Significance
Eukaryotic cells migrate through a gradient with a huge concentration range of chemoattractant stimuli by employing “adaptation,” in which cells no longer respond to the present stimuli, but remain sensitive to stronger stimuli. Many models agree on the “temporal adaptation”: a rapid “excitation” that triggers cellular responses and a temporally delayed “inhibition” that terminates the responses to reach adaptation. The inhibitory mechanism largely remains elusive, although many molecules of the excitatory signaling pathway have been identified. In the present study, we showed that GPCR-activated Ras negative regulator C2GAP1 locally inhibits Ras signaling for adaptation and long-range chemotaxis.
Keywords: chemotaxis, adaptation, G protein-coupled receptor, Ras activation, Ras GAP
Abstract
Eukaryotic cells chemotax in a wide range of chemoattractant concentration gradients, and thus need inhibitory processes that terminate cell responses to reach adaptation while maintaining sensitivity to higher-concentration stimuli. However, the molecular mechanisms underlying inhibitory processes are still poorly understood. Here, we reveal a locally controlled inhibitory process in a GPCR-mediated signaling network for chemotaxis in Dictyostelium discoideum. We identified a negative regulator of Ras signaling, C2GAP1, which localizes at the leading edge of chemotaxing cells and is activated by and essential for GPCR-mediated Ras signaling. We show that both C2 and GAP domains are required for the membrane targeting of C2GAP1, and that GPCR-triggered Ras activation is necessary to recruit C2GAP1 from the cytosol and retains it on the membrane to locally inhibit Ras signaling. C2GAP1-deficient c2gapA− cells have altered Ras activation that results in impaired gradient sensing, excessive polymerization of F actin, and subsequent defective chemotaxis. Remarkably, these cellular defects of c2gapA− cells are chemoattractant concentration dependent. Thus, we have uncovered an inhibitory mechanism required for adaptation and long-range chemotaxis.
Chemotaxis is a directional cell migration guided by chemoattractant gradients (1–3). This cellular behavior plays critical roles in many physiological processes, such as neuron patterning, immune responses, angiogenesis, metastasis of cancer cells, and the early development of the model organism Dictyostelium discoideum (4–6). Chemotactic cells detect and respond to a large range of concentrations of chemoattractants. For example, D. discoideum cells chemotax toward their chemoattractant cAMP gradients from 10−9 to 10−5 M (7). Chemoattractant sensing has several key features. First, in response to sustained stimuli, cells display a transient response, a process referred to as “adaptation” (8, 9). The critical nature of adaptation is that adaptive cells no longer respond to the continuing, existing stimuli but remain responsive to stimuli at higher concentrations. Second, cells translate extracellular cAMP gradients into polarized intracellular responses, a process called “spatial amplification” (9–12). Because of their capability for temporal adaptation and spatial amplification, the cells chemotax in a chemoattractant gradient over a large range of concentrations. To explain these features, many abstract models have been proposed over the years (9, 13, 14). All models agree on the temporal dynamics of adaptation: an increase in receptor occupancy activates two antagonistic signaling processes: a rapid “excitation” that triggers cell responses and a temporally delayed “inhibition” that terminates the responses to reach adaptation. The central debate focuses on the spatial distribution and the activation mechanism of the inhibition that balances excitation to achieve spatial amplification for gradient sensing (8, 9, 13, 15, 16). Although many of the molecular mechanisms of the excitation process have been discovered, those of the inhibitory processes are still largely elusive (17, 18).
In D. discoideum, cAR1 GPCR (cAMP receptor)-mediated PIP3 responses display all of the features of chemoattractant sensing. In response to uniformly applied cAMP stimulation, the signaling pathway leading to the PIP3 response contains four steps with different kinetics. First, cAMP binds to cAR1 receptor (19, 20). Second, activated cAR1 induces a persistent dissociation/activation of heterotrimeric G proteins (12, 21, 22), indicating that adaptation occurs downstream of heterotrimeric G protein activation. Third, Ras is activated by guanine nucleotide exchange factors (GEFs), which catalyze the exchange of RasGDP (inactive) to RasGTP (active); then GTPase-activating proteins (GAPs) inactivate RasGTP by converting it to RasGDP via stimulating its intrinsic GTPase activity (17, 18, 23, 24). A uniformly applied cAMP stimulation triggers a transient, robust Ras activation followed by a second, small activation associated with pseudopod protrusion (17, 24, 25). Fourth, PIP3 is generated via activation of PI3K and regulation of PTEN membrane localization. PI3K is activated by Ras and phosphorylates the phospholipid PIP2 to PIP3 in the membrane; in the meantime, PTEN transiently withdraws from the plasma membrane to allow accumulation of PIP3 and then returns to the membrane where it dephosphorylates PIP3 to PIP2 (24, 26–29). In conclusion, sustained cAMP stimulation induces persistent dissociation (activation) of heterotrimeric G protein as well as transient and adaptive responses of both Ras activation and PIP3 production (12, 24, 26, 28). Ras activation is the first step in the GPCR-mediated signaling pathway that displays adaptation behavior, indicating the involvement of an inhibitory process acting on Ras signaling and potential roles of Ras inhibitors in chemotaxis (17, 24) (Fig. 1A). There are 18 genes (Fig. S1) that encode potential Ras GAP proteins in the genome of D. discoideum. Disruption of RasGAP gene, nf1 (nf1− or axeB−) or ddnf1 (ddnf1− or nfaA−), results in enhanced Ras activation (18, 30). Ddnf1− cells display impaired chemotaxis toward the cAMP gradient (18), consistent with the pivotal role of Ras in GPCR-mediated chemotaxis. Interestingly, although nf1− cells also have increased Ras and PIP3 activation, they did not show clear chemotaxis defect, but instead, they displayed strong defects in micropinocytosis and axenic growth (30). Despite the potential roles of Ras inhibitors in chemotaxis, we still do not know the molecular mechanisms by which GPCR controls spatiotemporal activities of RasGAPs for chemoattractant sensing. We previously demonstrated the existence of a locally regulated inhibitory process that is upstream of PI3K/PTEN and is required for proper PIP3 responses (12, 14, 31). Thus, we propose that D. discoideum cells may require more than one GAP protein to regulate Ras activation in response to various stimuli and chemotaxis in different concentration gradients.
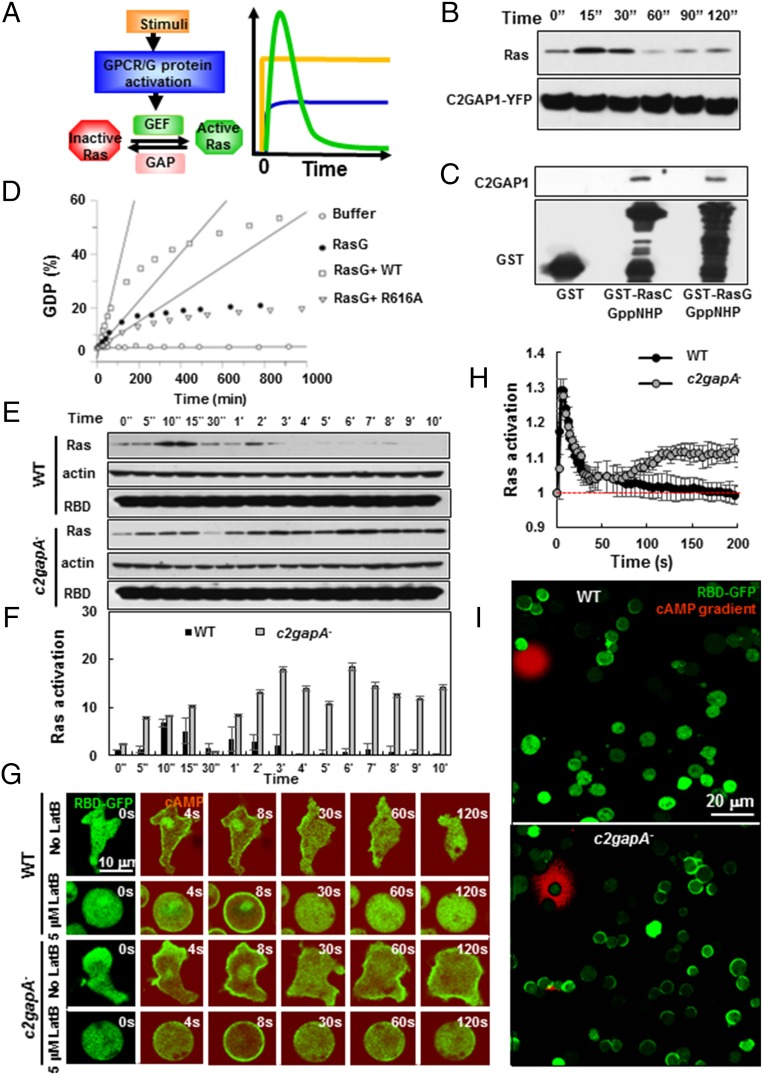
Fig. 1.
