′, 5′-gttcccatatctccgtggaatc-3′), at3g51895 (5′-ctatcaagctgccatccatctc-3′, 5′-ccaatctctacactgccgaatac-3′), at1g76180 (5′-cttgggaaagaagaaagacgaaac, 5′-gctccggctctgaaatatgaa-3′), at5g57560 (5′-gtctttggaacgctgatgattg'-3′, 5′- tgttgaaagccacggtaaga-3′), at3g50970 (5′-tcaccacgagaagaaaggaatg-3′, 5′-gtgaacaccaccagtgttagta-3′), at3g15450 (5′-acagtccggcttctcatttc-3′, 5′-tcgccgaagttcatagagaaag-3′), and ACTIN2 (5′-gacctttaactctcccgctatg-3′, 5′-gagacacaccatcaccagaat-3′).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This research was supported by the National Basic Research Program of China 973 Program (No. 2014CB954203); The National Natural Science Foundation of China (No. 31200298). We thank prof. Zhiguang Zhao for providing help in lethal temperature calculation.
References
- 1.Isshiki R, Galis I, Tanakamaru S. Farinose flavonoids are associated with high freezing tolerance in fairy primrose (Primula malacoides) plants. J Integr Plant Biol. 2014;56(2):181–8. doi: 10.1111/jipb.12145. [DOI] [PubMed] [Google Scholar]
- 2.Nicotra AB, Atkin OK, Bonser SP, Davidson AM, Finnegan EJ, Mathesius U, Poot P, Purugganan MD, Richards CL, Valladares F, et al.. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010;15(12):684–92. doi: 10.1016/j.tplants.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Albert KR, Boesgaard K, Ro-Poulsen H, Mikkelsen TN, Andersen SPilegaard K. Antagonism between elevated CO2, nighttime warming, and summer drought reduces the robustness of PSII performance to freezing events. Environ Exp Bot. 2013;93;1–12. doi: 10.1016/j.envexpbot.2013.03.008. [DOI] [Google Scholar]
- 4.Zanne AE, Tank DC, Cornwell WK, Eastman JM, Smith SA, FitzJohn RG, McGlinn DJ, O'Meara BC, Moles AT, Reich PB, et al.. Three keys to the radiation of angiosperms into freezing environments. Nature. 2014;506(7486):89-+. doi: 10.1038/nature12872. [DOI] [PubMed] [Google Scholar]
- 5.Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12(10):444–51. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Knight MR, Knight H. Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 2012;195(4):737–751. doi: 10.1111/j.1469-8137.2012.04239.x. [DOI] [PubMed] [Google Scholar]
- 7.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6(5):410–417. doi: 10.1016/S1369-5266(03)00092-X. [DOI] [PubMed] [Google Scholar]
- 8.Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol. 2009;12(2):133–9. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyko A, Kovalchuk I. Epigenetic control of plant stress response. Environ Mol Mutagen. 2008;49(1):61–72. doi: 10.1002/em.20347. [DOI] [PubMed] [Google Scholar]
- 10.Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. 2011;14(3):267–274. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Robertson AL, Wolf DE. The role of epigenetics in plant adaptation. Trends Evol Biol. 2012;4(1):19–25. doi: 10.4081/eb.2012.e4. [DOI] [Google Scholar]
- 12.Boyko A, Kovalchuk I. Genome instability and epigenetic modification–heritable responses to environmental stress? Curr Opin Plant Biol. 2011;14(3):260–6. doi: 10.1016/j.pbi.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Gutzat R, Mittelsten Scheid O. Epigenetic responses to stress: triple defense? Curr Opin Plant Biol. 2012;15(5):568–573. doi: 10.1016/j.pbi.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahu PP, Pandey G, Sharma N, Puranik S, Muthamilarasan M, Prasad M. Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep. 2013;32(8):1151–9. doi: 10.1007/s00299-013-1462-x. [DOI] [PubMed] [Google Scholar]
- 15.Zhu J, Jeong J C, Zhu Y, Sokolchik I, Miyazaki S, Zhu JK, Hasegawa PM, Bohnert HJ, Shi H, Yun DJ, et al.. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc Natl Acad Sci U S A. 2008;105(12):4945–50. doi: 10.1073/pnas.0801029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo M, Liu X, Singh P, Cui Y, Zimmerli L, Wu K. Chromatin modifications and remodeling in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819(2):129–36. [DOI] [PubMed] [Google Scholar]
