Significance
Toxin-antitoxin (TA) systems are small genetic modules known to modulate bacterial growth in response to stress. Their major contribution to the formation of persister cells and to the virulence of several important pathogens has highlighted them as promising new targets for therapy. TA-chaperone (TAC) modules are atypical TA systems tightly controlled by a third partner: a molecular chaperone that directly assists the antitoxin. Remarkably, TAC chaperones belong to the family of the canonical SecB chaperone known to facilitate protein secretion in bacteria, thus potentially connecting toxin activation and protein export. Herein, we have used the TAC system of the major human pathogen Mycobacterium tuberculosis to reveal how generic chaperones can rapidly evolve to become specialized for TA systems.
Keywords: HigB-HigA, SecA, trigger factor, DnaK, Rv1957
Abstract
SecB chaperones assist protein export in bacteria. However, certain SecB family members have diverged to become specialized toward the control of toxin-antitoxin (TA) systems known to promote bacterial adaptation to stress and persistence. In such tripartite TA-chaperone (TAC) systems, the chaperone was shown to assist folding and to prevent degradation of its cognate antitoxin, thus facilitating inhibition of the toxin. Here, we used both the export chaperone SecB of Escherichia coli and the tripartite TAC system of Mycobacterium tuberculosis as a model to investigate how generic chaperones can specialize toward the control of TA systems. Through directed evolution of SecB, we have identified and characterized mutations that specifically improve the ability of SecB to control our model TA system without affecting its function in protein export. Such a remarkable plasticity of SecB chaperone function suggests that its substrate binding surface can be readily remodeled to accommodate specific clients.
In bacteria, the SecB chaperone facilitates protein export via the general Sec pathway by binding to presecretory proteins and maintaining them in a nonnative state competent for translocation through the Sec translocon at the inner membrane (1). SecB bound to its presecretory protein client specifically interacts with the SecA motor subunit of Sec, which subsequently takes over the substrate and promotes its translocation by successive cycles of ATP hydrolysis (2). SecB is mainly present in α-, β- and γ-proteobacteria, but some secB sequences are sporadically found in other taxonomic groups, most of which are diderm bacteria (3). Remarkably, over 7% of these secB genes are clustered with genes encoding putative type II toxin-antitoxin (TA) systems (1). Type II TA systems are composed of two genes encoding a stable toxin and a less stable antitoxin, where both proteins form a complex in which the toxin is inactive. Under specific stress conditions, the antitoxin is degraded by stress proteases, releasing the active toxin which will act on its intracellular targets and inhibit bacterial growth (4). Such TA-mediated control of growth in response to stress has been involved in several major processes, including biofilm, formation of persister cells, and virulence of several important pathogens (5–8).
The human pathogen Mycobacterium tuberculosis possesses such a TA-associated SecB-like chaperone, named Mtb-SecBTA, which specifically controls the Mtb-HigB1(toxin)-HigA1(antitoxin) TA pair. This system was the first functional tripartite toxin-antitoxin-chaperone (TAC) system identified and is, so far, the most characterized (9, 10). We previously showed that Mtb-SecBTA directly interacts with the short carboxyl-terminal extension of Mtb-HigA1 antitoxin and protects it from both degradation and aggregation, allowing inactivation of the Mtb-HigB1 toxin (9, 11). The toxin Mtb-HigB1 of TAC severely inhibits Escherichia coli, Mycobacterium smegmatis, Mycobacterium marinum, and M. tuberculosis growth (10), and likely acts as a ribonuclease with a limited number of targets, most of which are involved in iron and zinc homeostasis (12). Despite significant specificity for the antitoxin, Mtb-SecBTA likely retains its export chaperone function, as judged by its ability to efficiently replace E. coli SecB both in vivo and in vitro (9). However, a direct role for Mtb-SecBTA in Sec-dependent protein export in M. tuberculosis remains to be demonstrated (a proposed model is illustrated in Fig. S1).
In this study, we have investigated how generic SecB chaperones can specialize toward a TAC antitoxin by recreating such a phenomenon in a directed evolution experiment. Remarkably, we found that single amino acid substitutions within distinct regions of the E. coli solitary export chaperone SecB are sufficient to significantly improve its ability to control the Mtb-HigBA1 TA pair without affecting its function in protein export.
Results
Directed Evolution of Solitary SecB Toward Control of a TA System.