C2GAP1 is a GPCR-activated RasGAP. (A) Scheme of GPCR-mediated Ras signaling pathway, containing stimulus; GPCR/G protein; GEF, which converts inactive Ras to active Ras; and GAP, which converts active Ras into inactive Ras. The graph represents the time course of the relative signaling level when a cell is suddenly exposed to uniformly applied chemoattractant stimulation. The stimulation (yellow) is applied at time 0. The kinetics of activation of GPCR/G protein (blue) and Ras (green) are shown as a function of time. (B) A coimmunoprecipitation analysis shows that cAMP stimulation induces the association between Ras and C2GAP1. Cells expressing C2GAP1-YFP were stimulated with 10 μM cAMP at time 0, and cells were collected and lysed at the indicated time points. Lysates were incubated with agarose beads coupled with anti-GFP antibody, and elutes were analyzed by immunoblotting to detect Ras and C2GAP1-YFP using anti-pan Ras (Top) and anti-GFP (Bottom) antibodies, respectively. (C) A coimmunoprecipitation analysis shows that C2GAP1 binds to active RasC and RasG. C2GAP1-YFP lysate was incubated with beads coupled to purified GST alone, or recombinant RasC or RasG loaded with GDP or the nonhydrolysable GTP analog GppNHp {guanosine-5′-[(β,γ)-imido]triphosphate}. Elutes were analyzed by immunoblotting to detect C2GAP1-YFP. (D) GAP activity of C2GAP1. GTP hydrolysis rate of RasG in the presence of equal amounts of the GAP domains of WT (squares, 0.33 ± 0.03% GDP/min), mutant R616A (point mutation at GAP domain, triangle, 0.05 ± 0.003% GDP/min) or buffer alone (filled circle, 0.09 ± 0.007% GDP/min) was measured as the GDP content (by percentage) plotted over time. Buffer alone (open circle) was used as a control. The linear regression was fitted using GraFit 5.0 (Erithacus software). The hydrolysis of GTP is promoted by a factor of 3.7 in the presence of GAP domain of C2GAP1. (E) cAMP-induced Ras activation in WT and c2gapA− cells determined by a pull-down assay. Upon stimulation with 10 μM cAMP at time 0, cells were collected and lysed at the indicated time points. Lysates were incubated with agarose beads coupled with RBD-GST (active Ras binding domain tagged with GST), and elutes were analyzed by immunoblotting with anti-pan Ras antibody (Top), anti-actin antibody (Middle), and RBD-GST (Bottom). (F) Normalized quantitative densitometry of the active Ras in E. The intensity ratio of the active Ras and actin intensities in WT at time 0 s was normalized to 1. Mean ± SD from three independent experiments is shown. (G) cAMP-induced membrane translocation of RBD-GFP in WT and c2gapA− cells with or without Lat B treatment. Cells expressing RBD-GFP (green) were treated with 5 μM Lat B 10 min before the experiment and stimulated with 10 μM cAMP (red) at 2 s. Images were captured at 2-s intervals and shown at selected time points. (Scale bar, 5 μm.) Also see Movies S1 and S2. (H) Measurement of Ras activation in Lat B-treated WT or c2gapA− cells in G. Ras activation was measured as the membrane translocation of RBD-GFP, and the fluorescence intensity on the membrane at time 0 was normalized to 1. Mean ± SD is shown, n = 6 or 5 for WT and c2gapA− cells, respectively. (I) WT and c2gapA− cells expressing RBD-GFP (green) were exposed to a 10-μM cAMP gradient (red). Active Ras polarization was measured as RBD-GFP accumulation in the membrane of Lat B-treated cells exposed to a 10-μM cAMP gradient. Cells were treated with 5 μM Lat B 10 min before the experiment. (Scale bar, 20 μm.) The experiments in Fig. 1 were repeated using two independent c2gapA− clones, with the same results.
In the present study, we studied the role of a C2 domain-containing Ras GAP protein (C2GAP1) in D. discoideum. Specifically, C2GAP1 is highly expressed in cAMP-chemotactic D. discoideum cells and localizes at the leading edge of chemotaxing cells. Cells without C2GAP1 failed in the GPCR-mediated adaptation of Ras activation and showed defects in gradient sensing, polarization, and chemotaxis in a chemoattractant concentration-dependent manner. Our findings uncover a molecular mechanism of the inhibitory process at the level of Ras activation, via which cells achieve the adaptation and effective chemotaxis in response to gradients of a chemoattractant over a broad range of concentrations.
Results
C2GAP1 Is a cAMP-Activated RasGAP.
It has been proposed that the cAR1/G protein machinery activates both an activator (RasGEF) and an inhibitor (RasGAP) to generate a transient Ras activation upon cAMP stimulation (14, 24, 31, 32). However, a cAR1-regulated inhibitor of Ras has yet to be identified. Our previous study suggested that there is a locally controlled inhibitory mechanism that is required for cAR1-mediated PIP3 responses and possibly acts on Ras signaling (14). This led us to identify membrane-targeting Ras GAPs and to study their roles in cAR1-mediated chemotactic responses. Among the 18 genes that encode potential GAP proteins (Fig. S1), we identified one putative RasGAP (DDB0220496, c2gapA), which possesses a C2 domain and is designated C2GAP1. Using a real-time PCR analysis to quantify mRNA levels, we found that the expression of the c2gapA gene highly increased in the cAMP chemotactic-competent stage during development of D. discoideum (Fig. S2). This temporal expression profile is similar to those of key components for cAMP-mediated chemotaxis, such as cAR1 and Gα2 (33). To evaluate the potential role of C2GAP1 in Ras signaling, we examined the association between YFP-tagged C2GAP1 (C2GAP1-YFP) and Ras upon cAMP stimulation, using a coimmunoprecipitation analysis (Fig. 1B). We found that the cAMP stimulation promoted a transient association between C2GAP1 and Ras proteins. Specifically, C2GAP1 interacted with RasC and RasG (Fig. 1C), which are two major Ras isoforms for cAMP chemotaxis in D. discoideum (34, 35). It is well known that Ras GAP proteins increase the intrinsic GTPase activity of Ras by providing a catalytic arginine called R finger (36). The R finger is conserved in C2GAP1 and corresponds to R616 (Fig. S3). To examine the enzymatic activity of C2GAP1, the GAP domains of wild-type (WT) and R-finger mutant (R616A) were purified from Escherichia coli and their GTPase activity was assessed by a HPLC-based reversed phase chromatography (Fig. 1D). The WT GAP domain stimulated the low intrinsic hydrolysis rate of RasG, while the R-finger mutant R616A completely lost its enzymatic activity. These results together indicate that C2GAP1 is a cAMP-mediated Ras GAP protein.
To investigate the function of C2GAP1, we generated c2gapA knockout (c2gapA−) cells using the homologous recombination technique and confirmed the disruption of the c2gapA gene using Southern blot analysis (Fig. S4A). All c2gapA− clones displayed similar phenotypes, which include impaired cAMP-mediated aggregation and fewer fruiting bodies during development. These phenotypes were rescued by the expression of C2GAP1 tagged with YFP (C2GAP1-YFP) in the cells (Fig. S4 B and C). c2gapA− cells showed normal Ras activation during micropinocytosis in a vegetative, growing stage (Fig. S4 D and E), consistent with the expressing profile of C2GAP1 only high in cAMP-chemotactic cells. To examine the possible role of C2GAP1 in cAMP-mediated Ras signaling, we used a pull-down assay to determine the temporal profiles of Ras activation in both WT and c2gapA− cells in response to cAMP stimulation (Fig. 1 E and F). WT and c2gapA− cells had similar cAMP production in response to cAMP stimulation, which was completely blocked by 5 mM caffeine treatment (Fig. S5). Hence, 5 mM caffeine was always presented to the cells in all of the following experiments to eliminate cAMP signal relay and its effect on cell response (37) (Fig. S5). After cells were stimulated with cAMP, cell lysates were incubated with agarose beads conjugated with GST-tagged RBD (the active Ras-binding domain of Raf1) to pull down active Ras proteins, which were detected by an anti-pan Ras antibody (Fig. 1 E and F). In contrast with the low basal Ras activation in WT cells, resting c2gapA− cells had a slightly higher Ras activation, indicating a role of C2GAP1 in keeping the basal Ras activity at a low level. As previously reported (17, 24), cAMP induced a large, transient activation of Ras followed by a small activation of Ras in WT cells. In c2gapA− cells, the stimulation also triggered an initial, transient Ras activation, unlike in WT cells, followed by a robust and prolonged second Ras activation, indicating that Ras activation in c2gapA− cells does not adapt properly.
We further monitored the spatiotemporal dynamics of Ras activation in WT and c2gapA− cells (Fig. 1G). Cells expressing an active Ras probe, RBD-GFP (active Ras-binding domain of Raf1 tagged with GFP, green) were imaged in time lapse by fluorescent microscopy before and after cAMP stimulation (red). In response to uniformly applied cAMP stimulation, RBD-GFP translocated to the membrane and then mostly returned to the cytoplasm followed by a second localization in the protrusion sites in WT cells, while more RBD-GFP remained on the membrane after the initial translocation in c2gapA− cells. To examine the F-actin–independent Ras adaptation profile, we then monitored RBD-GFP dynamics in latrunculin B (Lat B)-treated, immobile cells. Treatment with 5 μM latrunculin completely abolished preexisting F actin and blocked cAMP-triggered actin polymerization (Fig. S6A). In Lat B-treated, immobile WT cells, uniform cAMP stimulation also triggered membrane translocation of RBD-GFP, which peaked at about 6 s, then gradually decreased, and returned to the cytoplasm at about 30 s (Movie S1), indicating a typical Ras adaptation as previously reported (18, 24, 32). This result also confirms that the second, relatively small Ras activation in F-actin intact cells associates with the pseudopod and is F-actin dependent (17, 25, 32). In c2gapA− cells, the stimulation induced a similar transient membrane translocation of RBD-GFP followed by a return to cytosol at about 30 s. However, in contrast to WT cells, RBD-GFP translocated to the membrane again at 60 s (see 60 s and 120 s), indicating that Ras activation fails to persistently adapt in c2gapA− cells in an F-actin–independent manner (Movie S2). Interestingly, the second membrane translocation of RBD-GFP in c2gapA− cells was polarized, but random in direction (Fig. S6B). Quantitative analysis of Ras activation from multiple Lat B-treated cells is also shown (Fig. 1H and Fig. S6C). We then monitored Ras activation in WT and c2gapA− cells exposed to a 10-μM cAMP gradient (Fig. 1I and Fig. S6D). Only those WT cells that were relatively close to the cAMP source, displayed RBD-GFP accumulation in the front facing the cAMP source, while most c2gapA− cells, even those far away from the cAMP source, displayed RBD-GFP membrane accumulation but often in the wrong direction, suggesting hyperactivation of Ras and impaired gradient sensing in c2gapA− cells. Taken together, these results indicate that C2GAP1 is a GPCR-activated RasGAP that suppresses Ras activation in response to various fields of chemoattractant stimuli in an F-actin–independent fashion.
