Significance
In order for HIV to proliferate, viral proteins and genomic dimers are assembled at host cell membranes and released as immature virions. Disrupting this key intermediate step in viral replication is a potential target for treatment. However, a detailed molecular view of this process remains lacking. Here, we elucidate a network of constitutive interactions that regulate viral assembly dynamics through a combined computational and experimental approach. Specifically, our analysis reveals the active roles of nucleic acid and the membrane as scaffolds that promote the multimerization of Gag polyprotein, which proceeds through multistep and self-correcting nucleation. Our findings also illustrate the functional importance of the N-terminal, C-terminal, and spacer peptide 1 protein domains.
Keywords: self-assembly, coarse-grained molecular dynamics, HIV packaging and budding, Gag, CA–SP1 junction
Abstract
The packaging and budding of Gag polyprotein and viral RNA is a critical step in the HIV-1 life cycle. High-resolution structures of the Gag polyprotein have revealed that the capsid (CA) and spacer peptide 1 (SP1) domains contain important interfaces for Gag self-assembly. However, the molecular details of the multimerization process, especially in the presence of RNA and the cell membrane, have remained unclear. In this work, we investigate the mechanisms that work in concert between the polyproteins, RNA, and membrane to promote immature lattice growth. We develop a coarse-grained (CG) computational model that is derived from subnanometer resolution structural data. Our simulations recapitulate contiguous and hexameric lattice assembly driven only by weak anisotropic attractions at the helical CA–SP1 junction. Importantly, analysis from CG and single-particle tracking photoactivated localization (spt-PALM) trajectories indicates that viral RNA and the membrane are critical constituents that actively promote Gag multimerization through scaffolding, while overexpression of short competitor RNA can suppress assembly. We also find that the CA amino-terminal domain imparts intrinsic curvature to the Gag lattice. As a consequence, immature lattice growth appears to be coupled to the dynamics of spontaneous membrane deformation. Our findings elucidate a simple network of interactions that regulate the early stages of HIV-1 assembly and budding.
HIV type 1 (HIV-1) spreads infection through a robust replication cycle. A critical step during this life cycle is the assembly and packaging of viral proteins and RNA at the plasma membrane interface, followed by the budding and release of immature virions (1–4). Understanding the molecular mechanisms that regulate this process may highlight opportunities for antiretroviral treatment strategies.
The main structural constituent throughout HIV-1 replication is the group-specific antigen (Gag) polyprotein (5, 6), which comprises around 50% of the viral particle mass (7). Gag contains several functional domains: the matrix (MA) domain for membrane association, the capsid (CA) domain for protein–protein association, the nucleocapsid (NC) domain for RNA association, the p6 domain for endosomal sorting complex required for transport (ESCRT) machinery recruitment, and the spacer peptide 1 (SP1) and 2 (SP2) domains. Cryo-electron tomography (cryo-ET) experiments have demonstrated that Gag polyproteins form hexameric lattices that are highly pleomorphic within immature virions and arrested budding sites (7–11). While largely contiguous, the immature polyprotein shell of HIV-1 remains incomplete—a notable difference from icosahedral viruses—such that only 40–70% coverage is observed in immature virions (8, 9). The large gap-like defects throughout the immature lattice are believed to accommodate the strain from lattice curvature (10), in direct contrast to the incorporation of pentameric units that permit curvature in the closed mature capsid core (12–17). We further hypothesize that the structural variability of the immature lattice may be functionally significant for viral maturation.
Resolving the structure of multimerized Gag constructs within immature virions and virus-like particles (VLPs) has provided many insights into the relationship between polyprotein structure and supramolecular assembly. Most notably, recent advances in cryo-electron microscopy (cryo-EM) and cryo-ET have revealed an unprecedented subnanometer resolution of Gag–Gag interfaces (18–21). From these studies, important interactions have been inferred from close protein–protein contacts, such as at the CACTD helix 9 interface for dimerization and the helical CA–SP1 junction interface for hexameric bundling. Biochemical experiments have corroborated these findings; for example, the importance of the SP1 domain and nearby motifs, including the major homology region and type II β-turn, was demonstrated by the production of aberrant virions following mutations to associated residues (21–23). In fact, the helical transition of the CA–SP1 junction has been proposed as a conformational molecular switch that regulates immature lattice assembly (21–24).
For efficient virion production, concerted interactions between Gag and both RNA and lipids in the cell membrane appear to be necessary. In vitro assembly of VLPs with immature-like spherical geometries requires, at a minimum, Gag binding to oligonucleotides (25, 26) or through leucine zipper motifs (27) while binding to inositol derivatives appears to adjust particle sizes (28). Recent studies have suggested that Gag assembly at the cell membrane can promote selective RNA dimerization and packaging (29, 30); competitor tRNA is also observed to positively regulate viral RNA selectivity (30). In contrast, overexpression of nonsilencing micro-RNA is suggested to negatively regulate viral production by impeding Gag assembly (31). The composition of RNA within virions has also been connected to particle size dispersity (32). Furthermore, lipid domains in the plasma membrane may mediate Gag targeting and assembly (33–35), while membrane curvature can initiate nucleation of ESCRT machinery for scission (36). It is therefore clear that a network of interactive processes between Gag, RNA, and the plasma membrane may coordinate viral replication. Nonetheless, the underlying mechanisms that dictate the interfacial assembly dynamics remain unclear.
Molecular-dynamics (MD) simulations may afford insight into the dynamics of these cellular processes. As opposed to very computationally demanding all-atom MD simulations, coarse-grained (CG) MD models are particularly attractive as a means to access biological phenomena that typically occur on very large length and timescales. Notably, understanding of the self-assembly process for icosahedral capsids has emerged from CG MD simulations of simplified capsomer subunits, in which idealized shapes (e.g., trapezoidal, triangular, and pentagonal) can be assumed due to the high symmetry of the closed polyhedral shell (37–44). These studies have revealed rich phenomenology that relate capsomer [and polyelectrolyte (42, 43)] interaction strengths, concentrations, and temperature to the thermodynamics and kinetics of capsid assembly. The competition between thermodynamic driving forces and multiple kinetic pathways has consequently been shown to result in capsid polymorphism (38–42). In principle, such insights may be broadly applicable to other viral systems. However, only a few CG MD studies have been reported on HIV-1 as the viral constituents tend to be structurally and chemically complex, for example, as demonstrated by the aforementioned network of interactions, while also requiring thousands of copies of Gag for complete virions (7, 9). For example, CG simulations have recently predicted fundamental mechanisms in mature CA assembly (15, 17, 45, 46) and MA domain anchoring to the lipid membrane (47), while the importance of CACTD interactions to stabilize hexameric order in the immature virion has also been demonstrated (48).
Emerging fluorescence techniques, such as total internal reflection fluorescence (TIRF) and single-particle tracking photoactivated localization microscopy (spt-PALM), also enable the probing of protein mobility during live-cell imaging with high specificity, especially in crowded and heterogeneous environments such as viral buds (49, 50). Recently, spt-PALM analysis revealed a notable suppression in mobility for Gag clustered within viral puncta compared with membrane-bound or cytosolic Gag (31, 49). Hence, cooperative understanding from computer simulations and fluorescence experiments may help elucidate assembly dynamics at the cell membrane interface.
In this work, we present a CG computational study with our modeling developed with the help of recent experimental structures. We also present spt-PALM experimental data of Gag self-assembly during the early stages of immature lattice growth. Our intention here is to shed critical light on the network of constituents and interactions required for viable self-assembly and budding. First, we use CG MD simulations to investigate the assembly of ΔCANTD–CACTD–SP1 Gag and find that nucleation proceeds through six-helical bundling at the CACTD–SP1 junction contingent on scaffolding from RNA and the membrane. In particular, we focus on the role of RNA through analysis of Gag diffusion and clustering based on spt-PALM and CG MD trajectories. Interestingly, these lattices, while contiguous and hexameric, are predicted to be flat and, hence, unlikely to instigate budding. Next, we introduce the CANTD domain and find that the Gag lattice exhibits an intrinsic curvature. Simulations of these Gag constructs reveal that spontaneous curvature of the membrane promotes efficient nucleation, while gradual membrane deformation is essential for extended CA–SP1 lattice growth. Taken together, these findings provide insight into molecular-scale mechanisms that regulate the packaging and budding of immature HIV-1 virions.
