Significance
Arginine methylation is a common posttranslational modification serving as an epigenetic regulator of gene transcription, pre-mRNA splicing, and PIWI-interacting RNA (piRNA) biogenesis. Methylarginine recognition is mediated by the aromatic cage of the Tudor domain. TDRD2–PIWI interactions are essential for piRNA biogenesis, but the biochemical and structural basis whereby TDRD2 recognizes PIWI proteins is not clear. We used crystallography and biochemical studies to show that TDRD2 binds to PIWI-like protein 1 (PIWIL1) in an arginine methylation-independent manner. Our complex structures revealed a binding mode by which the extended Tudor domain of TDRD2 recognizes PIWIL1 distinct from the canonical Tudor recognition mode utilizing an aromatic cage. Our results provide a paradigm for how Tudor proteins harboring an incomplete aromatic cage bind to PIWI proteins.
Keywords: TDRD2, TDRKH, PIWI, piRNA, methylation-independent
Abstract
The P-element–induced wimpy testis (PIWI)-interacting RNA (piRNA) pathway plays a central role in transposon silencing and genome protection in the animal germline. A family of Tudor domain proteins regulates the piRNA pathway through direct Tudor domain–PIWI interactions. Tudor domains are known to fulfill this function by binding to methylated PIWI proteins in an arginine methylation-dependent manner. Here, we report a mechanism of methylation-independent Tudor domain–PIWI interaction. Unlike most other Tudor domains, the extended Tudor domain of mammalian Tudor domain-containing protein 2 (TDRD2) preferentially recognizes an unmethylated arginine-rich sequence from PIWI-like protein 1 (PIWIL1). Structural studies reveal an unexpected Tudor domain-binding mode for the PIWIL1 sequence in which the interface of Tudor and staphylococcal nuclease domains is primarily responsible for PIWIL1 peptide recognition. Mutations disrupting the TDRD2–PIWIL1 interaction compromise piRNA maturation via 3′-end trimming in vitro. Our work presented here reveals the molecular divergence of the interactions between different Tudor domain proteins and PIWI proteins.
The Tudor domain belongs to the “Royal family” protein domains, which also include PWWP, MBT, Chromo, and plant Agenet domains (1). Tudor domains recognize methylated arginine or lysine residues on target proteins and mediate protein–protein interactions (2–4). Characteristic of the Tudor domain is a barrel-like fold, consisting of four or five antiparallel β-strands, which normally harbors an aromatic cage capable of recognizing methylation marks. Tudor domain proteins can be classified as either methyllysine or methylarginine “readers” (2). Survival motor neuron protein (SMN), Splicing factor 30 (SPF30), Tudor domain-containing protein 3 (TDRD3), and the germline-specific Tudor domain-containing proteins (TDRDs) have been characterized as methylarginine-binders (5–8). Compared with the canonical Tudor domains of SMN and SPF30, the TDRD subfamily of Tudor proteins contains an extended Tudor (eTudor) domain. The eTudor domain contains flanking regions [i.e., a staphylococcal nuclease (SN) domain] extending the canonical Tudor domain, which together contribute to the recognition of methylarginine-containing ligands (9–11). Previous studies also suggest that a complete four-residue aromatic cage is essential for binding of methylarginine marks (8).
P-element–induced wimpy testis (PIWI) proteins are an evolutionarily conserved subclade of Argonaute family proteins that are predominantly expressed in animal germ cells (2, 12). PIWI-interacting RNAs (piRNAs) are typically 24–32 nt in length with essential roles in gametogenesis (12). PIWI proteins associated with piRNAs silence transposable elements and thereby maintain genome integrity. The N terminus of the PIWI protein harbors RG/RA-rich clusters (GAR motif), which are the substrates of the methyltransferase PRMT5 (13, 14). TDRD proteins are found to interact with arginine methylated PIWI proteins and to regulate piRNA biogenesis (2, 5, 11, 15). Mutations of TDRD1, TDRD2, TDRD4, TDRD9, or TDRD12 in mice have been shown to cause defects in piRNA production and spermatogenesis (16–19).