Ras Is Required for Membrane Targeting of C2GAP1.
To understand how cAR1 GPCR controls the function of C2GAP1, we examined possible molecular mechanisms underlying membrane targeting of C2GAP1 (Fig. 2). Using confocal microscopy, we first examined the spatiotemporal localization of C2GAP1 in live cells in response to cAMP stimulation (Fig. 2A). A cAMP stimulation (red) triggered robust membrane translocation of C2GAP1-YFP (green) in the cells without or with Lat B treatment. In addition, PI3K inhibitor did not block the cAMP-induced membrane translocation of C2GAP1 (Fig. S7). These results indicate that the cAR1-mediated membrane translocation of C2GAP1 is independent of F actin and PI3K-mediated PIP3 production. In a cAMP gradient, C2GAP1-YFP was recruited to the leading edge, where active Ras accumulates, in a chemotaxing cell (24) (Fig. 2B and Fig. S8). We next measured the dynamics of cAMP-induced Ras activation and membrane translocation of C2GAP1 in the cells expressing both RBD-RFP and C2GAP1-YFP (Fig. 2C). In nonpolarized, Lat B-treated cells, cAMP stimulation induced the membrane translocation and colocalization of RBD-RFP and C2GAP1-YFP. Our quantitative analysis showed similar membrane-translocation dynamics of C2GAP1-YFP and RBD-RFP (Fig. 2D), suggesting that Ras on the membrane might contribute to the recruitment of C2GAP1 from cytosol to the membrane. We then examined the membrane translocation of C2GAP1-GFP in the cells lacking both RasC and RasG. It has been previously shown that the rasC−:rasG− cells express a low level of cAR1 (35). Thus, we epigenetically expressed cAR1 in rasC−:rasG− cells (rasC−:rasG−:cAR1) (Fig. S9A) and then determined Ras activation and membrane translocation of C2GAP1 in the cells. We found that cAMP stimulation induced negligible membrane translocation of either RBD-GFP (Fig. S9B) or C2GAP1-YFP in rasC−:rasG−:cAR1 cells (Fig. 2 E and F), suggesting that RasC and/or RasG are required for the membrane recruitment of C2GAP1. In addition, cells expressing dominant negative RasC(S17N) or RasG(S17N) showed significantly reduced cAMP-mediated membrane translocation of C2GAP1, while the expression of constitutively active RasC(G12V) significantly enhanced the membrane translocation of C2GAP1 upon cAMP stimulation (Fig. S10 A and B). We also found that C2GAP1 interacts with RasC/G independent of the nucleotide-bound state (Fig. S10C) and the overexpression of the Ras mutants in the cells did not significantly affect the membrane localization of C2GAP1 in unstimulated cells (Fig. S10 D–F). These results together suggest that cAMP-mediated membrane translocation of C2GAP1 is Ras dependent, and requires an additional Ras-dependent signal. To further understand the domain requirement for C2GAP1 membrane targeting, we generated truncation mutants of C2GAP1 lacking either the GAP domain (∆GAP-YFP) or the C2 domain (∆C2-YFP) (Fig. 2G) and tested their membrane-targeting ability in response to cAMP stimulation. We found that both ∆GAP-YFP and ∆C2-YFP mutants failed to translocate to the membrane (Fig. 2 H and I). These results indicate that cAMP-induced membrane translocation of C2GAP1 is Ras dependent and requires both C2 and GAP domains.
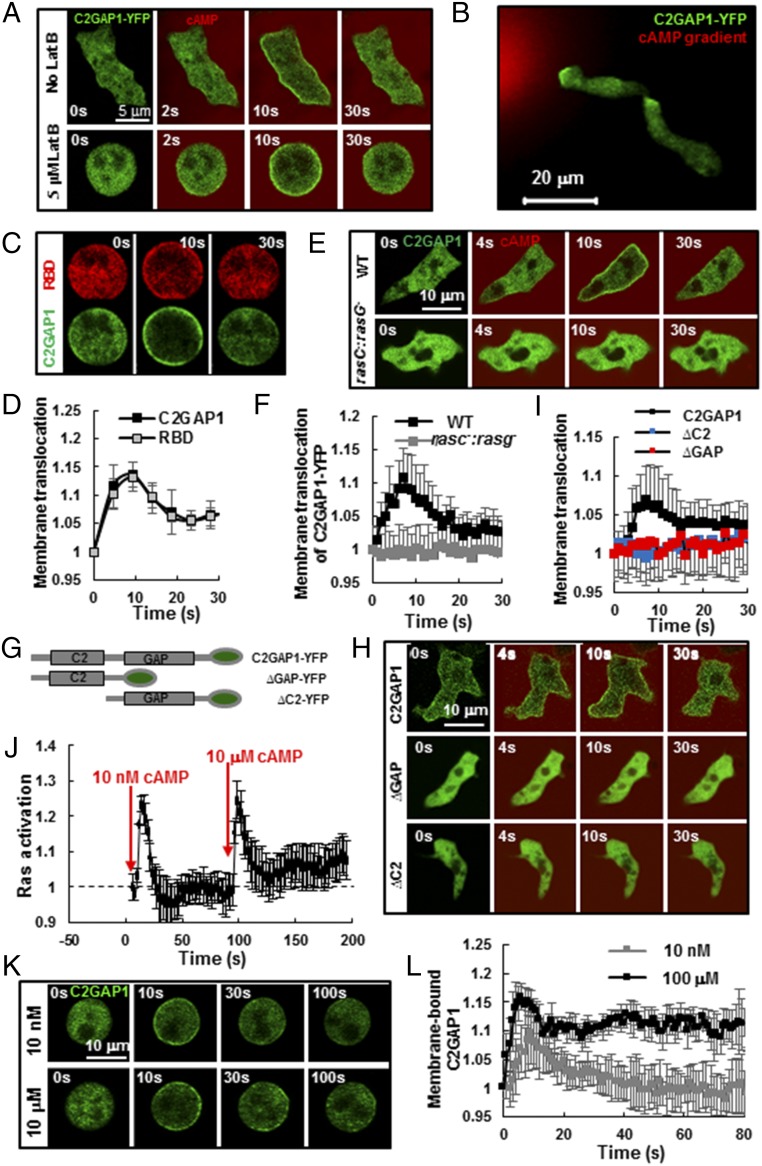
Fig. 2.
Active Ras is required for membrane targeting of C2GAP1. (A) Uniformly applied cAMP induces membrane translocation of C2GAP1-YFP in live cells without (No Lat B) or with (5 μM Lat B) Lat B treatment. Cells expressing C2GAP1-YFP (green) were imaged in time lapse. A total of 1 μM cAMP was mixed with the fluorescent dye Alexa 594 (red) and applied at 2 s. (Scale bar, 5 μm.) (B) C2GAP1 localizes in the leading edge of chemotaxing cells. (Scale bar, 20 μm.) Cells expressing C2GAP1-YFP (green) chemotaxed in a 1-μM cAMP gradient (red). (C) cAMP stimulation induces colocalization of C2GAP1-YFP and RBD-RFP in the cells. A cell expressing C2GAP1-YFP (green) and RBD-RFP (red) was treated with 5 μM Lat B 10 min before the experiment and stimulated with 1 μM cAMP at time 0 s. (Scale bar, 5 μm.) (D) Measurement of the membrane-translocation dynamics of C2GAP1 and RBD-RFP in response to uniform cAMP stimulation in C. Mean ± SD is shown, n = 4. The fluorescence intensity of each fluorescent protein on the membrane at time 0 s was normalized to 1. (E) cAMP-induced membrane translocation of C2GAP1 in WT and rasC−:rasG−:cAR1 cells. Cells expressing C2GAP1-YFP (green) were stimulated with a mixture of 1 μM cAMP and 1 μg/mL Alexa 594 (red) at time 0 s. (Scale bar, 10 μm.) (F) Measurement of the C2GAP1 membrane translocation in WT and c2gapA− cells in E. Mean ± SD is shown (n = 5 or 6 for WT and c2gapA− cells, respectively). The fluorescence intensity on the plasma membrane at time 0 s was normalized to 1. (G) Scheme shows YFP-tagged full-length (C2GAP1) or deletion mutants without the GAP domain (∆GAP) or the C2 domain (∆C2). C2 and GAP domains are between 90–217 aa and 524–860 aa, respectively. (H) Membrane translocation of C2GAP1, ∆GAP, and ∆C2 in response to cAMP stimulation. WT cells expressing C2GAP1-YFP, ∆GAP-YFP, or ∆C2-YFP (green) were stimulated with a mixture of 1 μM cAMP and 1 μg/mL Alexa 594 (red) at time 0 s. (Scale bar, 10 μm.) (I) Measurement of membrane translocation of C2GAP1 or its deletion mutants upon uniform cAMP stimulation in H. Mean ± SD is shown (n = 4, 4, or 5 for cells expressing C2GAP1-YFP, ∆GAP-YFP, or ∆C2-YFP, respectively). The fluorescence intensity on the plasma membrane at time 0 s was normalized to 1. (J) Ras activation in WT cells upon sequentially applied 10-nM and 10-μM cAMP stimulations. Ras activation was measured as the membrane translocation of RBD-GFP in the nonpolarized, Lat B-treated cells. Mean ± SD is shown (n = 5). The fluorescence intensity on the membrane at time 0 s was normalized to 1. (K) Membrane translocation of C2GAP1-YFP upon 10-nM or 10-μM cAMP stimulation. Cells expressing C2GAP1-YFP were treated with 5 μM Lat B 10 min before the experiments and stimulated with cAMP at time 0 s. (L) Measurement of C2GAP1 membrane translocation upon 10-nM or 10-μM uniform cAMP stimulation in K. Mean ± SD is shown, (n = 7 or 8 cells for 10-nM or 10-μM cAMP stimuli, respectively). The fluorescence intensity on the plasma membrane at time 0 s was normalized to 1.