Results
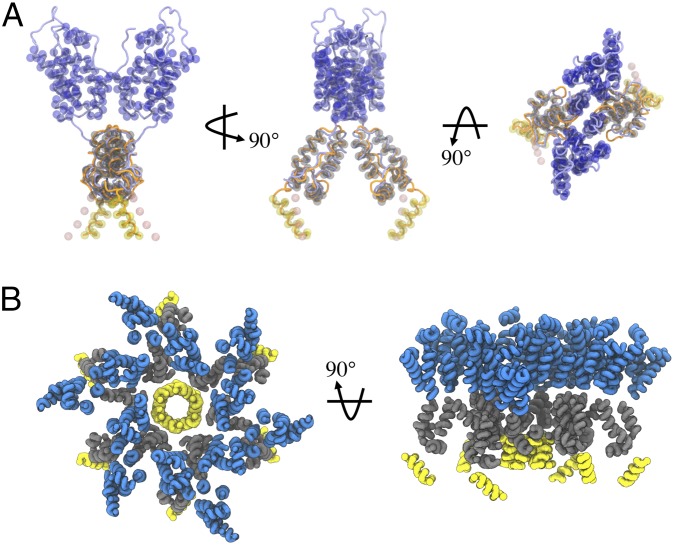
As depicted in Fig. 1A, we have developed a CG model of the CA–SP1 dimer that is directly derived from experimental structural data of the immature CANTD–CACTD [Protein Data Bank (PDB) ID code 4USN (19)] and CACTD–SP1 [PDB ID code 5I4T (21)] domains. The CG model is designed to recapitulate the internal structure of the polyprotein, relevant protein–protein interfaces, and quaternary structures (Fig. 1B) through a protocol adapted from previous work on the mature CA (17). Importantly, the collective α-helices within each of the three protein domains are maintained as rigid bodies (i.e., six total per dimer) with excluded volume interactions, while interdomain flexibility is maintained with a soft elastic network model. The dimerization is preserved through weak harmonic bonds at the helix 9–helix 9 (H9–H9) interface in the CACTD domain. Finally, the only attractive Gag–Gag interaction occurs between SP1 domains through virtual particles that serve as binding sites (Fig. 1A). In addition, we model RNA and the cell membrane as a linear chain and hexagonal mesh of CG particles, respectively. Attractive interactions are included between the topmost bead of the CANTD domain and the membrane and the bottommost bead of the SP1 domain and RNA to mimic MA–membrane and NC–RNA interactions, respectively. Full details of the model are described in Methods.
Fig. 1.
CG representation of CA–SP1 polyprotein. (A) Schematic of all CG sites of the CA–SP1 dimer shown as beads and reference experimental structures [refs. 19 (blue) and 21 (orange)] shown as tubes. The CANTD, CACTD, and SP1 particles are depicted as transparent blue, gray, and yellow spheres, respectively. The pink spheres depict virtual particles, which act as tethering sites for adjacent SP1 helices. (B) Schematic of a hexameric bundle that has assembled from six Gag dimers during our simulations. Each helix is shown as a tube with blue, gray, and yellow denoting CANTD, CACTD, and SP1 domains, respectively; this convention is used for all subsequent molecular snapshots.
Assembly Through SP1 Helical Bundling Is Activated by RNA and Membrane Scaffolds.
We first investigate the factors that enable self-assembly of CACTD–SP1 into an immature lattice at the plasma membrane interface. Recent high-resolution structural characterization has revealed that a 14-residue sequence in the CA–SP1 junction adopts an α-helical conformation that may mediate Gag–Gag interactions during assembly (20, 21, 24). Indeed, mutations in this region tend to abrogate viral particle formation (21–23). Therefore, our intention is to ascertain the assembly competence of CACTD–SP1 as facilitated by interactions at the CA–SP1 junction and the presence of both RNA and the cell membrane.
We prepared simulations with 144 CA–SP1 dimers initially arranged in an evenly spaced 12 × 12 grid (8.5-nm separation) along the xy plane; final xy lengths fluctuated around 102 nm such that the total surface coverage of Gag monomers was around 46 nmol/m2. The membrane and a 3,500-nt RNA were, respectively, placed 10 nm above and 20 nm below the layer of Gag dimers; here, the length of the RNA chain is deliberately long enough to provide an excess of binding sites. We also maintained membrane–CANTD interactions, which were included to implicitly represent Gag binding to the lipid bilayer, for example, through exposed myristyl and the MA domain; the excluded volume interactions of the CANTD domain were removed to emulate its deletion. Initially, a repulsive barrier was placed between the RNA and CA–SP1 dimers to independently evolve disordered configurations of the RNA and membrane-bound Gag over 5 × 105 CG MD time steps (τ) at 310 K. The barrier was subsequently removed and simulations were performed for 7.5 × 107 τ with trajectories saved every 2.5 × 104 τ for analysis. We note that the minimum SP1–SP1 binding strength (ESP1 = 2.35 kcal/mol) required to initiate Gag association was used for these simulations.
As seen in Fig. 2A, membrane-bound CACTD–SP1 dimers readily self-assemble into a contiguous hexagonal lattice (e.g., the orientation distribution shown in the Bottom Inset of Fig. 2A exhibits sixfold symmetry) once tethered to RNA. However, in the absence of binding to either membrane or RNA (e.g., Top Inset of Fig. 2A for the latter case), Gag multimerization was not observed throughout the entirety of our simulation trajectory (Fig. S1). This suggests that the presence of both RNA and membrane scaffolds may be necessary for efficient immature lattice assembly. As an additional test, we subsequently removed both the RNA and membrane and found that the extended lattice persisted indefinitely. The stability of the lattice suggests that the CA–SP1 junction interactions are sufficient for multimerization while RNA and cell membranes serve as catalysts for immature lattice assembly. One may interpret the roles of membrane and RNA as scaffolds that restrict the diffusive degrees of freedom for Gag (as discussed below), encourage favorable Gag orientations for binding (as discussed in a later section), and facilitate Gag oligomerization through increased colocalization. By simply increasing the surface coverage of Gag monomers to around 100 nmol/m2 (which is comparable to lower-end surface coverages within immature virions), Gag self-assembly is observed even in the absence of RNA (Fig. S1). In this case, we find that the growth of the primary cluster proceeds at a comparable rate to that of the RNA-present case, which further suggests that these scaffolds promote contiguous assembly through localized aggregation of Gag. Note that in vitro self-assembly of Gag in solution is possible, although additional agents such as nucleic acid or inositol derivatives are necessary to efficiently grow VLPs with spherical morphologies (25, 26, 28).
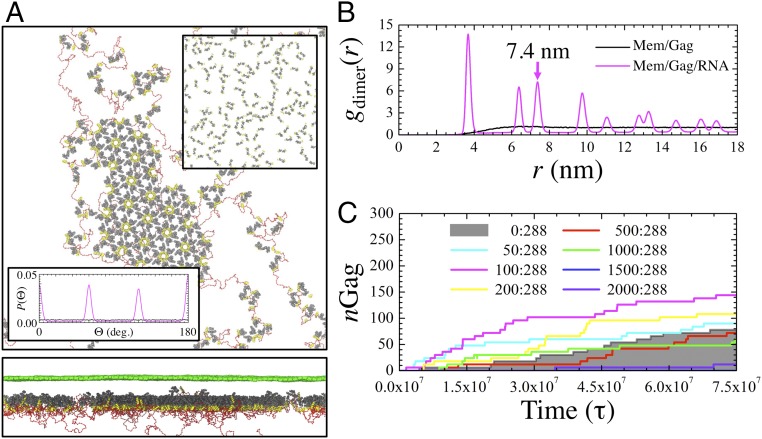
Fig. 2.
Nucleic acid modulates self-assembly of membrane-bound CACTD–SP1. (A) Top- and side-view depictions of the flat immature CACTD–SP1 lattice (gray and yellow tubes) at the membrane interface (green mesh) with bound RNA (red chain). The Top Inset shows the final configuration of CACTD–SP1 in the absence of RNA (and removal of the membrane yields comparably disperse Gag). The Bottom Inset shows the probability distribution of CACTD orientation relative to dimers within a radial distance of 8.5 nm while in the presence of RNA (purple; exhibiting sixfold symmetry) and without RNA (black). (B) Two-dimensional pair distribution function between the center of mass of Gag dimers (gdimer) as a function of radial distance (r). The arrow indicates the calculated lattice spacing of the hexagonal lattice. (C) Evolution in the number of Gag monomers (nGag) within completed hexamers as a function of MD time steps (τ). The legend lists the ratio of 20-nt miRNA to Gag monomers (i.e., [miRNA]:[Gag]) within the simulation domain. Trajectories within the shaded region exhibit hindered assembly relative to the miRNA-absent case.
Remarkably, the assembled lattice derived from our simple CG model is predicted to preserve a high degree of crystallinity. The lattice tends to be completely planar, which is in good agreement with the observed flat assemblies from purified CACTD–SP1 proteins (21). To characterize the crystallinity, we computed a 2D (e.g., in-plane) pair distribution function [gdimer(r)] for the dimer center of mass, which is shown in Fig. 2B. The observation of distinct peaks is indicative of crystallinity; by contrast, the gdimer(r) in the absence of RNA gradually increases toward unity, which indicates the absence of an ordered structure. Given that the repeating unit in the lattice is composed of six subunits of CA–SP1 dimers, the lattice constant can be approximated from the position of the third gdimer(r) peak (refer to Fig. S2). Our calculated lattice spacing of 7.4 nm, which is in excellent agreement with previous cryo-EM studies of tubular Gag constructs (∼7.25 nm) (18), and reproduction of hexagonal symmetry further support the validity of our CG model.
Visualization of the simulation trajectories (Movie S1) reveals that extended Gag oligomerization proceeds through a multistage process. Interestingly, Gag polyproteins continuously undergo cyclical association and dissociation behavior until a hexamer is formed (Fig. S1). In turn, these hexamers remain associated indefinitely. Hence, the extended lattice grows from both the nucleation of additional hexamers around a stable hexamer and the assimilation of hexamer-containing oligomers. Complementary evidence is provided by the time series profile (i.e., gray line) of the number of Gag (nGag) within hexamers shown in Fig. 2C; here, nGag evolves in a stepwise and monotonically increasing fashion. Similarly, fluorescence studies have suggested that Gag recruitment at membrane puncta is irreversible once complete (51). The preferential stability of hexamers points to its importance as a building block of the immature lattice, which we will discuss further in a later section.
