Significance
Transition state theory (TST) is the most popular theory to calculate the rates of enzymatic reactions. However, in some cases TST could fail due to the violation of the nonrecrossing hypothesis at the transition state. In the present work we show that even for one of the most controversial enzymatic reactions—the hydride transfer catalyzed by dihydrofolate reductase—the error associated to TST represents only a minor correction to the reaction rate. Moreover, this error is actually larger for the reaction in solution than in the enzymatic active site. Based on this finding and on previous studies we propose an “enzymatic shielding” hypothesis which encompasses various aspects of the catalytic process.
Keywords: enzymatic catalysis, transition state theory, dynamic effects, dihydrofolate reductase, transmission coefficient
Abstract
While being one of the most popular reaction rate theories, the applicability of transition state theory to the study of enzymatic reactions has been often challenged. The complex dynamic nature of the protein environment raised the question about the validity of the nonrecrossing hypothesis, a cornerstone in this theory. We present a computational strategy to quantify the error associated to transition state theory from the number of recrossings observed at the equicommittor, which is the best possible dividing surface. Application of a direct multidimensional transition state optimization to the hydride transfer step in human dihydrofolate reductase shows that both the participation of the protein degrees of freedom in the reaction coordinate and the error associated to the nonrecrossing hypothesis are small. Thus, the use of transition state theory, even with simplified reaction coordinates, provides a good theoretical framework for the study of enzymatic catalysis.
The origin of the enormous catalytic power of enzymes has been the subject of intensive research during recent decades. Many successful studies, including both interpretation and predictions, have been carried out in the framework of transition state theory (TST) (1–7). However, because of the complex dynamic nature of the enzymatic environment (8–10), it has been continuously questioned whether statistical rate theories, such as TST, can capture the whole impact of protein motions in the rate of the chemical reaction. The so-called “dynamic effect” hypothesis stresses the role of these motions in the interpretation of several phenomena. Strong temperature dependence of kinetic isotope effects (KIEs) in H-transfer reactions was interpreted as the result of enzymatic promoting vibrations that compress the donor–acceptor distance (11, 12). Correlation between the frequency of conformational changes and reactive events was seen as a signal of the existence of protein motions essential for the reaction (9, 13). The decrease of the catalytic efficiency observed after the introduction of mutation of residues placed far from the active site was explained as a consequence of the perturbation of some essential protein vibrations that expand beyond the active site (14, 15). Simulations were also performed for various enzymes to identify protein vibrational modes that would increase the probability of barrier crossing (16–18), although the methods used for this identification have been the subject of debate (19, 20). According to some models, the role of these promotion motions could be relevant in early and late stages of the reaction and not during barrier crossing (21, 22). In any case, the participation of protein motions in the chemical event could be the origin of practical or fundamental limitations for the application of TST to enzymatic catalysis. However, the dynamic effects hypothesis bears a serious shortcoming: There is no consensus in the field regarding the definition of what these effects are and how to quantify them. As a consequence, all of the interpretations based on these effects remain somewhat speculative (3–5, 23–25).
The term dynamic effect itself can be misleading (24, 26, 27), because TST is in fact a dynamical theory that accounts for the reactive flux, using an equilibrium description. There is nothing in TST saying that the dynamics of the system are unimportant to determine the reaction rate. The basic assumption in TST is that there is a hypersurface in the configurational space—the transition state (TS)—separating reactant and product basins that is never recrossed by trajectories coming from the reactants side. Then, the discussion about the validity of TST in enzymatic catalysis can be recast as a question about the validity of this nonrecrossing hypothesis and quantified by the transmission coefficient (κ), the ratio between the true rate constant and that obtained from the application of TST,
| [1] |
where κ is smaller or equal to unity. When κ = 1, there are no recrossings of the TS and the TST rate is exact. The nonrecrossing assumption also implies that the distribution formed at the TS by the reactive flux is the equilibrium one (28). According to this, the rate constant can be evaluated from the free energy difference between the reactants and the TS.
Quantification of the inherent error of TST is not a straightforward task because the TST rate constant and then the transmission coefficient depend on the choice of the dividing surface. This choice is typically made by selecting a function—a reaction coordinate (RC)—that describes the progress of the reaction and assigning the TS to some value of this function. In other words, the transmission coefficient is not an intrinsic property of the system. If barrier crossing involves changes in other degrees of freedom not included in a putative RC, there will be a friction (mean force) acting against the RC, causing recrossings and thus decreasing κ (24, 29). For example, enzymatic compression along the donor–acceptor distance could assist a transfer reaction (30). As a result, some of those trajectories that cross the TS surface defined along a simplified RC (e.g., the antisymmetric stretch in a transfer reaction) eventually fall back to reactants, violating the nonrecrossing assumption of TST. Then, a better definition of the dividing surface (with higher κ) can be obtained by modifying the initial RC to include protein degrees of freedom. This is the motivation for the variational TST (VTST), the application of the variational principle in the space of RCs to maximize κ (31–34). However, there is no guarantee that an ideal RC, providing a value for κ equal to unity, exists, even if all environmental degrees of freedom are considered in its definition. That would result in a limit for the exactness of TST.
