Significance
Psoriasis is a chronic skin disease characterized by the infiltration of inflammatory T cells to the skin in response to injury. When inflammatory T cells and dendritic cells are recruited to the skin by CCL20 and other chemokines, they release cytokines that contribute to psoriatic inflammation. We engineered a molecule derived from the natural CCL20 protein that adopts a unique dimeric structure, partially activates its G-protein receptor, blocks T cell homing, and prevents the signs of psoriasis in a mouse model of this common human skin disease. Our remarkable findings reveal the potential of engineered-CCL20 molecules as therapeutic agents for psoriasis and the general utility of chemokine engineering for treating inflammatory diseases.
Keywords: CCL20, Th17, psoriasis, protein engineering, X-ray
Abstract
Psoriasis is a chronic inflammatory skin disease characterized by the infiltration of T cell and other immune cells to the skin in response to injury or autoantigens. Conventional, as well as unconventional, γδ T cells are recruited to the dermis and epidermis by CCL20 and other chemokines. Together with its receptor CCR6, CCL20 plays a critical role in the development of psoriasiform dermatitis in mouse models. We screened a panel of CCL20 variants designed to form dimers stabilized by intermolecular disulfide bonds. A single-atom substitution yielded a CCL20 variant (CCL20 S64C) that acted as a partial agonist for the chemokine receptor CCR6. CCL20 S64C bound CCR6 and induced intracellular calcium release, consistent with G-protein activation, but exhibited minimal chemotactic activity. Instead, CCL20 S64C inhibited CCR6-mediated T cell migration with nominal impact on other chemokine receptor signaling. When given in an IL-23–dependent mouse model for psoriasis, CCL20 S64C prevented psoriatic inflammation and the up-regulation of IL-17A and IL-22. Our results validate CCR6 as a tractable therapeutic target for psoriasis and demonstrate the value of CCL20 S64C as a lead compound.
Psoriasis affects up to 3% of the world’s populations, depending on geographic and genetic factors, and is one of the most common autoimmune diseases that affects the skin (1). There is abundant evidence that the chemokine–receptor pair, CCL20/CCR6, plays a key role in psoriatic skin inflammation by regulating dendritic and T cell trafficking to sites of injury or infection (2). In psoriatic inflamed skin, CCL20, an ∼8-kDa chemokine, is abundantly produced by psoriatic keratinocytes and endothelial cells where it binds and activates CCR6, its cognate seven-transmembrane G-protein–coupled receptor (GPCR), which is expressed on the surface of migratory immune cells (3–6). Mice injected intradermally with IL-23 had high expression levels of CCL20 in the epidermis and increased recruitment of CCR6+, γδ-low T cells (GDL T cells) that expressed the IL-23 receptor (7, 8). Moreover, CCR6-deficient mice were highly resistant to IL-23–associated psoriasiform dermatitis compared with wild-type (WT) mice (7). Recruited GDL T cells expressed substantial levels of critical Th17 cytokines, IL-17A and IL-22, which are recognized biomarkers of psoriatic inflammation (8). Treatment with proven TNF-α inhibitors is highly effective but risk reactivation of TB (9). Similarly, new IL-17 inhibitors are effective but are accompanied by increased risk for fungal infection such as mucosal candidiasis (10). Because a neutralizing anti-CCL20 monoclonal antibody reduced psoriasis-like inflammation in mice (11), inhibition of CCL20/CCR6-mediated T cell recruitment as a therapeutic strategy in humans may offer a therapeutic option.
Chemokines engage their receptors via an extensive protein–protein interface that encompasses domains at the extracellular surface and a deep pocket within the transmembrane domain (the orthosteric site) where the chemokine N terminus binds (12). Native chemokines are balanced GPCR agonists that elicit a combination of intracellular responses, including activation of heterotrimeric G proteins and β-arrestin–mediated signal transduction that culminate in directional cell migration. Inhibition of chemokine signaling by small molecule or peptide antagonists typically blocks GPCR signaling by binding the orthosteric site and preventing activation by the chemokine agonist (13, 14). However, partial or biased agonists of chemokine receptors, which are characterized by a selective loss of efficacy in certain types of signaling, can also function as potent inhibitors. For example, AOP-RANTES, a chemically modified form of RANTES/CCL5, induces CCR5-mediated calcium signaling but lacks promigratory signaling and has altered receptor recycling (15, 16). The greater anti-HIV potency of AOP-RANTES relative to native RANTES demonstrated the utility of engineered chemokines with partial agonist activity as alternatives to small-molecule GPCR antagonists for therapeutic development (17).
Manipulation of a chemokine’s oligomeric state can also change its signaling profile in useful ways. Chemokine self-association enhances binding to extracellular matrix glycosaminoglycans, which is essential for in vivo chemokine function (18), but full GPCR activation is generally attributed only to the monomeric state (19, 20). NMR and X-ray structures have revealed two conserved types of chemokine dimerization corresponding to the CC and CXC subfamilies (21). Additionally, cell-based assays have shown that CC dimers prevent receptor binding by sequestering residues of the N terminus within the dimer interface (22). In contrast, the N terminus of a typical CXC dimer remains fully accessible because the dimer interface forms along the β1 strand of opposing subunits. As a result of the CXC dimer conformation, an engineered CXCL1 dimer can bind and activate its receptor CXCR2 with nanomolar potency (23). We discovered that monomeric and dimeric forms of CXCL12 can each bind and activate the CXCR4 receptor, inducing profoundly different cellular responses (24). Whereas the CXCL12 monomer is a balanced agonist that promotes chemotactic cell migration, the disulfide-locked CXCL12 dimer is a biased agonist that potently inhibits chemotaxis in vitro and blocks the CXCR4-mediated spread of metastatic cancer cells in vivo (25–27).
Since typical CC chemokine dimers are incompatible with high-affinity receptor binding, CCL20 would seem to be an unlikely candidate for engineered dimerization. However, unlike most other CC chemokines, the two published crystal structures of CCL20 show the protein in a CXC-type dimeric arrangement. Based on our work with the disulfide-locked CXCL12 dimer, we speculated that a similar approach could convert CCL20 into a T cell-specific, antiinflammatory agent. Herein, we present the design and construction of a disulfide-linked CCL20 dimer, which binds and activates the chemokine receptor CCR6 but inhibits T cell chemotaxis. We show that the engineered CCL20 dimer reduces disease severity in an IL-23–dependent mouse model of psoriasis. This constitutively dimeric CCL20 molecule may have utility in other types of Th17-mediated inflammation, including autoimmune diseases like rheumatoid arthritis or multiple sclerosis.