While viral RNA seems to orchestrate Gag multimerization, the influence of nonviral RNA on the self-assembly process remains an open question. Nonsilencing micro-RNAs (miRNAs) are particularly interesting as a previous study (31) demonstrated their negative regulatory effect on viral infectivity. We proceeded to include short (20-nt) RNA into the CG model to represent typical nonsilencing miRNA lengths [20–25 nt (52)] and repeated the same simulation procedure as above. Here, we assume no differences between the Gag binding affinities of the viral RNA and miRNA. Hence, the length and concentration of the two RNA species are the only distinctions; the miRNA remains sufficiently long to bind to multiple diffuse Gag (around 1–3) and across Gag hexamers. The time series profiles of nGag for varying concentrations of miRNA are shown in Fig. 2C. Interestingly, the profiles for small concentrations of miRNA (less than a ratio of 200:288:1 miRNA:Gag:RNA) remain consistently above the shaded region, which indicates that the rate of Gag oligomerization is enhanced compared with the miRNA-absent case. These miRNA appear to aid in the initial nucleation of hexamers and other small oligomers. Throughout assembly, these miRNA-tethered oligomers are condensed into the largest cluster that is organized primarily by the viral RNA (Fig. S3), which then anneal as dissociated Gag dimers remain available. However, beyond a critical concentration of miRNA (more than a ratio of 1,500:288:1), lattice assembly appears virtually stagnant. Our trajectories show that miRNA-bound Gag impede viral RNA binding through competitive inhibition (Fig. S3), thereby preventing the tethering activity of viral RNA.
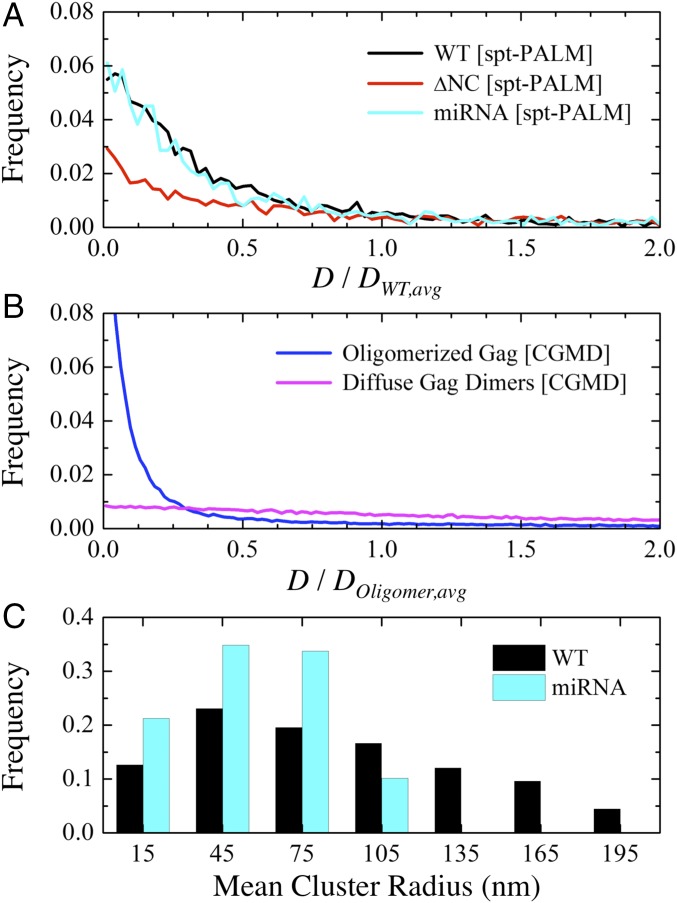
Our CG simulation results are supported by evidence from spt-PALM. We are specifically interested in changes to the diffusive behavior of Gag under different RNA environments since these can manifest due to changes in the multimerized Gag cluster size. We prepared wild-type (WT) and miRNA-overexpressing (has-miR-146a with 22 nt) HEK293 cells transfected with a full-length HIV-1 proviral clone and its derivative tagged with mEOS2, which were expressed in a 3:1 ratio to ensure viral particle formation (Methods). To remove RNA influence on Gag multimerization (53–55), WT cell lines with NC-deleted Gag (ΔNC) mutants were similarly prepared. Fig. 3A displays the distribution of Gag diffusivities (D) from the WT, miRNA, and ΔNC lines with calculated mean values of 0.092, 0.144, and 0.438 μm2/s, respectively. Similarly, a distribution of D for multimerized Gag and diffuse Gag dimers as computed from our CG MD simulations are shown in Fig. 3B with calculated mean values of 570 and 2,950 μm2/s, respectively. We normalize these two distributions to establish a basis for comparison to account for the well-known separation between CG and real-time dynamics (56, 57); although the CG dynamics are several orders of magnitude faster than reality, the relative ratio of the mean D for ΔNC to WT lines (∼4.8) is comparable to that of CG predictions for oligomerized to diffuse Gag (∼5.2). It is also notable that the distributions of D for the WT and miRNA Gag (Fig. 3A) and that of oligomerized Gag (Fig. 3B) exhibit a qualitatively similar preference for slow D (<25% of the mean) while the distributions of both ΔNC and diffuse Gag show comparably broad (i.e., heavy-tail) behavior. These shared features indirectly suggest that Gag can oligomerize in both miRNA-rich and miRNA-absent environments, while ΔNC-Gag expectedly remain largely dissociated.
Fig. 3.
Dynamics and clustering behavior of Gag. (A) Histogram of Gag diffusivities (D) from spt-PALM trajectories in WT, ΔNC, and miRNA lines (from 3,732, 3,619, and 2,373 tracks, respectively). These results are normalized by the mean diffusivity of WT Gag (DWT,avg = 0.092 μm2/s). (B) Histogram of D from CG MD trajectories in simulations with oligomerized Gag and diffuse (i.e., noninteracting) Gag. These results are normalized by the mean diffusivity of oligomerized Gag (DOligomer,avg = 570 μm2/s). (C) Histogram of mean cluster radii from spt-PALM trajectories in WT and miRNA lines. Clustered Gag (Methods) account for 54.2% and 50.9% of tracks, respectively. Bin sizes of 0.0025 μm2/s, 10 μm2/s, and 30 nm were used in A, B, and C, respectively.
We next used PALM cluster analysis (Methods) to quantify the distribution of Gag cluster sizes in the WT and miRNA lines. The distributions shown in Fig. 3C confirm that Gag can form tightly clustered regions in both WT and miRNA lines. However, it is apparent that Gag in WT lines tend to form much larger clusters (approaching 150 nm) compared with miRNA lines. Therefore, our combined simulation and experimental results suggest that, while viral RNA instigates Gag multimerization, an excess of miRNA can limit the extent of multimerization and thereby reduce viral particle production (31).
Weak Gag–Gag Interaction Strength Regulates Against Kinetic Trapping.
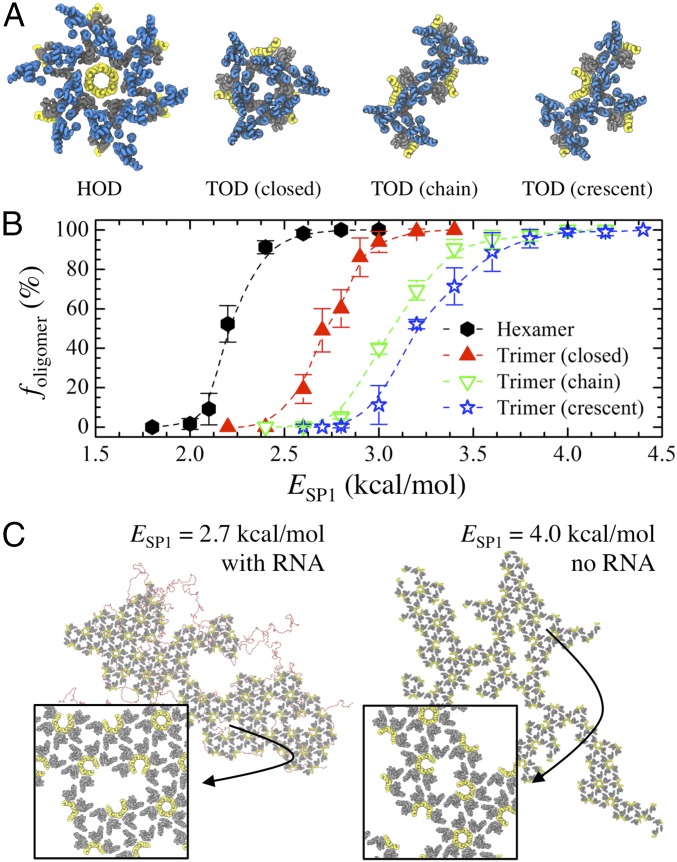
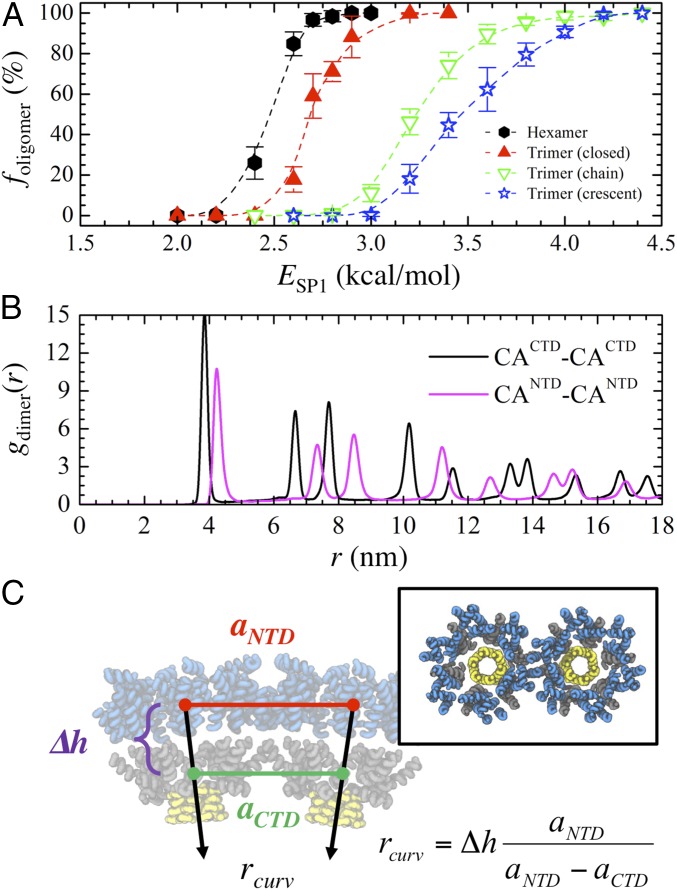
To this point, we have established that CACTD–SP1 is assembly competent and nucleates through six-helical bundling at the CA–SP1 junction interface. Importantly, RNA and the plasma membrane also appear to catalyze assembly, which may be a consequence of the weak protein–protein interactions. We next sought to determine whether this inherent interaction strength is important for successful assembly by modulating ESP1 (i.e., the junction binding affinity). First, it is informative to analyze the stability of different Gag oligomers, each observed during general assembly, as ESP1 is adjusted. To do so, we approximate the probability (foligomer) that each of four representative oligomers—a hexamer of dimers (HOD), a closed trimer of dimers (TOD), a chain of TODs, and a crescent fan of TODs (depicted in Fig. 4A)—resists dissociation due to thermal fluctuations at 310 K within a constant period of time (e.g., 1 × 106 τ); larger foligomer values imply greater kinetic stability and longer lifetimes. For each type of oligomer, we initialized systems of evenly spaced 5 × 5 × 5 CA–SP1 oligomers separated by 25 nm in all directions within a 125 × 125 × 125-nm3 simulation domain. Simulations were performed in the canonical ensemble (constant NVT) over production runs of 1 × 106 τ with trajectories saved every 2.5 × 104 τ for analysis. The reported foligomer is averaged over four independent trajectories for each ESP1 that is varied between 1.5 and 4.5 kcal/mol.