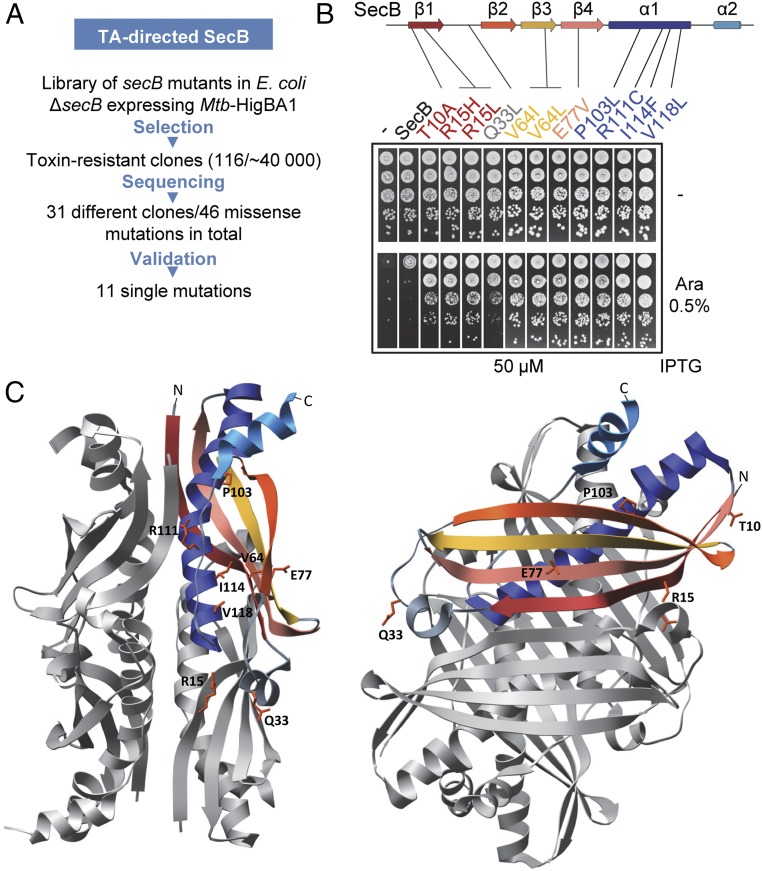
In contrast to Mtb-SecBTA, a robust overexpression of the E. coli solitary SecB is necessary to suppress the toxicity induced by Mtb-HigBA1 (9). We took advantage of this limited suppression by SecB to investigate how a generic export chaperone can specialize toward the control of TA systems. A plasmid library of randomly mutagenized E. coli secB was first transformed in the W3110 ∆secB strain expressing the Mtb-HigBA1 under conditions in which expression of wild-type SecB does not prevent growth inhibition by the toxin, and clones that formed viable colonies were selected for further analysis (Fig. 1A). This approach allowed the identification of 11 single amino acid substitutions in SecB that improve its ability to specifically control Mtb-HigBA1 (Fig. 1B). Steady-state expression of these mutants was comparable to that of wild-type SecB after 2 h of induction with 50 μM isopropyl β-d-1-thiogalactopyranoside (IPTG) (Fig. S2). To confirm that the improved TA control by the suppressors, herein named SecBTA, was antitoxin-dependent and not related to an indirect effect on the toxin alone, we coexpressed the Mtb-HigB1 toxin and the SecBTA variants in the absence of the antitoxin and show that, indeed, none of the SecB variants inactivate Mtb-HigB1 toxicity without Mtb-HigA1 (Fig. S2).
Fig. 1.
TA-directed SecB mutants. (A) Genetic selection to identify SecB variants with improved ability to control Mtb-HigBA1. (B) Suppression of Mtb-HigBA1 toxicity by SecBTA variants. The position of the suppressors is indicated on SecB secondary structures, with strand β1 in red, strand β2 in orange, strand β3 in yellow, strand β4 in salmon, helix α1 in dark blue, and helix α2 in light blue. W3110 ΔsecB containing plasmids pK6-Mtb-HigBA1 and p29SEN(−), p29-SecB, or p29-SecBTA variant was serially diluted; spotted on agar plates containing arabinose (Ara) and IPTG inducers; and incubated at 37 °C. (C) Localization of the mutations isolated on the E. coli SecB tetramer (1QYN) in front (Left) and side (Right) views. Residues are displayed as orange sticks on one SecB monomer, which is colored according to secondary structure elements from B.
The localization of the mutations carried by the SecBTA suppressors on the available structure of the E. coli SecB tetramer (13) is presented in Fig. 1C. The substitutions identified herein are located on strands β1 (T10A, R15H/L), β3 (V64I/L), and β4 (E77V); on the cross-over loop between strands β1 and β2 (Q33L); and in helix α1 (P103L, R111C, I114F, V118L). Residues T10, Q33, V64, and E77 were shown to be involved in substrate binding, and T10 and E77 were shown to additionally contact SecA (14–16). These mutations could directly affect SecB substrate binding specificity, thus increasing functional interaction with Mtb-HigA1. Interestingly, mutations at position E77, including the E77V substitution identified here, were previously found to inactivate SecB export function and interaction with SecA (17). However, the well-characterized E77K mutation known to affect SecB export function and interaction with SecA (18) does not improve TA control (Fig. S2). This indicates that export defect through an alteration of SecA binding by SecB is not sufficient to generate TA-specialized SecB and that the E77V substitution may facilitate TA control mainly via modulation of substrate binding.
Among the positions mutated in SecBTA variants that were not previously shown to interact with client proteins, R15, I114, and V118, are in close vicinity to the primary substrate binding site, suggesting that they could also act by modulating the affinity of SecB for Mtb-HigA1. The two remaining positions, R111 and P103, are more buried in the structure of the protein, suggesting that mutations of these residues might affect the conformation of SecB to improve antitoxin binding. A detailed structural location of the mutated residues is depicted in Fig. S3. The I114F mutation was largely overrepresented in our genetic selection (i.e., 73 of 116 suppressors carried this substitution), thus suggesting a main role for this position in specialization toward the antitoxin. This residue is located in helix α1 of SecB, in a region that contains a motif highly conserved between SecB family members (Fig. S4), including TACs (3). Positioned directly underneath the main substrate binding surface, I114 was identified by NMR spectroscopy to be in the vicinity of the substrate (either PhoA or MBP), but showed no direct interaction (16). This suggests that residue I114 might be important for the proper structure of the substrate binding site, and mutations at this position could destabilize or somehow remodel the binding site on SecB, thus modulating substrate binding properties (16).
SecBI114Y Is a Bona Fide Specialized SecBTA.