Thus, there are two different issues to be quantified regarding the applicability of TST in the context of enzymatic catalysis: (i) How important is the inclusion of protein degrees of freedom in the definition of the RC? (ii) How important is the error associated to the nonrecrossing hypothesis for the best possible RC (one that includes all of the relevant degrees of freedom)? While the first question addresses a practical problem during the application of TST to realistic systems, the second one refers to a more basic issue in TST: Does a nonrecrossing surface exist for enzymatic reactions? And, if not, does this have important consequences in the evaluation of the rate constant?
To give an unambiguous answer to these two questions one has to look for intrinsic characteristics of the system, independent of arbitrary choices. If for a particular system a nonrecrossing surface exists, it must coincide with the 0.5 isosurface of the committor function pB—the probability for a random trajectory initiated at some point on the dividing surface to end up in the products basin. If all of the trajectories that cross a given surface are reactive, exactly half of the equilibrium flux (the one coming from the reactants side) will end up in products, while another half (the one coming from the products side) will commit to the reactants basin. The 0.5 isocommittor surface (the equicommittor) can be recrossed only either by trajectories that recross the equicommittor an even number of times (reactive recrossings) or by the so-called recrossing pairs: a reactants → reactants and a products → products trajectory pair that share a common configuration (35). If recrossing pairs exist, then the distribution formed by reactive flux at the dividing surface is different from the equilibrium one (36). This nonequilibrium effect is an intrinsic property of the system: There is no surface that can avoid the recrossing pairs if they exist, so they cannot be eliminated by RC optimization and thus the equicommittor provides an upper limit for κ. Therefore, the value of the transmission coefficient at the equicommittor answers the two questions raised in the previous paragraph. The first question, “How important is the inclusion of protein degrees of freedom in the definition of the RC?”, is answered by measuring the importance of these degrees of freedom in the improvement of the transmission coefficient up to the value reached at the equicommittor. The answer to the second one, “How important is the error associated to the nonrecrossing hypothesis for the best possible RC, one that includes all of the relevant degrees of freedom?”, is provided by the deviation of κ from unity at the equicommittor.
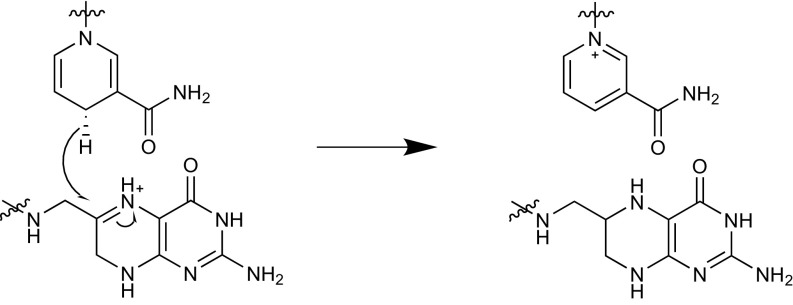
Analytic expressions of the committor are not available for realistic systems and thus cannot be directly used in combination with enhanced sampling methods to characterize the TS and to obtain the transmission coefficient. However, it can be calculated indirectly using a reweighting scheme (37) (Methods), provided that a good-overlapping ensemble can be adequately sampled. Such ensemble can be obtained using the generalized hyperplanar TS (GHTS) optimization technique, recently developed in our group, which provides the dividing surface with highest κ, using a linear combination of a set of collective variables (38). We here applied this methodology to analyze a hydride transfer reaction catalyzed by the human dihydrofolate reductase (hsDHFR). hsDHFR catalyzes the reduction of dihydrofolate to tetrahydrofolate by NADPH (Fig. 1). The κ values obtained for enzymatic hydride transfer reactions using a simple antisymmetric combination of breaking and forming bonds are typically around 0.5 or lower (39, 40), while higher values can be obtained with more collective RCs based in an empirical valence bond description (41). Many geometric degrees of freedom are coupled to the hydride transfer because the reaction involves changes in the aromaticity of the two reacting fragments and then it should not be surprising that a simple antisymmetric RC was not able to provide a good dividing surface. Using transition path sampling techniques on this enzyme, Masterson and Schwartz (18) recently proposed that fast protein motions might be dynamically coupled to the RC, increasing the likelihood of barrier crossing. However, an increase of the mass of hsDHFR by isotopic substitution had no detectable effects on the KIEs for the hydride transfer, which led Kohen and coworkers (42) to the conclusion that protein motions promoting the hydride transfer are hardly affected by the mass change. Therefore, the hydride transfer in hsDHFR is a representative and challenging system for the study of the limits of TST applied to enzymatic catalysis. Our results demonstrate that the intrinsic error associated to TST is small and that the participation of the environment in the definition of the best possible RC is reduced in the catalyzed reaction with respect to the counterpart process in solution.
Fig. 1.
Hydride transfer in DHFR.