Fig. 4.
Weak interactions ensure preferential oligomerization into hexamers. (A) Schematic of four representative CA–SP1 oligomers [hexamer of dimers (HOD) and trimer of dimers (TOD)] observed during CG MD simulations; blue, gray, and yellow tubes represent helices in the CANTD, CACTD, and SP1 domains, respectively. (B) The probability (foligomer) that the listed CACTD–SP1 oligomer remains associated after 1 × 106 MD time steps as the SP1–SP1 interaction energy (ESP1) is varied. (C) Snapshots of final CACTD–SP1 lattices (gray and yellow tubes) assembled at the membrane interface when (Left) ESP1 = 2.7 kcal/mol and (Right) ESP1 = 4.0 kcal/mol. In the former (latter) case, RNA (red beads) is still (not) required for lattice assembly to occur. The Insets show a zoomed view of the indicated regions, which highlight kinetically frustrated configurations.
Fig. 4B shows the foligomer profiles for each oligomer type as a function of ESP1. In every case, a sigmoidal profile was obtained in which below (above) a certain ESP1, all oligomers spontaneously dissociated (remained associated) while both associated and dissociated states existed in the intermediate ESP1 regime (i.e., near the inflection point); we should note that the previously used ESP1 (=2.35 kcal/mol) is within the intermediate regime for HODs. We find that HODs are considerably more kinetically stable than closed TODs, which in turn are more stable than the open forms of TODs (i.e., chain and crescent forms); note that the tethering of small RNA (20 nt) to each oligomer yielded comparable foligomer profiles. Additional discussion on the factors contributing to oligomer stability can be found in Supporting Information. The preferential stability (i.e., lifetime) of HODs over other oligomers is notable as it further supports the idea of the aforementioned multistage nucleation process in which individual HODs must first assemble through helical bundling at the CA–SP1 junction before the extended lattice is grown; the growth proceeds in a cascading fashion, either from additional HODs nucleating on the edges of the initial HOD or from assimilation of independently formed HODs (Movie S1).
Next, we investigate the impact of increasing ESP1 to 2.7 and 4.0 kcal/mol, which should increase the stability of closed and open TODs (Fig. 4B). As depicted in Fig. 4C, the resultant lattice growth exhibits notable anisotropy. This dendritic growth can be attributed to the increased stability of oligomers at the edges of the lattice, which in turn present additional nucleating sites for CA–SP1 consolidation. Furthermore, the degree of anisotropy appears to scale directly with increasing oligomer stability (e.g., the ESP1 = 4.0 kcal/mol case), while concurrently mitigating the necessity of RNA to promote CA–SP1 association. Vacancy-like defects (i.e., incomplete hexamers) can also form throughout the lattice (Fig. 4C, Insets). These defects likely arise due to the extended lifetime of nonhexameric oligomers that are subsequently incorporated as kinetically frustrated configurations. Therefore, unlike our previous simulations (i.e., ESP1 = 2.35 kcal/mol), the absence of a self-regulating mechanism to heal lattice defects through cyclic CA–SP1 association and dissociation can result in locally defective immature lattices.
CANTD Domain Introduces Natural Curvature Through Volumetric Occlusion.
In this section, we describe the impact of the CANTD domain on the structure of the immature CA–SP1 lattice. First, we reexamine the kinetic stability of CA–SP1 oligomers once the explicit volume occupied by the CANTD domain is introduced. The foligomer profiles depicted in Fig. 5A exhibit sigmoidal profiles similar to that of the CACTD–SP1 cases (Fig. 4B). Importantly, the CANTD–CACTD–SP1 profiles are shifted to the right (e.g., the inflection point of each profile is higher by as much as 0.3 kcal/mol) in comparison with the previous CACTD–SP1 cases. This indicates that the steric occlusion by the globular CANTD region tends to destabilize each oligomer. The volume occupied by the CANTD domain has an important influence on the immature lattice structure. We assembled a hexameric lattice of CANTD–CACTD–SP1 (described in a later section) and computed gdimer(r) profiles between the center of masses of the CACTD domains and the CANTD domains for comparison. From Fig. 5B, the sharp peaks associated with the CACTD and CANTD layers are indicative of crystallinity. However, one notable difference is their respective lattice spacings, that is, the position of their third peaks; the CACTD sublattice (∼7.7 nm) is more tightly packed than the CANTD sublattice (∼8.5 nm). Given that the lattice spacing of the CACTD domain is reduced to around 7.4 nm when the CANTD domain is deleted, these results indicate that the space filled by the CANTD domain introduces lateral strain to the assembled complex. As the two domains are connected, this strain has the important consequence of introducing an inherent lattice curvature. Following the geometric relationship depicted in Fig. 5C, we approximate the natural radius of curvature (rcurv) to be 39.6 ± 1.3 nm. For comparison, experimental VLPs constructed with ΔMA–Gag can have radii as small as 40–45 nm (58, 59), while WT virions, which are composed of full-length Gag proteins, typically have larger radii that vary between 50 and 100 nm (7, 9, 11).
Fig. 5.
Strain from the CANTD domain imparts intrinsic curvature. (A) The probability (foligomer) that the listed CANTD–CACTD–SP1 oligomers remain associated after 1 × 106 MD time steps as the SP1–SP1 interaction energy (ESP1) is varied. (B) Two-dimensional pair distribution function between the center of masses of the listed Gag domain (gdimer) as a function of radial distance (r). (C) Schematic of two adjacent CANTD–CACTD–SP1 hexameric bundles depicted as blue, gray, and yellow tubes, respectively. The intrinsic radius of curvature (rcurv) is estimated from the lattice spacing between the center of masses of the CANTD (aNTD) and CACTD (aCTD) domains and from the separation distance (∆h) between domains. Our calculations estimate rcurv to be 39.6 ± 1.3 nm.
Spontaneous Membrane Curvature Triggers Extended Assembly.
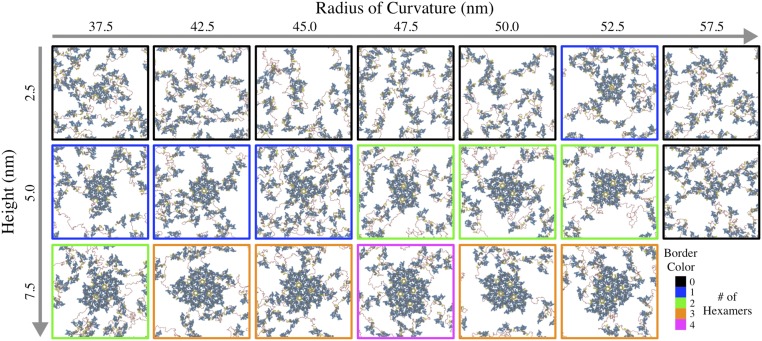
Surprisingly, we found that CANTD–CACTD–SP1 dimers remained largely dissociated when following our previous simulation procedure (with ESP1 = 2.6 kcal/mol). This remained true even when ESP1 was enhanced to 3.5 kcal/mol in an attempt to promote assembly (Fig. S4); although small oligomers readily formed, extended assembly was not observed. We attribute the impeded assembly to the competition between the natural curvature of the CA–SP1 lattice and the resistance to curvature from the membrane. In other words, our simulations suggest that the weak binding affinity between Gag–Gag and membrane–Gag may be insufficient to overcome the free-energy penalty for membrane bending. As a result, the misalignment in the binding interface of adjacent CA–SP1 helical junction may be conceived as an additional entropic barrier. Nonetheless, we found that inducing localized curvature (i.e., small punctas) on the membrane, for example, from lateral phase segregation of lipids or membrane rafts (60, 61), allowed extended lattice formation to proceed. To further investigate the influence of spontaneous membrane curvature, we instigated local puncta of variable size with a weakly repulsive spherical potential (Methods) that was slowly pushed (over 5 × 106 τ) into the membrane with 64 bound dimers and without RNA (with lateral dimensions of 70 × 70 nm2); we limited system sizes to study a wide range of puncta heights (with respect to the base of the membrane) and radii of curvature. Then, a 2,500-nt chain of RNA was introduced and the systems were allowed to equilibrate for 1.5 × 108 τ at 310 K; it is notable that simulations required much longer trajectories than the CACTD–SP1 cases as the CANTD domain introduces additional barriers associated with reorganization that slow HOD formation.