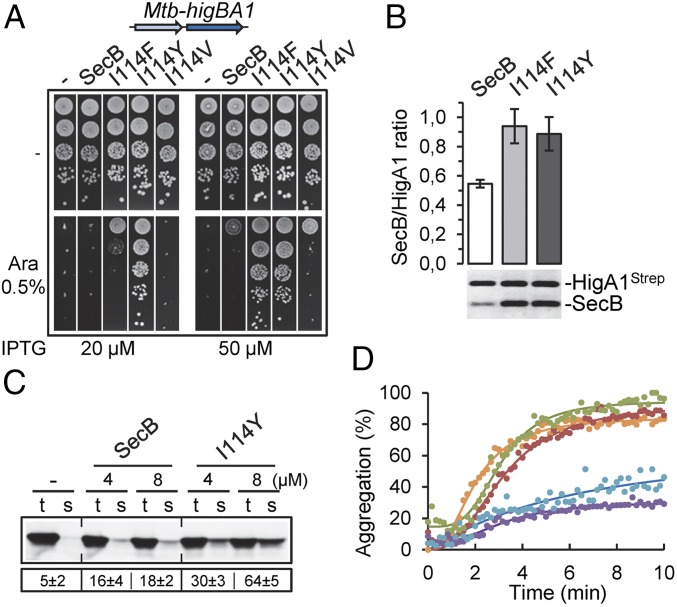
Position I114 of SecB likely corresponds to V145 in the sequence of Mtb-SecBTA (Fig. S4). However, mutation I114V in the conserved helix α1 motif of E. coli SecB does not improve the TA control (Fig. 2A). However, residue V145 of Mtb-SecBTA is framed by two tyrosines (Fig. S4), and we thus reasoned that introduction of an aromatic residue in this region of SecB could be responsible for the increased TA control function of the SecBI114F variant. Accordingly, mutation I114Y further improves TA control (Fig. 2A). Fig. S4 shows the molecular environment of residue I114 in the structures of SecB alone and in complex with PhoA, and illustrates predictions of the structural impact of the mutation to a tyrosine residue at this position. Note that the fact that substitution for a tyrosine was not obtained during the genetic selection procedure is not surprising because I114Y substitution requires two nucleotide changes within the same codon, while all of the mutations isolated resulted from single nucleotide changes. The SecBI114Y mutant, which turned out to be an efficient specialized SecBTA variant, was used to further characterize the mechanism of specialization of SecB toward TA systems.
Fig. 2.
Specialized SecBI114Y is directed toward Mtb-HigA1. (A) Suppression of Mtb-HigBA1 toxicity. W3110 ΔsecB containing plasmids pK6-Mtb-HigBA1 and p29SEN(−), p29-SecB, p29-SecB(I114F), p29-SecB(I114Y), or p29-SecB(I114V) was serially diluted, spotted on agar plates with or without inducers, and incubated at 37 °C. (B) In vivo interaction between Mtb-HigA1 and SecBTA variants. W3110 ΔsecB containing plasmids pK6-Mtb-HigA1 and p29-SecB or p29-SecBTA variant was grown to midlog phase, SecB expression was induced with 50 μM IPTG, and Mtb-HigA1 was induced with 0.5% arabinose (Ara). Crude cell extracts were mixed with streptactin Sepharose resin, and protein complexes were eluted with desthiobiotin. Protein quantification from elution fractions was carried out after migration on SDS/PAGE, followed by coloration with SYPRO orange using Multi Gauge software. The ratio of SecB/Mtb-HigA1 was calculated for each SecBTA. The results are given as fold change normalized to wild-type SecB, and are the mean of three independent experiments. Error bars indicate SD. (C) Improved solubilization of newly translated Mtb-HigA1 by SecBI114Y. Mtb-HigA1 was expressed in a cell-free translation system with or without SecB or SecBI114Y tetramer at 4 μM and 8 μM. After translation, the total (t) and soluble (s) fractions were separated on SDS/PAGE and Mtb-HigA1 was revealed by Western blot. The numbers below the electrophoretic pattern represent the mean solubility values (%), calculated by the ratio of the amount of translation products in the s and t fractions obtained from three different translation experiments. The SD is indicated. A representative result of three different experiments is shown. (D) Aggregation kinetics of denatured Mtb-HigA1 (4 μM) were followed at 30 °C by light scattering at 350 nm without (orange) or with 0.5 μM or 1 μM purified SecB (green and red, respectively) or SecBI114Y (blue and violet, respectively) tetramer.
SecBI114Y Exhibits Enhanced Chaperone Activity Toward Mtb-HigA1.
We next asked whether evolved SecB had an increased affinity for the antitoxin at steady state, using an in vivo pull-down assay with coexpressed SecBTA variants and strep-tagged Mtb-HigA1 antitoxin as bait (9). We found that both SecBI114F and SecBI114Y indeed show an increased interaction with Mtb-HigA1, compared with SecB wild type. This is in full agreement with the more robust inhibition of Mtb-HigB1 toxicity observed for both TA-directed mutants (Fig. 2B). SecBI114Y was then purified and characterized in vitro. Gel filtration analysis confirmed that, as observed for SecB, SecBI114Y forms a tetramer in vitro (Fig. S5). In addition, no difference could be detected between purified SecB and SecBI114Y proteins by circular dichroism or following partial α-chymotrypsin proteolysis (Fig. S5). The ability of purified SecBI114Y and SecB to solubilize Mtb-HigA1 in vitro was first investigated using a cell-free coupled transcription/translation assay, as performed previously (11). In this case, a significant fraction of the newly synthesized antitoxin was solubilized by SecBI114Y (up to 64% in the presence of 8 μM chaperone), and to a lesser extent by SecB wild type (up to 18% at the same concentration; Fig. 2C). Note that solubilization by SecBI114Y in this assay is not as robust as that observed with the native Mtb-SecBTA chaperone, which fully solubilized Mtb-HigA1 under the same conditions (11). We next monitored the ability of SecBI114Y and SecB to directly prevent aggregation of denatured Mtb-HigA1 in vitro. In this assay, we found that SecBI114Y significantly prevents Mtb-HigA1 aggregation, while SecB wild type showed no detectable effect (Fig. 2D). In this case, SecBI114Y was nearly as efficient as Mtb-SecBTA to prevent the aggregation of Mtb-HigA1 (Fig. S6). Together, these results demonstrate that the specialized SecB mutants have been efficiently redirected toward the antitoxin.