TDRD2 (TDRKH), a protein containing Tudor and K homology (KH) domains with enriched expression in the testis, is essential for spermatogenesis and male fertility. TDRD2-null mice are sterile and show meiotic arrest at the zygotene stage (17). Previous studies show that TDRD2 interacts directly with PIWI proteins [PIWI-like protein 1 (PIWIL1), PIWIL2, and PIWIL4) and is an important component of the piRNA pathway (5, 14, 17). Mutation of TDRD2 leads to the reduction of mature piRNAs and the accumulation of piRNA intermediates, indicating that TDRD2 has an essential role in piRNA precursor trimming (17). Most recently, the 3′–5′ exonuclease PNLDC1 (called “Trimmer” in silkworms) was identified as a pre-piRNA trimming enzyme (20) whose activity is dependent on BmPAPI (the TDRD2 ortholog in silkworms). BmPAPI assists pre-piRNA trimming via recruitment of pre-piRNA-loaded SIWI (silkworm PIWI) to Trimmer (20, 21). Overexpression of BmPAPI, but not of Trimmer, accelerates the trimming reaction, indicating that BmPAPI is a limiting factor for piRNA maturation in silkworm cells.
Here we systematically characterized the binding ability of the eTudor domains of various human TDRD members with the GAR motif peptide of PIWIL1. Intriguingly, we found that TDRD2 preferentially recognizes the unmethylated peptide with high affinity. We then determined both the apo structure of the extended Tudor domain of TDRD2 and its complex with a GAR peptide of PIWIL1. Structural analysis demonstrates that TDRD2 binds to the unmethylated PIWIL1 through multivalent interactions involving both the Tudor–SN interface and the incomplete aromatic cage. However, the TDRD2–PIWIL1 interaction is primarily mediated by the Tudor–SN interface, which is also in line with our piRNA 3′-end trimming results.
Results
Arginine Methylation-Independent TDRD2–PIWIL1 Interaction.
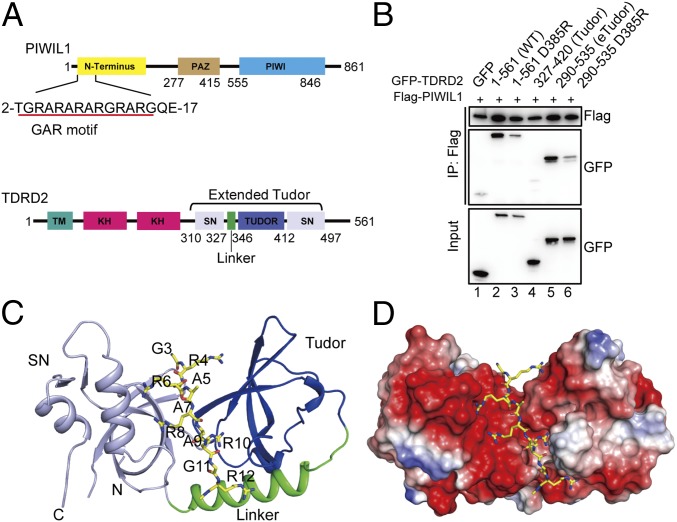
To characterize the binding affinity and specificity of the eTudor domains of human TDRD members, we cloned and expressed the eTudor domains of all human TDRD members (TDRD1–TDRD12) in Escherichia coli (Fig. S1) and were able to obtain soluble and well-behaved proteins for most of these eTudor domains (Table S1). We then measured the binding affinities of these eTudor domains against a well-characterized PIWIL1 peptide in different methylation states (Fig. 1A) (5, 11). As expected, the results showed that eTudor domains harboring a four-residue aromatic cage display selectivity toward the dimethylated arginine (Rme2) peptide over the monomethylated arginine (Rme1) and unmodified arginine (Rme0) peptides (Fig. S1 and Table S1). However, all eTudor domains with an incomplete aromatic cage (one to four substitutions of the aromatic cage residues) except TDRD2 exhibited weak or no binding for the PIWIL1 peptides. Surprisingly, unlike the binding preference of other reported eTudor domains, TDRD2 showed a binding preference for unmethylated over methylated peptides (Table S1). The binding affinity of TDRD2 to the unmethylated GAR peptide of PIWIL1 (R4me0) was 0.2 μM, 20-fold stronger than to the R4me2s peptide. Our isothermal titration calorimetry (ITC) data further confirmed that TDRD2 preferentially recognizes the unmethylated GAR motif (Fig. S2A). To test whether an intact eTudor domain of TDRD2 is required for the binding of PIWIL1, we performed ITC binding experiments using truncated eTudor proteins and the PIWIL1 peptide and found that the canonical Tudor domain of TDRD2 does not show any interaction with the PIWIL1 peptide (Fig. S2B). We further confirmed the requirement of the intact eTudor domain in PIWIL1 binding by coimmunoprecipitation experiments in HEK293T cells. We found that the eTudor domain but not the canonical Tudor domain efficiently coprecipitates with FLAG-tagged PIWIL1 (Fig. 1B, lanes 4 and 5). Taken together, these results demonstrate that the intact eTudor domain of TDRD2 is essential for binding to PIWIL1, with a preference for unmethylated peptide.