Results
Construction of the CCL20 S64C Variant.
For chemokines that self-associate, dimerization is essential for biological function in vivo, and, as a result, engineered monomers and dimers of various CC and CXC chemokines have been used to probe their respective roles in chemokine signaling (18). The CXC-type dimer observed in two different CCL20 X-ray crystal structures [Protein Data Bank (PDB) ID codes 2HCI and 1M8A] is unusual for a member of the CC chemokine family, and NMR studies indicate that CCL20 self-association is relatively weak and pH dependent (28–30). To probe the functional activity of the crystallographic CCL20 dimer, we used the protein engineering algorithm Disulfide by Design and visual inspection of the X-ray crystal structure (PDB ID code 2HCI) to design a panel of cysteine substitutions predicted to form intermolecular disulfide bonds (31). The five CCL20 variants selected for initial experimental testing included two single-cysteine and three double-cysteine substitutions in either the first β-strand (V21C/T24C, G22C/T24C, and F23C) or the α-helix (V60C/V67C and S64C) (Fig. S1A). Each variant was expressed in Escherichia coli and refolded. Only one variant, CCL20 S64C, which is predicted to make a symmetric, intermolecular disulfide bond between opposing α-helices (Fig. S1B), yielded properly folded protein that behaved as a dimer as assessed by SDS/PAGE.
Coinjection of CCL20 WT and CCL20 S64C proteins onto reverse-phase HPLC produced a chromatogram with two distinct peaks (Fig. S1C). Injection of CCL20 WT or CCL20 S64C alone revealed that the retention times for each protein matched those in the coinjection, and purity of CCL20 S64C was estimated to exceed 99.5%. The intact mass of CCL20 S64C was determined by mass spectrometry to be 16,080.4 Da (Fig. S2), consistent with the presence of a disulfide bond linking two CCL20 S64C molecules. The oligomeric state of CCL20 S64C was confirmed by SDS/PAGE of disulfide-reduced and nonreduced samples, which migrated at ∼10 and 20 kDa, respectively (Fig. S1D). A 2D 1H–15N heteronuclear single-quantum coherence spectrum of CCL20 S64C shows 79 well-dispersed peaks consistent with a homogeneous, folded protein in a pattern that is distinct from CCL20 WT (Fig. S1E). Based on the biophysical data, we concluded that replacement of serine 64 with cysteine yielded a disulfide-linked symmetric CCL20 homodimer.
X-Ray Crystal Structure of CCL20 S64C.
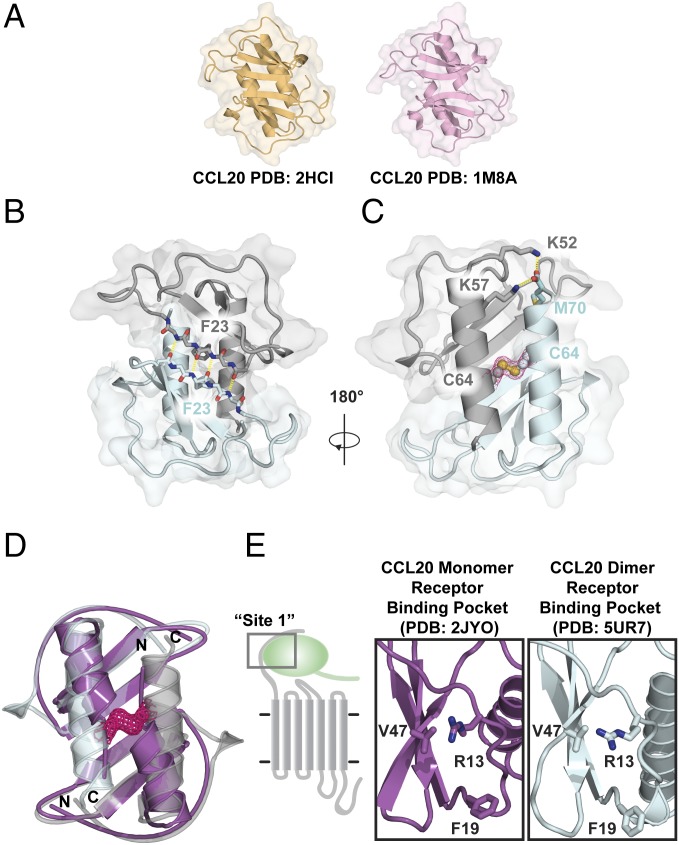
In two previous X-ray structures, CCL20 adopts a CXC-like dimer conformation (29, 30) (Fig. 1A). We solved the CCL20 S64C structure at 2.0-Å resolution and observed a CXC-type dimer in the asymmetric unit (PDB ID code 5UR7) (Table S1). Except for the first four residues of each polypeptide, complete electron density was present throughout, including M70, the C-terminal residue, which was not observed in previous structures. The dimer interface is composed of various intermolecular contacts, including a characteristic pattern of intermolecular hydrogen bonds involving V21, F23, and R25, which link the β1 strands of each subunit to create a six-stranded sheet (Fig. 1B). The intermolecular disulfide, readily discernable in the electron density, is augmented by specific hydrogen bond and electrostatic contacts between subunits (Fig. 1C). For example, intermolecular hydrogen bonds are formed between K57 Nζ and K52 Nζ and the backbone carbonyl oxygen from M70 in the opposing subunit.
Fig. 1.
Crystal structure of CCL20 S64C shows a CXC dimer conformation. (A) Previously reported X-ray crystal structures of CCL20 WT (PDB ID codes 2HCI and 1M8A) exhibited the dimeric arrangement common to most CXC chemokines instead of the typical CC dimer interface. (B) Hydrogen bonds linking backbone atoms of the β1 strands define the CXC-type dimer interface and form an intermolecular β-sheet centered at F23. (C) Strong electron density (pink mesh) confirms the formation of the engineered disulfide bond. The carboxyl terminus is stabilized by hydrogen bonds between the K52 and K57 sidechain of one subunit with the backbone carbonyl of M70 of the opposing subunit. (D) Two copies of the CCL20 NMR monomer structure (purple; PDB ID code 2JYO) superimposed on the CCL20 S64C structure (gray/cyan; PDB ID code 5UR7). (E) Canonical receptor binding epitopes (site 1) are structurally conserved between the monomer structure of CCL20 (purple; PDB ID code 2JYO) and dimeric structure of CCL20 S64C (cyan; PDB ID code 5UR7).