Finally, all of the other SecBTA variants identified in this work were also tested for interaction with Mtb-HigA1 by pull-down in vivo. As observed for SecBI114Y and SecBI114F, eight of 11 variants show an increased affinity for Mtb-HigA1 in vivo (Fig. S7). Variants T10A and R15H were comparable to SecB wild type, and R111C showed reduced binding, somehow suggesting a more transient interaction that may not be detected under the steady-state conditions tested. Accordingly, we purified both SecBT10A and SecBR15H variants and found that they could efficiently prevent Mtb-HigA1 aggregation in vitro (Fig. S7), in a manner comparable to that of SecBI114Y (Fig. 2D).
Evolved SecBTA Interacts with the Chaperone Addiction Extension of Mtb-HigA1.
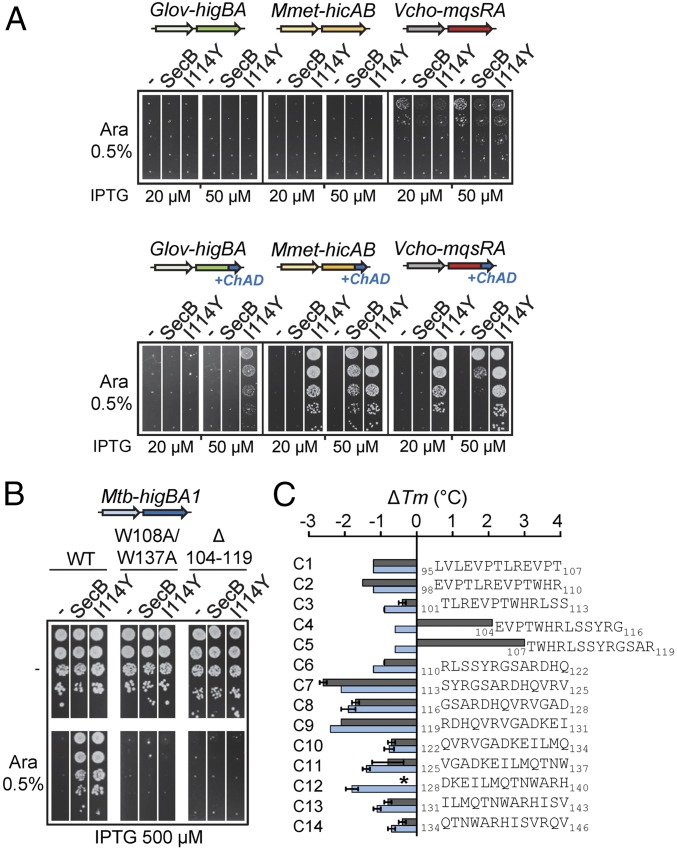
Antitoxins of TAC systems possess a carboxyl-terminal extension named chaperone addiction (ChAD), which is responsible for interaction with the dedicated chaperone and renders TAC antitoxins chaperone-dependent (11). ChAD regions are highly variable in length and amino acid composition, and they present high specificity for their respective SecBTA (11). We took advantage of these findings to investigate whether evolved SecBI114Y mutant is specifically directed toward the ChAD region of Mtb-TAC, like Mtb-SecBTA. Growth inhibition assays were performed in the presence of SecBI114Y coexpressed with chaperoneless TA pairs of three different TAC systems: Mmet-TAC from Methylomonas methanica, Vcho-TAC from Vibrio cholerae, and Glov-TAC from Geobacter lovleyi (3, 11). Remarkably, SecBI114Y was not capable of assisting the three chaperoneless TA pairs of these systems unless their original ChAD sequences were replaced by the one of Mtb-TAC (Fig. 3A, compare top and bottom spot tests; control plates of this experiment with no arabinose inducer added are shown in Fig. S8). Furthermore, assistance to chimeric TA pairs by the evolved SecBI114Y mutant is much more robust than that observed with SecB wild type (11). In support of such specific targeting of SecBI114Y to Mtb-ChAD, we found that the Mtb-HigA1 harboring the double W108A/W137A mutation in ChAD, previously shown to affect interaction with Mtb-SecBTA (11), also inhibits the ability of SecBI114Y to neutralize Mtb-HigBA1 (Fig. 3B).
Fig. 3.
SecBTA is directed toward the primary ChAD region. (A) Functional transfer of the SecBI114Y/Mtb-ChAD-specific pair. W3110 ΔsecB containing the pK6-Glov-HigBA, pK6-Mmet-HicAB, or pK6-Vcho-MqsRA native TA pair (Top) or their chimeric derivatives with their respective ChAD sequences being replaced by Mtb-ChAD (Bottom) was cotransformed with p29SEN-based SecB and SecBI114Y and analyzed by spot tests as in Fig. 1B. (B) Mutations in ChAD affect suppression by SecBI114Y. W3110 ΔsecB containing plasmid pK6-Mtb-HigBA1, pK6-Mtb-HigA1W108A/W137A, or pK6-Mtb-HigA1Δ(104–119) and p29SEN(−), p29-SecB, or p29-SecBI114Y was spotted on agar plates containing the indicated concentrations of arabinose (Ara) and IPTG inducers, and incubated at 37 °C. (C) Thermal stability of SecB and SecBI114Y was monitored by DSF in the presence of each peptide in a 60-fold excess. Tm values were deduced from the fluorescence curves recorded using a temperature gradient from 20 to 90 °C. Shifts in melting temperature are shown for SecB (blue bars) and SecBI114Y (gray bars). Mean and SEM of three replicates are shown. An asterisk (*) indicates no accurate Tm determination. Note that, in most cases, there was no detectable difference between replicates.