It has been proposed that a stronger receptor activation induces a higher level of inhibition to produce adaptive responses in a cell (14, 32). To explore the role of C2GAP1 in the response to stimulation with different concentrations of cAMP, we first determined the dynamics of Ras activation in response to stimulation with low (10 nM) and high (10 μM) concentrations of cAMP. We sequentially stimulated Lat B-treated WT cells expressing RBD-GFP with 10 nM cAMP at time 0 s and then with 10 μM at 90 s, and measured the membrane translocation of RBD-GFP, which monitors dynamics of Ras activation (Fig. 2J). We found that each of the stimuli triggered a transient Ras activation. However, after the transient activation, Ras adapted to different levels: a complete return to the prestimulus level, designated a perfect adaptation, in response to 10 nM cAMP; and an incomplete return to a higher level than the prestimulus baseline, designated an imperfect adaptation, in response to 10 μM cAMP. Thus, more Ras proteins remain active after adaptation in response to a stronger stimulation. We next monitored the membrane translocation of C2GAP1-YFP in response to stimulation with 10 nM or 10 μM cAMP (Fig. 2K). We found that in response to 10 nM cAMP stimulation, C2GAP1 rapidly translocated to the membrane and then returned to the cytosol at close to the prestimulus level. At the same time, in response to 10 μM cAMP stimulation, C2GAP1-YFP also quickly translocated to the membrane, and more C2GAP1-YFP remained on the membrane instead of returning to the cytosol (Fig. 2L). Taken together, these results suggest that a stronger cAMP stimulus induces more active Ras during the adaptation, and that more C2GAP1 proteins are subsequently retained on the plasma membrane.
c2gapA− Cells Fail to Generate a Persistent Adaptive PIP3 Response upon Uniform cAMP Stimulation in a Concentration-Dependent Manner.
The cAR1/G protein machinery regulates Ras activation for proper PIP3 responses, such as the adaptation to uniform cAMP stimuli (6, 24). To examine the function of C2GAP1 in the cAMP-controlled PIP3 response, we examined cAMP-induced membrane translocation of pleckstrin homology domain of CRAC tagged with GFP (PHcrac-GFP) and PKBR1 phosphorylation (6, 38). In WT cells, we observed biphasic PIP3 production and a transient PKBR phosphorylation in response to uniform stimulation, consistent with previous reports (38–40) and the Ras activation profile in WT cells (Fig. 1E). In c2gapA− cells, we detected prolonged, nonadaptive PIP3 production and PKBR1 phosphorylation (Fig. 3 A and B and Fig. S11). To examine the F-actin–independent PIP3 response, we next monitored PHcrac-GFP membrane translocation in nonpolarized, Lat B-treated WT and c2gapA− cells (Fig. 3 C and D). In WT cells, stimulation with a saturating dose of cAMP (10 μM) triggered a robust membrane translocation of PHcrac-GFP, which peaked at about 6 s, then gradually decreased, and finally returned to its basal level at about 20 s in WT cells, a typical adaptation of PIP3 response as previously reported (6). This result also indicates that the second PIP3 production observed in Fig. 3A is F-actin dependent. In c2gapA− cells, the same stimulus induced an initial transient membrane translocation of PHcrac-GFP followed by a return to cytosol at about 20 s. However, PHcrac-GFP translocated to the membrane again at 120 s, indicating a failure of the persistent adaptation of the PIP3 response in c2gapA− cells in a F-actin–independent manner. Interestingly, we found that stimulation with a low dose of cAMP (10 nM) induced adaptive PIP3 responses in both WT and c2gapA− cells. We quantitatively measured PIP3 responses in WT and c2gapA− cells and confirmed the defect in PIP3 adaptation in c2gapA− cells when they were stimulated with 10 μM cAMP (Fig. 3D). We next determined PIP3 responses to stimulation with different concentrations of nonhydrolysable cAMP, Sp-cAMPS, (10 nM, 100 nM, 1 μM, and 10 μM) (41). Consistent with results in Fig. 3 A and B, WT cells displayed PIP3 adaptation to all concentrations of cAMP, while c2gapA− cells often failed to adapt to the stimuli at high concentrations (Fig. S12 and Movies S3 and S4). The result shows a correlation between the strength of the cAMP stimuli and the percentage of c2gapA− cells that fail in the PIP3 adaptation (Fig. 3E). To understand the relationship between Ras activation and PIP3 production, we simultaneously monitored the membrane translocation of RBD-GFP and PHcrac-RFP in the same cells. We found that the dynamics of PIP3 production closely followed those of Ras activation in the cells (Fig. S13). All results demonstrate that C2GAP1-mediated Ras signaling is essential for the proper adaptation of the PIP3 response in a chemoattractant concentration-dependent, F-actin–independent manner.
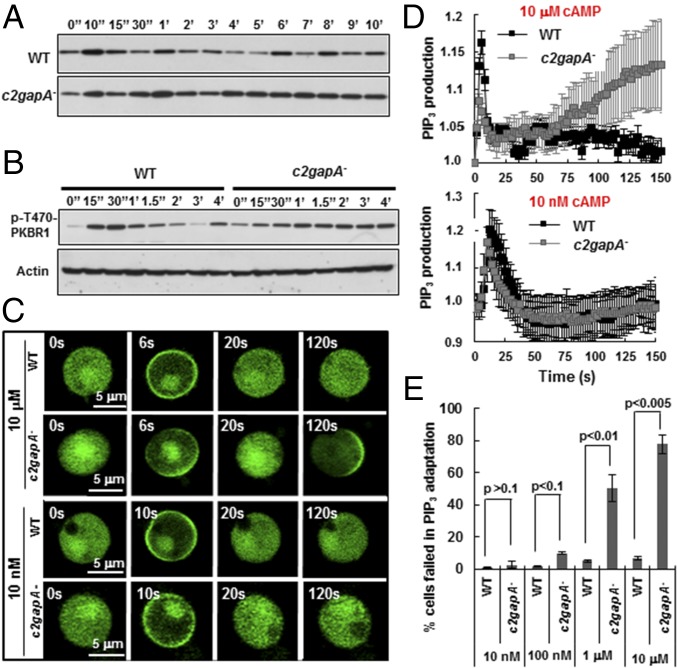
Fig. 3.
c2gapA− cells fail in persistent adaptation of PIP3 production upon uniform stimulation in a cAMP concentration-dependent fashion. (A) Nonadaptive PIP3 response of c2gapA− cells measured by detecting PHcrac-GFP membrane translocation in response to 10-μM cAMP stimulation using filter assay. Aliquots of cells at the indicated time points after cAMP stimulation were immediately filter lysed. The membrane fraction was collected by centrifugation at 16,000 × g for 2 min, then mixed with SDS loading buffer, and subjected to Western blot detection of PHcrac-GFP with anti-GFP antibody. (B) Prolonged PKBR phosphorylation in c2gapA− cells upon cAMP stimulation. WT or c2gapA− cells were stimulated with 10 μM cAMP and sampled at the indicated time points. The phosphorylation of PKBR1 (Top) was detected with antibody recognizing phospho-HM of PKBR1. The amount of actin (Bottom) was detected by anti-actin antibody and used as the loading control. (C) cAMP-induced PIP3 production visualized by the membrane translocation of PHcrac-GFP in nonpolarized, Lat B-treated WT and c2gapA− cells upon uniformly applied 10-nM or 10-μM cAMP stimulation. Cells expressing PHcrac-GFP (green) were treated with 5 μM Lat B 10 min before the experiments and stimulated with cAMP at time 0 s. (D) Measurement of PIP3 kinetics in Lat B-treated WT and c2gapA− cells upon 10-nM or 10-μM cAMP stimulation in A. The fluorescence intensity on the membrane at time 0 was normalized to 1. Mean ± SD is shown (for 10 nM cAMP stimulation, n = 7 or 8 in WT and c2gapA− cells, respectively; for 10-μM cAMP stimulation, n = 6 or 7 in WT and c2gapA− cells, respectively). (E) Percentage of WT and c2gapA− cells failed in PIP3 adaptation upon stimulation with cAMP at various concentrations. Mean ± SD from three sets of independent experiments is shown. Student’s t test, P values are as indicated in each group. Also see Movies S3 and S4. Equivalent results were obtained using two independent c2gapA− clones.
c2gapA− Cells Display Impaired Gradient Sensing.