Fig. 6 summarizes the results of our simulations for CANTD–CACTD–SP1 assembly as a function of puncta height and radius of curvature. Here, Gag self-assembly proceeds at the center of the domain where the membrane puncta is located. Interestingly, the efficiency of Gag association, as characterized by the total number of completed hexamers at the end of our simulations, appears to be sensitive to both the height and curvature of the puncta. It is intuitive that the final number of hexamers increases with height as the available surface area of the deformed region also increases. However, unexpectedly, an optimal radius of curvature also appears at each discrete height, decreasing from around 52.5 to 47.5 nm as the height increases from 2.5 to 7.5 nm. In instances both above and below this critical radius, Gag multimerization appears to be hindered. These results suggest that the alignment between local membrane and intrinsic Gag curvature becomes increasingly important as the lattice grows to promote Gag binding onto the cluster periphery. Time-series profiles of hexamer formation (Fig. S5) suggest that the rate of lattice assembly follows a qualitatively similar trend. We therefore speculate that the extent of lattice growth is coupled to the dynamics of membrane deformation (i.e., bud formation).
Fig. 6.
Matrix of final top-view configurations of CANTD–CACTD–SP1 dimers, depicted as blue, gray, and yellow tubes, with RNA (red spheres) at the membrane interface after 1.5 × 108 CG MD time steps; each snapshot represents an independent simulation. The membrane was spontaneously deformed with a weak spherical indenter (applied centrally) at the listed height and radius of curvature. The border color indicates the number of completed hexamers.
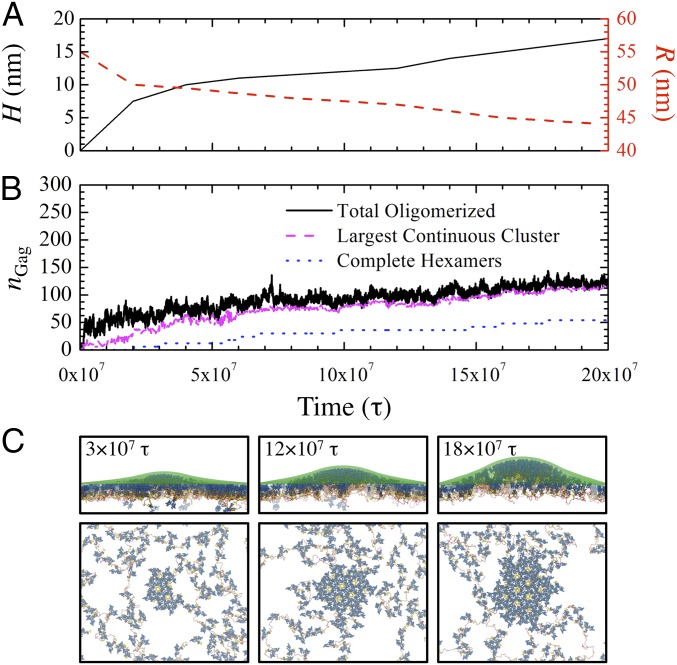
To emulate initial budding, we performed a nonequilibrium MD simulation (Movie S2) of 144 dimers with RNA and a gradually deforming membrane (initial lateral domain size of 100 × 100 nm2) over 2 × 108 τ. We monotonically increased the height and radius of curvature of the puncta following the time series profiles shown in Fig. 7A. Fig. 7B presents nGag profiles according to three classifications: (i) total oligomerized, (ii) within the largest contiguous cluster, and (iii) within complete hexamers. Throughout the trajectory, we observe relatively rapid binding and unbinding of Gag, reined in by RNA, to the central multimerized cluster to complete hexamers at the lattice periphery (Fig. 7C). Meanwhile, hexamer creation increases monotonically, which is consistent with our observations from TIRF experiments (Fig. S6); note that the actual time required for hexamer formation can vary widely as evidenced by the plateaus in the hexamerized nGag profile. Analysis of the dynamic rate of membrane deformation and Gag lattice growth (Fig. S7) further suggests that, while the extent of Gag aggregation is proportional to the rate of membrane deformation, hexamer formation rates remain largely independent. The snapshots in Fig. 7C also show that the lattice grows isotropically such that hexamer formation mostly occurs in concentric layers around the center of the cluster. We can expect this behavior since the entropic burden for hexamer formation (through concerted Gag localization) is reduced for every adjacent hexamer, which provide tethered monomers for hexamer incorporation; the extent to which this isotropic growth persists as the lattice approaches virion-scale sizes remains an open question. Altogether, our simulations suggest that slow and self-correcting Gag assembly concurrent with gradual membrane deformation may be essential features of immature lattice growth.
Fig. 7.
Assembly dynamics of CA–SP1 with RNA at membrane puncta. Time series profiles as a function of MD time step (τ) of the (A) height (H) and radius of curvature (R) of the continuously deforming membrane site and (B) the total number of multimerized Gag (nGag) categorized in three contexts: anywhere within the simulation domain, within the largest continuous cluster of Gag, and within any completed hexamers. (C) Side and top views of assembled CANTD–CACTD–SP1 lattices, depicted as blue, gray, and yellow tubes, at the listed τ with RNA (red beads) and membrane (transparent green beads) also shown.
Discussion
We have developed a CG computational model from experimental structural data that recapitulates assembly of HIV-1 immature lattices having contiguous and hexameric structures, in general agreement with previous cryo-EM and cryo-ET experiments (7–11). Our joint CG MD simulations and spt-PALM experiments support recent fluorescence experiments (51, 62–64) that suggested Gag multimerization occurs at the cell membrane interface rather than through the delivery of small Gag oligomers preassembled within the cytoplasm. Importantly, we highlight the catalytic role of both membrane puncta and viral RNA to drive multimerization. Our simulations indicate that assembly proceeds through a slow, self-regulated, and multistage process in which Gag hexamers are gradually formed along the periphery of a growing cluster; the multistep nature of this assembly process may contribute to the pauses that have been observed during viral assembly (65). We should emphasize that our CG model is intentionally simplified to reconstitute the minimal features required for assembly and may not be directly comparable to the complex ecosystem within cells. As a result, some possible mechanistic nuances, such as molecular conformational switches (21, 22, 24, 30) or regulatory signals from viral RNA-specific sequences and structures (66–70), are not included at this juncture in the modeling. Nonetheless, the simplicity of the present CG model provides a platform to distinguish possible regulatory roles of Gag protein domains, RNA, and the cell membrane on the early stages of immature lattice assembly.
The immature lattice consists of hexagonally arranged building blocks which themselves are hexamers of Gag. Therefore, successful assembly is contingent on controlled hexagonal order both within the hexameric unit and throughout the extended lattice. Our simulations show that anisotropic attractive interactions at the helical CA–SP1 junction, which may arise from interactions between hydrophobic sidechains (21) that emerge due to the amphipathic character of the helix (22), can provide hexagonal order within the hexameric unit through six-helical bundling. The inherent weakness of this interaction may be significant for two reasons. First, hexamers are preferentially stable over other small oligomers, which subsequently allows nonideal oligomers to anneal into proper hexamers. Second, hexamer formation requires concerted localization and reorganization of Gag, which can be facilitated by the RNA scaffold that, in part, makes RNA indispensable (and ensures that RNA is packaged). Our predicted requirement for weak interprotein interactions to prevent kinetically frustrated assembly is also consistent with computational studies of icosahedral capsid assembly (38, 40–42, 71), but further suggests that only a subset of protein interfaces is needed to mediate assembly. Hence, based on our simulation results, we also hypothesize that enhancing the interactions at the CA–SP1 helical interface can either induce aberrant Gag multimerization or abolish RNA packaging requirements; for example, A366E and M367K mutations exhibit Gag localization on the plasma membrane (i.e., arrested virion release) or nonspherical virion phenotypes (22), which may be interpreted based on additional electrostatic attraction at the helical junction. In other words, perturbing the narrow window of weak interactions at the CA–SP1 junction, either through diminishing or heightening, may be a viable therapeutic target to disrupt immature virion production.