We also compared the CCL20 S64C crystal structure with the NMR structure of the CCL20 monomer. Superposition of the NMR structure (PDB ID code 2JYO) on each subunit of the CCL20 S64C dimer (Fig. 1D) shows close agreement for the β-sheet (Cα rmsd = 0.97 Å for residues 10–50). However, the orientation of the helix in the NMR structure is incompatible with the dimer configuration. The CCL20 helix must therefore pivot out of the way to permit dimerization, much like the CXCL12 conformational change described by Veldkamp et al. (32). In most chemokines, residues of the N-loop and β3 strand form a conserved receptor binding cleft (33–35), far from the intermolecular disulfide in the structure of the CCL20 S64C dimer. Side chains of R13, F19, and V47 of CCL20 are predicted to form an analogous binding pocket for the CCR6 N-terminal domain (36). These residues adopt similar orientations in the monomer and dimer structures, with the R13 amino group pointing into the pocket in close proximity to V47 (Fig. 1E). This suggests that the CCL20 S64C locked dimer preserves a functional CCR6 recognition site. Taken together, the comparisons with structures of WT CCL20 suggest that CCL20 S64C is likely to bind CCR6, but being locked into the dimeric state may alter its ability to activate the receptor.
CCL20 S64C Is a Partial Agonist for CCR6.
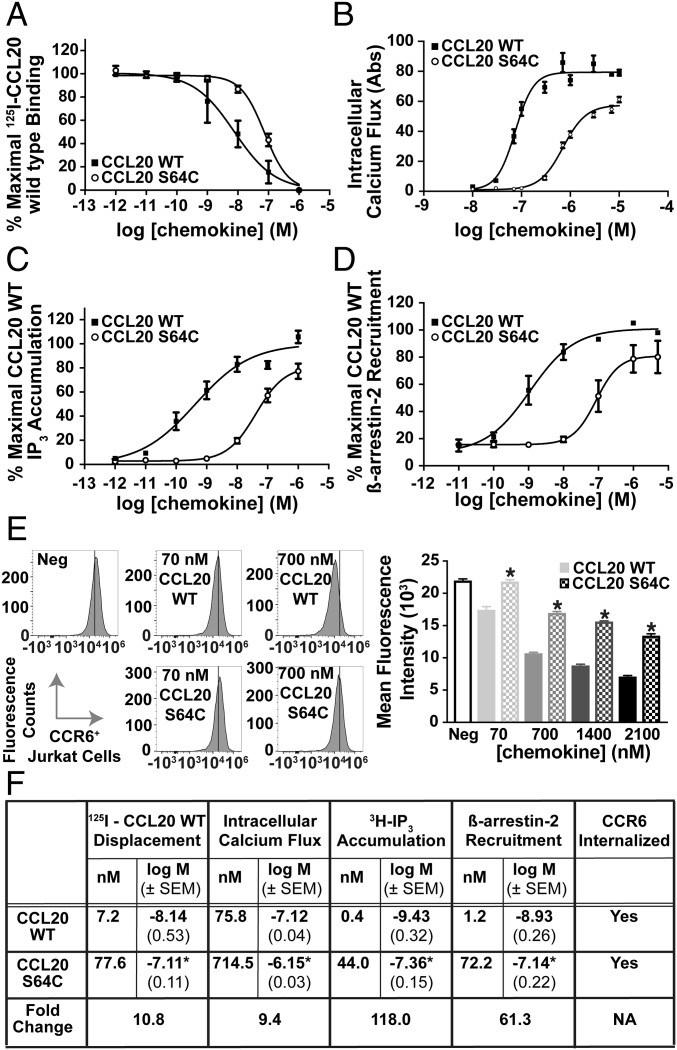
Some CXC chemokine dimers can bind and activate their receptors, but dimerization of CC chemokines typically blocks binding, rendering them nonfunctional as GPCR agonists (22, 23). To understand the biological properties of the CCL20 dimer, we compared CCL20 S64C and WT CCL20 for their ability to bind and activate CCR6. Radioligand displacement of 125I-CCL20 WT on CCR6+ COS7 cells showed a 10-fold change in affinity between CCL20 WT and CCL20 S64C with Kd values of 7.2 and 77.6 nM, respectively (Fig. 2A). Next, we evaluated the dimer’s ability to activate CCR6 relative to CCL20 WT by measuring intracellular calcium release. In CCR6+ Jurkat cells, both molecules induced CCR6-mediated calcium flux at nanomolar concentrations with CCL20 S64C being ∼10-fold less potent than the unmodified chemokine (Fig. 2B). As a way to test for Gαi subunit interactions with CCR6, inositol triphosphate (IP3) accumulation was measured in COS-7 cells transfected with the Gqi4myr chimeric G protein (37). Concentration-dependent administration of CCL20 WT and CCL20 S64C on COS-7 cells resulted in CCR6-mediated IP3 accumulation with EC50 values of 0.4 and 44.0 nM, respectively. CCL20 WT activation of CCR6 is known to recruit Gαi and is sensitive to pertussis toxin (38). In agreement with CCL20 WT, treatment of CCR6+ Jurkat cells with pertussis toxin completely blocked CCL20 S64C calcium flux (Fig. S3). In assays evaluating G-protein signaling, the CCL20 S64C retains the ability to activate CCR6, albeit at higher concentrations than CCL20 WT (Fig. 2C).
Fig. 2.
Biochemical characterization of CCL20 S64C activation of CCR6. (A) Binding of CCL20 proteins was observed by 125I-CCL20 WT displacement from CCR6+ transfected COS-7 cells. The Kd values for CCL20 WT and S64C binding to CCR6 were calculated as 7.2 nM (n = 4) and 77.6 nM, respectively (n = 3). (B) Administration of CCL20 WT and S64C on CCR6+ Jurkat cells promoted intracellular calcium release with EC50 values of 75.8 and 714.5 nM, respectively (n = 3). (C) Accumulation of 3H-IP3 was determined by radioactive measurements on transfected CCR6+ COS-7 cells in response to CCL20 WT and S64C with resulting EC50 values of 0.4 and 44.0 nM (n = 4). (D) Dose-dependent treatment of U2OS cells with CCL20 WT and S64C promoted β-arrestin-2 recruitment to CCR6 with EC50 values of 1.2 nM (n = 5) and 72.2 nM (n = 4). (E) Treatment with CCL20 S64C reduced CCR6 cell surface expression less efficiently than CCL20 WT. *P < 0.05 vs. same WT group (n = 2). (F) Table, summary of experimental EC50 values, corresponding logEC50 ± SEM, and receptor internalization results. *P < 0.05 vs. CCL20 WT group.