We next used differential scanning fluorimetry (DSF) to investigate the thermal stability of SecB and SecBI114Y, and to test whether SecBI114Y and Mtb-SecBTA share the same short region of interaction within the ChAD domain of Mtb-HigA1 (11). While thermal denaturation of SecB revealed a high melting temperature (Tm) of 71.7 ± 0.1 °C, the SecBI114Y variant exhibited a significantly reduced thermal stability (53.7 ± 0.2 °C; Fig. S8). This suggests that the I114Y mutation might indeed destabilize the substrate binding site of SecB, and thus potentially modulate interaction with the substrate (16). Notably, Mtb-SecBTA was also shown to have a significantly lower Tm than E. coli SecB (11). We then analyzed the effect of a set of 14 13-mer peptides encompassing the whole Mtb-HigA1 ChAD region (Fig. S8) on both SecB wild type and SecBI114Y, as previously described (11). Strikingly, only the C4 and C5 peptides, previously shown to contain the main site of interaction with Mtb-SecBTA, were able to induce a significant increase in the Tm of SecBI114Y but not of SecB wild type (Fig. 3C). In agreement with the in vivo results presented above, these data further show that SecBI114Y has indeed specialized toward a region within ChAD that is also recognized by the native Mtb-SecBTA. In support of such specificity, Mtb-HigA1 deleted for amino acids 104–119 comprising peptides C4 and C5 was not rescued by SecBI114Y or SecB (Fig. 3B).
Generic Export Functions of Evolved SecBTA.
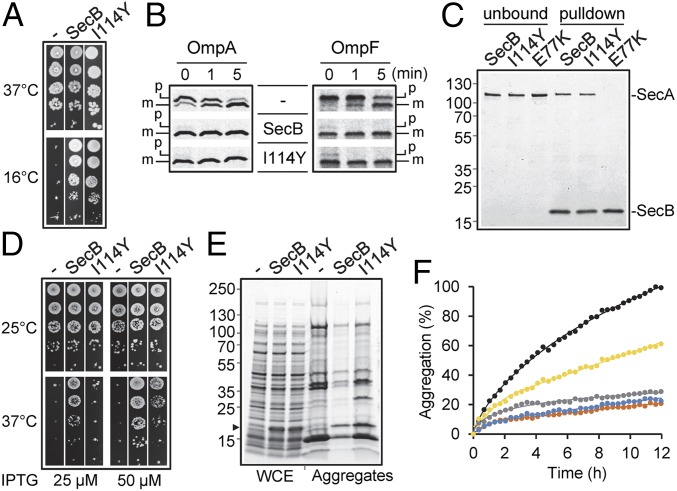
Improving chaperone activity toward a specific substrate generally results in a decreased generic chaperone function (19, 20). We thus asked whether specialized SecB variants fulfill their generic chaperone function at low temperature and in the presence of novobiocin antibiotic, with both reflecting a protein export defect (21, 22). Remarkably, SecBI114Y fully complements for the lack of SecB as efficiently as the wild type (Fig. 4A and Fig. S9, respectively). Note that this was also the case for all SecBTA variants isolated, with the exception of the previously described export-deficient variant SecBE77V (Fig. S9). We next followed the maturation kinetics of two SecB substrates by pulse-chase analyses in a secB mutant. In this case, expression of SecBI114Y fully restores export of proOmpA and proOmpF to levels comparable to those of wild-type SecB (Fig. 4B). Finally, we additionally confirmed that binding of SecBI114Y to SecA was not affected in vitro (Fig. 4C). Taken together, these data indicate that evolution toward TA control does not alter the export function of SecB.
Fig. 4.
Generic chaperone functions of TA-directed SecBI114Y. (A) W3110 ΔsecB containing plasmid p29SEN(−), p29-SecB, or p29-SecB(I114Y) was spotted on agar plates at indicated temperatures. (B) Pulse-chase analysis showing the processing of radiolabeled proOmpA and proOmpF in the strain W3110 ΔsecB containing plasmid p29SEN(−), p29-SecB, or p29-SecB(I114Y). Mature (m) and precursor (p) forms of both OmpA and OmpF are shown. (C) In vitro interaction of His-tagged SecB, SecBE77K ,and SecBI114Y with Flag-tagged SecA. Pull-down was performed using nickel-nitrilotriacetic acid resin. The unbound and elution (pull-down) fractions are shown. A protein ladder (in kilodaltons) is shown to the left of the gel. (D) MC4100 Δtig ΔdnaKdnaJ strain containing p29SEN(−), p29-SecB, or p29-SecB(I114Y) was grown to midlog phase at 25 °C; spotted on agar plates containing IPTG inducer as indicated; and incubated at 25 °C or 37 °C. (E) Prevention of aggregation in the MC4100 Δtig ΔdnaKdnaJ double-mutant strain harboring plasmid pSE380ΔNcoI(−), pSE-SecB, or p2SE-SecB(I114Y). Cells were grown to midlog phase at 25 °C, induced with 0.5 mM IPTG for 1 h, and switched at 33 °C for 1 h, and aggregates were extracted. Whole-cell extracts (WCE) and corresponding aggregate fractions (Aggregates) are shown. The position of SecB is shown with a black arrow. (F) In vitro aggregation of l-MDH. Aggregation kinetics of denatured l-MDH (2 μM) were followed at 30 °C by light scattering at 350 nm without (black) or with either 0.25 μM or 0.5 μM SecB (gray and orange, respectively) or SecBI114Y (yellow and blue, respectively) tetramers.
Cytosolic Chaperone Functions of SecBTA.
