ABSTRACT
Recent advances in fluorescence microscopy have opened up new possibilities to investigate chromosomal and nuclear 3D organization on the nanoscale. We here discuss their potential for elucidating topographical details of the nuclear lamina. Single molecule localization microscopy (SMLM) in combination with immunostainings of lamina proteins readily reveals tube-like invaginations with a diameter of 100–500 nm. Although these invaginations have been established as a frequent and general feature of interphase nuclei across different cell types, their formation mechanism and function have remained largely elusive. We critically review the current state of research, propose possible connections to lamina associated domains (LADs), and revisit the discussion about the potential role of these invaginations for accelerating mRNA nuclear export. Illustrative studies using 3D super-resolution imaging are shown and will be instrumental to decipher the physiological role of these nanoscale invaginations.
KEYWORDS: chromatin, DNA repair, heterochromatin, lamina associated domains, lamin, mechanics, nuclear architecture, nuclear envelope, nuclear structures, nuclear transport, topography
The nuclear lamina delineates the inner nuclear membrane (INM) and provides structural integrity to the nucleus. Moreover, for a long time it has been implicated in assisting gene regulation. Peripheral regions of the nucleus close to the nuclear lamina are commonly enriched in densely packed heterochromatin, which is often (but not always) transcriptionally inactive. The interactions mediating these specific associations are a field of intense study. Lamina-associated domains (LADs) have been identified and shown to interact directly with INM components such as the Lamin B Receptor (LBR) and some LEM family proteins (emerin, LAP2α/β).1,2 They further associate with repressive histone modifications (H3L27me3, H3K9me2), RNA polymerase II and transcription factors (MOK2, c-Fos, BAF) that can also mediate interactions between the nuclear lamina and chromatin.3 Loss of the integrity of the nuclear lamina arising from mutations on lamins or other lamina proteins (laminopathic cells) leads to disorganized peripheral heterochromatin.4 Moreover, these interactions are dynamically regulated in the sense that they can form or disassemble as the methylation and/or phosphorylation state of the involved interaction partners changes.5 Importantly, LADs significantly change during development, in particular upon embryonic stem cell differentiation into different lineages.3,6 Together, these and other observations support the view that the biochemical composition and structural organization of the nuclear lamina co-regulates gene transcription,7 and more recent findings imply that even mechanical forces add an additional layer of regulation.8-11
Many of these functional insights into chromatin-nuclear lamina interactions have been obtained by biochemical and sequencing technology (i.e. by DamID and ChIP).3,7,12 Recently developed super-resolution microscopy techniques offer new opportunities to directly visualize the fine-structure of the nuclear lamina and the precise localization of specific genomic loci in single nuclei with molecular specificity and down to the nanometer scale. Three-dimensional structured illumination microscopy (3D SIM) has been most widely used to study the nuclear envelope (NE), and in particular the distribution of lamins, with an about 2-fold increased axial and lateral resolution compared with confocal microscopy.13-15 Higher resolution can in principle be reached by STimulated Emission Depletion (STED) microscopy or, even more, by single molecule localization microscopy (SMLM) techniques comprising 3D PhotoActivated Localization Microscopy (PALM), 3D STochastic Optical Reconstruction Microscopy (3D-STORM) or 3D DNA-PAINT. STED and STORM successfully have visualized the localization of proteins and subunits within nuclear pore complexes (NPCs).16,17 3D STORM together with multiplexed FISH has recently succeeded in revealing the compaction of individual chromosomes or chromosomal sub-regions based on the localization of topologically associating domain (TAD) sequences.18 Despite some studies in which the distribution of lamins was depicted by 3D PALM,19 its resolution has not been exploited to systematically look into nanoscale features of the nuclear lamina until very recently.20
We here investigated the suitability of 3D SMLM to study the topography of the nuclear lamina, focusing on features that are below the optical diffraction limit and are thus not fully resolved by the most commonly performed confocal microscopy. Immunofluorescence stainings for lamins in nuclei of fixed fibroblasts show a predominantly homogenous sheet-like distribution, with notable exceptions of thin invaginations that penetrate into the nucleus (Fig. 1). These invaginations are lined by both A- and B-type lamins (Fig. 1a) and form hollow tubes, which emanate perpendicular to the basal (Fig. 1b,c) or apical (Fig. 1d) NE. Their diameters range from 0.1–0.5 µm (Fig. 1e), they occur relatively frequently (Fig. 1f), and they contain NPCs (Fig. 1g). These invaginations are well-known to many researchers who have imaged the nuclear lamina, and have been termed type II nucleoplasmic reticulum (NR).21 Already in 1979, transmission electron microscopy (TEM) of different cell types revealed hollow finger-like intrusions that seemingly ended at nucleoli and contained NPCs.22 In an extensive paper on this subject, Fricker and coworkers found that 0.2–0.5 µm diameter thick invaginations occurred with an average frequency of 1–2 per interphase nucleus and ended at nucleoli or sometimes transversed the nucleus in the basal-apical direction.23 These invaginations contained inner and outer nuclear membrane, as well as NPCs and were surrounded by lamins.23 By combining ultrastructural methods with confocal microscopy and also STORM, Auer and coworkers recently showed that tunnel-like invaginations occurred in 30–50% of nuclei of human mammary epithelial cells in 3D culture (similar to the frequency in fibroblasts, see Fig. 1f), that these invaginations contained lamin B1 and NPCs, were rich in SUN-1, associated with heterochromatin, and engulfed cytoskeletal actin and keratin filaments.20 Thus, SMLM readily yields structural biology information that previously could only be obtained by a combination of fluorescence and electron microscopy techniques, and typically achieves a much higher throughput than EM, such that statistics about structural characteristics (Fig. 1e, f) can be obtained more easily.