Upon agonist activation of chemokine receptors, G-protein receptor kinases (GRKs) phosphorylate the GPCR cytoplasmic tail and recruit multiple β-arrestin isotypes to the intracellular face of the receptor (39). We tested the dimer’s ability to recruit β-arrestin-2 using a β-galactosidase luminescence assay on transfected U2OS cells (Fig. 2D). β-Arrestin-2 recruitment to CCR6 was measured in response to CCL20 WT and CCL20 S64C and produced EC50 values of 1.2 and 72.2 nM, respectively. Similar to IP3 accumulation, both CCL20 ligands recruit β-arrestin-2 but with large differences in potency. We also evaluated CCR6 internalization in response to CCL20 WT and CCL20 S64C. Jurkat cells were incubated with each protein at concentrations ranging from 70 to 2,100 nM for 30 min, and CCR6 expression at the cell surface was quantified by flow cytometry. Whereas CCL20 WT produced a concentration-dependent change in cell surface CCR6 with a reduction of nearly 70% at the highest dose, CCL20 S64C-induced CCR6 internalization was significantly less efficient at all tested concentrations (∼40% maximum attenuation of cell surface CCR6) (Fig. 2E).
The CCL20 Locked Dimer Blocks Promigratory Function.
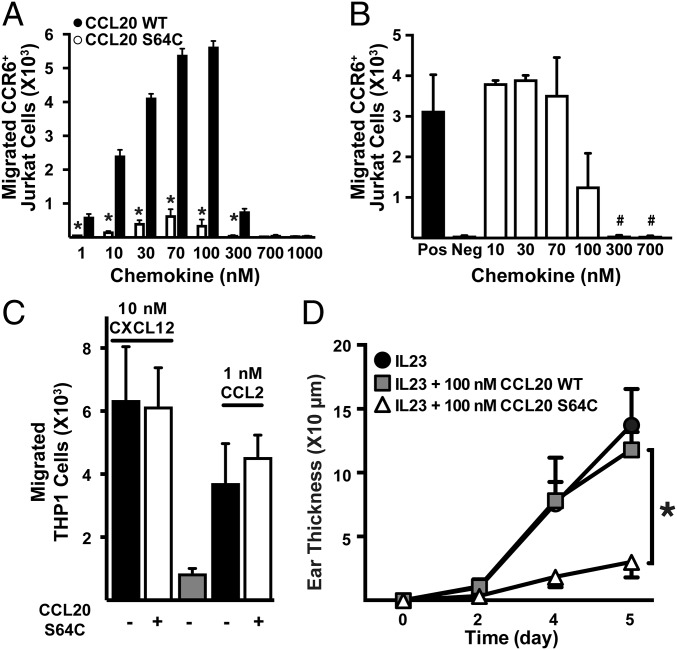
Dissociation of G proteins and recruitment of β-arrestin at the chemokine receptor intracellular face produce cellular changes that lead to actin cytoskeleton rearrangement and cell migration (40). To further measure the dimer’s activation of CCR6, we compared the abilities of CCL20 WT and CCL20 S64C to promote cell migration. In a filter-based cell migration assay, CCL20 induced CCR6+ Jurkat cell migration with a typical biphasic concentration dependence and a maximal response observed at 100 nM (EC50 ∼15 nM) (Fig. 3A). In contrast, the cell migration response to CCL20 S64C was attenuated by ∼90% with a similar concentration-dependent profile. These results suggest that the disulfide-linked CCL20 dimer retains the ability to bind CCR6 and induce Gαi and β-arrestin-2 signaling but fails to stimulate the full repertoire of CCL20-induced signaling pathways required for efficient cellular chemotaxis.
Fig. 3.
CCL20 S64C blocks CCL20 WT-dependent cell migration and is a preventative therapeutic for IL-23–induced psoriasis. (A) Migration of CCR6+ Jurkat cells after 2-h incubation was measured by flow cytometry (n = 2). *P < 0.05 vs. equal concentration CCL20 WT. (B) Inhibition of CCR6+ Jurkat cell migration by CCL20 S64C in the presence of 30 nM CCL20 WT. #P < 0.07 vs. CCL20 WT positive control. (C) CCL20 S64C inhibition of CCL2 (1 nM)- or CXCL12 (10 nM)-dependent THP-1 cell migration (n = 2). (D) Time course of ear swelling measured as the difference in ear thickness from day 0. *P < 0.05 vs. CCL20 WT group (n = 6). Three mice were injected in both ears for each experiment. Similar results were obtained in two independent experiments; plot is representative of one independent experiment.
We hypothesized that CCL20 S64C could function as an inhibitor of CCL20-mediated chemotaxis and, therefore, monitored the effect of CCL20 S64C on CCR6+ Jurkat cell migration induced by 30 nM CCL20 WT. CCL20 S64C dose-dependently inhibited chemotaxis with an IC50 ∼100 nM (Fig. 3B). Of note, CCL20 S64C did not inhibit THP-1 cell migration mediated by CXCR4 and CCR2, the cognate receptors for chemokines CXCL12 and CCL2, respectively (Fig. 3C). Taken together, these results indicated that CCL20 S64C inhibits CCL20-induced chemotaxis by binding specifically to the CCR6 receptor rather than interacting directly with CCL20 or by blocking other downstream cell migration signaling pathways.
CCL20 S64C Ameliorates IL-23–Induced Psoriasiform Inflammation.
Based on its ability to block CCL20-mediated cell migration in vitro in a CCR6-specific manner, we speculated that CCL20 S64C could attenuate the inflammation associated with psoriasiform dermatitis in vivo. IL-23 is critical for Th17 cell differentiation, activates Th17 cytokine expression, and stimulates the migration of T cells and other inflammatory cells into the skin following intradermal injections, resulting in psoriasiform epidermal hyperproliferation and dermal inflammation (7, 8). We injected IL-23 with CCL20 S64C, IL-23 with CCL20 WT, or IL-23 alone into the ear skin of mice and measured ear thickness every other day for 5 d. As expected, mice injected with IL-23 and CCL20 WT or IL-23 alone displayed reproducible ear swelling. Coinjection of CCL20 S64C with IL-23 resulted in a significant reduction in total ear thickness compared with the other groups (Fig. 3D). The lack of prevention by equimolar amounts of CCL20 WT indicates that the observed effects are due to the disulfide-linked dimeric conformation of CCL20 S64C and not to increased levels of CCL20 in general. Thus, CCL20 S64C effectively prevents IL-23–induced psoriasiform inflammation.