SecB overexpression partially suppresses the temperature-sensitive growth and prevents cytosolic protein aggregation in the absence of both cytosolic TF and DnaK chaperones (23). Under these conditions, suppression by SecB is independent of its export function (23). We took advantage of the severe growth phenotype of the tig dnaKJ mutant to test our SecBTA variants in vivo and found that none of the SecB mutants identified could suppress the growth defect better than SecB wild type (Fig. S9). On the contrary, seven of 12 SecBTA variants, including SecBI114Y, showed a significantly reduced ability to replace TF and DnaK compared with SecB wild type (Fig. 4D and Fig. S9). Accordingly, prevention of cytosolic protein aggregation by SecBI114Y in the absence of TF and DnaK was also less efficient (Fig. 4E). These results mirror the previously observed weak suppression of the tig dnaK mutant phenotypes by Mtb-SecBTA (9). Although the reasons for this are unknown, we cannot exclude the possibility that subtle changes within the substrate binding site of the TA-directed SecB could affect its ability to support bacterial growth in such a stringent context or that the absence of the major DnaK/TF pathway could affect the stability and conformation of certain SecB variants. Notably, all of our previous attempts to improve SecB function in the absence of TF and DnaK using the same SecB mutant library and a selection for growth above 37 °C were unsuccessful. This suggests that improvement of SecB toward cytosolic protein substrates of the TF/DnaK chaperone pathway may not be achievable under the conditions tested. We next asked whether SecBI114Y was also affected in its ability to prevent in vitro aggregation of the model substrate l-malate dehydrogenase (MDH). In this case, SecBI114Y exhibits reduced ability to prevent the aggregation of MDH compared with wild-type SecB (Fig. 4F). This is in sharp contrast to the increased aggregation prevention observed for the Mtb-HigA1 antitoxin, and indicates that the mutation I114Y has distinct effects on the interaction with different substrates.
Interplay Between Export Function and TA Control.
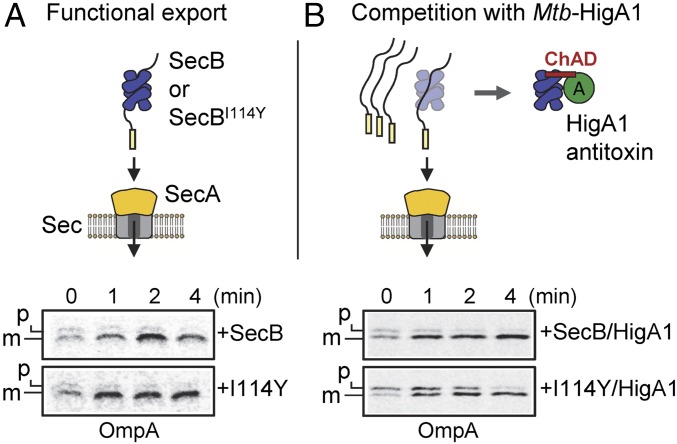
All SecB-like proteins of TAC systems tested so far have retained their ability to efficiently suppress the export defect of an E. coli secB mutant, while being specialized toward their cognate ChAD extension (3, 11). This suggests that competitive binding of SecBTA to the antitoxin or to preprotein clients could underlie the mechanism of activation of TAC systems, as proposed (3, 9). We took advantage of the evolved SecBI114Y, which retained its full ability to assist protein export, to test whether the antitoxin and preprotein clients could indeed compete for binding to SecB. We thus asked whether overexpression of the Mtb-HigA1 antitoxin could interfere with the export function of TA-specialized SecB mutant. Both SecB and SecBI114Y were coexpressed with the antitoxin in E. coli ΔsecB (9), and export kinetics of proOmpA were measured by pulse-chase analysis (Fig. 5A). In the absence of antitoxin, no difference could be detected between SecB and SecBI114Y in the rescuing of OmpA export, confirming that the mutant’s export function is kept intact. Strikingly, expression of Mtb-HigA1 in these conditions leads to a severe delay of maturation of proOmpA in the case of the TA-directed SecBI114Y mutant, while it has no detectable effect on SecB wild type (Fig. 5B). The same result was found with proOmpF substrate (Fig. S10). This indicates that Mtb-HigA1 can indeed compete with the export function of specialized SecBTA more than it does with SecB wild type, suggesting that under some circumstances, SecBTA chaperone function might be recruited either by the antitoxin or by accumulation of preproteins.
Fig. 5.
Mtb-HigA1 can compete with the export function of specialized SecB. Pulse-chase analysis showing the processing of the radiolabeled OmpA precursor (p) or mature (m) form in the strain W3110 ΔsecB expressing SecB or SecBI114Y without (A) or with (B) coexpressed Mtb-HigA1. Cells were grown at 30 °C to midlog phase, and were induced with 50 μM IPTG and with 0.002% arabinose 30 min later. After 30 min, cultures were switched to 18 °C, pulse-labeled, and chased for the indicated times. Samples were immunoprecipitated with anti-OmpA antibodies, separated by SDS/PAGE, and analyzed by autoradiography.
Discussion
In this work, we have isolated single amino acid substitutions within SecB that significantly improve TA control, thus illustrating the remarkable adaptability of this chaperone. The fact that all SecB-like chaperones of TAC tested so far have kept the ability to perform generic SecB export functions (3, 11) and that a directed evolution toward TA specialization does not impair the export capacity of SecB suggests that SecBTA chaperones might have undergone subfunctionalization, with optimization of a preexisting TA control function and a generic export function unaffected (24). Whether comparable chaperone plasticity can be found in other chaperone families remains unknown. Notably, the HscA member of the Hsp70 chaperone family, which is involved in the biogenesis of iron-sulfur proteins, has specialized toward a conserved “LPPVK” motif within its IscU substrate and, as a consequence, has lost its ability to bind a broad range of substrates and to functionally replace the multifunctional DnaK/Hsp70 homolog (25, 26). Similarly, a directed evolution of GroEL (Hsp60) toward heterologous GFP identified mutations in the chaperone that markedly improved GFP folding but that, in turn, significantly altered its ability to fold natural GroEL substrates and to replace GroEL in vivo (19). Therefore, in contrast to the unaffected SecB export function, specialization toward specific substrates severely impacted the main generic chaperone properties of these chaperones.