- 17.Chen LT, Luo M, Wang YY, Wu K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot. 2010;61(12):3345–53. doi: 10.1093/jxb/erq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo M, Wang YY, Liu X, Yang S, Lu Q, Cui Y, Wu K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot. 2012;63(8):3297–306. doi: 10.1093/jxb/ers059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steward N, Ito M, Yamaguchi Y, Koizumi N, Sano H. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J Biol Chem. 2002;277(40):37741–37746. doi: 10.1074/jbc.M204050200. [DOI] [PubMed] [Google Scholar]
- 20.Liang D, Zhang Z, Wu H, Huang C, Shuai P, Ye CY, Tang S, Wang Y, Yang L, Wang J, et al.. Single-base-resolution methylomes of Populus trichocarpa reveal the association between DNA methylation and drought stress. BMC Genet. 2014;15 Suppl 1, S9. doi: 10.1186/1471-2156-15-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim H, Kim KC, Son J, Shin Y, Yoon CH, Kang C, Choi BS. Synergistic reactivation of latent HIV-1 provirus by PKA activator dibutyryl-cAMP in combination with an HDAC inhibitor. Virus Res. 2016;227;1–5. [DOI] [PubMed] [Google Scholar]
- 22.Staberg M, Michaelsen SR, Rasmussen RD, Villingshoj M. Poulsen H SHamerlik P, Inhibition of histone deacetylases sensitizes glioblastoma cells to lomustine. Cell Oncol (Dordr). 2017;40(1):21–32. doi: 10.1007/s13402-016-0301-9. [DOI] [PubMed] [Google Scholar]
- 23.Tiper IV, Webb TJ. Histone deacetylase inhibitors enhance CD1d-dependent NKT cell responses to lymphoma. Cancer Immunol Immunother. 2016;65(11):1411–1421. doi: 10.1007/s00262-016-1900-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee K, Park OS, Jung SJ, Seo PJ. Histone deacetylation-mediated cellular dedifferentiation in Arabidopsis. J Plant Physiol. 2016;191;95–100. doi: 10.1016/j.jplph.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Cui Y, Ling Y, Zhou J, Li X. Interference of the Histone Deacetylase Inhibits Pollen Germination and Pollen Tube Growth in Picea wilsonii Mast. PLoS One. 2015;10(12):e0145661. doi: 10.1371/journal.pone.0145661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang P, Zhao L, Hou H, Zhang H, Huang Y, Wang Y, Li H, Gao F, Yan S, Li L. Epigenetic Changes are Associated with Programmed Cell Death Induced by Heat Stress in Seedling Leaves of Zea mays. Plant Cell Physiol. 2015;56(5):965–76. doi: 10.1093/pcp/pcv023. [DOI] [PubMed] [Google Scholar]
- 27.Hou H, Wang P, Zhang H, Wen H, Gao F, Ma N, Wang Q, Li L. Histone Acetylation is Involved in Gibberellin-Regulated sodCp Gene Expression in Maize Aleurone Layers. Plant Cell Physiol. 2015;56(11):2139–49. [DOI] [PubMed] [Google Scholar]
- 28.Tyagi T, Treas JN, Mahalingaiah P KSingh KP. Potentiation of growth inhibition and epigenetic modulation by combination of green tea polyphenol and 5-aza-2′-deoxycytidine in human breast cancer cells. Breast Cancer Res Treat. 2015;149(3):655–68. doi: 10.1007/s10549-015-3295-5. [DOI] [PubMed] [Google Scholar]
- 29.Zhao C, Zhang Z, Xie S, Si T, Li Y, Zhu JK. Mutational Evidence for the Critical Role of CBF Transcription Factors in Cold Acclimation in Arabidopsis. Plant Physiol. 