Results
We have analyzed the hydride transfer reaction catalyzed by hsDHFR, using the GHTS method (38) to optimize the TS on multidimensional free energy surface described by quantum mechanics/molecular mechanics (QM/MM) Hamiltonian both in solution and in the enzyme (Methods). Three different approaches to the RC were employed (Fig. 2). We first used as RC the antisymmetric combination of hydride–donor and hydride–acceptor distances. We then optimized the RC, using a basis set of 62 collective variables (CVs) that basically includes all of the relevant degrees of freedom of the substrate and the cofactor (distances and hybridization coordinates) (Methods and RC Optimization and Analysis for the Enzymatic Reaction). The resulting RC is called here “the chemical RC” because it is assumed to include all of the degrees of freedom of the chemical system (substrate and cofactor) that might contribute to the RC. Finally, the ensemble generated in this way was used to characterize the properties of the equicommittor surface, using the reweighting procedure (Methods). The equicommittor as a coordinate involves all of the degrees of freedom of the system (chemical subsystem and environment) and provides the best possible RC, minimizing the error associated to TST.
Fig. 2.
Representation of the studied dividing surfaces and the origin of the differences among them. Note that the equicommittor can be recrossed only by recrossing pairs (Top) and reactive recrossings (Bottom).
Transmission Coefficient at the Equicommittor Surface.
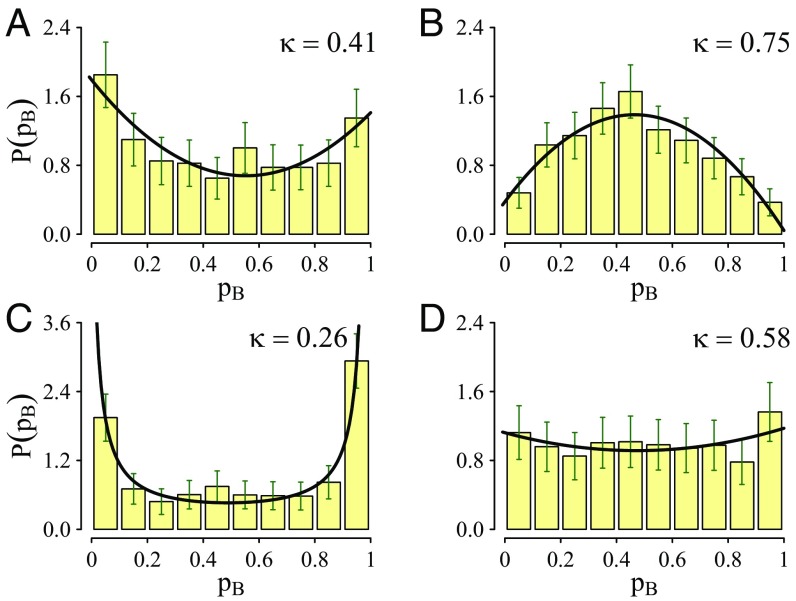
The transmission coefficients and committor histograms obtained for the dividing surfaces defined by the three different RCs (antisymmetric, chemical, and equicommittor) are given in Table 1 and Fig. 3, respectively. The transmission coefficients obtained with the antisymmetric coordinate in solution and in hsDHFR are similar to those obtained in previous works using the same coordinate (40, 43, 44). The committor histograms clearly show that this is not a high-quality coordinate and that an additional free energy barrier associated to other degrees of freedom must be overcome. Then the highest probability for a trajectory initiated at the TS defined with this coordinate is to recross to the reactants or products basins (the histograms peak at 0 and 1). However, the fact that the antisymmetric stretching coordinate alone does not provide a committor histogram peaked at 0.5 does not imply a strong participation of solvent or protein degrees of freedom in the RC.
Table 1.
Transmission coefficients for different dividing surfaces
| System | Antisymmetric RC | Chemical RC | Equicommittor |
| Enzyme | 0.41 (±0.02) | 0.75 (±0.02) | 0.94 (±0.02) |
| Water | 0.26 (±0.02) | 0.58 (±0.03) | 0.88 (±0.03) |
Fig. 3.
The committor histograms for the transfer and chemical dividing surfaces for enzymatic and aqueous solution reactions. (A) Enzyme, antisymmetric RC; (B) enzyme, chemical RC; (C) water, antisymmetric RC; (D) water, chemical RC.
A chemical RC was optimized in a space formed by 62 CVs that includes only degrees of freedom of the substrate and the cofactor. An importance analysis of the different CVs in the definition of the chemical RC is presented in Fig. S3 and discussed in RC Optimization and Analysis for the Enzymatic Reaction. Using the chemical RC, the transmission coefficient increases substantially, becoming close to the value reached at the equicommittor (second and third columns in Table 1), while the committor histogram (Fig. 3B) is clearly peaked at 0.5, stressing the quality of the coordinate. This finding solves the first of the two questions raised in the Introduction: A good RC for the enzymatic process can be defined without including explicitly protein degrees of freedom. A similar conclusion was reached in a theoretical analysis of the reaction in Escherichia coli DHFR that showed that the reaction involves a small group of atoms that are dynamically uncoupled from motions of the protein environment (45). These findings also agree with the results of recent experiments showing that the alteration of the protein mass by isotopic substitution (40, 42, 46) had a very small effect on the hydride transfer step in DHFRs (42).
Interestingly, the committor histogram obtained in aqueous solution using the chemical RC is not peaked at 0.5 and the transmission coefficient is significantly lower than in the enzyme, indicating a larger participation of environmental degrees of freedom for the reaction in solution (see RC Optimization and Analysis for the Reaction in Solution for a discussion of the reaction in solution). This is related to a larger reorganization of the environment coupled to the reaction progress in aqueous solution (47, 48).