Analysis of TA-directed SecB revealed that the isolated mutations either affect residues previously known to directly interact with substrate or more buried residues that had not been characterized yet. Substitutions at these buried positions could alter the structure of the tetramer in a way that specifically enhances the productive interaction with the Mtb-HigA1 antitoxin, as our results suggest in the case of SecBI114Y. Notably, destabilized variants of the Spy periplasmic chaperone selected on the basis of their ability to stabilize a poorly folded immunity protein 7 mutant have also been isolated (27). However, in this case, the chaperone activity of Spy variants was also improved for all of the other substrates tested. In sharp contrast, SecBI114Y seems to only assist a certain type of substrate, as we observed reduced chaperone activity toward cytosolic substrates in vivo and in vitro and no chaperone activity toward other antitoxins tested in this work. This shows that discreet changes in SecB can readily remodel the chaperone to specifically accommodate very diverse ChAD amino acid extensions that are present in TAC antitoxins. The fact that SecB can bind long stretches of polypeptides on its remarkably large substrate binding surface (over 7,600 Å2) indeed suggests that subtle changes within this surface could modulate the interaction with specific substrates without affecting the binding to presecretory proteins (16, 28).
Directed evolution of molecular chaperones has been successfully employed to enhance the folding of aggregation-prone proteins involved in neurodegenerative diseases (29). For example, mutations in the middle domain of the yeast Hsp104 disaggregase improve its ability to rescue proteotoxicity in models of Parkinson’s disease and amyotrophic lateral sclerosis (30). The remarkably adjustable substrate selectivity of SecB suggests that this chaperone might be easily genetically manipulated to accommodate new substrates, including aggregation-prone disease proteins or other heterologous proteins for biotechnology purposes.
Materials and Methods
Bacterial Strains and Culture Conditions.
Genetic experiments were performed in E. coli strains W3110 ΔsecB::CmR (21) and MC4100 Δtig::CmR ΔdnaKdnaJ::KanR (31). Bacteria were grown in LB or M9 medium supplemented with ampicillin (100 or 50 μg/mL when specified), chloramphenicol (15 μg/mL), or kanamycin (40 μg/mL) when necessary.
Genetic Selection.
To construct a SecB mutant library, the secB gene was amplified by error-prone PCR, cloned as an EcoRI/HindIII fragment into plasmid p29SEN under an IPTG-inducible promoter, and transformed in E. coli DH5α. Approximately 15,000 transformants were pooled, and plasmids were extracted. Sequencing of random clones from the library indicates that the rate of mutations in secB was two (eight mutations for 467 bp). To select for TA-specific SecB mutants, the W3110 ΔsecB::CmR strain containing pK6-Mtb-HigBA1 was transformed with the p29SEN-based library of SecB mutants and grown at 37 °C on LB ampicillin/kanamycin agar plates supplemented with 0.5% arabinose and 50 μM IPTG to induce expression of the TA system and SecB, respectively. Note that under these conditions, transformation with the plasmid p29-SecB expressing SecB wild type did not produce any viable colony on plates. A control aliquot of transformants plated on LB ampicillin/kanamycin agar plates without inducer indicates that the number of transformants tested during the selection was ∼40,000. Plasmids that confer bacterial growth in the presence of inducer were independently extracted from a total of 120 colonies, retransformed in E. coli DH5α strain, and selected on LB ampicillin agar plates. Kanamycin-sensitive colonies were selected, grown in LB ampicillin, and subjected to plasmid extraction. Plasmids were retransformed in W3110 ΔsecB::CmR cells containing pK6-Mtb-HigBA1 and tested for suppression in the presence of 0.5% arabinose and 50 μM IPTG inducers. The suppressors that passed the second round of selection (n = 116) were sequenced using primers pSE-For (5′-gtgtggaattgtgagcggat-3′) and pSE-Rev (5′-gatttaatctgtatcaggctg-3′).
Bacterial Growth Assays.
E. coli strain W3110 ΔsecB::CmR was cotransformed with pK6-Mtb-HigBA1 and p29SEN, p29-SecB, or p29-SecBTA variant on LB ampicillin/kanamycin agar plates containing 0.2% glucose at 37 °C. Fresh transformants were grown in LB ampicillin/kanamycin to midlog phase, serially diluted, and spotted on LB ampicillin/kanamycin agar plates with or without IPTG and arabinose inducers as indicated. Plates were incubated at 37 °C overnight. For complementation of SecB chaperone activity in vivo, fresh transformants of W3110 ΔsecB::CmR containing p29SEN-based constructs were grown at 37 °C to midlog phase in LB ampicillin, serially diluted, and spotted on LB ampicillin agar plates in the absence or presence of IPTG inducer. Plates were incubated for 1 d at 37 °C or for 5 d at 16 °C. For growth complementation using the MC4100 Δtig::CmR ΔdnaKdnaJ::KanR temperature-sensitive strain (32), midlog-phase cultures of fresh transformants grown at 25 °C in LB ampicillin (50 μg/mL) were serially diluted and spotted on LB ampicillin (50 μg/mL) agar plates at 25 °C and 37 °C with or without IPTG inducer.
Details about plasmid constructs, pull-down assays, pulse-chase analysis, protein purification, DSF, aggregation assays, circular dichroism, chymotrypsin proteolysis, and cell-free protein synthesis are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Charalampos Babis Kalodimos (University of Minnesota) for discussion and Gianluca Cioci and Bertrand Delahaye for technical assistance. The DSF equipment and the spectropolarimeter are part of the Integrated Screening Platform of Toulouse (PICT, IBiSA). This work was supported by the Ministère de l’Education Nationale de la Recherche et de la Technologie and the Fondation pour la Recherche Médicale Grants FDT20140930836 (to A.J.S.), UPS-2014-113 (to S.A.), ANR-13-BSV8-0010-01 (to L.M. and P.G.), and SNF CRSII3_160703 (to P.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710456114/-/DCSupplemental.