Fig. 1.
The eTudor domain of TDRD2 preferentially recognizes unmethylated PIWIL1. (A, Upper) Domain organization of PIWIL1. (Lower) Domain organization of TDRD2. PAZ, PIWI/Argonaute/Zwille; TM, transmembrane domain. (B) The coimmunoprecipitation assay of PIWIL1 with TDRD2. HEK293T cells were cotransfected with FLAG-PIWIL1 and different GFP-TDRD2 constructs. The complex was analyzed by Western blot with anti-GFP and anti-FLAG antibodies. (C) Overall structure of the eTudor domain of TDRD2 in complex with the PIWIL1 peptide. The color scheme is the same as in A. (D) Electrostatic surface representation of the PIWIL1-binding cleft of TDRD2. PIWIL1 is shown in a stick model.
Crystal Structure of the TDRD2 eTudor Domain.
To understand the preferential recognition of unmethylated PIWIL1 by TDRD2, we solved the crystal structure of the TDRD2 eTudor domain (Table S2). There are two TDRD2 molecules in each asymmetric unit of our crystal structure, although TDRD2 behaves as a monomer in solution (Fig. S3). The overall structure of the TDRD2 eTudor domain resembles other available eTudor domain structures, including those of Drosophila Tud11, murine TDRD1, and human SND1 (rmsd <2.04 Å) (9–11). The eTudor domain of TDRD2 consists of a canonical Tudor domain and a SN domain, which are linked by a long α-helix (Fig. S4A). The canonical Tudor domain adopts a four-stranded β-barrel topology (β3–β6), packing against a short α-helix (α2) (Fig. S4A). The SN domain, which is split by the intervening canonical Tudor domain in sequence, folds into a single domain of five antiparallel β-strands (β1, β2, β7, β9, and β10) surrounded by two α-helices (α3 and α4) and two β-strands (β8 and β11). The Tudor domain interacts with the SN domain through β-strands (β1, β2 and β5, β6) (Fig. S4A).
Crystal Structure of the TDRD2–PIWIL1 Complex.
We next determined the crystal structure of the TDRD2 eTudor domain in complex with an unmethylated PIWIL1 peptide. Extensive attempts to cocrystallize TDRD2 with different peptides were not successful, so we generated a fusion construct in which the N-terminal PIWIL1 peptide is fused to TDRD2 using a linker to increase the local peptide concentration and the stability of the TDRD2–PIWIL1 complex during crystallization. We were able to crystallize the fusion construct, and the structure was determined to a resolution of 1.9 Å (Table S2).
The complex structure is similar to the apo structure with an rmsd value of 0.85 Å. Inspection of electron density maps revealed positive mFo-DFc peaks, suggesting the presence of a peptide. The electron density allowed us to build the residues G3–R12 of PIWIL1 into the model (Fig. 1C and Fig. S4B). Structural comparisons with other eTudor–PIWIL1 complex structures revealed that the unmodified PIWIL1 peptide bound to TDRD2 in a similar orientation as the PIWIL1-R4me2s peptide bound to SND1 (Fig. S5) (13).