The value obtained for κ in the enzyme using the chemical RC (0.75) already reduces significantly the upper limit for possible nonequilibrium contributions in this process. However, one could still argue that these effects could be responsible for a 25% reduction of the reaction rate with respect to the TST result. Since the equicommittor region is well sampled with the chemical RC (Fig. 3), we applied the reweighting procedure (Methods) to characterize the properties of this dividing surface, which provides the best possible RC. For the reweighted TS ensembles formed by ∼500 structures that presented a committor value of 0.50 ± 0.01 the transmission coefficients are 0.94 and 0.88, for the reaction in the enzyme and in aqueous solution, respectively (third column in Table 1). As explained before, these transmission coefficients are an intrinsic characteristic of the system. An important conclusion can be obtained from these values: Undoubtedly some deviation from equilibrium is observed for the best possible RC, but the effect is negligible. A transmission coefficient of 0.94/0.88 translates to a 0.04/0.08 kcal·mol−1 underestimation of the activation free energy at 300 K, which is far below any other error present in state-of-the-art free energy calculations. We can now answer the second question raised in the Introduction. The error associated to the nonrecrossing hypothesis for the best possible TS is very small and is of essentially no importance for a quantitative estimation of the rate constant.
From the values of the transmission coefficient at the equicommittor we can conclude that the error due to recrossings of this hypersurface is actually smaller in the enzyme than in water. It is hard to find a physical interpretation for this fact, since it is not clear what features of the energy landscape are responsible for the appearance of recrossing pairs (35). The usage of the equicommittor surface ensures that there is no mean force acting on the system and then there is no bias for the system to commit to reactants or products due to the lack of some environmental rearrangement (24). Instead, the appearance of recrossing pairs is due to configurational changes that modify the forces acting on the system and thus the velocities along the RC. If the environment does not suffer significant changes at the timescale of the reactive event, as in the case of a preorganized protein active site (22, 46), nonequilibrium effects are reduced. The lower number of recrossing pairs in the enzyme with respect to the reaction in solution indicates that the chemical system is less sensitive to the rest of the environmental degrees of freedom in the former environment. The enzyme works as a protecting “cage,” shielding the impact of the environment on the reaction. This “enzymatic shielding” effect also includes a smaller participation of the environmental degrees of freedom in the RC and a smaller contribution of the environment reorganization to the free energy barrier (47, 48).
The present analysis provides a numerical estimate of nonequilibrium effects, measured as the total TST error due to equicommittor recrossings. From a practical point of view both reactive recrossings and recrossing pairs cause an overestimation of the reaction rate and should be considered together. However, from a more fundamental perspective, it might be more appropriate to consider as nonequilibrium effects only the contribution of those recrossings that result in nonreactive trajectories. Reactive recrossings do not alter the distribution at the TS if one considers the total flux (both positive and negative) through the dividing surface. Nonetheless, they contribute to the total TST error, since TST ignores the negative flux through the TS. In contrast, recrossing pairs do cause a deviation of the TS distribution from the equilibrium one, since the P → P trajectories are excluded from the distribution when only the flux originated from the reactants basin is considered. From this perspective, only the amount of flux forming the recrossing pairs should be considered as nonequilibrium effects.
Since the committor function is not explicitly known, it is not possible to find how many times a given trajectory will cross the equicommittor surface and then it is not possible to distinguish reactive recrossings from reactive trajectories that do not recross the committor. However, the number of times that a trajectory crossed the equicommittor can be approximated by the number of crossings of the isosurface of a good RC that intersects the equicommittor. The total flux through the equicommittor surface can be then trivially split into contributions from trajectories with different numbers of surface crossings (Table 2). The results show that, if only recrossing pairs are considered, nonequilibrium effects account for only 3% and 5% of the total flux at the equicommittor, for the reaction in enzyme and water, respectively.
Table 2.
Fraction of flux formed by trajectories with different number of recrossings at the equicommittor
| System | No recrossings | Reactive recrossings | Recrossing pairs |
| Enzyme | 0.94 (±0.02) | 0.03 (±0.01) | 0.03 (±0.01) |
| Water | 0.88 (±0.03) | 0.08 (±0.02) | 0.05 (±0.01) |
Geometry of the TS.
While in free energy terms the difference observed between the rate constants obtained with a naive and an optimized RC can be small, there is no guarantee that a lower-quality RC provides an adequate geometric description of the TS, which can be essential for the design of TS analogs or biocatalysts or the analysis of KIEs (49). To our knowledge, only one study explicitly addressed this question before for an enzymatic reaction (50). We then used simulations and the reweighting procedure to calculate the distributions of essential geometric properties at the three dividing surfaces analyzed above (Table 3 and Table S1).
Table 3.
Distribution of the donor–acceptor distance for the studied dividing surfaces
| Enzyme | Water | |||
| Transition state | μ | σ | μ | σ |
| Antisymmetric | 2.645 (±0.006) | 0.059 (±0.005) | 2.675 (±0.006) | 0.067 (±0.005) |
| Chemical | 2.652 (±0.005) | 0.062 (±0.004) | 2.663 (±0.006) | 0.064 (±0.004) |
| Equicommittor | 2.654 (±0.012) | 0.058 (±0.005) | 2.702 (±0.037) | 0.066 (±0.008) |
Errors for the mean values and the SDs correspond to 95% confidence intervals. All values are given in angstroms.