Figure 1.
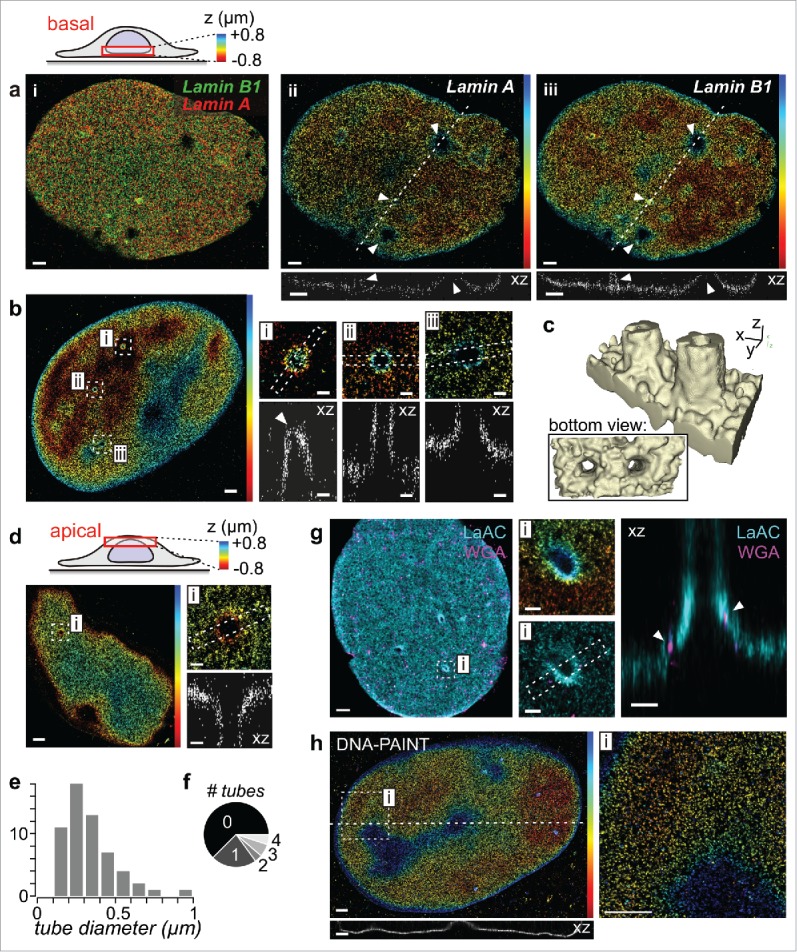
Investigating nanoscale nuclear envelope topography and invaginations by 3D SMLM. (a) A- and B-type lamins share the same topography. Lamin A and lamin B1 of fixed human foreskin fibroblasts were immunostained and imaged by dual-color 3D STORM at the basal side of the nucleus. Shown are (i) an overlay and (ii+iii) z-color coded images of individual lamins with line profiles underneath. Arrowheads indicate tube-like invaginations. Scale bars: 1 µm. (b) Tube-like invaginations at the basal side of a lamin A stained nucleus. Arrowhead: tip of an invagination. Scale bars: 1 µm (overview) or 0.2 µm (magnified insets i-iii). (c) 3D reconstruction of 2 nearby tubes. (d) Invagination at the apical side of a lamin A stained nucleus. Scale bars: 1 µm (overview) or 0.2 µm (inset i). (e) Pooled statistics of tube diameters (n = 57, from 30 cells). (f) Occurrence frequency of the number of tubes per imaged side of the nucleus (60 basal, 20 apical). As only one side could be imaged per nucleus, this represents a lower estimate. (g) Dual-color 3D STORM of A-type lamins (LaAc; cyan) and wheat germ agglutinin (WGA; magenta) as an NPC stain. Arrowheads: NPCs. Scale bars: 1 µm (overview) or 0.2 µm (insets).(h) 3D DNA-PAINT measurement of lamin B1. Scale bars: 1 µm.