2016;171(4):2744–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Hu G, Liu X, Zhou Y, Li Y, Zhang X, Yuan X, Zhang Q, Yang D, Wang T, et al.. Transcriptome Sequencing Identified Genes and Gene Ontologies Associated with Early Freezing Tolerance in Maize. Front Plant Sci. 2016;7;1477. doi: 10.3389/fpls.2016.01477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Y, Liu L, Feng Y, Wei Y, Yue X, He W, Zhang H, An L. Chilling- and Freezing- Induced Alterations in Cytosine Methylation and Its Association with the Cold Tolerance of an Alpine Subnival Plant, Chorispora bungeana. PLoS One. 2015;10(8):e0135485. doi: 10.1371/journal.pone.0135485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nah G, Lee M, Kim DS, Rayburn AL, Voigt T, Lee DK. Transcriptome Analysis of Spartina pectinata in Response to Freezing Stress. PLoS One. 2016;11(3):e0152294. doi: 10.1371/journal.pone.0152294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang S, Pikaard CS. Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J Biol Chem. 2005;280(1):796–804. doi: 10.1074/jbc.M409053200. [DOI] [PubMed] [Google Scholar]
- 34.Song Y, Wu K, Dhaubhadel S, An L, Tian L. Arabidopsis DNA methyltransferase AtDNMT2 associates with histone deacetylase AtHD2s activity. Biochem Biophys Res Commun. 2010;396(2):187–92. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Yu CW, Duan J, Luo M, Wang K, Tian G, Cui Y, Wu K. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 2012;158(1):119–29. doi: 10.1104/pp.111.184275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuler MA. The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol. 1996;112(4):1411–9. doi: 10.1104/pp.112.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cummins I, Dixon DP, Freitag-Pohl S, Skipsey MEdwards R. Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metab Rev. 2011;43(2):266–80. doi: 10.3109/03602532.2011.552910. [DOI] [PubMed] [Google Scholar]
- 38.Strommer J. The plant ADH gene family. Plant J. 2011;66(1):128–42. doi: 10.1111/j.1365-313X.2010.04458.x. [DOI] [PubMed] [Google Scholar]
- 39.Solomonson LP, Barber MJ. Assimilatory nitrate reductase: Functional properties and regulation. A nnu Rev Plant Physiol Plant Mol Biol. 1990;41;225–253. [Google Scholar]
- 40.Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM. The Molecular-Genetics of Nitrogen Assimilation into Amino Acids in Higher Plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47;569–593. doi: 10.1146/annurev.arplant.47.1.569. [DOI] [PubMed] [Google Scholar]
- 41.Sechley KA, Oaks A, Bewley JD. Enzymes of Nitrogen Assimilation Undergo Seasonal Fluctuations in the Roots of the Persistent Weedy Perennial Cichorium intybus. Plant Physiol. 1991;97(1):322–9. doi: 10.1104/pp.97.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miflin BJ. The location of nitrite reductase and other enzymes related to amino Acid biosynthesis in the plastids of root and leaves. Plant Physiol. 1974;54(4):550–5. doi: 10.1104/pp.54.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu Y, Zhang L, Zhao L, Li J, He S, Zhou K, Yang F, Huang M, Jiang L, Li L. Trichostatin A selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLoS One. 2011;6(7):e22132. doi: 10.1371/journal.pone.0022132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Plant Physiol. 1962;15;473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 45.Rohde P, Hincha DK, Heyer AG. Heterosis in the freezing tolerance of crosses between two Arabidopsis thaliana accessions (Columbia-0 and C24) that show differences in non-acclimated and acclimated freezing tolerance. Plant J. 2004;38(5):790–9. doi: 10.1111/j.1365-313X.2004.02080.x. [DOI] [PubMed] [Google Scholar]
- 46.Song Y, Liu L, Wei Y, Li G, Yue X, An L. Metabolite Profiling of adh1 Mutant Response to Cold Stress in Arabidopsis. Front Plant Sci. 2016;7;2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7;252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al.. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hosack DA, Dennis G Jr., Sherman BT, Lane H CLempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al.. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]