In the complex structure, the PIWIL1 peptide displays an extended conformation (Fig. 1C). The binding cleft is predominantly negatively charged, in accordance with the positively charged nature of the ligand, indicating that the electrostatic interactions between TDRD2 and PIWIL1 play a major role in binding (Fig. 1D). Consistent with this observation, TDRD2 displayed a nanomolar affinity (20 nM) for the PIWIL1 peptide at a lower salt concentration (25 mM NaCl) but suffered a 700-fold reduction in binding affinity (∼14 μM) at a higher salt concentration (500 mM NaCl) (Fig. S2B).
Recognition Mechanism of PIWIL1 by TDRD2.
The available eTudor-ligand structures demonstrate that the binding of eTudor domains to methylated GAR peptides shares similar structural features and that the aromatic cages of these eTudor domains play a key role in binding (9–11). Surprisingly, the PIWIL1 peptide bound to TDRD2 in a distinct manner: The N terminus of the PIWIL1 peptide (G3–R8) is located in a unique groove created by the Tudor and SN domains, and the remainder of the PIWIL1 peptide (A9–R12) lies in the aromatic cage region (Fig. 2A).
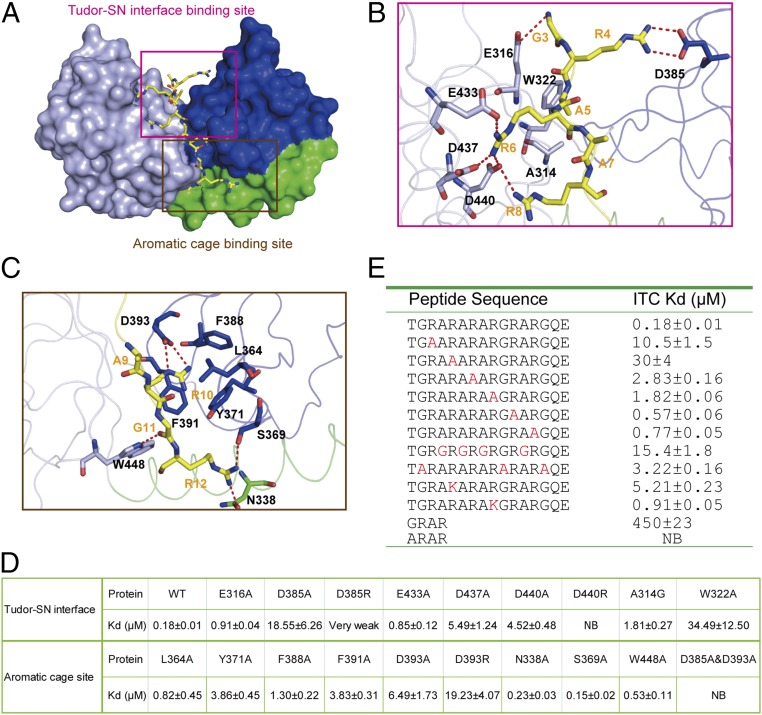
Fig. 2.
Recognition mechanism of PIWIL1 by TDRD2. (A) Recognition of the PIWIL1 peptide by TDRD2 via multivalent interactions (Tudor–SN interface binding site and aromatic cage binding site). The bound PIWIL1 peptide is shown in yellow. (B) Close-up view of the TDRD2–PIWIL1 interactions involved in the Tudor–SN interface. (C) Close-up view of the TDRD2–PIWIL1 interactions involved in the aromatic cage. (D) Quantification of the binding affinity between the unmethylated PIWIL1 peptide (amino acids 290–535) and indicated WT and mutant TDRD2 by ITC. (E) Dissociation constants of TDRD2 with WT and mutant PIWIL1 peptides by ITC.
Residues G3–R8 of the PIWIL1 peptide are sandwiched between the Tudor and SN domains. Specifically, G3 forms a hydrogen bond with the side chain of Glu316 of TDRD2 (Fig. 2B). The guanidinium moiety of R4 forms a salt bridge with Asp385 of TDRD2. A5 is buried in a hydrophobic pocket formed by Ala314 and Trp322 of TDRD2. R6 and R8 form a network of salt bridges with Glu433, Asp437, and Asp440 of TDRD2. Therefore, residues from both the Tudor and SN domains contribute to the recognition of PIWIL1, explaining why the extended regions of the eTudor domain are required for binding of PIWIL1.