One key feature of gradient sensing is that a cell is able to translate an extracellular gradient into a polarized intracellular response, such as a sharp accumulation of PHcrac-GFP (PIP3 crescent) in the front of the cell facing the source of a chemoattractant (9). To understand the consequence of Ras hyperactivation observed in c2gapA− cells (Fig. 1I), we monitored the distribution of PHcrac-GFP in WT and c2gapA− cells in response to a cAMP gradient (Fig. 4A). Cells expressing PHcrac-GFP were treated with 5 μM Lat B and exposed to 10 μM cAMP gradients. WT cells showed PHcrac-GFP accumulation in a small and restricted area of the plasma membrane facing the cAMP source. However, c2gapA− cells displayed significantly broadened PHcrac-GFP accumulation in the front (Fig. 4 A and B). This result indicates that c2gapA− cells fail to restrict the PIP3 response to form a sharp PIP3 polarization.
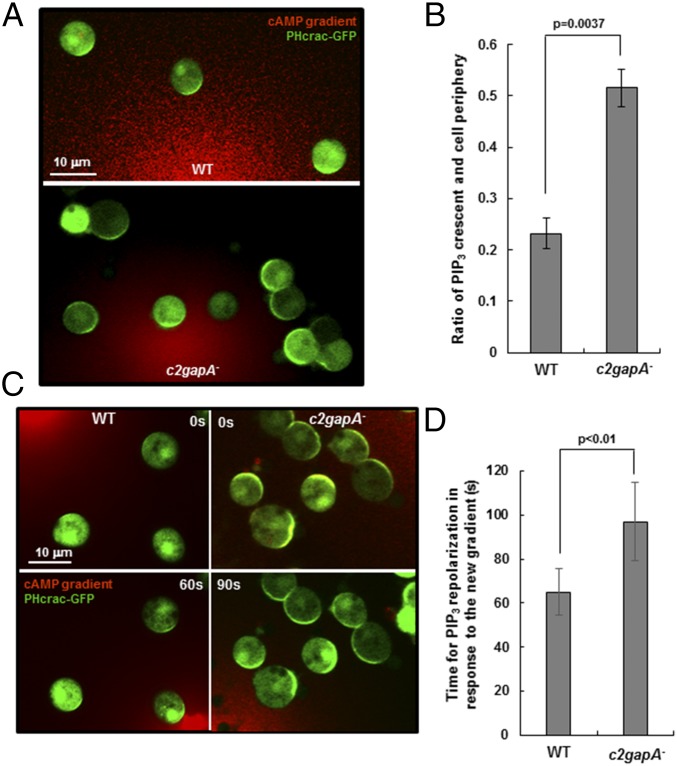
Fig. 4.
c2gapA− cells display impaired gradient sensing capability. (A) Montages show gradient sensing of Lat B-treated WT and c2gapA− cells as PHcrac-GFP accumulation (PIP3 crescents or PIP3 polarization) in the membrane of cells facing a 10-μM cAMP gradient. Cells expressing PHcrac-GFP (green) were treated with 5 μM Lat B and exposed to a 10-μM cAMP gradient (red). To visualize the gradient, cAMP was mixed with 1 μg/μL Alexa 594 (red) in A and C. (B) Ratio of the length of PIP3 crescent versus circumference of the cell in WT and c2gapA− cells. Mean ± SD is shown (n = 11 or 8 for WT and c2gapA− cells, respectively). Student’s t test, P < 0.01. (C) Montages show PIP3 polarization in Lat B-treated WT and c2gapA− cells exposed to the “old” gradient at time 0 s and PIP3 repolarization in the “new” gradient at the indicated time. Cells expressing PHcrac-GFP were treated with 5 μM Lat B for 10 min before the experiments. The new gradient was introduced to the cells at ∼180°. (D) Quantitative analysis of the time to reestablish PIP3 polarization in WT and c2gapA− cells in response to the new gradient shown in C. Cells that displayed the first and the second PIP3 polarization in response to the old and new gradients with ∼180° direction change were measured. Mean ± SD is shown (n = 11 or 12 in WT and c2gapA− cells, respectively). Experiments were repeated using two independent c2gapA− clones, with similar results.
Another feature of gradient sensing is that cells are able to reorient their directional responses toward a changing gradient (22). To examine whether c2gapA− cells are able to reestablish PIP3 polarization efficiently in response to a new gradient, we performed the “changing-direction” experiment, in which an original cAMP gradient was removed from cells that had established PIP3 polarization, followed by the application of a “new” gradient in the opposite direction (Fig. 4C). In response to the change in gradient direction, WT cells rapidly cleared existing PIP3 crescents and quickly made new ones facing the new gradient at about 60 s. However, c2gapA− cells took a significantly longer time (∼90 s) to discard the original PIP3 crescents and to generate new ones facing the new gradient (Fig. 4D). Some c2gapA− cells even failed to generate PIP3 crescents facing the new direction (Fig. 4C). Our results indicated that c2gapA− cells have an impaired gradient sensing machinery that is unable to generate proper responses to gradients, especially to changing ones.
Excess Actin Polymerization Impairs Chemotaxis in c2gapA− Cells.
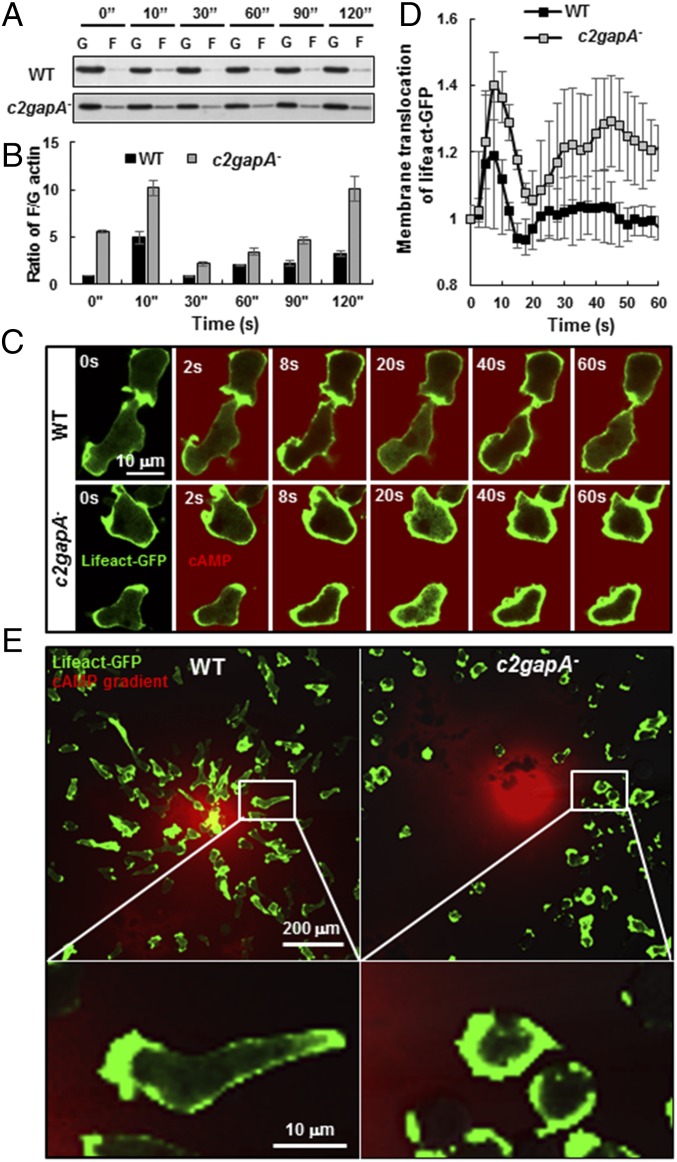
To understand the function of C2GAP1 in cAR1-mediated actin responses, we first measured cAMP-induced actin polymerization by quantifying the ratio of globular (G) actin and actin filaments (F) over a time course (Fig. 5 A and B). In WT cells, stimulation with a high concentration of cAMP (10 μM) induced an actin polymerization process that had an initial, rapid increase followed by an elongated, small second peak, as previously reported (17, 38). On the other hand, in c2gapA− cells, the stimulation triggered an excess actin polymerization that had a larger initial increase followed by a much bigger and persistent elevation. Next, we imaged the cAMP-induced actin response in live cells expressing an F-actin probe (lifeact-GFP) using confocal microscopy (42). Upon cAMP stimulation at 2 s (red), lifeact-GFP translocated to the cell cortex, indicating an increase in F actin in both wild-type and c2gapA− cells (Fig. 5C). Quantitative analysis showed that after the initial response, c2gapA− cells displayed much stronger second membrane translocation of lifeact-GFP than WT cells did (Fig. 5D), indicating that cAMP stimulation induces excess actin polymerization in cells lacking C2GAP1. We then visualized F-actin distribution in chemotaxing cells in a cAMP gradient (Fig. 5E). WT cells chemotaxed effectively toward the micropipette releasing 10 μM cAMP, while c2gapA− cells did not migrate close to the micropipette. Migrating WT cells were highly polarized, with narrow leading fronts and trailing ends, which were marked by lifeact-GFP. However, c2gapA− cells were less polarized, with significantly broadened fronts or massive accumulations of lifeact-GFP on the periphery. Taken together, our results show that saturating cAMP stimuli induce excess actin polymerization and subsequent impairment of chemotaxis in c2gapA− cells.