The results are strikingly similar for the three TSs. The differences between the mean values in all cases are on the order of 0.01 Å and are barely statistically significant. It was shown above that the transmission coefficient is significantly different for the three TSs. So, it seems that κ is extremely sensitive even to tiny changes in the geometry of the dividing surface. Turning this finding upside down, we reach an important conclusion: The TS geometry has low sensitivity to the quality of the RCs tested. As a result, when one is interested only in the geometry of the TS, a simple coordinate (such as the transfer coordinate) can provide a reasonable result. This finding is in line with the tremendous success of free energy-based computational methods in enzymology: Although most of the studies used simple geometric RCs, the results are generally in very good agreement with experiments (2–4, 6, 7, 51). If the nature of the TS were very sensitive to the choice of RC, that probably would not be the case. However, we stress that this result is obtained by comparing the best possible dividing surface to others with already reasonable quality: For the worst RC analyzed here (the antisymmetric transfer coordinate) the transmission coefficient is 0.41, which in free energy terms means an underestimation of the activation free energy <1 kBT (at room temperature). Probably, in cases where the RC (and thus the dividing surface) deviates strongly from the correct one (κ << 1) this conclusion does not apply and, therefore, should not be abused to make claims about the TS structure without checking the value of κ.
Discussion
The lack of practical approaches to unambiguously quantify the error of TST has been one of the reasons for ongoing debates regarding the importance of this factor, frequently attributed to so-called dynamic effects, in enzymatic catalysis. Here we have shown how a combination of RC optimization and ensemble reweighting can solve this issue. Using this methodological combination, we have been able to address two important concerns often mired in this debate: the environmental participation in the RC and the inherent error associated to the use of statistical rate theories. The last one can be quantified from the transmission coefficient at the equicommittor surface. The results for the hydride transfer catalyzed by hsDHFR clearly show that these two factors have a negligible effect on the evaluation of the enzymatic reaction rate. Therefore, despite existing claims (18), it is hardly credible that such small effects could have practical consequences, for example in the improvement of the catalytic efficiency of enzymes during natural or directed evolution. Moreover, deviation from the equilibrium assumption in the enzyme is smaller than in aqueous solution. We have introduced the “enzymatic shielding” concept that summarizes this and other differences between enzymatic reaction and reaction in solution, resulting in an increased reaction rate: less reorganization of the environment, smaller participation of environmental degrees of freedom during the barrier crossing, and finally reduced nonequilibrium effects. Although further research may be required to generalize these conclusions, some of the results presented here have been already observed in systems involving larger charge redistributions. In particular, the methyl transfer reaction catalyzed by catechol O-methyl transferase (COMT) proceeds from charged reactants (catecholate and S-adenosyl methionine) to neutral products. In that case it was found that the transmission coefficient for the enzymatic TS defined using a simple antisymmetric transfer coordinate was quite high (52). In addition, the quality of the RC, determined from the committor histogram, was not noticeably improved when optimized, including also a collective environmental coordinate. Instead, the quality of the RC was noticeably improved when the same procedure was followed for the counterpart process in solution.
The analysis of the TS geometry reveals an interesting and useful feature: For the studied reaction the antisymmetric RC provides an average TS geometry that is only slightly different from the one obtained with optimized and even ideal (committor function) RCs. While the quality of a RC should always be checked by computing the associated transmission coefficient before extracting any conclusion, it seems that for most practical tasks, such as the calculation of activation free energies or the design of TS mimics, expensive RC optimization is usually not required. However, RC optimization might be necessary for reactions with poorly understood mechanisms, where dividing surfaces with large values of κ are challenging to obtain (53).
The results presented here also offer some guidelines for future research work. First, since TST is able to calculate successfully the rate of enzymatic reactions, the main effort should be focused on the improvement of free energy calculation techniques. This includes sampling efficiency, the quality of Hamiltonians, and the RC definition. Second, our analysis is based on a classical description of nuclear motion and the consequences of a quantum description of some degrees of freedom are unclear. A rigorous theoretical background and practical approaches with affordable computational cost are needed to address this issue, which could be important in reactions with strong tunneling contribution. We hope that the present work will stimulate advances in these directions.
Methods
The simulation system was prepared based on the structure 4M6K (54) from Protein Data Bank (www.rcsb.org/). The enzyme was embedded in a TIP3P (55) water box of size 58 × 55 × 67 Å3. The QM/MM scheme was used with Amber ff12SB (56) forcefield and AM1 (57) semiempirical Hamiltonian with specific AM1 parameters for DHFR catalyzed reaction taken from Doron et al. (58) for the MM and QM regions, respectively (Fig. S1). Langevin dynamics at 300 K were used in all cases with periodic boundary conditions, a time step of 0.5 fs, and particle mesh Ewald (59) to treat long-range electrostatic interactions. The cutoff to limit the direct space sum was set to 8 Å. For the committor test and κ calculation the velocities for each trajectory were sampled from the Maxwell–Boltzmann distribution at 300 K and the dynamics were propagated deterministically both forward and backward in time.