To study the nucleus as a whole, 3D SMLM variants with increased axial range are needed beyond imaging of a single plane. To this end, illumination by a (lattice) light sheet19 can be used but requires sophisticated instrumentation. Fig. 1h demonstrates that DNA-PAINT24 can be used as an alternative to STORM. The higher brightness of dyes in DNA-PAINT (no blinking) and the unlimited measurement duration (steady renewal of imager strands) are ideally suited to image multiple planes sequentially, even on commercial spinning disk microscopes (R. Jungmann, personal communication), and thus enable 3D whole cell imaging. Another potential advantage is that contextual information might be obtained in sequential imaging rounds by exchange-PAINT24 or PAINT with DNA-binding dyes,19,25 thus opening up multiplexing beyond the limited 2 channels in STORM. In this way, the localization of NPCs (Fig. 1g) or functional DNA binders26 relative to interchromatin compartments27 could be studied. To maximize labeling density without introducing labeling artifacts, either careful design of CRISPR-Cas gene knock-in28 or small affinity binders such as nanobodies29 could be implemented. Such improvements would allow to study the distribution of lamins, NPCs, INM proteins, and methylated histones along tube-like invaginations. While SMLM techniques offer compelling advantages i.e., in resolution (in our experiments it was between 20–25 nm according to an established measure30 and thus about fourfold higher than in 3D-SIM) they are not well suited to study live cell dynamics. They thus should be primarily regarded as a tool for structural biology. To obtain dynamic information about the NE, 3D-SIM and its variants31,32 currently are the best choice.
Despite substantial evidence that type II NR invaginations are a common feature of many cell types21 and that they are present and dynamic in living cells,20,23 their formation has remained largely elusive. From a conceptual point of view, tubular membrane structures in cells often arise as a consequence of mechanical force. One possibility are pushing forces exerted by the polymerization of cytoskeletal filaments, e.g., during the formation of filopodia by actin polymerization. Another possibility are pulling forces, e.g., exerted by (cytoplasmic or nuclear33,34) molecular motors35 that give rise to the tube-like appearance of the endoplasmic reticulum.31 Both mechanisms yield tubes with diameters in the range of few hundred nanometers,31 very similar to the dimensions of type II NR (Fig. 1e). Upon isolation of nuclei, tube-like invaginations disappeared36 which supports that they are stabilized mechanically. This finding also speaks against a third potential formation mechanism involving dedicated engulfment machinery37 (which might play a role for type I NR; see also review38). So what roles could pushing or pulling forces play for the formation of type II NR invaginations?
The ‘pushing’ hypothesis is supported by the presence of cytoskeletal filaments such as microtubules,39 actin40 or keratin20 in the interior of NE invaginations which is continuous with the cytoplasm.41 Their rather loose organization without apparent bundling20 (as opposed to filopodia or cilia), however, implies weak mechanical strength and questions whether pushing forces were sufficiently high to penetrate the nucleus, notwithstanding they might contribute to the deepening of pre-formed invaginations by their polymerization. In contrast, these filaments have been suggested to transmit contractile forces to the nucleus by a physical linkage through SUN proteins20 and thus could play a role in nuclear tensional homeostasis.
The ‘pulling’ hypothesis requires a handle for tearing at the NE and a pulling force, for instance chromatin-lamin interactions and large-scale rearrangements of chromatin. In this respect, several studies confirmed that invaginations were associated tightly with chromatin.20,42 Invaginations contained lamin A,43 emerin,44 and the latent binding protein LAP2α (but not LAP2β),45 and their occurrence positively correlated with overexpression of LBR46 or lamin B itself.47 These pieces of information suggest that tube-like NE invaginations rely on interactions between NE proteins with chromatin (Fig. 2). Natural candidates for genomic loci involved in such interactions are LADs. The often reported association of NE tubes with nucleoli22,23 supports this idea because LADs share sequence similarities with nucleolus-associated domains (NADs).3,48 Hence, the same chromosomal regions can either attach to the lamina or to nucleoli, and some ‘facultative’ LADs have indeed been found at either location.49 The topographical rearrangement of LADs is thought to happen during mitosis before NE re-assembly or before the chromatin-NE connection has been re-established50 (Fig. 2b). Interestingly, many NE invaginations appear during telophase46 or early G1 phase50 when chromosomal organization is being re-established51 and large movements of chromatin (up to 4 µm) are still common.52 If chromosomal reorganizations occurred in an NE-attached state, the NE could be pulled into the nucleus together with the LAD and thus form an invagination (Fig. 2c). Chromosomal sites that had made contact with the lamina have indeed been observed ∼1 µm away from the nuclear border50 and thus in principle fulfill the requirements for the formation of NE tubes. On the other hand, NE invaginations also arise during interphase.23,38,46 Translocations of genomic loci within the interphase nucleus are typically limited to submicron scales.51 Larger movements were occasionally observed in cultured cells,53 as well as systematically upon chromosome condensation,54 transcriptional activation of peripherally located chromosome sites,53 or in response to environmental stimuli or double strand breaks.55 Of note, NE invaginations have been found near sites of DNA repair after gamma-irradiation,56 which suggests that they might have formed in conjunction with a potential rearrangement triggered by the damage response. Chromosomal movements may be driven by different forces, with physical contributions from a shrinkage by chromatin condensation, nucleosome remodeling,52 or nuclear motor proteins. While the existence of nuclear motors has become an accepted fact,33,34 and it has been shown that interphase translocations of genomic loci were sensitive to perturbations of actomyosin activity,57 the linkage between motor activity and these motions remains controversial. In summary, the ‘pulling’ hypothesis could explain the observation that the number of NE tubes increases with the ‘stickiness’ of the lamina for LADs (by increased LBR or lamin B levels46,47). It further predicts that their number should correlate with the frequency of chromosomal rearrangements in the nucleus – irrespective of the mechanical origin of these movements – which could offer a way to test it.