CCL20 S64C Reduces the Recruitment of Inflammatory γδ-Low T Cells.
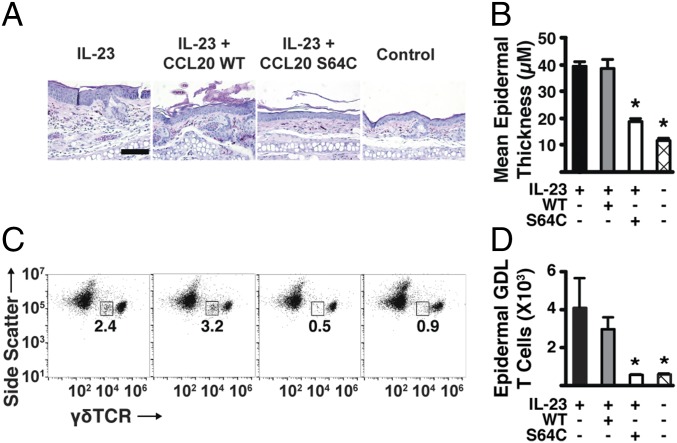
In the IL-23–induced psoriasis model, γδ-low T cells migrate to the epidermis, produce the cytokines IL-17A and IL-22, and lead to the development of psoriasis symptoms (8). Histologically, injection with IL-23 alone (or with CCL20 WT protein) resulted in marked epidermal hyperplasia and a dense mixture of mononuclear inflammatory infiltrate (Fig. 4A). Similar to the reduction in total ear thickening, administration of CCL20 S64C significantly reduced epidermal thickness at the injection site compared with the IL-23–treated mice (Fig. 4B). In contrast, mice treated with IL-23 and CCL20 S64C showed reduced epidermal hyperplasia and decreased inflammatory cell infiltration, which was comparable to the non–IL-23 control. Thus, CCL20 S64C treatment markedly reduced epidermal hyperplasia and dermal inflammation, two key signatures of psoriasiform dermatitis.
Fig. 4.
The CCL20 locked dimer blocks the recruitment of γδ-low–expressing T cells to the epidermis. (A) H&E staining after treatment for 6 d. (Scale bar: 100 µm.) Data are representative of at least three mice. Similar results were obtained in three independent experiments. (B) Mean epidermal thickness measured per mouse for each group (n = 3). Plotted values are averaged from two pooled mouse ears. Similar results were obtained in two independent experiments. (C) Representative flow cytometry results of epidermal cell suspensions from each mouse ear stained with mAbs against γδ-TCR. The numbers indicate the proportion of side-scattermed γδlow GDL T cells. Similar results were obtained in three independent experiments. (D) Total number of epidermal GDL T cells per mouse for each group (n = 3). Plotted values are averaged from two pooled mouse ears. Similar results were obtained in two independent experiments. All experimental data are expressed as mean ± SEM. *P < 0.05 vs. IL-23–alone groups for all experiments.
Next, we evaluated the specific effects of CCL20 treatments on T cell trafficking and biomarker expression in the epidermis. Epidermal cell suspensions were stained with monoclonal antibodies against the γδ-T cell receptor (TCR). In murine skin, two distinct populations of γδ-T cells are found in normal, noninflammatory conditions. Under steady-state conditions, cells expressing high levels of the γδ-TCR (dendritic epidermal T cells) are found in much higher numbers relative to low-expressing γδ-TCR cells (41). In our previous report, we showed that the mice treated with intradermal IL-23 alone showed an increase in epidermal thickness with an accompanying influx of CCR6-positive GDL cells expressing high levels of IL-17A and IL-22, whereas resident γδ-high dendritic T cells in the epidermis expressed low levels of CCR6 (8). Similarly, the mice treated herein with IL-23 showed an increase in epidermal GDL T cell populations and significant epidermal thickening compared with the non–IL-23 control (P < 0.05) (Fig. 4 C and D). While injection of 100 nM CCL20 WT resulted in a marked increase in GDL T cells, CCL20 S64C-treated mice showed no increase in epidermal GDL T cell accumulation (Fig. 4 C and D). These results confirmed our hypothesis that CCL20 S64C, but not CCL20 WT, can block the trafficking of GDL T cells to the epidermis.
To further examine relevant psoriasis biomarker expression levels, we performed RT-PCR analysis of IL-17A and IL-22, which are known to be elevated in the affected skin of human psoriasis (42). Epidermal cell suspensions from ears treated with IL-23 and CCL20 S64C showed a significant reduction in the expression of IL-22 and IL-17A mRNA compared with IL-23–only treatment or IL-23 and CCL20 WT coadministration (Fig. S4). Thus, CCL20 S64C blocked the accumulation of GDL T cells in the epidermis and reduced the expression of key Th17 effector cytokines in the epidermis of treated mice.
Discussion
CCL20 plays an important role in mucosal immune surveillance by orchestrating the trafficking of CCR6-expressing dendritic and T cells. Inappropriate or prolonged CCL20-mediated recruitment of T cells contributes to various diseases of the skin, including atopic dermatitis (43) and psoriasis, where CCL20 is the most highly up-regulated chemokine in active lesions (5) and IL-17A+, CCR6+ T cells are present in greater numbers compared with normal patient samples (44, 45). Recent studies suggest that CCL20 activation of CCR6 also drives Th17-mediated pathology in nondermatologic contexts, including spinal cord injury (46) and cervical cancer (47). Because the major clinical need for new therapeutic agents that block CCL20/CCR6 signaling remains unmet, we used the tools of protein design to create an engineered CCL20 molecule with an altered oligomeric state and functional profile. Whereas WT CCL20 promotes chemotaxis of CCR6-expressing cells, the CCL20 locked dimer inhibits chemotaxis and our in vivo studies revealed its striking effects on T cell recruitment. Remarkably, selective functional engineering of CCL20 converts it from a balanced GPCR agonist with promigratory properties to a partial agonist of CCR6 that blocks chemotactic migration and is antiinflammatory in vivo.