For the reaction in water NADPH and DHF were solvated in a water box of size 36 × 48 × 41 Å3 and were kept close to their orientation in the Michaelis complex by applying weak harmonic restraints (K = 2 kcal⋅mol−1⋅Å−2) to the positions of four heavy atoms, one on the adenine base of NADPH and three on DHF (Fig. S2). Five sodium atoms were added to neutralize the system. The rest of the simulation protocol, including the definition of the QM part and the Hamiltonian, was identical to that of the enzymatic system.
For the GHTS method (38) we used an active space defined by 62 CVs involving only degrees of freedom of the chemical system to localize the optimum dividing surface (chemical RC). We stress that while a large number of CVs were used to optimize the dividing surface, the RC itself is one dimensional and, therefore, provides a valid 3N-1 dimensional TS. Essentially all of the bond distances and hybridization coordinates of the two conjugated systems were considered (Fig. S2). The hybridization coordinate was defined as a point-plane distance from the central atom (carbon or nitrogen) to the plane formed by the three substituents in the sp2 state (60). However, no protein degrees of freedom were included. It was done on purpose—since the active space covers all of the relevant degrees of freedom of the substrate, the difference between the true TS (equicommittor) and the hyperplane should roughly correspond to the degree of participation of environment in the TS crossing. The GHTS optimization protocol and an analysis of the resulting chemical RC are given in RC Optimization and Analysis for the Enzymatic Reaction.
The equilibrium properties (including κ) on the equicommittor surface were calculated using the ensemble reweighting (37):
| [2] |
Here the averaging is done over structures obtained using a harmonic bias that maintains the system close to the hyperplane (“b” subindex) and having pB = 0.5 ± 0.01 at the same time (Dirac’s delta function). The reweighting factor compensates for the biasing potential and recovers the surface integral over pB = 0.5 from one defined using Dirac’s delta function . The latter is approximated as
| [3] |
where is the directional derivative of pB along which was evaluated numerically.
For the reweighting procedure to converge, one needs the two ensembles to be as similar as possible. If not, the reweighting factors become too large, increasing dramatically the statistical error. The optimized chemical RC did not perform so well for the reaction in water; then an extended space of trial coordinates including environmental CVs was used to obtain a better RC with κ = 0.65 (RC Optimization and Analysis for the Reaction in Solution). A total of 20,000 structures were generated for surfaces in protein and water. For each structure, 100 trajectories were integrated forward and backward in time, resulting in 2·106 trajectories for each system. For both systems, among the 20,000 structures sampled ∼500 structures had the value of committor function = 0.5 ± 0.01. The importance analysis was performed as in ref. 38 and details are given in RC Optimization and Analysis for the Enzymatic Reaction.
All of the error bars provided here represent 95% confidence intervals and were calculated by statistical bootstrapping with 10,000 bootstrap samples.
Supplementary Material
Acknowledgments
The authors acknowledge computational facilities of the Servei d’Informàtica de la Universitat de València in the “Tirant” supercomputer. The authors gratefully acknowledge financial support from Ministerio de Economía y Competitividad and Fondo Europeo de Desarrollo Regional funds (Project CTQ2015-66223-C2-2-P). K.Z. acknowledges a Formación Profesorado Universitario fellowship from Ministerio de Educación.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710820114/-/DCSupplemental.
References
- 1.Åqvist J, Warshel A. Simulation of enzyme reactions using valence bond force fields and other hybrid quantum/classical approaches. Chem Rev. 1993;93:2523–2544. [Google Scholar]
- 2.Martí S, et al. Theoretical insights in enzyme catalysis. Chem Soc Rev. 2004;33:98–107. doi: 10.1039/b301875j. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Viloca M, Gao J, Karplus M, Truhlar DG. How enzymes work: Analysis by modern rate theory and computer simulations. Science. 2004;303:186–195. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- 4.Gao J, et al. Mechanisms and free energies of enzymatic reactions. Chem Rev. 2006;106:3188–3209. doi: 10.1021/cr050293k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamerlin SCL, Warshel A. At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis? Proteins. 2010;78:1339–1375. doi: 10.1002/prot.22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glowacki DR, Harvey JN, Mulholland AJ. Taking Ockham’s razor to enzyme dynamics and catalysis. Nat Chem. 2012;4:169–176. doi: 10.1038/nchem.1244. [DOI] [PubMed] [Google Scholar]