Once the formation of the initial hexamer unit is complete, additional hexamers nucleate in concentric rings around this seed. Our results suggest that this process is enabled by both Gag dimerization and Gag recruitment driven by RNA. Recall that Gag dimerization, which occurs through the H9–H9 interface in the CACTD domain, is preferred in the cytoplasm (72), although the relative orientation of these helices differs in mature CA dimers and lattices (18, 73–75). Hence, hexamer formation utilizes six distinct dimers but only one-half of each; the other half of each dimer remains flexibly bound on each of six facets. These dangling dimers serve as a nucleating template by reducing the number of Gag that must be recruited through RNA scaffolding; as a result, the immature lattice can expand in a largely contiguous fashion (8, 9). Based on our findings, we suggest that efficient particle release may require RNA that is sufficiently long to continuously recruit free Gag into the lattice. Previous experiments have demonstrated that viral RNA as short as 3 kb can generate viral particles (76), although the efficacy of much shorter RNA remains to be explored. Our joint experiments and simulations on ultrashort (20-nt) miRNA, and related observations from previous experiments using miRNA (31) and tRNA (30), suggest that an excess of short competitor RNA can negatively regulate the ability of viral RNA to orchestrate Gag multimerization by restricting the availability of Gag–RNA binding sites.
The dynamics of membrane deformation during budding appear to further regulate the assembly process. This is related to our finding that the CANTD domain imparts intrinsic curvature to the immature lattice. There has been some debate on the role and relevance of the CANTD domain since ΔCANTD Gag has been reported to successfully produce VLPs (27, 59), remain aggregated at the plasma membrane (77), or assemble into flat sheets in vitro (21). Our findings show that both CANTD–Gag and ΔCANTD–Gag are assembly competent, but the former may require spontaneous membrane curvature to initiate nucleation while extended growth is contingent on gradual membrane deformation. Beyond simple thermal fluctuations, curvature may be passively promoted by the heterogeneous compositions of membranes, such as through lipid raft domains (44, 78, 79) or transmembrane proteins (80–82). In fact, the binding of MA domains (and associated myristyl anchors) to the membrane may be important to initiate PIP2 enrichment into nanodomains (47, 83) and possibly membrane bending. In addition, the actin cytoskeleton has been proposed to expedite viral assembly and budding (84), although its role is dispensable as shown by recent fluorescence experiments (85, 86). We further speculate that membrane deformation dynamics can influence the formation of previously observed defects throughout the immature lattice (8–10), especially since immature virions tend to be highly pleomorphic. While local hexameric defects may be similar to the vacancy-like defects observed in this work, extended gap-like defects might also arise from anisotropic lattice growth at the leading edges, which might be instigated by the strain imposed from the mismatch between the local curvature of the Gag lattice and membrane. Hence, the presence of these possible curvature generation factors is likely important to understand differences in observed virion phenotypes, which may be a future direction for study. Beyond incorporation of these potential factors, computational challenges remain for virion-scale assembly and budding. Given that immature virions are composed of an average of 2,400 Gag (9), the increased system sizes and anticipated trajectory lengths would make CG simulations (in the mold of this work) exceptionally expensive. Nonetheless, we are encouraged by the rapid development of enhanced sampling techniques such as metadynamics (87), which we foresee as one promising approach for future simulations in this direction.
To summarize, our findings reveal that RNA and the cell membrane are active participants of viral assembly that both initiate and coordinate Gag multimerization. Deformations along the membrane can regulate the location and size of the assembling Gag cluster, which can only proceed when RNA drives the localization and reorganization of multiple free Gag toward the cluster periphery. Our results suggest that immature lattice assembly and budding are coupled and proceed concurrently (88). These insights reveal a network of coordinated interactions that are likely to be essential for viral replication.
Methods
CA–SP1 CG Model.
To reproduce the internal structure of the CA–SP1 dimer, we only considered the structured portions within each protein domain. As depicted in Fig. 1A, the relative position of each CG site was obtained from the average position of each C-α throughout the α-helices within the CANTD (seven helices), CACTD (four helices), and SP1 (one helix) domains, which were calculated over all monomer chain configurations. We used two experimental datasets [PDB ID codes 4USN (19) and 5I4T (21)] that were superposed by minimizing the root-mean-squared deviation of the C-α positions in the CACTD domain. As NMR spectroscopy has suggested that each protein domain remains folded in solution and reorients semiindependently from each other (72), the collection of α-helices within each domain was considered as a rigid body with rigid-body dynamics (89). The tertiary structure of CA–SP1 was maintained with an elastic network model (ENM) that connected CG particles in adjacent domains (e.g., between helix 10 of CACTD and the SP1 helix) through flexible harmonic bonds; the potential energy of the bonds (Ebond) was given by the following:
where Kbond = 0.5 kcal⋅mol−1⋅Å−2, r is the separation distance, and r0 is the computed average distance. To maintain the shape of the polyprotein, each CG particle had an excluded volume through a soft cosine potential (Eexcl) given by the following:
when r < rexcl. The excluded volume separation (rexcl) between CG particles in adjacent helices was set to their average separation distances if less than 10 Å or, otherwise, set to 10 Å (i.e., CG particle radii are 5 Å by default); in all cases, A = 25 kcal/mol. The mass of every CG site was set to 150 Da.
The CG model also incorporated two important protein–protein interfaces that have previously been identified. First, we used a flexible ENM (Kbond = 0.05 kcal⋅mol−1⋅Å−2) between adjacent H9 in the CACTD domains to dimerize CA–SP1 based on the helical orientations observed in the aforementioned experimental data sets; our ENM represents the strong preference for CA proteins to exist as homodimers in the cytoplasm (72, 75), although the relative tilt between H9 can vary (73, 74). In addition, we assume that the CA–SP1 junction forms an ordered helix that assembles into a high-density bundle (8, 10, 18–21). To recapitulate this interface, we used the relative positions of four residues (P356, A360, A364, S368) in an adjacent SP1 domain when confined within a hexamer configuration to assign the positions of virtual particles (pink spheres in Fig. 1A); as a result, each CG monomer consisted of 157 residue beads and four virtual particles (a total of 161 CG beads per monomer). Each of these virtual particles were allowed to tether to any CG particle of the same residue through an attractive Gaussian potential (Etether) given by the following:
where ESP1 is the well depth (varied throughout this manuscript), B is the inverse interaction range (=0.2 Å−2), and r is the distance between the virtual and tether site. This interaction was the only attractive nonbonding interaction between CA–SP1 dimers used throughout this work and effectively represents the net contributions from the SP1 helical interactions, the major homology region, and the nearby type II β-turn (19–21).
By combining the excluded volume and structural ENM throughout the CA–SP1 polyprotein, the flexible ENM at the H9–H9 dimer interface, and the anisotropic tethering potential at the CA–SP1 junction, the tertiary and quaternary structure of assembled CA–SP1 recapitulated the expected hexamer structure (Fig. 1B).
RNA and Membrane Model.
Simplified CG models of RNA and the membrane were used throughout this work. RNA was represented as a linear chain of CG particles that were linked with flexible harmonic bonds (Kbond = 0.5 kcal⋅mol−1⋅Å−2; see Ebond above) and excluded volume with rexcl = 7.0 Å (i.e., particle radius of 3.5 Å; see Eexcl above). The mass of each CG site was set to 330 Da. To implicitly include binding between RNA and the NC domain, each RNA particle was allowed to tether to the final SP1 particle with a well depth of 7.5 kcal/mol and inverse interaction range of 0.1 Å−2; as this interaction is, in part, due to electrostatic attraction between the negatively charged phosphate groups in RNA and positively charged Zn2+ fingers in the NC domain (66, 90), the chosen potential energy is intentionally both stronger and wider-ranged than that of the hydrophobic SP1–SP1 interaction (21, 22).
The simplified membrane was represented by an elastic mesh of particles (with a mass of 1 kDa) arranged in a hexagonal lattice with a lattice constant of 0.85 nm. Bonds between particles are maintained with a Morse potential (Emorse) given by the following:
where D = 450.0 kcal/mol, α = 0.17 Å−1, and rEM = 5.0 Å; each particle was connected to their three neighbors. Particles also interacted through a long-range 12-6 Lennard Jones potential with a modified soft core (Esclj) (91) given by the following:
where n = 2, αLJ = 0.5, λ = 0.6, ε = 0.03 kcal/mol, and σ = 20 Å. These two potentials, at minimum, were found to stabilize membrane planarity even without angular or dihedral terms. These parameters have been adjusted to reproduce a fluctuation spectrum (Fig. S8) with bending modulus of around 12 kBT (92). Our intention was simply to emulate the membrane as a substrate that reduces the dimensionality of CA–SP1 assembly (44). To implicitly include binding between the membrane and Gag through exposed myristyl and the MA domain (93, 94), each elastic mesh particle interacted with the top-most particle of the CANTD domain (i.e., in helix 6, which is furthest from the CA–SP1 junction) through Esclj, where n = 2, αLJ = 0.5, λ = 0.6, ε = 1.5 kcal/mol, and σ = 12 Å; note that weaker interaction strengths prevent Gag binding, while stronger interaction strengths prevent extended lattice growth (Fig. S4). In addition, spontaneous curvature in the membrane was induced through a spherical indentation force (Find) given by the following:
where Kind = 0.001 kcal⋅mol−1⋅Å−3 and Rind is the radius of the indenter; the force is repulsive when r < Rind and zero otherwise. In these cases, an opposing planar force (Kind = 0.0001 kcal⋅mol−1⋅Å−3) was weakly applied to the outermost edges of the mesh (with thickness around 15 Å) to prevent vertical drift.
Unless otherwise specified, default excluded volume interactions (e.g., radius of 5 Å) were maintained between CA–SP1, RNA, and the membrane.
MD Simulations.