While engineered CC-type chemokine dimers [e.g., CCL2 (22)] cannot bind their cognate GPCRs, disulfide-locked CXCL1 (23), CXCL8 (48), and CXCL12 (25) dimers retain high affinity for their receptors with varying types of agonist activity. The CXCL12 locked dimer, which activates G-protein signaling but blocks cell migration, is a prototype for new inhibitors that use biased agonism to disrupt pathologic chemokine activity (25). Like the engineered CXC chemokine dimers studied previously, the CCL20 locked dimer is also a functional GPCR agonist with a CCR6 activation profile that differs from CCL20 WT. While both ligands trigger Ca2+ mobilization, IP3 accumulation, β-arrestin-2 recruitment, and ligand-induced receptor internalization, CCL20 S64C exhibited lower efficacy for Ca2+ mobilization (Fig. 2B) and less efficient CCR6 internalization at all concentrations tested (Fig. 2E). Despite the relatively modest differences in their CCR6 activation profiles, CCL20 S64C is minimally chemotactic and inhibits CCR6-mediated migration.
Closely related GPCR ligands with distinct signaling profiles are hypothesized to stabilize different active receptor conformations. For example, Corbisier et al. (49) found that both enantiomers of a small-molecule CCR2 agonist had equal potency and efficacy for Gαoa activation while only one enantiomer induced β-arrestin-2 recruitment. In the context of CCR6 activation, CCL20 S64C acts as a partial agonist for intracellular calcium release (Fig. 2B), and in the context of cellular migration it is a potent inhibitor. The pattern of phosphorylation by GRKs and subsequent internalization through β-arrestin and clathrin recruitment is now recognized as a means of modulating the downstream signaling response to a chemokine ligand (50, 51). The CCL20 locked dimer may stimulate a unique GRK-mediated CCR6 phosphorylation profile that recruits β-arrestin-2 but fails to stimulate efficient receptor internalization or chemotactic cell migration. While our results show that CCL20 S64C displaces CCL20 WT from CCR6 (Fig. 2A), we acknowledge that in vivo effects could also arise from indirect mechanisms such as functional cross talk observed in the chemokine (52) and GPCR families (53).
In conclusion, we find that manipulation of the CCL20 oligomeric state via protein engineering subtly shifts its pharmacological profile as a CCR6 agonist with profound consequences for cell migration and inflammation. As first observed for the engineered CXCL12 dimer (24), CCL20 self-association converts the promigration chemokine to an inhibitor of CCR6-mediated chemotaxis. The engineered CCL20 dimer blocked recruitment of CCR6+ T cells to the epidermis, thereby reducing the clinical signs of IL-23–induced psoriasiform dermatitis in mice (11). Future work will test the engineered CCL20 dimer in a therapeutic model of psoriasis to evaluate its ability to treat established disease. Consequently, we speculate that CCR6-directed intervention using a recombinant protein inhibitor is a promising approach for treatment of psoriasis. Biologic drugs derived from the CCL20 dimer described in this report could also find broad utility in a wide range of Th17-mediated inflammatory and autoimmune diseases such as rheumatoid arthritis, inflammatory bowel disease, and multiple sclerosis. Guided by the utility of engineered CCL20 and CXCL12 dimers, other T cell-specific CXC chemokines (e.g., CXCL10) may be suitable candidates for the development of novel anti-inflammatory agents.
Materials and Methods
Molecular Biology.
CCR6 cDNA was cloned into the pcDNA3.1(+) vector (Invitrogen) using sticky end ligation. The CCR6 gene was amplified by PCR without stop codons and ligated into the pCMV-ProLink1 (PK1) vector (DiscoveRx) directly upstream of the PK1 tag for the β-arrestin-2 recruitment assays. See details in SI Materials and Methods.
Protein Crystallization.
Purified CCL20 S64C was resuspended in water at a concentration of 16 mg/mL. Crystallization was conducted at 19 °C by sitting drop vapor diffusion by mixing equal volumes of the protein with well solution containing 200 mM ammonium acetate, 100 mM Na-HEPES (pH 7.5), and 25% isopropanol (vol/vol). See details in SI Materials and Methods.
Radioligand Displacement Assay.
125I-CCL20 WT was produced by oxidative iodination using ChloramineT (PerkinElmer), and labeled chemokine was purified and verified by reverse-phase HPLC chromatography, as previously described (54). See details in SI Materials and Methods.
Calcium Flux Assay.
Transfected CCR6+ Jurkat cells were washed twice in a Hanks’ buffered saline solution (HBSS), 25 mM Hepes (pH 7.4), and 0.2% (wt/vol) BSA buffer, and 2 × 105 cells per well were incubated with FLIPR Calcium 4 dye at 37 °C, 5% CO2 for 1 h in a 384-well HTS plate (Greiner Bio-One). See details in SI Materials and Methods.
β-Arrestin-2 Recruitment Assay.
CCR6-PK1 transfected U2OS-A2 cells were seeded into white, 96-well plates at 35,000 cells per well overnight. After 24 h, ligands were added to the cells, and the PathHunter β-arrestin GPCR assay was carried out according to the manufacturer’s instructions (DiscoveRx).
Chemotaxis Assay.
Chemokine dilutions were prepared as indicated in a RPMI 1640, 25 mM Hepes (pH 7.4), and 0.2% BSA buffer and added to the lower well of a Corning HTS Transwell 96-well plate (5-µm pore size). See details in SI Materials and Methods.
Cytokine Injections into Mouse Ears.
All animal experiment protocols were approved by the Medical College of Wisconsin Institutional Animal Care and Use Committee. Intradermal injections of 500 ng of recombinant mouse IL-23 (BioLegend) in 20 µL of PBS were performed into both ears of anesthetized mice every other day for 6 d as described previously (8). See details in SI Materials and Methods.
Processing of Epidermal Cells from Mouse Ears.
After recovery of mouse ears, skin sheets were separated from cartilage and incubated in PBS containing 0.5% trypsin (Affymetrix) for 40 min at 37 °C to separate epidermal sheets from dermal sheets. To obtain cell suspensions, epidermal sheets were treated in DMEM (Invitrogen) containing 0.05% DNase I (Sigma-Aldrich) as described (11).
Supplementary Material
Acknowledgments
We thank Drs. J. C. Fox, I. Roy, and M. B. Dwinell, as well as A. B. Kleist for fruitful discussion and help with manuscript preparation, and D. R. Jensen for collection and analysis of mass spectrometry data. This work was supported in part by the Uehara Foundation (to Y.I.) as well as a National Psoriasis Foundation translational grant and National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant 1R01AR063091-01A1 (to S.T.H.). This work was supported in part by National Institutes of Health Grants R01 AI058072 and R01 GM097381 (to B.F.V.).
Footnotes
Conflict of interest statement: B.F.V. and F.C.P. have ownership interests in Protein Foundry, LLC. The other authors state no conflict of interest. The technology presented is protected under provisional patent filing no. 62/170,347.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5UR7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704958114/-/DCSupplemental.