The C-terminal portion of the PIWIL1 peptide (A9–R12) is anchored in the aromatic cage of TDRD2 through salt bridges between R10 of PIWIL1 and Asp393, as well as the cation–π interactions between R10 of PIWIL1 and Tyr371, Phe388, and Phe391 of TDRD2 (Fig. 2C). Additionally, the hydrogen-bonding interactions (G11–Trp448, R12–Asn338, and R12–Ser369) also contribute to the interaction between PIWIL1 and TDRD2. Taken together, the arginine-mediated salt bridge interactions between the GAR motif of PIWIL1 and TDRD2 play a significant role in the TDRD2–PIWI interactions.
The Tudor–SN Interface of TDRD2 Is Primarily Responsible for Binding of PIWIL1.
To confirm the validity of the interactions observed from the complex structure, we introduced point mutations into TDRD2 for binding studies (Fig. 2D). Substitution of the G3-interacting Glu316 with alanine reduced the binding affinity by fivefold. Mutating the R4-interacting Asp385 to alanine diminished the binding affinity by 100-fold. Mutating the A5-interacting Ala314 to glycine or Trp322 to alanine diminished the binding affinity by 10- and 190-fold, respectively. Substitution of R6-interacting residues (Glu433, Asp437, and Asp440) with alanine led to fourfold to 30-fold loss in binding. Likewise, mutating the R10-interacting residues (Asp393, Tyr371, Phe388, Phe391) to alanine resulted in a sevenfold to 36-fold reduction in binding affinity. However, substitution of G11- or R12-interating residues (Asn338, Ser369, and Trp448) with alanine had very little or no impact on binding. Taken together, the interaction between TDRD2 and PIWIL1 is more significantly compromised by the Tudor–SN interface mutations than by the aromatic cage mutations. On the other hand, the double mutant D385A/D393A exhibited totally abolished binding, suggesting that both binding sites are important for the TDRD2–PIWIL1 interaction (Fig. 2D).
In addition, mutating any of the residues (Asp385, Asp393, or Asp440) to arginine, which form salt bridges with the PIWIL1 arginine residues, significantly reduced or disrupted the TDRD2 binding to the PIWIL1 peptides (Fig. 2D). Substitution to arginine would reverse the charge of the negatively charged binding groove of TDRD2 and cause electrostatic repulsion with the PIWIL1 peptide, reinforcing the crucial role of the electrostatic interactions in the recognition of PIWIL1.
To explore the importance of the interacting residues within the PIWIL1 peptide, we synthesized a series of PIWIL1-derived mutant peptides and measured their binding affinity to TDRD2 by ITC (Fig. 2E). Consistent with the TDRD2 mutation data, replacement of arginine in the Tudor–SN interface (R4, R6, or R8) with alanine significantly weakened the binding. Substitution of R10 with ananine also reduced the binding affinity by 10-fold. Mutating these arginine residues would disrupt its salt-bridge interactions with TDRD2. Consistently, methylation of these arginine residues would also disrupt its salt-bridge interactions with TDRD2, in agreement with our findings that arginine methylation of the GAR peptide reduces its binding to TDRD2 (Fig. S2A). However, replacement of arginine with lysine was better tolerated than its replacement with alanine (Fig. 2E), further confirming that electrostatic contacts mediate the interaction between PIWIL1 and TDRD2 and also explaining why mutation of the potential methylarginine sites to lysine on SIWI does not fully abolish the BmPAPI–SIWI binding (20–22).
The methyl group of A5 of PIWIL1 fits tightly into a narrow hydrophobic pocket formed by Ala314 and Trp322 (Fig. 2B). Therefore, residues bulkier than alanine would not be favored here. On the other hand, glycine substitution of A5 of PIWIL1, with the potential to lose the hydrophobic interaction, showed substantially diminished binding affinity for TDRD2 (Fig. 2E). Introduction of bulky residues at G3 and G11 would also weaken the interaction due to steric constraints or changes in protein stability. Replacement of these glycines with alanines (G3A/G11A/G15A) resulted in a 16-fold reduction of affinity (Fig. 2E).