Figure 2.
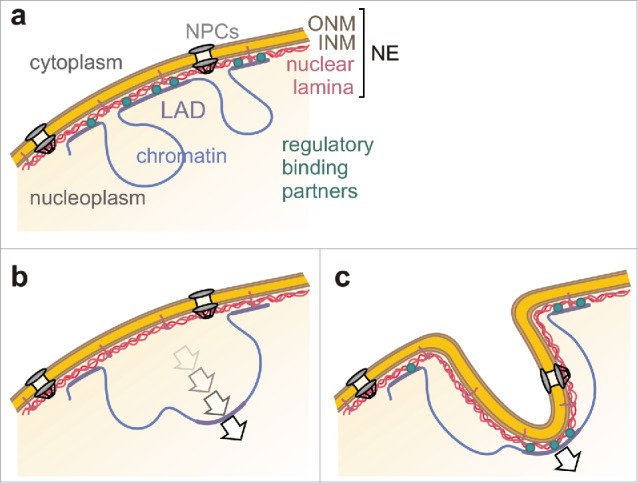
Proposed formation mechanism of tube-shaped NE invaginations. (a) Chromatin regions (i.e., LADs) build physical connections with the NE. These interactions are dynamically regulated by proteins of the nuclear lamina and chromatin binding partners. ONM: outer nuclear membrane; INM: inner nuclear membrane; NPCs: nuclear pore complexes. (b) During genomic translocations, LADs detach from the NE, move into the nucleus, and eventually bind to the nucleolar periphery. (c) Alternatively, if the connection between LADs and the NE is strong enough to withstand detachment, the NE might be pulled inward along with the LAD and form a tube-like invagination.
With the advent of these novel super-resolution light imaging modalities to study nanoscale NE invaginations, the old question about the function of NE tubes can be revived. Several authors have suggested a functional role for nuclear import/export.22,23,38,46 As many tubular membrane structures are involved in intracellular transport, it has been speculated that NE invaginations shorten diffusional transport of transcribed mRNA to the nuclear boundary. Following a simplified argumentation, the presence of invaginations shortens the distance from a nucleolus to the nearest boundary, resulting in diffusion time shortening. However, as diffusion is not one-dimensional and the dimensions of a tube are rather small such that the chance to hit it is smaller than hitting the outer membrane, this simplified argument needs more careful consideration. To this end, we performed Monte Carlo simulations using typical nuclear dimensions and intra-nuclear diffusion coefficients for mRNA (see Supplementary Information) and found that the presence of a tubular invagination shortens intra-nuclear diffusional transport to the NE (Fig. 3a,b). This effect is strongly dependent on the relative location within the nucleus (Fig. 3c). A significant reduction of diffusion times is observed locally around the invagination whereas the global average was decreased by only -10 % (Fig. 3d). This result suggests that a potential advantage of NE tubes – if there is any – would require a controlled positioning of NPCs (cf Fig. 1g), interchromatin compartments26,27,58,59 that facilitate diffusion, and of active gene transcription sites along them. Especially the complex and heterogeneous structure of chromatin around NE tubes with respect to compacted regions, interchromatin channels, and potentially transcriptionally active loops, as it has been studied in the Barr body,26 is expected to distinctly affect the local transport of mRNA.59 Existing labeling schemes for LADs50 and methodologies that are currently being developed, e.g., within the 4D Nucleome program (http://www.4dnucleome.org), will be important tools to specifically look into chromatin architecture, its relation to LADs, and dynamical changes of both. Combining these new tools with multiplexed imaging of NE invaginations could further help to test whether invaginations are positioned close to actively transcribed chromosomes. Ultimately, super-resolved live cell imaging31 could allow to study the dynamic formation of NE invaginations and the driving forces, i.e., LAD rearrangements50 or cytoskeletal dynamics in detail. Efforts on several fronts will be needed to elucidate the relation between NE invaginations and the physiological60,61 or pathological21,62 regulation of cellular phenotype.