Fig. 5.
Excess F-actin polymerization upon cAMP stimulation impairs chemotaxis in c2gapA− cells. (A) The amount of globular actin (G) and actin filaments (F) in WT and c2gapA− cells was determined by centrifugation F-actin assay. Cells were stimulated with 10 μM cAMP at time 0, and samples were analyzed at various time points. (B) Normalized quantitative densitometry of the F/G-actin ratio in the WT and c2gapA− cells in A. Mean ± SD from three independent experiments is shown. The F/G ratio of WT cells at 0 s was normalized to 1. (C) cAMP-induced actin polymerization in WT and c2gapA− cells using confocal microscopy. Cells expressing F-actin probe, lifeact-GFP (green), were stimulated with a final concentration of 10 μM cAMP (red) at time 0 s. (Scale bar, 10 μm.) (D) Normalized actin polymerization in WT and c2gapA− cells measured as the membrane translocation of lifeact-GFP in C. Mean ± SD is shown (n = 4 or 5 for WT and c2gapA− cells, respectively). The fluorescence intensity on the plasma membrane at time 0 s was normalized to 1. (E) F-actin distribution in WT and c2gapA− cells exposed to a cAMP gradient. Cells expressing lifeact-GFP (green) were exposed to a 10-μM cAMP (red) gradient for 10 min. (Scale bar, 10 or 200 μm as indicated.)
Chemotaxis Defects in c2gapA− Cells Are cAMP-Concentration Dependent.
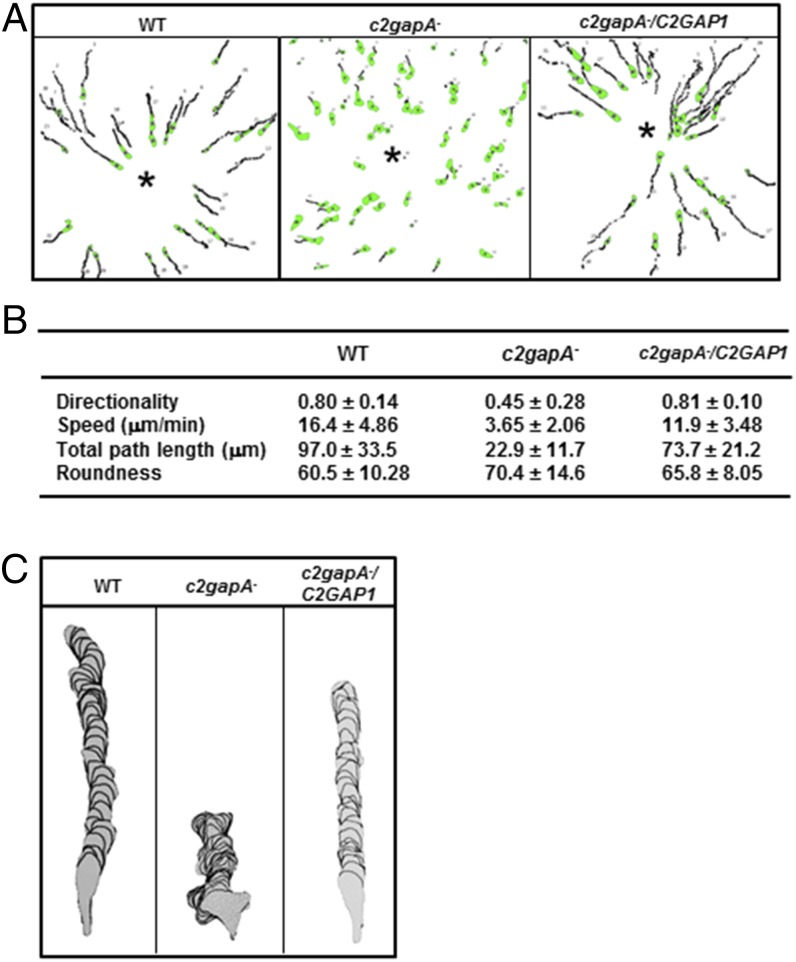
To understand the role of C2GAP1 in chemotaxis, we imaged cell movement toward a micropipette releasing cAMP (10 μM), traced the cell migration, and quantitatively determined four chemotaxis parameters for WT, c2gapA−, and c2gapA−/C2GAP1-YFP cells using the Dynamic Imaging Analysis System (DIAS) (43) (Fig. 6A). We found that c2gapA− cells exhibited poorer directionality, lower speed, shorter travel path length, and much reduced cell polarization (Fig. 6B). Consistent with the significant accumulations of active Ras and PIP3 on the front (Figs. 2 and 4), migrating c2gapA− cells showed significantly broadened leading fronts and often generated random pseudopods (Fig. 6C). c2gapA− cells were also much less polarized and less mobile than WT cells. These defects of c2gapA− cells were rescued by expressing C2GAP1-YFP. The behavior of c2gapA− cells resembles the chemotaxis defects observed in the cells expressing constitutively active Ras (38). Taken together, our results demonstrate that C2GAP1 plays a key role in GPCR cAR1-mediated chemotaxis in D. discoideum cells.
Fig. 6.
C2GAP1 is required for chemotaxis in a 10 μM cAMP gradient. (A) Quantitative analysis of WT, c2gapA−, and c2gapA−/C2GAP1 cells migrating toward a micropipette releasing 10 μM cAMP. * shows the position of the micropipette from which a cAMP gradient was generated. (B) Chemotaxis behaviors measured from A using DIAS software and described as four parameters: directionality is “total” directionality measurement, where 0 represents random movement and 1 represents straight movement to the micropipette; speed, defined as the distance that a cell’s centroid moves as a function of time; total path length the total distance a cell has traveled; and roundness, an indication of cell polarization, where 0 represents a straight line, perfect polarization; and 100% represents a circle, nonpolarization. (C) Typical morphologies of WT, c2gapA−, and c2gapA−/C2GAP1 cells migrating to the micropipette. Experiments in were repeated using two independent c2gapA− clones, with similar results.
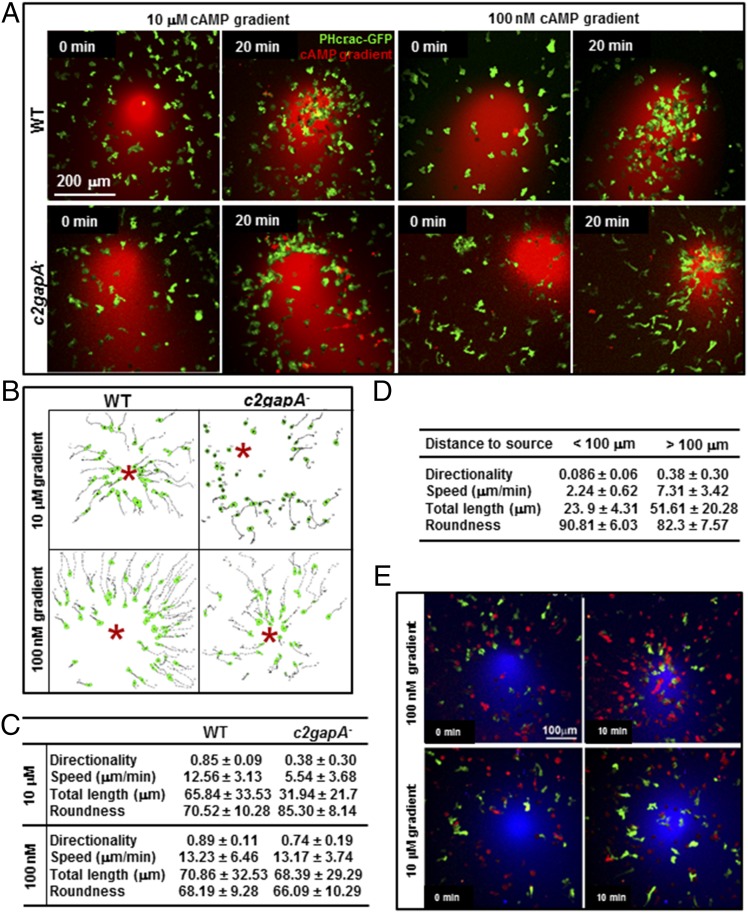
It has been proposed that cells require inhibition processes to adapt to existing stimuli while maintaining sensitivity to stronger stimulation for long-range chemotaxis (9). In this context, we last examined the chemotaxis behavior of WT and c2gapA− cells in cAMP gradients at high (10 μM) or low (100 nM) concentrations. A micropipette releasing cAMP (red) was placed next to the cells (green), and cell migration toward the gradient was imaged in time lapse by confocal microscopy. At 20 min, many WT cells had accumulated near the micropipette generating either 10 μM or 100 nM cAMP gradient (Fig. 7A). The c2gapA− cells accumulated near the micropipette releasing 100 nM cAMP, but not near the one releasing 10 μM cAMP. Using the DIAS software, we traced the chemotaxing cells in time lapse and measured the chemotaxis parameters of WT and c2gapA− cells in both 10 μM and 100 nM cAMP gradients (Fig. 7B). We found that c2gapA− cells not only displayed a more severe chemotaxis defect in a 10 μM cAMP gradient, including poor directionality, low speed, short path, and less polarization (Fig. 7C), but also, the severity of their chemotaxis defects inversely correlated with the distance from the cAMP source (Fig. 7D). Furthermore, we carried out a “mixing” experiment: WT (green) and c2gapA− (red) cells were mixed and allowed to chemotax in the same 100-nM or 10-μM cAMP gradient. We verified that c2gapA− cells chemotaxed as well as WT cells did in the 100 nM cAMP gradient but did poorly in a 10-μM cAMP gradient (Fig. 7E). These results show that C2GAP1 is especially important for cells to chemotax in a higher-concentration cAMP gradient.