TDRD2 Specifically Binds to a GRAR Consensus Sequence.
To generalize the binding motif of TDRD2, a random dodecapeptide phage-displayed library (2 × 1010 unique members) was used to perform panning against TDRD2. The resultant phage pools were analyzed by sequencing, which yielded 20 unique phage clones (Fig. S6A). Interestingly, the 20 unique peptides share a conserved GRAR motif (Fig. S6B), which is in agreement with the N-terminal portion of the PIWIL1 sequence involved in TDRD2 binding in our complex structure and further confirms the importance of the PIWIL1–Tudor–SN interface interaction. The two top-ranked peptides were synthesized for ITC binding studies, which showed binding affinities comparable to those of the PIWIL1 peptide we used in this study (Fig. S6C). Moreover, the selected phage clones showed significant cross-reactivity toward eTudor but not toward the single Tudor domain (Fig. S6D), further demonstrating that intact eTudor domain is required for ligand binding.
Mechanistic Similarity and Diversity of Ligand Recognition Between Mammalian TDRD2 and Silkworm BmPAPI.
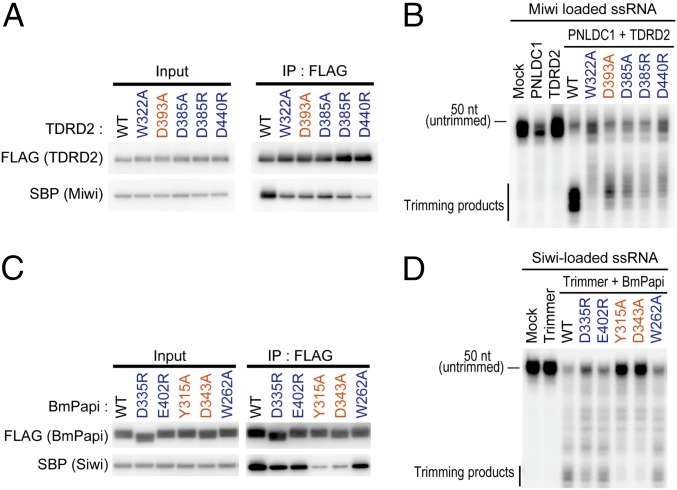
Our structural and in vitro binding assay revealed that the Tudor–SN interface of TDRD2 primarily contributes to the arginine methylation-independent interaction with PIWIL1. To validate this TDRD2–PIWIL1 interaction mode in cells, we generated full-length mouse TDRD2 mutants harboring a mutation in the Tudor–SN interface or the aromatic cage. We expressed these TDRD2 mutants together with mouse PIWIL1 (MIWI) in HEK293T cells and examined their interactions. In line with the in vitro binding assay (Fig. 2D), all the mutants exhibited decreased interaction with MIWI (Fig. 3A). Among these, D440R, a mutation in one of salt-bridging residues, significantly reduced the TDRD2–PIWIL1 interaction (Fig. 3A), verifying the importance of the electrostatic interactions in the PIWIL1 binding.
Fig. 3.
Recognition diversity in arginine methylation between mammalian TDRD2 and BmPAPI. (A) The coimmunoprecipitation assay of MIWI with WT and mutant TDRD2. The residues in the Tudor–SN interface are shown in blue, and the residue in the aromatic cage is shown in orange. (B) In vitro trimming assay for MIWI-loaded ssRNAs. HEK293T whole-cell lysate coexpressing mouse PNLDC1 and WT or mutant mouse TDRD2 was used as trimming lysate. (C) The coimmunoprecipitation assay of SIWI with WT and mutant BmPAPI. The residues in the Tudor–SN interface are shown in blue, and the residues in the aromatic cage are shown in orange. (D) In vitro trimming assay for SIWI-loaded ssRNAs. S2 cells were cotransfected with Trimmer (silkworm PNLDC1) and WT or mutant BmPAPI, and the 1,000 × g pellet fraction was used as trimming lysate.