Figure 3.
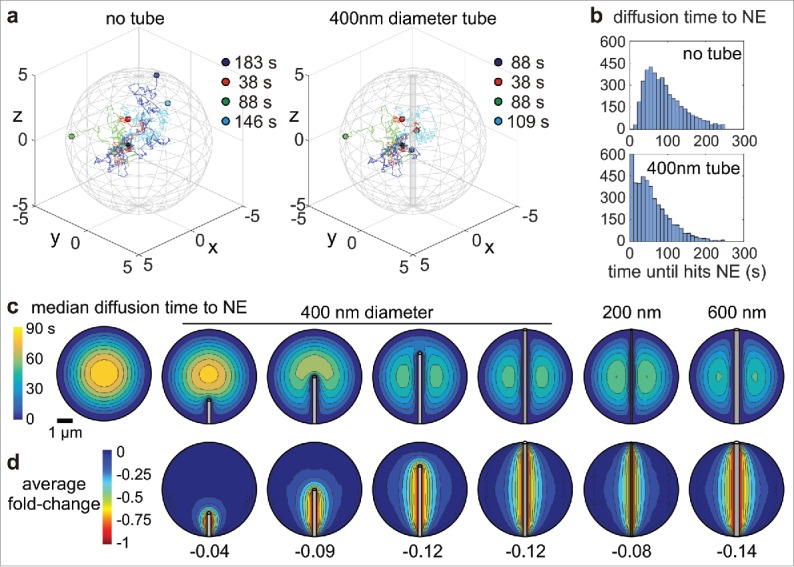
Theoretical shortening of intra-nuclear diffusive transport to the nuclear envelope by NE invaginations. (a) Three-dimensional diffusive paths of 4 exemplary particles starting in the point (1,0,0) and ending at the NE. Indicated are the times and locations when/where the particles hit the outer boundary (left) and/or the tubular invagination (right). The nucleus was modeled as a sphere with 10 µm diameter. The diffusion constant was set to 0.04 µm2s−1;63 anomalous diffusion or directed transport were neglected. (b) Distribution of diffusion times to the NE for 4000 particles starting at the point (1,0,0) in a nucleus without (top) or with (bottom) a tubular invagination. (c) Median diffusion times to the NE (color coded) as a function of the starting position in the nucleus, as well as of the length and diameter of a tubular invagination. (d) Average fold-change of diffusion times to the NE (color coded) in the presence of a NE tube for the same cases as in (c). A fold-change of -0.5 indicates a reduction of diffusion times by 50% relative to the nucleus without invagination.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Nikhil Jain for critical comments on the manuscript.
Funding
This work was supported by the Holcim Science Foundation (I.S.), the Swiss National Science Foundation (SNSF) through project CR32I3–156931 (V.V.) and by ETH Zurich.
References
- [1].Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear Lamins. Cold Spring Harb Perspect Biol 2010; 2:a000547-a000547; https://doi.org/ 10.1101/cshperspect.a000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Barrales RR, Forn M, Georgescu PR, Sarkadi Z, Braun S. Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev 2016; 30:133-48; PMID:26744419; https://doi.org/ 10.1101/gad.271288.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Collas P, Lund EG, Oldenburg AR. Closing the (nuclear) envelope on the genome: How nuclear lamins interact with promoters and modulate gene expression. BioEssays 2014; 36:75-83; PMID:24272858; https://doi.org/ 10.1002/bies.201300138 [DOI] [PubMed] [Google Scholar]
- [4].Taimen P, Pfleghaar K, Shimi T, Möller D, Ben-Harush K, Erdos MR, Adam SA, Herrmann H, Medalia O, Collins FS, et al.. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc Natl Acad Sci U S A 2009; 106:20788-93; https://doi.org/ 10.1073/pnas.0911895106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al.. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008; 453:948-51; PMID:18463634; https://doi.org/ 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- [6].Shivashankar G V. Mechanosignaling to the Cell Nucleus and Gene Regulation. Annu Rev Biophys 2011; 40:361-78; https://doi.org/ 10.1146/annurev-biophys-042910-155319 [DOI] [PubMed] [Google Scholar]
- [7].Lemaître C, Bickmore WA. Chromatin at the nuclear periphery and the regulation of genome functions. Histochem Cell Biol 2015; 144:111-22; PMID:26170147; https://doi.org/ 10.1007/s00418-015-1346-y [DOI] [PubMed] [Google Scholar]
- [8].Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, Wang N. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater 2016; 15:1287-96; PMID:27548707; https://doi.org/ 10.1038/nmat4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Le HQ, Ghatak S, Yeung C-YC, Tellkamp F, Günschmann C, Dieterich C, Yeroslaviz A, Habermann B, Pombo A, Niessen CM, et al.. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol 2016; 18:1-23; https://doi.org/ 10.1038/ncb3387 [DOI] [PubMed] [Google Scholar]
- [10].Ihalainen TO, Aires L, Herzog FA, Schwartlander R, Moeller J, Vogel V. Differential basal-to-apical accessibility of lamin A/C epitopes in the nuclear lamina regulated by changes in cytoskeletal tension. Nat Mater 2015; 14:1252-61. http://www.nature.com/doifinder/10.1038/nmat4389; https://doi.org/ 10.1038/nmat4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, Burridge K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol 2014; 16:376-81; PMID:24609268; https://doi.org/ 10.1038/ncb2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kubben N, Voncken JW, Misteli T. Mapping of protein- and chromatin-interactions at the nuclear lamina. Nucleus 2010; 1:460-71. https://doi.org/10.4161/nucl.1.6.13513; https://doi.org/ 10.4161/nucl.1.6.13513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard D a, Gustafsson MGL, et al.. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 2008; 320:1332-6. PMID:2916659; https://doi.org/ 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell 2015; 26:4075-86; https://doi.org/ 10.1091/mbc.E15-07-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Markaki Y, Smeets D, Fiedler S, Schmid VJ, Schermelleh L, Cremer T, Cremer M. The potential of 3D-FISH and super-resolution structured illumination microscopy for studies of 3D nuclear architecture. BioEssays 2012; 34:412-26; https://doi.org/ 10.1002/bies.201100176 [DOI] [PubMed] [Google Scholar]
- [16].Göttfert F, Wurm CA, Mueller V, Berning S, Cordes VC, Honigmann A, Hell SW. Coaligned Dual-Channel STED Nanoscopy and Molecular Diffusion Analysis at 20 nm Resolution. Biophys J 2013; 105:L01-3; https://doi.org/ 10.1016/j.bpj.2013.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Loschberger A, van de Linde S, Dabauvalle M-C, Rieger B, Heilemann M, Krohne G, Sauer M. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J Cell Sci 2012; 125:570-5; PMID:22389396; https://doi.org/ 10.1242/jcs.098822 [DOI] [PubMed] [Google Scholar]
- [18].Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, Fudenberg G, Imakaev M, Mirny LA, Wu C, Zhuang X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 2016; 529:418-22. https://doi.org/ 10.1038/nature16496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Legant WR, Shao L, Grimm JB, Brown TA, Milkie DE, Avants BB, Lavis LD, Betzig E. High-density three-dimensional localization microscopy across large volumes. Nat Methods 2016; 13:1-9; PMID:26950745; https://doi.org/ 10.1038/nmeth.3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jorgens DM, Inman JL, Wojcik M, Robertson C, Palsdottir H, Tsai W-T, Huang H, Bruni-Cardoso A, López CS, Bissell MJ, et al.. Deep nuclear invaginations linked to cytoskeletal filaments: Integrated bioimaging of epithelial cells in 3D culture. J Cell Sci 2016; 130(1):177-189; https://doi.org/ 10.1242/jcs.190967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Malhas A, Goulbourne C, Vaux DJ. The nucleoplasmic reticulum: Form and function. Trends Cell Biol 2011; 21:362-73; https://doi.org/ 10.1016/j.tcb.2011.03.008 [DOI] [PubMed] [Google Scholar]
- [22].Bourgeois CA, Hemon D, Bouteille M. Structural Relationship Between the Nucleolus and the Nuclear-Envelope. J Ultrastruct Res 1979; 68:328-40; PMID:490761; https://doi.org/ 10.1016/S0022-5320(79)90165-5 [DOI] [PubMed] [Google Scholar]
- [23].Fricker M, Hollinshead M, White N, Vaux D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol 1997; 136:531-44; PMID:9024685; https://doi.org/ 10.1083/jcb.136.3.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jungmann R, Avendaño MS, Woehrstein JB, Dai M, Shih WM, Yin P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods 2014; 11:313-8; PMID:24487583; 4108088https://doi.org/10.1038/nmeth.2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schoen I, Ries J, Klotzsch E, Ewers H, Vogel V. Binding-Activated Localization Microscopy of DNA Structures. Nano Lett 2011; 11:4008-11; https://doi.org/ 10.1021/nl2025954 [DOI] [PubMed] [Google Scholar]
- [26].Smeets D, Markaki Y, Schmid VJ, Kraus F, Tattermusch A, Cerase A, Sterr M, Fiedler S, Demmerle J, Popken J, et al.. Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenetics Chromatin 2014; 7:8; PMID:4108088; https://doi.org/ 10.1186/1756-8935-7-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Żurek-Biesiada D, Szczurek AT, Prakash K, Mohana GK, Lee H-K, Roignant J-Y, Birk UJ, Dobrucki JW, Cremer C. Localization microscopy of DNA in situ using Vybrant® DyeCycle™ Violet fluorescent probe: A new approach to study nuclear nanostructure at single molecule resolution. Exp Cell Res 2016; 343:97-106; https://doi.org/ 10.1016/j.yexcr.2015.08.020 [DOI] [PubMed] [Google Scholar]