References
- 1.Sala A, Bordes P, Genevaux P. Multitasking SecB chaperones in bacteria. Front Microbiol. 2014;5:666. doi: 10.3389/fmicb.2014.00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Randall LL, Hardy SJ. SecB, one small chaperone in the complex milieu of the cell. Cell Mol Life Sci. 2002;59:1617–1623. doi: 10.1007/PL00012488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sala A, Calderon V, Bordes P, Genevaux P. TAC from Mycobacterium tuberculosis: A paradigm for stress-responsive toxin-antitoxin systems controlled by SecB-like chaperones. Cell Stress Chaperones. 2013;18:129–135. doi: 10.1007/s12192-012-0396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 5.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 6.Van Melderen L. Toxin-antitoxin systems: Why so many, what for? Curr Opin Microbiol. 2010;13:781–785. doi: 10.1016/j.mib.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Helaine S, et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soo VW, Wood TK. Antitoxin MqsA represses curli formation through the master biofilm regulator CsgD. Sci Rep. 2013;3:3186. doi: 10.1038/srep03186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bordes P, et al. SecB-like chaperone controls a toxin-antitoxin stress-responsive system in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2011;108:8438–8443. doi: 10.1073/pnas.1101189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sala A, Bordes P, Genevaux P. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) 2014;6:1002–1020. doi: 10.3390/toxins6031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bordes P, et al. Chaperone addiction of toxin-antitoxin systems. Nat Commun. 2016;7:13339. doi: 10.1038/ncomms13339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuessler DL, et al. Induced ectopic expression of HigB toxin in Mycobacterium tuberculosis results in growth inhibition, reduced abundance of a subset of mRNAs and cleavage of tmRNA. Mol Microbiol. 2013;90:195–207. doi: 10.1111/mmi.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekker C, de Kruijff B, Gros P. Crystal structure of SecB from Escherichia coli. J Struct Biol. 2003;144:313–319. doi: 10.1016/j.jsb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Crane JM, et al. Mapping of the docking of SecA onto the chaperone SecB by site-directed spin labeling: Insight into the mechanism of ligand transfer during protein export. J Mol Biol. 2005;353:295–307. doi: 10.1016/j.jmb.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 15.Lilly AA, Crane JM, Randall LL. Export chaperone SecB uses one surface of interaction for diverse unfolded polypeptide ligands. Protein Sci. 2009;18:1860–1868. doi: 10.1002/pro.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C, Rossi P, Saio T, Kalodimos CG. Structural basis for the antifolding activity of a molecular chaperone. Nature. 2016;537:202–206. doi: 10.1038/nature18965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimsey HH, Dagarag MD, Kumamoto CA. Diverse effects of mutation on the activity of the Escherichia coli export chaperone SecB. J Biol Chem. 1995;270:22831–22835. doi: 10.1074/jbc.270.39.22831. [DOI] [PubMed] [Google Scholar]
- 18.Fekkes P, et al. Preprotein transfer to the Escherichia coli translocase requires the co-operative binding of SecB and the signal sequence to SecA. Mol Microbiol. 1998;29:1179–1190. doi: 10.1046/j.1365-2958.1998.00997.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang JD, Herman C, Tipton KA, Gross CA, Weissman JS. Directed evolution of substrate-optimized GroEL/S chaperonins. Cell. 2002;111:1027–1039. doi: 10.1016/s0092-8674(02)01198-4. [DOI] [PubMed] [Google Scholar]
- 20.Aponte RA, Zimmermann S, Reinstein J. Directed evolution of the DnaK chaperone: Mutations in the lid domain result in enhanced chaperone activity. J Mol Biol. 2010;399:154–167. doi: 10.1016/j.jmb.2010.03.060. [DOI] [PubMed] [Google Scholar]
- 21.Ullers RS, Ang D, Schwager F, Georgopoulos C, Genevaux P. Trigger factor can antagonize both SecB and DnaK/DnaJ chaperone functions in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:3101–3106. doi: 10.1073/pnas.0608232104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichols RJ, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ullers RS, et al. SecB is a bona fide generalized chaperone in Escherichia coli. Proc Natl Acad Sci USA. 2004;101:7583–7588. doi: 10.1073/pnas.0402398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahi C, et al. Sequential duplications of an ancient member of the DnaJ-family expanded the functional chaperone network in the eukaryotic cytosol. Mol Biol Evol. 2013;30:985–998. doi: 10.1093/molbev/mst008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hesterkamp T, Bukau B. Role of the DnaK and HscA homologs of Hsp70 chaperones in protein folding in E.coli. EMBO J. 1998;17:4818–4828. doi: 10.1093/emboj/17.16.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoff KG, Ta DT, Tapley TL, Silberg JJ, Vickery LE. Hsc66 substrate specificity is directed toward a discrete region of the iron-sulfur cluster template protein IscU. J Biol Chem. 2002;277:27353–27359. doi: 10.1074/jbc.M202814200. [DOI] [PubMed] [Google Scholar]
- 27.Quan S, et al. Super spy variants implicate flexibility in chaperone action. Elife. 2014;3:e01584. doi: 10.7554/eLife.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crane JM, et al. Sites of interaction of a precursor polypeptide on the export chaperone SecB mapped by site-directed spin labeling. J Mol Biol. 2006;363:63–74. doi: 10.1016/j.jmb.2006.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackrel ME, Shorter J. Protein-remodeling factors as potential therapeutics for neurodegenerative disease. Front Neurosci. 2017;11:99. doi: 10.3389/fnins.2017.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackrel ME, et al. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156:170–182. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruel N, et al. Hsp33 controls elongation factor-Tu stability and allows Escherichia coli growth in the absence of the major DnaK and trigger factor chaperones. J Biol Chem. 2012;287:44435–44446. doi: 10.1074/jbc.M112.418525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genevaux P, et al. In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep. 2004;5:195–200. doi: 10.1038/sj.embor.7400067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakr S, et al. Lon protease quality control of presecretory proteins in Escherichia coli and its dependence on the SecB and DnaJ (Hsp40) chaperones. J Biol Chem. 2010;285:23506–23514. doi: 10.1074/jbc.M110.133058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol. 2001;40:397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- 35.Bordoli L, et al. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.