References
- 1.Parisi R, Symmons DP, Griffiths CE, Ashcroft DM. Identification and Management of Psoriasis and Associated ComorbidiTy (IMPACT) Project Team Global epidemiology of psoriasis: A systematic review of incidence and prevalence. J Invest Dermatol. 2013;133:377–385. doi: 10.1038/jid.2012.339. [DOI] [PubMed] [Google Scholar]
- 2.Harper EG, et al. Th17 cytokines stimulate CCL20 expression in keratinocytes in vitro and in vivo: Implications for psoriasis pathogenesis. J Invest Dermatol. 2009;129:2175–2183. doi: 10.1038/jid.2009.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba M, et al. Identification of CCR6, the specific receptor for a novel lymphocyte-directed CC chemokine LARC. J Biol Chem. 1997;272:14893–14898. doi: 10.1074/jbc.272.23.14893. [DOI] [PubMed] [Google Scholar]
- 4.Liao F, et al. CC-chemokine receptor 6 is expressed on diverse memory subsets of T cells and determines responsiveness to macrophage inflammatory protein 3 alpha. J Immunol. 1999;162:186–194. [PubMed] [Google Scholar]
- 5.Homey B, et al. Up-regulation of macrophage inflammatory protein-3 alpha/CCL20 and CC chemokine receptor 6 in psoriasis. J Immunol. 2000;164:6621–6632. doi: 10.4049/jimmunol.164.12.6621. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y, Li J. Expression of CC chemokine ligand 20 and CC chemokine receptor 6 mRNA in patients with psoriasis vulgaris. J Huazhong Univ Sci Technol Med Sci. 2004;24:297–299. doi: 10.1007/BF02832019. [DOI] [PubMed] [Google Scholar]
- 7.Hedrick MN, et al. CCR6 is required for IL-23-induced psoriasis-like inflammation in mice. J Clin Invest. 2009;119:2317–2329. doi: 10.1172/JCI37378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mabuchi T, Takekoshi T, Hwang ST. Epidermal CCR6+ γδ T cells are major producers of IL-22 and IL-17 in a murine model of psoriasiform dermatitis. J Immunol. 2011;187:5026–5031. doi: 10.4049/jimmunol.1101817. [DOI] [PubMed] [Google Scholar]
- 9.Xie X, Li F, Chen JW, Wang J. Risk of tuberculosis infection in anti-TNF-α biological therapy: From bench to bedside. J Microbiol Immunol Infect. 2014;47:268–274. doi: 10.1016/j.jmii.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Blauvelt A. Safety of secukinumab in the treatment of psoriasis. Expert Opin Drug Saf. 2016;15:1413–1420. doi: 10.1080/14740338.2016.1221923. [DOI] [PubMed] [Google Scholar]
- 11.Mabuchi T, et al. CCR6 is required for epidermal trafficking of γδ-T cells in an IL-23-induced model of psoriasiform dermatitis. J Invest Dermatol. 2013;133:164–171. doi: 10.1038/jid.2012.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kufareva I, et al. Stoichiometry and geometry of the CXC chemokine receptor 4 complex with CXC ligand 12: Molecular modeling and experimental validation. Proc Natl Acad Sci USA. 2014;111:E5363–E5372. doi: 10.1073/pnas.1417037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crump MP, et al. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenkilde MM, et al. Molecular mechanism of AMD3100 antagonism in the CXCR4 receptor: Transfer of binding site to the CXCR3 receptor. J Biol Chem. 2004;279:3033–3041. doi: 10.1074/jbc.M309546200. [DOI] [PubMed] [Google Scholar]
- 15.Rodríguez-Frade JM, et al. Similarities and differences in RANTES- and (AOP)-RANTES-triggered signals: Implications for chemotaxis. J Cell Biol. 1999;144:755–765. doi: 10.1083/jcb.144.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mack M, et al. Aminooxypentane-RANTES induces CCR5 internalization but inhibits recycling: A novel inhibitory mechanism of HIV infectivity. J Exp Med. 1998;187:1215–1224. doi: 10.1084/jem.187.8.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons G, et al. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 18.Proudfoot AE, et al. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci USA. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleist AB, et al. New paradigms in chemokine receptor signal transduction: Moving beyond the two-site model. Biochem Pharmacol. 2016;114:53–68. doi: 10.1016/j.bcp.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stone MJ, Hayward JA, Huang C, Huma ZE, Sanchez J. Mechanisms of regulation of the chemokine-receptor network. Int J Mol Sci. 2017;18:E342. doi: 10.3390/ijms18020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clore GM, Gronenborn AM. Three-dimensional structures of alpha and beta chemokines. FASEB J. 1995;9:57–62. doi: 10.1096/fasebj.9.1.7821760. [DOI] [PubMed] [Google Scholar]
- 22.Tan JH, et al. Design and receptor interactions of obligate dimeric mutant of chemokine monocyte chemoattractant protein-1 (MCP-1) J Biol Chem. 2012;287:14692–14702. doi: 10.1074/jbc.M111.334201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravindran A, Sawant KV, Sarmiento J, Navarro J, Rajarathnam K. Chemokine CXCL1 dimer is a potent agonist for the CXCR2 receptor. J Biol Chem. 2013;288:12244–12252. doi: 10.1074/jbc.M112.443762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veldkamp CT, et al. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci Signal. 2008;1:ra4. doi: 10.1126/scisignal.1160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drury LJ, et al. Monomeric and dimeric CXCL12 inhibit metastasis through distinct CXCR4 interactions and signaling pathways. Proc Natl Acad Sci USA. 2011;108:17655–17660. doi: 10.1073/pnas.1101133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takekoshi T, Ziarek JJ, Volkman BF, Hwang ST. A locked, dimeric CXCL12 variant effectively inhibits pulmonary metastasis of CXCR4-expressing melanoma cells due to enhanced serum stability. Mol Cancer Ther. 2012;11:2516–2525. doi: 10.1158/1535-7163.MCT-12-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy I, et al. Pancreatic cancer cell migration and metastasis is regulated by chemokine-biased agonism and bioenergetic signaling. Cancer Res. 2015;75:3529–3542. doi: 10.1158/0008-5472.CAN-14-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan DI, Hunter HN, Tack BF, Vogel HJ. Human macrophage inflammatory protein 3alpha: Protein and peptide nuclear magnetic resonance solution structures, dimerization, dynamics, and anti-infective properties. Antimicrob Agents Chemother. 2008;52:883–894. doi: 10.1128/AAC.00805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoover DM, et al. The structure of human macrophage inflammatory protein-3alpha/CCL20. Linking antimicrobial and CC chemokine receptor-6-binding activities with human beta-defensins. J Biol Chem. 2002;277:37647–37654. doi: 10.1074/jbc.M203907200. [DOI] [PubMed] [Google Scholar]