- [28].Ratz M, Testa I, Hell SW, Jakobs S. CRISPR/Cas9-mediated endogenous protein tagging for RESOLFT super-resolution microscopy of living human cells. Sci Rep 2015; 5:9592; PMID:25892259; https://doi.org/ 10.1038/srep09592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mikhaylova M, Cloin BMC, Finan K, van den Berg R, Teeuw J, Kijanka MM, Sokolowski M, Katrukha E a, Maidorn M, Opazo F, et al.. Resolving bundled microtubules using anti-tubulin nanobodies. Nat Commun 2015; 6:7933; https://doi.org/ 10.1038/ncomms8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nieuwenhuizen RPJ, Lidke K a, Bates M, Puig DL, Grünwald D, Stallinga S, Rieger B. Measuring image resolution in optical nanoscopy. Nat Methods 2013; 10:557-62; PMID:23624665; https://doi.org/ 10.1038/nmeth.2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nixon-Abell J, Obara CJ, Weigel A V, Li D, Legant WR, Xu CS, Pasolli HA, Harvey K, Hess HF, Betzig E, et al.. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 2016; 354:aaf3928; https://doi.org/ 10.1126/science.aaf3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, Liu Z, et al.. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science 2014; 346:1257998-1257998; PMID:25342811; https://doi.org/ 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].de Lanerolle P, Serebryannyy L. Nuclear actin and myosins: life without filaments. Nat Cell Biol 2011; 13:1282-8; PMID:22048410; https://doi.org/ 10.1038/ncb2364 [DOI] [PubMed] [Google Scholar]
- [34].Lee YM, Lee S, Lee E, Shin H, Hahn H, Choi W, Kim W. Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. Biochem J 2001; 360:549-56. https://doi.org/ 10.1042/bj3600549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Koster G, VanDuijn M, Hofs B, Dogterom M. Membrane tube formation from giant vesicles by dynamic association of motor proteins. Proc Natl Acad Sci U S A 2003; 100:15583-8; PMID:14663143; https://doi.org/ 10.1073/pnas.2531786100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Avedanian L, Jacques D, Bkaily G. Presence of tubular and reticular structures in the nucleus of human vascular smooth muscle cells. J Mol Cell Cardiol 2011; 50:175-86. https://doi.org/ 10.1016/j.yjmcc.2010.10.005 [DOI] [PubMed] [Google Scholar]
- [37].von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull M-T, Banterle N, Parca L, Kastritis P, et al.. In situ structural analysis of the human nuclear pore complex. Nature 2015; 526:140-3; https://doi.org/ 10.1038/nature15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Drozdz MM, Vaux DJ. Shared mechanisms in physiological and pathological nucleoplasmic reticulum formation. Nucleus 2017; 8:34-45; https://doi.org/ 10.1080/19491034.2016.1252893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Seisenberger G, Ried MU, Endress T, Büning H, Hallek M, Bräuchle C. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science 2001; 294:1929-32; PMID: 11729319; https://doi.org/ 10.1126/science.1064103 [DOI] [PubMed] [Google Scholar]
- [40].Clubb BH, Locke M. 3T3 cells have nuclear invaginations containing F-actin. Tissue Cell 1998; 30:684-91; https://doi.org/ 10.1016/S0040-8166(98)80087-6 [DOI] [PubMed] [Google Scholar]
- [41].Lui PPY, K SK, Kw TT, Lee CY. The Nucleus of HeLa Cell Contains Tubular Structures for Ca2+ Signalling. Biochem Biophys Res Commun 1998; 93:88-93; https://doi.org/ 10.1006/bbrc.1998.8649 [DOI] [PubMed] [Google Scholar]
- [42].Park PC, Boni UDE. Nuclear Membrane Modifications in Polytene Nuclei of Drosophila rnelanogaster : Serial Reconstruct ion and Cytochemistry. Anat Rec 1992; 234:15-26; https://doi.org/ 10.1002/ar.1092340103 [DOI] [PubMed] [Google Scholar]
- [43].Broers JL, Machiels BM, van Eys GJ, Kuijpers HJ, Manders EM, van Driel R, Ramaekers FC. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci 1999; 112:3463-75; http://www.ncbi.nlm.nih.gov/pubmed/10504295 [DOI] [PubMed] [Google Scholar]
- [44].Manilal S, Nguyen TM, Morris GE. Colocalization of emerin and lamins in interphase nuclei and changes during mitosis. Biochem Biophys Res Commun 1998; 249:643-7; PMID:9731189; https://doi.org/ 10.1006/bbrc.1998.9209 [DOI] [PubMed] [Google Scholar]
- [45].Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R, Ashery-Padan R, Weiss AM, Feinstein N, Gruenbaum Y, et al.. Lamina-associated polypeptide 2alpha binds intranuclear A-type lamins. J Cell Sci 2000; 113:3473-84. http://www.ncbi.nlm.nih.gov/pubmed/10984438 [DOI] [PubMed] [Google Scholar]