Fig. 7.
c2gapA− cells display cAMP concentration-dependent chemotaxis deficiency. (A) WT and c2gapA− cells expressing PHcrac-GFP (green) were exposed to a 10-μM or 100-nM cAMP gradient (red) at 0 min and allowed to chemotax for 20 min. Images at time 0 and 20 min are shown. (Scale bar, 200 μm.) (B) Quantitative analysis of WT and c2gapA− cells migrating toward a micropipette filled with 10 μM or 100 nM cAMP. The travel paths of WT and c2gapA− cells in cAMP gradient at the indicated concentrations are traced. * indicates the position of the micropipette. (C) Chemotaxis behaviors measured from B and described as four parameters: directionality, where 0 represents random movement and 1 represents straight movement to the micropipette; speed, defined as the distance that a cell’s centroid moves as a function of time; total path length, the total distance a cell has traveled; and roundness, an indication of cell polarization, where 0 represents a straight line, perfect polarization, and 100% represents a circle, nonpolarization. (D) Comparison of chemotaxis parameters from the cells either close (<50 μm) or far (>200 μm) from the 10 μM cAMP source in B. (E) WT (green) and c2gapA− (red) cells were exposed to the same 100 nM or 10 μM cAMP gradient (blue) at time 0 min and allowed to chemotax for 10 min. WT cells expressed RBD-GFP (green), and c2gapA− cells expressed RBD-RFP (red). To visualize the cAMP gradient, cAMP stimuli were mixed with 1 μg/mL Alexa 633 (blue). (Scale bar, 100 μm.)
Discussion
The existence of an inhibitory process and its function in GPCR-mediated gradient sensing and chemotaxis has been proposed for 40 years. Adaptation behavior of GPCR-mediated Ras activation indicates the involvement of an inhibitory process. The regulation of Ras adaptation through DDNF1 and PKB/PKBR1-mediated negative feedback mechanism has been reported (17, 18). However, the inhibitors are still poorly understood. Here, we reveal a negative regulator of Ras signaling, C2GAP1. We show that C2GAP1 is mediated by GPCR activation and recruited from the cytosol in a Ras-dependent manner to inhibit Ras signaling for its adaptation. We also show that C2GAP1-regulated Ras adaptation is essential for chemoattractant gradient sensing and long-range chemotaxis.
GAPs are negative regulators of Ras proteins that are key components of gradient sensing and chemotaxis in both neutrophils and D. discoideum (13, 18, 23, 24, 44). The D. discoideum genome encodes 18 GAP-like proteins. Disruption of one gap gene, ddnf1 (ddnf1−), results in prolonged Ras activation in response to cAMP stimulation and impaired chemotaxis toward a cAMP gradient (18). However, whether cAR1 GPCR regulates DdNF1’s function is still not clear. Our previous study suggested that a chemoattractant GPCR must control negative regulators that locally suppress Ras signaling for effective gradient sensing and efficient chemotaxis (14). In the present study, we discovered C2GAP1 that translocates from the cytosol to the cell membrane upon cAMP stimulation and localizes at the leading edge of a chemotaxing cell, indicating its involvement in a cAR1-mediated inhibitory mechanism acting locally on Ras signaling. As previously reported, GPCR-mediated Ras activation is a complex, including F-actin–dependent positive and negative feedback loops (14, 17, 24, 25). Actin polymerization inhibitors eliminate F-actin–mediated positive and negative feedback loops and thus provide a simplified system for dissecting the F-actin–independent signaling pathway from the F-actin–dependent one. Using inhibitor-treated cells, different Ras activation/adaptation behaviors were observed in WT, ddnf1−, and c2gapA− cells. In response to uniform cAMP stimulation, WT cells show a transient Ras activation, while ddnf1− cells exhibit prolonged Ras activation (18). Unexpectedly, c2gapA− cells display the same initial, transient Ras activation as WT cells upon a persistent cAMP stimulation (24), but fail to adapt persistently to the stimulation (Fig. 1). Based on the Ras activation profiles in WT, ddnf1−, and c2gapA− cells, we suggest that the F-actin–independent adaptation of Ras signaling includes two sequential steps: an initial, transient activation, followed by a persistent adaptation. Both DdNF1 and C2GAP1 are required for the adaptation process, but act at different steps. DdNF1 appears to be responsible for the quick turnoff of Ras activation at the initial step, and C2GAP1 plays a crucial role in persistent, long-term adaptation at the second step in the adaptation process. Moreover, F-actin–independent adaptation is involved in and contributes to the adaptation mechanism with cytoskeletal activity.
How a chemoattractant GPCR regulates inhibitors to achieve adaptation and chemotaxis in eukaryotic cells is still not clear. A theoretical study demonstrated that there are two classes of simple network for receptor-induced adaptation: an incoherent feedforward loop with a proportional node (IFFLP) or a negative feedback loop with a buffer node (NFBLB) (32, 45). The difference between these two classes of networks is how the negative regulator is activated and consequently terminates the cell response to reach adaptation. In the IFFLP model, GPCR signaling via something other than the product of the response activates the negative regulator, while in the NFBLB model, the response itself triggers the activation of the negative regulator (8, 45). Several models have proposed that the chemoattractant GPCR-induced inhibitions are likely IFFLP type (9, 31, 32). In the present study, we have shown that several GPCR-mediated responses, such as PIP3 production or actin polymerization, are not required for membrane targeting of C2GAP1 (Fig. 2A and Fig. S7). The spatiotemporal distribution of C2GAP1 relies on and closely follows Ras activation (Fig. 2 and Fig. S10). In addition to the GAP domain, the C2 domain is required for the membrane targeting of the C2GAP1 (Fig. 2 H and I) as it does in many other proteins in D. discoideum, such as PTEN and PI3K (26, 28, 29). Recently, it was shown that the C2 domain of RASAL, a mammalian C2 domain-containing RasGAP protein, regulates its membrane targeting through calcium and lipids (46). Interestingly, the basal levels of C2GAP1 membrane of all Ras mutants is similar to that in wild-type cells (Fig. S10 A–E), and the in vitro binding assay indicates that C2GAP1 binds both GTP- and GDP-bound Ras protein (Fig. S10A). Taking these results together, we propose a two-step membrane-translocation mechanism of C2GAP1, which is dependent on Ras and Ras activation and requires both the C2 and GAP domains of C2GAP1.
The inhibitory process of GPCR-mediated gradient sensing and chemotaxis has been suggested by many models (9, 13, 14). The molecular mechanisms of inhibition required for adaptation during chemotaxis of eukaryotic cells are still a focal point for debates. A GPCR-mediated signaling network for chemotaxis consists of many signaling events, including activation of GPCR, G protein dissociation, Ras activation, PIP3 production, and other events linked to actin reorganization. Theoretically, receptor-mediated adaptation could be as simple as the following: activation of receptor induces a fast excitation to generate a response, and a slow inhibition terminates the response to reach adaptation. However, the cAR1-induced adaptation occurs at many signaling steps, including Ras activation and PIP3 production. Thus, we propose that eukaryotic cells utilize various inhibitory mechanisms at different steps of the signaling pathway for chemotaxis. In response to a cAMP gradient, cells generate different spatiotemporal responses at each signaling event that eventually lead to a directional biochemical response (gradient sensing) for chemotaxis. Activation of cAR1 induces persistent G protein dissociation/activation (12, 21). It triggers a transient Ras activation that quickly returns to low levels around the cell membrane, but only those cells exposed to a strong and steep gradient displayed polarized Ras activation. The gradient also induces a polarized PIP3 response through activation of PI3K in the cell membrane and redistribution of PTEN (26, 28). Our study reveals one of the inhibitors, C2GAP1, that controls the Ras signaling event for directional sensing and long-range chemotaxis, especially in a high-concentration gradient. A similar high concentration-dependent defect in gradient sensing and directed cell migration has been reported in gip1 null cells (47). G protein-interacting protein 1 (GIP1) binds and sequesters G proteins in the cytosolic pool, and subsequently regulates GPCR-mediated G protein shuttling between the cytosol and the membrane. It will be important to know whether the recycling of heterotrimeric G protein plays any role in the adaptation process of downstream effectors. Many other inhibitory mechanisms are yet to be identified in the GPCR-mediated network for chemotaxis, and future studies are needed to reveal additional negative inhibitors essential for the chemotaxis of eukaryotic cells.
Materials and Methods
Chemotaxis Assay.
As was previously described (12), cells were placed in a one-well chamber (Nalge Nunc International) and imaged with a Zeiss laser scanning microscope, LSM 780 or LSM 880, with a 63×, 1.4 N.A. Plan-Neofluar objective lens. A chemoattractant gradient was generated with a microinjector (Eppendorf) connected to a micropipette filled with cAMP solution. Cell migration was recorded at 10-s intervals. Computer analysis was performed using DIAS software (43).
GAP Activity Measurement of C2GAP1.