All CG MD simulations were performed using the large-scale atomic/molecular massively parallel simulator (LAMMPS) (95). Three different classes of simulations were performed throughout this manuscript; the general details of each are described below. In all cases, a temperature of 310 K was maintained using Langevin dynamics (96) with a damping constant of 100 ps. A Nosé–Hoover chain barostat (97, 98) in the coupled xy (i.e., lateral) dimensions was used to maintain zero tension with a damping constant of 250 ps, unless otherwise specified. A CG MD time step (τ) of 200 fs was used; we chose the largest τ that was possible without noticeable energy drift in the microcanonical (constant NVE) ensemble. All pair potentials used a 25-Å radial cutoff with an additional 5-Å buffer for particle neighbor lists. All simulations were periodic in all three directions, although in all cases, unless otherwise specified, the z direction was bound by repulsive walls and essentially nonperiodic. Several different simulations were performed throughout this manuscript; specific details are described within the appropriate subsections in Results. All simulations were executed on either 64 or 96 AMD Bulldozer CPUs (2.3 GHz) with execution speeds around 1.2 × 107 and 1.7 × 107 τ/d, respectively. Further details on trajectory analysis are provided in SI Methods.
Preparation of Cells and Plasmids.
WT HEK293 cells and HEK293 cells overexpressing exogenous human miRNA has–miR–146a were cultured and maintained in phenol red-free DMEM supplemented with 10% (vol/vol) FBS (Invitrogen) and 2 mM glutamine (Cellgro). Designs of miRNA expression plasmids and viral constructs used for expressing Gag (pNL4-3ΔPΔE, pNL4-3-imEOS2-ΔPΔE, and pNL4-3-iEGFP-ΔPΔE) and generation of miRNA cell line have been described before (31).
PALM Imaging.
Live-cell spt-PALM experiments were carried out with an Elyra PS.1 using a 100×, 1.46 N.A. oil-immersion objective (Carl Zeiss). Cells were plated in cleaned 25-mm #1.5 coverslips (Warner Instruments) and transfected with pNL4-3ΔPΔE and pNL4-3-imEOS2-ΔPΔE plasmids as described before (31). Live-cell spt-PALM experiments (31, 49) were performed with cells placed in imaging medium (phenol red-free DMEM containing 25 mM Hepes and 1% FBS) maintained at 37 °C. During PALM image acquisition, mEOS2 probes were photoconverted using a 561-nm laser (readout-induced activation mode), and images were collected at 20 frames per s. The 561-nm laser power was calibrated to maintain a sparse distribution of single mEOS2 fluorescent spots at every image frame of the time series.
PALM images of live cells were collected using illumination conditions where the average distance between single-molecule fluorescent spots was significantly larger than the localization precision of single Gag molecules. Single mEOS2-tagged Gag molecules were identified and fit with a 2D symmetric Gaussian point spread function using either commercially available software (Zeiss Zen Black super resolution module; Carl Zeiss) or custom code written in MATLAB. Further details on PALM image analysis are provided in SI Methods.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under Award P50-GM082545 (to G.A.V. and M.Y.). The J.L.-S. laboratory acknowledges financial support from the Howard Hughes Medical Institute. The J.A.G.B. laboratory acknowledges financial support from the European Molecular Biology Laboratory, Chica und Heinz Schaller Stiftung, and Deutsche Forschungsgemeinschaft Grant BR 3536/2-1 (to J.A.G.B.). Computational resources were provided by the Blue Waters sustained-petascale computing project, which is supported by National Science Foundation Awards OCI-0725070 and ACI-1238993 and the State of Illinois. Blue Waters is a joint effort of the University of Illinois at Urbana–Champaign and its National Center for Supercomputing Applications. This work is part of the “Ultra–Coarse-Grained Simulations of Biomolecular Processes at the Petascale” Petascale Computing Resource Allocation support to G.A.V. by National Science Foundation Award OCI-1440027.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706600114/-/DCSupplemental.
References
- 1.Briggs JAG, Kräusslich HG. The molecular architecture of HIV. J Mol Biol. 2011;410:491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 2.Sundquist WI, Kräusslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lingappa JR, Reed JC, Tanaka M, Chutiraka K, Robinson BA. How HIV-1 Gag assembles in cells: Putting together pieces of the puzzle. Virus Res. 2014;193:89–107. doi: 10.1016/j.virusres.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freed EO. HIV-1 assembly, release and maturation. Nat Rev Microbiol. 2015;13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freed EO. HIV-1 gag proteins: Diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 6.Bell NM, Lever AML. HIV Gag polyprotein: Processing and early viral particle assembly. Trends Microbiol. 2013;21:136–144. doi: 10.1016/j.tim.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Briggs JAG, et al. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 8.Wright ER, et al. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson LA, et al. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs JAG, et al. Structure and assembly of immature HIV. Proc Natl Acad Sci USA. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson LA, et al. Cryo electron tomography of native HIV-1 budding sites. PLoS Pathog. 2010;6:e1001173. doi: 10.1371/journal.ppat.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 13.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 14.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grime JMA, Voth GA. Early stages of the HIV-1 capsid protein lattice formation. Biophys J. 2012;103:1774–1783. doi: 10.1016/j.bpj.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao G, et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grime JMA, et al. Coarse-grained simulation reveals key features of HIV-1 capsid self-assembly. Nat Commun. 2016;7:11568. doi: 10.1038/ncomms11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bharat TAM, et al. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc Natl Acad Sci USA. 2014;111:8233–8238. doi: 10.1073/pnas.1401455111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schur FKM, et al. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature. 2015;517:505–508. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 20.Schur FKM, et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016;353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 21.Wagner JM, et al. Crystal structure of an HIV assembly and maturation switch. Elife. 2016;5:e17063. doi: 10.7554/eLife.17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta SAK, et al. On the role of the SP1 domain in HIV-1 particle assembly: A molecular switch? J Virol. 2011;85:4111–4121. doi: 10.1128/JVI.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datta SAK, et al. Dimerization of the Sp-1 region of HIV-1 Gag induces a helical conformation and association into helical bundles: Implications for particle assembly. J Virol. 2015;90:1773–1787. doi: 10.1128/JVI.02061-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayro MJ, Ganser-Pornillos BK, Zadrozny KK, Yeager M, Tycko R. Helical conformation in the CA-SP1 junction of the immature HIV-1 lattice determined from solid-state NMR of virus-like particles. J Am Chem Soc. 2016;138:12029–12032. doi: 10.1021/jacs.6b07259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross I, Hohenberg H, Kräusslich HG. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 26.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Accola MA, Strack B, Göttlinger HG. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol. 2000;74:5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell S, et al. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc Natl Acad Sci USA. 2001;98:10875–10879. doi: 10.1073/pnas.191224698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, et al. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc Natl Acad Sci USA. 2016;113:E201–E208. doi: 10.1073/pnas.1518572113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlson L-A, Bai Y, Keane SC, Doudna JA, Hurley JH. Reconstitution of selective HIV-1 RNA packaging in vitro by membrane-bound Gag assemblies. Elife. 2016;5:e14663. doi: 10.7554/eLife.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen AK, et al. MicroRNA binding to the HIV-1 Gag protein inhibits Gag assembly and virus production. Proc Natl Acad Sci USA. 2014;111:E2676–E2683. doi: 10.1073/pnas.1408037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faivre-Moskalenko C, et al. RNA control of HIV-1 particle size polydispersity. PLoS One. 2014;9:e83874. doi: 10.1371/journal.pone.0083874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dick RA, Vogt VM. Membrane interaction of retroviral Gag proteins. Front Microbiol. 2014;5:187. doi: 10.3389/fmicb.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yandrapalli N, Muriaux D, Favard C. Lipid domains in HIV-1 assembly. Front Microbiol. 2014;5:220. doi: 10.3389/fmicb.2014.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mücksch F, Laketa V, Müller B, Schultz C, Kräusslich H-G. Synchronized HIV assembly by tunable PIP2 changes reveals PIP2 requirement for stable Gag anchoring. Elife. 2017;6:25287. doi: 10.7554/eLife.25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee I-H, Kai H, Carlson L-A, Groves JT, Hurley JH. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc Natl Acad Sci USA. 2015;112:15892–15897. doi: 10.1073/pnas.1518765113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rapaport DC. Self-assembly of polyhedral shells: A molecular dynamics study. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70:051905. doi: 10.1103/PhysRevE.70.051905. [DOI] [PubMed] [Google Scholar]