- [46].Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol 1997; 138:1193-206; PMID:9298976; https://doi.org/ 10.1083/jcb.138.6.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Volkova EG, Kurchashova SY, Polyakov VY, Sheval E V. Self-organization of cellular structures induced by the overexpression of nuclear envelope proteins: A correlative light and electron microscopy study. J Electron Microsc (Tokyo) 2011; 60:57-71; PMID:20926432; https://doi.org/ 10.1093/jmicro/dfq067 [DOI] [PubMed] [Google Scholar]
- [48].van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-Resolution Whole-Genome Sequencing Reveals That Specific Chromatin Domains from Most Human Chromosomes Associate with Nucleoli. Mol Biol Cell 2010; 21:3735-48; https://doi.org/ 10.1091/mbc.E10-06-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 2015; 16:245-57; https://doi.org/ 10.1038/nrm3965 [DOI] [PubMed] [Google Scholar]
- [50].Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-Cell Dynamics of Genome-Nuclear Lamina Interactions. Cell 2013; 153:178-92; https://doi.org/ 10.1016/j.cell.2013.02.028 [DOI] [PubMed] [Google Scholar]
- [51].Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J. Global Chromosome Positions Are Transmitted through Mitosis in Mammalian Cells. Cell 2003; 112:751-64; https://doi.org/ 10.1016/S0092-8674(03)00189-2 [DOI] [PubMed] [Google Scholar]
- [52].Dion V, Gasser SM. Chromatin movement in the maintenance of genome stability. Cell 2013; 152:1355-64; https://doi.org/ 10.1016/j.cell.2013.02.010 [DOI] [PubMed] [Google Scholar]
- [53].Tumbar T, Belmont AS. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat Cell Biol 2001; 3:134-9; https://doi.org/ 10.1038/35055033 [DOI] [PubMed] [Google Scholar]
- [54].Bauer CR, Hartl TA, Bosco G. Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes. PLoS Genet 2012; 8:1-12; https://doi.org/ 10.1371/journal.pgen.1002873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. Spatial dynamics of chromosome translocations in living cells. Science 2013; 341:660-4. PMID:23929981; https://doi.org/ 10.1126/science.1237150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Legartová S, Stixová L, Laur O, Kozubek S, Sehnalová P, Bártová E. Nuclear structures surrounding internal lamin invaginations. J Cell Biochem 2014; 115:476-87; PMID:24123263; https://doi.org/ 10.1002/jcb.24681 [DOI] [PubMed] [Google Scholar]
- [57].Bridger JM. Chromobility: the rapid movement of chromosomes in interphase nuclei. Biochem Soc Trans 2011; 39:1747; PMID:22103519; https://doi.org/ 10.1042/BST20110696 [DOI] [PubMed] [Google Scholar]
- [58].Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Küpper K, Joffe B, Thormeyer T, von Hase J, et al.. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosom Res 2006; 14:707-33. https://doi.org/ 10.1007/s10577-006-1086-x [DOI] [PubMed] [Google Scholar]
- [59].Rouquette J, Genoud C, Vazquez-Nin GH, Kraus B, Cremer T, Fakan S. Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: A novel electron-microscopic approach to reconstructing nuclear architecture. Chromosom Res 2009; 17:801-10; https://doi.org/ 10.1007/s10577-009-9070-x [DOI] [PubMed] [Google Scholar]
- [60].Johnson N, Krebs M, Boudreau R, Giorgi G, LeGros M, Larabell C. Actin-filled nuclear invaginations indicate degree of cell de-differentiation. Differentiation 2003; 71:414-24. https://doi.org/ 10.1046/j.1432-0436.2003.7107003.x [DOI] [PubMed] [Google Scholar]
- [61].Popken J, Graf A, Krebs S, Blum H, Schmid VJ, Strauss A, Guengoer T, Zakhartchenko V, Wolf E, Cremer T. Remodeling of the nuclear envelope and lamina during bovine preimplantation development and its functional implications. PLoS One 2015; 10:1-22; https://doi.org/ 10.1371/journal.pone.0124619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Malhas AN, Vaux DJ. Nuclear Envelope Invaginations and Cancer. In: Cancer Biology and the Nuclear Envelope 2014; page 523-35; https://doi.org/ 10.1007/978-1-4899-8032-8 [DOI] [PubMed] [Google Scholar]
- [63].Braga J, McNally JG, Carmo-Fonseca M. A reaction-diffusion model to study RNA motion by quantitative fluorescence recovery after photobleaching. Biophys J 2007; 92:2694–703. https://doi.org/ 10.1529/biophysj.106.096693 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