- 7.Hammes-Schiffer S. Catalytic efficiency of enzymes: A theoretical analysis. Biochemistry. 2013;52:2012–2020. doi: 10.1021/bi301515j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henzler-Wildman KA, et al. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 9.Henzler-Wildman KA, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 10.Nashine VC, Hammes-Schiffer S, Benkovic SJ. Coupled motions in enzyme catalysis. Curr Opin Chem Biol. 2010;14:644–651. doi: 10.1016/j.cbpa.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Goodey NM, Benkovic SJ, Kohen A. Coordinated effects of distal mutations on environmentally coupled tunneling in dihydrofolate reductase. Proc Natl Acad Sci USA. 2006;103:15753–15758. doi: 10.1073/pnas.0606976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hay S, Scrutton NS. Good vibrations in enzyme-catalysed reactions. Nat Chem. 2012;4:161–168. doi: 10.1038/nchem.1223. [DOI] [PubMed] [Google Scholar]
- 13.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watney JB, Agarwal PK, Hammes-Schiffer S. Effect of mutation on enzyme motion in dihydrofolate reductase. J Am Chem Soc. 2003;125:3745–3750. doi: 10.1021/ja028487u. [DOI] [PubMed] [Google Scholar]
- 15.Benkovic SJ, Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz SD, Schramm VL. Enzymatic transition states and dynamic motion in barrier crossing. Nat Chem Biol. 2009;5:551–558. doi: 10.1038/nchembio.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz SD. Protein dynamics and the enzymatic reaction coordinate. In: Klinman J, Hammes-Schiffer S, editors. Dynamics in Enzyme Catalysis. Springer; Berlin: 2013. pp. 189–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masterson JE, Schwartz SD. Evolution alters the enzymatic reaction coordinate of dihydrofolate reductase. J Phys Chem B. 2015;119:989–996. doi: 10.1021/jp506373q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters B. Comment on “Toward identification of the reaction coordinate directly from the transition state ensemble using the kernel PCA method”. J Phys Chem B. 2011;115:12671–12673. doi: 10.1021/jp206156m. [DOI] [PubMed] [Google Scholar]
- 20.Antoniou D, Schwartz SD. Response to “Comment on ‘Toward identification of the reaction coordinate directly from the transition state ensemble using the kernel PCA method.”. J Phys Chem B. 2011;115:12674–12675. doi: 10.1021/jp207463g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters B. Transition-state theory, dynamics, and narrow time scale separation in the rate-promoting vibrations model of enzyme catalysis. J Chem Theory Comput. 2010;6:1447–1454. doi: 10.1021/ct100051a. [DOI] [PubMed] [Google Scholar]
- 22.García-Meseguer R, Martí S, Ruiz-Pernía JJ, Moliner V, Tuñón I. Studying the role of protein dynamics in an SN2 enzyme reaction using free-energy surfaces and solvent coordinates. Nat Chem. 2013;5:566–571. doi: 10.1038/nchem.1660. [DOI] [PubMed] [Google Scholar]
- 23.Kamerlin SCL, Warshel A. An analysis of all the relevant facts and arguments indicates that enzyme catalysis does not involve large contributions from nuclear tunneling. J Phys Org Chem. 2010;23:677–684. doi: 10.1002/poc.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuñón I, Laage D, Hynes JT. Are there dynamical effects in enzyme catalysis? Some thoughts concerning the enzymatic chemical step. Arch Biochem Biophys. 2015;582:42–55. doi: 10.1016/j.abb.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warshel A, Bora RP. Perspective: Defining and quantifying the role of dynamics in enzyme catalysis. J Chem Phys. 2016;144:180901. doi: 10.1063/1.4947037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandler D. Statistical mechanics of isomerization dynamics in liquids and the transition state approximation. J Chem Phys. 1978;68:2959–2970. [Google Scholar]
- 27.Pu J, Gao J, Truhlar DG. Multidimensional tunneling, recrossing, and the transmission coefficient for enzymatic reactions. Chem Rev. 2006;106:3140–3169. doi: 10.1021/cr050308e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson JB. Statistical theories of chemical reactions. Distributions in the transition region. J Chem Phys. 1973;58:4684–4692. [Google Scholar]
- 29.Ruiz-Pernía JJ, Tuñón I, Moliner V, Hynes JT, Roca M. Dynamic effects on reaction rates in a Michael addition catalyzed by chalcone isomerase. Beyond the frozen environment approach. J Am Chem Soc. 2008;130:7477–7488. doi: 10.1021/ja801156y. [DOI] [PubMed] [Google Scholar]
- 30.Antoniou D, Schwartz SD. Phase space bottlenecks in enzymatic reactions. J Phys Chem B. 2016;120:433–439. doi: 10.1021/acs.jpcb.5b11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wigner E. Calculation of the rate of elementary association reactions. J Chem Phys. 1937;5:720–725. [Google Scholar]
- 32.Juro H. On the statistical mechanical treatment of the absolute rate of chemical reaction. Bull Chem Soc Jpn. 1938;13:210–216. [Google Scholar]
- 33.Truhlar DG, Garrett BC. Variational transition state theory. Annu Rev Phys Chem. 1984;35:159–189. [Google Scholar]
- 34.Vanden-Eijnden E, Tal FA. Transition state theory: Variational formulation, dynamical corrections, and error estimates. J Chem Phys. 2005;123:184103. doi: 10.1063/1.2102898. [DOI] [PubMed] [Google Scholar]
- 35.Mullen RG, Shea J-E, Peters B. Communication: An existence test for dividing surfaces without recrossing. J Chem Phys. 2014;140:041104. doi: 10.1063/1.4862504. [DOI] [PubMed] [Google Scholar]