In piRNA biogenesis, TDRD2 plays a critical role in the pre-piRNA trimming to generate mature piRNAs with optimal lengths (17, 20, 21). To evaluate the functional importance of the arginine methylation-independent interactions between TDRD2 and PIWIL1, we performed a trimming assay using the above-described TDRD2 mutants. To this end, we coexpressed the TDRD2 mutants and mouse PNLDC1 in HEK293T cells in which endogenous TRDR2 was depleted by RNAi. The cell lysate was incubated with MIWI-loaded ssRNAs for the trimming assay (20). Compared with wild-type TDRD2, all the TDRD2 mutations attenuated pre-piRNA trimming activity (Fig. 3B). Consistent with our ITC data and structural analysis, the Tudor–SN interface mutants (W322A, D385A/R, D440R) showed more severe impairment in pre-piRNA trimming than the aromatic cage mutant (D393A) (Fig. 3B), suggesting that the Tudor–SN interface is more important for the recognition of PIWI proteins and efficient piRNA 3′-end trimming and maturation.
To determine if the TDRD2 recognition mode is conserved in other species, we next analyzed the interaction of BmPAPI (silkworm TDRD2) with SIWI (a silkworm PIWI protein). Previous studies reported that the BmPAPI recognizes the GAR motif of SIWI and facilitates pre-piRNA trimming by bridging SIWI and Trimmer (silkworm PNLDC1) (20, 21). We performed ITC using the eTudor domain of BmPAPI and a GAR motif-containing SIWI peptide. Unexpectedly, BmPAPI had a modest preference for the Rme2s peptide over the Rme0 peptide (Fig. S7B). Sequence alignment of the TDRD2 proteins from different species revealed the evolutionary conservation of the eTudor domains and some species-specific differences between human TDRD2 and BmPAPI (Fig. S7C). The major difference is observed in the aromatic cage, where BmPAPI harbors a complete four-residue aromatic cage, suggesting that BmPAPI is an Rme2s binder. In agreement with this notion, the aromatic cage mutants of BmPAPI dramatically decreased the SIWI binding (Fig. 3C). To confirm the above observations, we examined the ability of the BmPAPI mutants in the Tudor–SN interface and the aromatic cage to promote pre-piRNA trimming. While the aromatic cage mutations (Y315A and D343A) significantly impaired piRNA trimming, the Tudor–SN mutations (D335R, E402R and W262A) had less impact (Fig. 3D). Thus, unlike mammalian TDRD2, the aromatic cage of BmPAPI is crucial for SIWI recognition and piRNA trimming. Nevertheless, mutation of the Tudor–SN interface also modestly impeded SIWI binding and piRNA trimming (Fig. 3 C and D), suggesting that the Tudor–SN interface of BmPAPI plays a role in piRNA biogenesis as well. These results are consistent with the notion that arginine methylation is not a prerequisite for the recognition of SIWI by BmPAPI and piRNA trimming (20–22) and suggest that Rme2s would fine-tune the SIWI binding and piRNA trimming.
Molecular Basis of the Recognition of the Unmethylated PIWIL1 Peptide by the TDRD2 Aromatic Cage.