A total of 1 µM of RasG with and without an equal amount of C2GAP or mutant was incubated with 50 µM of GTP at 20 °C in 50 mM Tris pH 7.5, 50 mM NaCl, and 5 mM MgCl2. After different times, the GDP content of the samples was analyzed by a HPLC (Themo Ultimate 3000): a reversed phase C18 column was employed to detect GDP and GTP content (by percentage) as previously described by Eberth and Ahmadian (48). Linear rates of GDP production were plotted (first four to eight time points) using GraFit 5.0 (Erithacus software). Full materials and methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Xinzhuan Su and Joseph Brzostowski for their critical reading of the article; Wei Quan for his help with chemotaxis video analysis; and Dr. Peter van Haastert, Susanne Terheyden, and Kavya Prasad for assistance with the biochemical experiments. This work was supported by the intramural fund of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and the American Heart Association (AHA0930127N).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703208114/-/DCSupplemental.
References
- 1.Iijima M, Huang YE, Devreotes P. Temporal and spatial regulation of chemotaxis. Dev Cell. 2002;3:469–478. doi: 10.1016/s1534-5807(02)00292-7. [DOI] [PubMed] [Google Scholar]
- 2.Van Haastert PJ, Devreotes PN. Chemotaxis: Signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 3.Zigmond SH. Chemotaxis by polymorphonuclear leukocytes. J Cell Biol. 1978;77:269–287. doi: 10.1083/jcb.77.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condeelis J, Singer RH, Segall JE. The great escape: When cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- 5.Murphy PM. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 6.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 7.Janssens PM, Van Haastert PJ. Molecular basis of transmembrane signal transduction in Dictyostelium discoideum. Microbiol Rev. 1987;51:396–418. doi: 10.1128/mr.51.4.396-418.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoeller O, Gong D, Weiner OD. How to understand and outwit adaptation. Dev Cell. 2014;28:607–616. doi: 10.1016/j.devcel.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parent CA, Devreotes PN. A cell’s sense of direction. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 10.Chung CY, Funamoto S, Firtel RA. Signaling pathways controlling cell polarity and chemotaxis. Trends Biochem Sci. 2001;26:557–566. doi: 10.1016/s0968-0004(01)01934-x. [DOI] [PubMed] [Google Scholar]
- 11.Servant G, et al. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, Meier-Schellersheim M, Jiao X, Nelson LE, Jin T. Quantitative imaging of single live cells reveals spatiotemporal dynamics of multistep signaling events of chemoattractant gradient sensing in Dictyostelium. Mol Biol Cell. 2005;16:676–688. doi: 10.1091/mbc.E04-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houk AR, et al. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 2012;148:175–188. doi: 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Meier-Schellersheim M, Yan J, Jin T. Locally controlled inhibitory mechanisms are involved in eukaryotic GPCR-mediated chemosensing. J Cell Biol. 2007;178:141–153. doi: 10.1083/jcb.200611096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: Distinctions between directional sensing and polarization. J Biol Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima A, Ishihara S, Imoto D, Sawai S. Rectified directional sensing in long-range cell migration. Nat Commun. 2014;5:5367. doi: 10.1038/ncomms6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charest PG, et al. A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev Cell. 2010;18:737–749. doi: 10.1016/j.devcel.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Charest PG, Firtel RA. Spatiotemporal regulation of Ras activity provides directional sensing. Curr Biol. 2008;18:1587–1593. doi: 10.1016/j.cub.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein PS, et al. A chemoattractant receptor controls development in Dictyostelium discoideum. Science. 1988;241:1467–1472. doi: 10.1126/science.3047871. [DOI] [PubMed] [Google Scholar]
- 20.Van Haastert PJ. Down-regulation of cell surface cyclic AMP receptors and desensitization of cyclic AMP-stimulated adenylate cyclase by cyclic AMP in Dictyostelium discoideum. Kinetics and concentration dependence. J Biol Chem. 1987;262:7700–7704. [PubMed] [Google Scholar]
- 21.Janetopoulos C, Jin T, Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. 2001;291:2408–2411. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- 22.Jin T, Zhang N, Long Y, Parent CA, Devreotes PN. Localization of the G protein betagamma complex in living cells during chemotaxis. Science. 2000;287:1034–1036. doi: 10.1126/science.287.5455.1034. [DOI] [PubMed] [Google Scholar]
- 23.Insall RH, Borleis J, Devreotes PN. The aimless RasGEF is required for processing of chemotactic signals through G-protein-coupled receptors in Dictyostelium. Curr Biol. 1996;6:719–729. doi: 10.1016/s0960-9822(09)00453-9. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Haastert PJ, Keizer-Gunnink I, Kortholt A. Coupled excitable Ras and F-actin activation mediates spontaneous pseudopod formation and directed cell movement. Mol Biol Cell. 2017;28:922–934. doi: 10.1091/mbc.E16-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 27.Funamoto S, Milan K, Meili R, Firtel RA. Role of phosphatidylinositol 3′ kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in dictyostelium. J Cell Biol. 2001;153:795–810. doi: 10.1083/jcb.153.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 29.Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem. 2004;279:16606–16613. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- 30.Bloomfield G, et al. Neurofibromin controls macropinocytosis and phagocytosis in Dictyostelium. Elife. 2015;4:e04940. doi: 10.7554/eLife.04940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier-Schellersheim M, et al. Key role of local regulation in chemosensing revealed by a new molecular interaction-based modeling method. PLOS Comput Biol. 2006;2:e82. doi: 10.1371/journal.pcbi.0020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeda K, et al. Incoherent feedforward control governs adaptation of activated ras in a eukaryotic chemotaxis pathway. Sci Signal. 2012;5:ra2. doi: 10.1126/scisignal.2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Devreotes PN. G protein-linked signaling pathways control the developmental program of Dictyostelium. Neuron. 1994;12:235–241. doi: 10.1016/0896-6273(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 34.Kae H, Lim CJ, Spiegelman GB, Weeks G. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 2004;5:602–606. doi: 10.1038/sj.embor.7400151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolourani P, Spiegelman GB, Weeks G. Rap1 activation in response to cAMP occurs downstream of ras activation during Dictyostelium aggregation. J Biol Chem. 2008;283:10232–10240. doi: 10.1074/jbc.M707459200. [DOI] [PubMed] [Google Scholar]
- 36.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 37.Brenner M, Thoms SD. Caffeine blocks activation of cyclic AMP synthesis in Dictyostelium discoideum. Dev Biol. 1984;101:136–146. doi: 10.1016/0012-1606(84)90124-6. [DOI] [PubMed] [Google Scholar]
- 38.Cai H, et al. Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J Cell Biol. 2010;190:233–245. doi: 10.1083/jcb.201001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Postma M, et al. Uniform cAMP stimulation of Dictyostelium cells induces localized patches of signal transduction and pseudopodia. Mol Biol Cell. 2003;14:5019–5027. doi: 10.1091/mbc.E03-08-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang YE, et al. Receptor-mediated regulation of PI3Ks confines PI(3,4,5)P3 to the leading edge of chemotaxing cells. Mol Biol Cell. 2003;14:1913–1922. doi: 10.1091/mbc.E02-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Haastert PJ, Van der Heijden PR. Excitation, adaptation, and deadaptation of the cAMP-mediated cGMP response in Dictyostelium discoideum. J Cell Biol. 1983;96:347–353. doi: 10.1083/jcb.96.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukujin F, Nakajima A, Shimada N, Sawai S. Self-organization of chemoattractant waves in Dictyostelium depends on F-actin and cell-substrate adhesion. J R Soc Interface. 2016;13:20160233. doi: 10.1098/rsif.2016.0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wessels D, et al. A computer-assisted system for reconstructing and interpreting the dynamic three-dimensional relationships of the outer surface, nucleus and pseudopods of crawling cells. Cell Motil Cytoskeleton. 1998;41:225–246. doi: 10.1002/(SICI)1097-0169(1998)41:3<225::AID-CM4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 44.Wang MJ, Artemenko Y, Cai WJ, Iglesias PA, Devreotes PN. The directional response of chemotactic cells depends on a balance between cytoskeletal architecture and the external gradient. Cell Rep. 2014;9:1110–1121. doi: 10.1016/j.celrep.2014.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sot B, Behrmann E, Raunser S, Wittinghofer A. Ras GTPase activating (RasGAP) activity of the dual specificity GAP protein Rasal requires colocalization and C2 domain binding to lipid membranes. Proc Natl Acad Sci USA. 2013;110:111–116. doi: 10.1073/pnas.1201658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamimura Y, Miyanaga Y, Ueda M. Heterotrimeric G-protein shuttling via Gip1 extends the dynamic range of eukaryotic chemotaxis. Proc Natl Acad Sci USA. 2016;113:4356–4361. doi: 10.1073/pnas.1516767113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eberth A, Ahmadian MR. In vitro GEF and GAP assays. Curr Protoc Cell Biol. 2009;Chapter 14:Unit 14.19. doi: 10.1002/0471143030.cb1409s43. [DOI] [PubMed] [Google Scholar]
- 49.Bosgraaf L, van Haastert PJ, Bretschneider T. Analysis of cell movement by simultaneous quantification of local membrane displacement and fluorescent intensities using Quimp2. Cell Motil Cytoskeleton. 2009;66:156–165. doi: 10.1002/cm.20338. [DOI] [PubMed] [Google Scholar]
- 50.Veltman DM, Akar G, Bosgraaf L, Van Haastert PJ. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 2009;61:110–118. doi: 10.1016/j.plasmid.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Snaar-Jagalska BE, Van Haastert PJ. G-protein assays in Dictyostelium. Methods Enzymol. 1994;237:387–408. doi: 10.1016/s0076-6879(94)37077-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.