- 38.Rapaport DC. Role of reversibility in viral capsid growth: A paradigm for self-assembly. Phys Rev Lett. 2008;101:186101. doi: 10.1103/PhysRevLett.101.186101. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen HD, Reddy VS, Brooks CL., 3rd Deciphering the kinetic mechanism of spontaneous self-assembly of icosahedral capsids. Nano Lett. 2007;7:338–344. doi: 10.1021/nl062449h. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen HD, Reddy VS, Brooks CL., 3rd Invariant polymorphism in virus capsid assembly. J Am Chem Soc. 2009;131:2606–2614. doi: 10.1021/ja807730x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagan MF, Elrad OM, Jack RL. Mechanisms of kinetic trapping in self-assembly and phase transformation. J Chem Phys. 2011;135:104115. doi: 10.1063/1.3635775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perlmutter JD, Perkett MR, Hagan MF. Pathways for virus assembly around nucleic acids. J Mol Biol. 2014;426:3148–3165. doi: 10.1016/j.jmb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perlmutter JD, Qiao C, Hagan MF. Viral genome structures are optimal for capsid assembly. Elife. 2013;2:e00632. doi: 10.7554/eLife.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz-Herrero T, Hagan MF. Simulations show that virus assembly and budding are facilitated by membrane microdomains. Biophys J. 2015;108:585–595. doi: 10.1016/j.bpj.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen B, Tycko R. Simulated self-assembly of the HIV-1 capsid: Protein shape and native contacts are sufficient for two-dimensional lattice formation. Biophys J. 2011;100:3035–3044. doi: 10.1016/j.bpj.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiao X, Jeon J, Weber J, Zhu F, Chen B. Construction of a novel coarse grain model for simulations of HIV capsid assembly to capture the backbone structure and inter-domain motions in solution. Data Brief. 2015;5:506–512. doi: 10.1016/j.dib.2015.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charlier L, et al. Coarse-grained simulations of the HIV-1 matrix protein anchoring: Revisiting its assembly on membrane domains. Biophys J. 2014;106:577–585. doi: 10.1016/j.bpj.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ayton GS, Voth GA. Multiscale computer simulation of the immature HIV-1 virion. Biophys J. 2010;99:2757–2765. doi: 10.1016/j.bpj.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 50.Baumgärtel V, Müller B, Lamb DC. Quantitative live-cell imaging of human immunodeficiency virus (HIV-1) assembly. Viruses. 2012;4:777–799. doi: 10.3390/v4050777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 53.Berkowitz RD, Ohagen A, Höglund S, Goff SP. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burniston MT, Cimarelli A, Colgan J, Curtis SP, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. 1999;73:8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell S, Vogt VM. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Izvekov S, Voth GA. Modeling real dynamics in the coarse-grained representation of condensed phase systems. J Chem Phys. 2006;125:151101. doi: 10.1063/1.2360580. [DOI] [PubMed] [Google Scholar]
- 57.Davtyan A, Dama JF, Voth GA, Andersen HC. Dynamic force matching: A method for constructing dynamical coarse-grained models with realistic time dependence. J Chem Phys. 2015;142:154104. doi: 10.1063/1.4917454. [DOI] [PubMed] [Google Scholar]
- 58.Gross I, et al. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borsetti A, Ohagen A, Göttlinger HG. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol. 1998;72:9313–9317. doi: 10.1128/jvi.72.11.9313-9317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simons K, Gerl MJ. Revitalizing membrane rafts: New tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 61.Lorizate M, Kräusslich H-G. Role of lipids in virus replication. Cold Spring Harb Perspect Biol. 2011;3:a004820. doi: 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivanchenko S, et al. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jouvenet N, et al. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kutluay SB, Bieniasz PD. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010;6:e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ku PI, et al. Identification of pauses during formation of HIV-1 virus like particles. Biophys J. 2013;105:2262–2272. doi: 10.1016/j.bpj.2013.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu D, Searles MA, Klug A. Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature. 2003;426:96–100. doi: 10.1038/nature02088. [DOI] [PubMed] [Google Scholar]
- 67.Jalalirad M, Laughrea M. Formation of immature and mature genomic RNA dimers in wild-type and protease-inactive HIV-1: Differential roles of the Gag polyprotein, nucleocapsid proteins NCp15, NCp9, NCp7, and the dimerization initiation site. Virology. 2010;407:225–236. doi: 10.1016/j.virol.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 68.Abd El-Wahab EW, et al. Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat Commun. 2014;5:4304. doi: 10.1038/ncomms5304. [DOI] [PubMed] [Google Scholar]
- 69.Kutluay SB, et al. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell. 2014;159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Comas-Garcia M, et al. Dissection of specific binding of HIV-1 Gag to the “packaging signal” in viral RNA. Elife. 2017;6:27055. doi: 10.7554/eLife.27055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hagan MF, Chandler D. Dynamic pathways for viral capsid assembly. Biophys J. 2006;91:42–54. doi: 10.1529/biophysj.105.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deshmukh L, Ghirlando R, Clore GM. Conformation and dynamics of the Gag polyprotein of the human immunodeficiency virus 1 studied by NMR spectroscopy. Proc Natl Acad Sci USA. 2015;112:3374–3379. doi: 10.1073/pnas.1501985112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Worthylake DK, Wang H, Yoo S, Sundquist WI, Hill CP. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr D Biol Crystallogr. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- 74.Byeon IJL, et al. Structural convergence between cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deshmukh L, et al. Structure and dynamics of full-length HIV-1 capsid protein in solution. J Am Chem Soc. 2013;135:16133–16147. doi: 10.1021/ja406246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nikolaitchik OA, et al. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 2013;9:e1003249. doi: 10.1371/journal.ppat.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jalaguier P, Turcotte K, Danylo A, Cantin R, Tremblay MJ. Efficient production of HIV-1 virus-like particles from a mammalian expression vector requires the N-terminal capsid domain. PLoS One. 2011;6:e28314. doi: 10.1371/journal.pone.0028314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shlomovitz R, Schick M. Model of a raft in both leaves of an asymmetric lipid bilayer. Biophys J. 2013;105:1406–1413. doi: 10.1016/j.bpj.2013.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meinhardt S, Vink RLC, Schmid F. Monolayer curvature stabilizes nanoscale raft domains in mixed lipid bilayers. Proc Natl Acad Sci USA. 2013;110:4476–4481. doi: 10.1073/pnas.1221075110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brannigan G, Brown FLH. Contributions of Gaussian curvature and nonconstant lipid volume to protein deformation of lipid bilayers. Biophys J. 2007;92:864–876. doi: 10.1529/biophysj.106.094953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marsh D. Lateral pressure profile, spontaneous curvature frustration, and the incorporation and conformation of proteins in membranes. Biophys J. 2007;93:3884–3899. doi: 10.1529/biophysj.107.107938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aimon S, et al. Membrane shape modulates transmembrane protein distribution. Dev Cell. 2014;28:212–218. doi: 10.1016/j.devcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yandrapalli N, et al. Self assembly of HIV-1 Gag protein on lipid membranes generates PI(4,5)P2/cholesterol nanoclusters. Sci Rep. 2016;6:39332. doi: 10.1038/srep39332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gladnikoff M, Shimoni E, Gov NS, Rousso I. Retroviral assembly and budding occur through an actin-driven mechanism. Biophys J. 2009;97:2419–2428. doi: 10.1016/j.bpj.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rahman SA, et al. Investigating the role of F-actin in human immunodeficiency virus assembly by live-cell microscopy. J Virol. 2014;88:7904–7914. doi: 10.1128/JVI.00431-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stauffer S, et al. The nucleocapsid domain of Gag is dispensable for actin incorporation into HIV-1 and for association of viral budding sites with cortical F-actin. J Virol. 2014;88:7893–7903. doi: 10.1128/JVI.00428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tribello GA, Giberti F, Sosso GC, Salvalaglio M, Parrinello M. Analyzing and driving cluster formation in atomistic simulations. J Chem Theory Comput. 2017;13:1317–1327. doi: 10.1021/acs.jctc.6b01073. [DOI] [PubMed] [Google Scholar]
- 88.Zhang R, Nguyen TT. Model of human immunodeficiency virus budding and self-assembly: Role of the cell membrane. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;78:051903. doi: 10.1103/PhysRevE.78.051903. [DOI] [PubMed] [Google Scholar]
- 89.Miller TF, et al. Symplectic quaternion scheme for biophysical molecular dynamics. J Chem Phys. 2002;116:8649–8659. [Google Scholar]
- 90.De Guzman RN, et al. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 91.Beutler TC, Mark AE, van Schaik RC, Gerber PR, van Gunsteren WF. Avoiding singularities and numerical instabilities in free energy calculations based on molecular simulations. Chem Phys Lett. 1994;222:529–539. [Google Scholar]
- 92.Brandt EG, Braun AR, Sachs JN, Nagle JF, Edholm O. Interpretation of fluctuation spectra in lipid bilayer simulations. Biophys J. 2011;100:2104–2111. doi: 10.1016/j.bpj.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saad JS, et al. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fledderman EL, et al. Myristate exposure in the human immunodeficiency virus type 1 matrix protein is modulated by pH. Biochemistry. 2010;49:9551–9562. doi: 10.1021/bi101245j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Plimpton S. Fast parallel algorithms for short-range molecular dynamics. J Comput Phys. 1995;117:1–19. [Google Scholar]
- 96.Schneider T, Stoll E. Molecular-dynamics study of a three-diemensional one-component model for distortive phase transitions. Phys Rev B. 1978;17:1302–1322. [Google Scholar]
- 97.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 98.Shinoda W, Shiga M, Mikami M. Rapid estimation of elastic constants by molecular dynamics simulation under constant stress. Phys Rev B. 2004;69:134103. [Google Scholar]
- 99.Hoshen J, Kopelman R. Percolation and cluster distribution. I. Cluster multiple labeling technique and critical concentration algorithm. Phys Rev B. 1976;14:3438–3445. [Google Scholar]
- 100.Dellago C, Bolhuis PG, Csajka Fl S, Chandler D. Transition path sampling and the calculation of rate constants. J Chem Phys. 1998;108:1964–1977. [Google Scholar]
- 101.Faradjian AK, Elber R. Computing time scales from reaction coordinates by milestoning. J Chem Phys. 2004;120:10880–10889. doi: 10.1063/1.1738640. [DOI] [PubMed] [Google Scholar]
- 102.Tiwary P, Parrinello M. From metadynamics to dynamics. Phys Rev Lett. 2013;111:230602. doi: 10.1103/PhysRevLett.111.230602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.