- 30.Malik ZA, Tack BF. Structure of human MIP-3alpha chemokine. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:631–634. doi: 10.1107/S1744309106006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dombkowski AA. Disulfide by design: A computational method for the rational design of disulfide bonds in proteins. Bioinformatics. 2003;19:1852–1853. doi: 10.1093/bioinformatics/btg231. [DOI] [PubMed] [Google Scholar]
- 32.Veldkamp CT, et al. Monomeric structure of the cardioprotective chemokine SDF-1/CXCL12. Protein Sci. 2009;18:1359–1369. doi: 10.1002/pro.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanpain C, et al. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J Biol Chem. 2003;278:5179–5187. doi: 10.1074/jbc.M205684200. [DOI] [PubMed] [Google Scholar]
- 34.Baly DL, et al. A His19 to Ala mutant of melanoma growth-stimulating activity is a partial antagonist of the CXCR2 receptor. J Immunol. 1998;161:4944–4949. [PubMed] [Google Scholar]
- 35.Bondue A, Jao SC, Blanpain C, Parmentier M, LiWang PJ. Characterization of the role of the N-loop of MIP-1 beta in CCR5 binding. Biochemistry. 2002;41:13548–13555. doi: 10.1021/bi026087d. [DOI] [PubMed] [Google Scholar]
- 36.Ziarek JJ, Heroux MS, Veldkamp CT, Peterson FC, Volkman BF. Sulfotyrosine recognition as marker for druggable sites in the extracellular space. Int J Mol Sci. 2011;12:3740–3756. doi: 10.3390/ijms12063740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenkilde MM, et al. Molecular pharmacological phenotyping of EBI2. An orphan seven-transmembrane receptor with constitutive activity. J Biol Chem. 2006;281:13199–13208. doi: 10.1074/jbc.M602245200. [DOI] [PubMed] [Google Scholar]
- 38.Vongsa RA, Zimmerman NP, Dwinell MB. CCR6 regulation of the actin cytoskeleton orchestrates human beta defensin-2- and CCL20-mediated restitution of colonic epithelial cells. J Biol Chem. 2009;284:10034–10045. doi: 10.1074/jbc.M805289200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aramori I, et al. Molecular mechanism of desensitization of the chemokine receptor CCR-5: Receptor signaling and internalization are dissociable from its role as an HIV-1 co-receptor. EMBO J. 1997;16:4606–4616. doi: 10.1093/emboj/16.15.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norgauer J, Metzner B, Czech W, Schraufstatter I. Reconstitution of chemokine-induced actin polymerization in undifferentiated human leukemia cells (HL-60) by heterologous expression of interleukin-8 receptors. Inflamm Res. 1996;45:127–131. doi: 10.1007/BF02265165. [DOI] [PubMed] [Google Scholar]
- 41.Girardi M. Immunosurveillance and immunoregulation by gammadelta T cells. J Invest Dermatol. 2006;126:25–31. doi: 10.1038/sj.jid.5700003. [DOI] [PubMed] [Google Scholar]
- 42.Sutton CE, et al. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–341. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama T, et al. Inducible expression of a CC chemokine liver- and activation-regulated chemokine (LARC)/macrophage inflammatory protein (MIP)-3 alpha/CCL20 by epidermal keratinocytes and its role in atopic dermatitis. Int Immunol. 2001;13:95–103. doi: 10.1093/intimm/13.1.95. [DOI] [PubMed] [Google Scholar]
- 44.Laggner U, et al. Identification of a novel proinflammatory human skin-homing Vγ9Vδ2 T cell subset with a potential role in psoriasis. J Immunol. 2011;187:2783–2793. doi: 10.4049/jimmunol.1100804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kagami S, Rizzo HL, Lee JJ, Koguchi Y, Blauvelt A. Circulating Th17, Th22, and Th1 cells are increased in psoriasis. J Invest Dermatol. 2010;130:1373–1383. doi: 10.1038/jid.2009.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu J, Yang Z, Li X, Lu H. C-C motif chemokine ligand 20 regulates neuroinflammation following spinal cord injury via Th17 cell recruitment. J Neuroinflammation. 2016;13:162. doi: 10.1186/s12974-016-0630-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walch-Rückheim B, et al. Stromal fibroblasts induce CCL20 through IL6/C/EBPβ to support the recruitment of Th17 cells during cervical cancer progression. Cancer Res. 2015;75:5248–5259. doi: 10.1158/0008-5472.CAN-15-0732. [DOI] [PubMed] [Google Scholar]
- 48.Nasser MW, et al. Differential activation and regulation of CXCR1 and CXCR2 by CXCL8 monomer and dimer. J Immunol. 2009;183:3425–3432. doi: 10.4049/jimmunol.0900305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corbisier J, Huszagh A, Galés C, Parmentier M, Springael JY. Partial agonist and biased signaling properties of the synthetic enantiomers J113863/UCB35625 at chemokine receptors CCR2 and CCR5. J Biol Chem. 2017;292:575–584. doi: 10.1074/jbc.M116.757559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tarrant TK, et al. G protein-coupled receptor kinase-3-deficient mice exhibit WHIM syndrome features and attenuated inflammatory responses. J Leukoc Biol. 2013;94:1243–1251. doi: 10.1189/jlb.0213097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh V, Raghuwanshi SK, Smith N, Rivers EJ, Richardson RM. G protein-coupled receptor kinase-6 interacts with activator of G protein signaling-3 to regulate CXCR2-mediated cellular functions. J Immunol. 2014;192:2186–2194. doi: 10.4049/jimmunol.1301875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mellado M, et al. Chemokine receptor homo- or heterodimerization activates distinct signaling pathways. EMBO J. 2001;20:2497–2507. doi: 10.1093/emboj/20.10.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jordan BA, Devi LA. G-protein-coupled receptor heterodimerization modulates receptor function. Nature. 1999;399:697–700. doi: 10.1038/21441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hatse S, et al. AMD3465, a monomacrocyclic CXCR4 antagonist and potent HIV entry inhibitor. Biochem Pharmacol. 2005;70:752–761. doi: 10.1016/j.bcp.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 55.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.