- 36.Garrett BC, Truhlar DG. Generalized transition state theory. Classical mechanical theory and applications to collinear reactions of hydrogen molecules. J Phys Chem. 1979;83:1052–1079. [Google Scholar]
- 37.Zwanzig RW. High‐temperature equation of state by a perturbation method. I. Nonpolar gases. J Chem Phys. 1954;22:1420–1426. [Google Scholar]
- 38.Zinovjev K, Tuñón I. Transition state ensemble optimization for reactions of arbitrary complexity. J Chem Phys. 2015;143:134111. doi: 10.1063/1.4931596. [DOI] [PubMed] [Google Scholar]
- 39.Roca M, Oliva M, Castillo R, Moliner V, Tuñón I. Do dynamic effects play a significant role in enzymatic catalysis? A theoretical analysis of formate dehydrogenase. Chemistry. 2010;16:11399–11411. doi: 10.1002/chem.201000635. [DOI] [PubMed] [Google Scholar]
- 40.Luk LYP, et al. Unraveling the role of protein dynamics in dihydrofolate reductase catalysis. Proc Natl Acad Sci USA. 2013;110:16344–16349. doi: 10.1073/pnas.1312437110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agarwal PK, Billeter SR, Hammes-Schiffer S. Nuclear quantum effects and enzyme dynamics in dihydrofolate reductase catalysis. J Phys Chem B. 2002;106:3283–3293. [Google Scholar]
- 42.Francis K, Sapienza PJ, Lee AL, Kohen A. The effect of protein mass modulation on human dihydrofolate reductase. Biochemistry. 2016;55:1100–1106. doi: 10.1021/acs.biochem.5b00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Viloca M, Truhlar DG, Gao J. Reaction-path energetics and kinetics of the hydride transfer reaction catalyzed by dihydrofolate reductase. Biochemistry. 2003;42:13558–13575. doi: 10.1021/bi034824f. [DOI] [PubMed] [Google Scholar]
- 44.Luk LYP, et al. Chemical ligation and isotope labeling to locate dynamic effects during catalysis by dihydrofolate reductase. Angew Chem Int Ed Engl. 2015;54:9016–9020. doi: 10.1002/anie.201503968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boekelheide N, Salomón-Ferrer R, Miller TF., 3rd Dynamics and dissipation in enzyme catalysis. Proc Natl Acad Sci USA. 2011;108:16159–16163. doi: 10.1073/pnas.1106397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Antoniou D, Schwartz SD, Schramm VL. Hydride transfer in DHFR by transition path sampling, kinetic isotope effects, and heavy enzyme studies. Biochemistry. 2016;55:157–166. doi: 10.1021/acs.biochem.5b01241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warshel A. Electrostatic origin of the catalytic power of enzymes and the role of preorganized active sites. J Biol Chem. 1998;273:27035–27038. doi: 10.1074/jbc.273.42.27035. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz-Pernia JJ, et al. Increased dynamic effects in a catalytically compromised variant of Escherichia coli dihydrofolate reductase. J Am Chem Soc. 2013;135:18689–18696. doi: 10.1021/ja410519h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knott BC, et al. The mechanism of cellulose hydrolysis by a two-step, retaining cellobiohydrolase elucidated by structural and transition path sampling studies. J Am Chem Soc. 2014;136:321–329. doi: 10.1021/ja410291u. [DOI] [PubMed] [Google Scholar]
- 50.Doron D, Kohen A, Nam K, Major DT. How accurate are transition states from simulations of enzymatic reactions? J Chem Theory Comput. 2014;10:1863–1871. doi: 10.1021/ct5000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Kamp MW, Mulholland AJ. Combined quantum mechanics/molecular mechanics (QM/MM) methods in computational enzymology. Biochemistry. 2013;52:2708–2728. doi: 10.1021/bi400215w. [DOI] [PubMed] [Google Scholar]
- 52.García-Meseguer R, Zinovjev K, Roca M, Ruiz-Pernía JJ, Tuñón I. Linking electrostatic effects and protein motions in enzymatic catalysis. A theoretical analysis of catechol o-methyltransferase. J Phys Chem B. 2015;119:873–882. doi: 10.1021/jp505746x. [DOI] [PubMed] [Google Scholar]
- 53.Mullen RG, Shea J-E, Peters B. Transmission coefficients, committors, and solvent coordinates in ion-pair dissociation. J Chem Theory Comput. 2014;10:659–667. doi: 10.1021/ct4009798. [DOI] [PubMed] [Google Scholar]
- 54.Bhabha G, et al. Divergent evolution of protein conformational dynamics in dihydrofolate reductase. Nat Struct Mol Biol. 2013;20:1243–1249. doi: 10.1038/nsmb.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 56.Duan Y, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 57.Dewar MJS, Zoebisch EG, Healy EF, Stewart JJP. Development and use of quantum mechanical molecular models. 76. AM1: A new general purpose quantum mechanical molecular model. J Am Chem Soc. 1985;107:3902–3909. [Google Scholar]
- 58.Doron D, Major DT, Kohen A, Thiel W, Wu X. Hybrid quantum and classical simulations of the dihydrofolate reductase catalyzed hydride transfer reaction on an accurate semi-empirical potential energy surface. J Chem Theory Comput. 2011;7:3420–3437. doi: 10.1021/ct2004808. [DOI] [PubMed] [Google Scholar]
- 59.Darden T, York D, Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 60.Vardi-Kilshtain A, Major DT, Kohen A, Engel H, Doron D. Hybrid quantum and classical simulations of the formate dehydrogenase catalyzed hydride transfer reaction on an accurate semiempirical potential energy surface. J Chem Theory Comput. 2012;8:4786–4796. doi: 10.1021/ct300628e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.