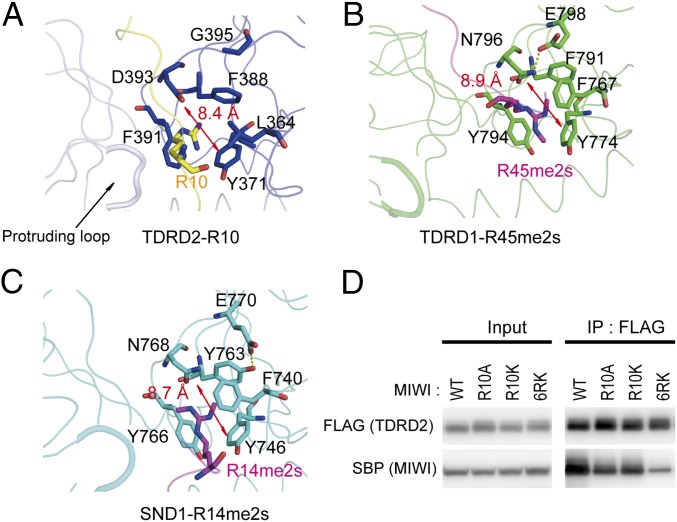
In the complex structure, R10 of PIWIL1 is inserted into the incomplete aromatic cage of TDRD2 formed by Tyr371, Phe388, and Phe391 of TDRD2. This interaction is further strengthened by a salt bridge between R10 of PIWIL1 and Asp393 of TDRD2 (Figs. 2C and 4A). ITC results showed that TDRD2 is involved in the arginine methylation-independent interaction. (Fig. S7A). Consistently, the TDRD2-R10me0 complex is slightly more stable, with a melting temperature of 65.9 °C compared with 65.1 °C for the TDRD2–R10me2s complex (Fig. S8A). This contrasts with the strong preference of TDRD1 and SND1 for GAR motif peptides bearing Rme2s modifications (10, 11). Structural comparisons reveal that the aromatic cage of TDRD2 is smaller (8.4 Å) than those of TDRD1 and SND1 (8.9 Å and 8.7 Å, respectively) (Fig. 4 A–C); thus steric hindrance within the aromatic cage of TDRD2 might disfavor methylarginine binding. To validate the role of the R10me2s in the TDRD2–PIWIL1 interaction, we constructed the R10A, R10K, and 6RK (replacement of all of the six arginine residues with lysine) mutants of PIWIL1 and tested their binding with TDRD2 in HEK293T cells by coimmunoprecipitation. R10A and R10K only slightly weakened the interaction, and even 6RK retained a detectable binding ability to TDRD2 (Fig. 4D). Thus, arginine methylation is not a prerequisite for the PIWIL1 recognition by TDRD2, whereas electrostatic interactions between TDRD2 and PIWIL1 play an important role in the PIWIL1–TDRD2 interaction.
Fig. 4.
Unmethylated arginine state-specific recognition by the incomplete aromatic cage of TDRD2. (A) The arginine-binding pocket in the TDRD2–PIWIL1 complex. The bound R10 residue is colored yellow, and the residues of the aromatic cage are colored blue. The protruding loop (light blue) and G395 of TDRD2 are shown in stick representations. (B) The arginine-binding pocket in the murine TDRD1–R45me2s complex [Protein Data Bank (PDB) ID code: 4B9W]. The loop of TDRD1 that is equivalent to the protruding loop of TDRD2 is shown as a thick line. (C) The arginine-binding pocket in the SND1–R14me2s complex (PDB ID code: 3OMG). The loop of SND1 that is equivalent to the protruding loop of TDRD2 is shown as a thick line. (D) Coimmunoprecipitation assays of TDRD2 with WT and mutant MIWI.
It has been demonstrated previously that the intact four-aromatic-residue cage of the eTudor domain is essential for its interaction with Rme2-containing peptides (8). Because TDRD2 has an incomplete aromatic cage due to Leu364 (Fig. S1B), we mutated Leu364 to phenylalanine, but we did not observe the expected change in binding preference (Table S3). Similarly, the L2058F mutation (aromatic cage residue) of Drosophila Tud9 does not restore methylarginine binding (23). These results imply that additional structural differences also contribute to the methylarginine mark recognition.
First, the SN domain of TDRD2 bears a protruding loop spanning amino acids His444–Pro450 and pointing toward the aromatic cage (Fig. 4A), suggesting that this loop might be involved in modulating the aromatic cage. In contrast, the corresponding loops in TDRD1 and SND1 are shorter and project away from the aromatic cage (Fig. 4 B and C). Second, a glutamate residue is positioned at the entrance of the aromatic cage in both TDRD1 (Glu798) and SND1 (Glu770) (Fig. 4 B and C) and forms hydrogen bonds with the aromatic cage, thus stabilizing the aromatic cage. However, the corresponding position in TDRD2 is Gly395, which does not interact with the aromatic cage, potentially resulting in a more unstable aromatic cage. Therefore, the preferential recognition of unmethylated ligand by TDRD2 may be attributed to all three of these structural features. Consistent with the structural analysis, a triple mutant involving these three structural features (L364F, deletion of the protruding loop, and G394E) was found to convert the ligand-binding preference from Rme0 to Rme2s (Table S3), together indicating that completion of a four-residue aromatic cage, a shortened loop, and a cage-stabilizing residue are determinants of selectivity for methylarginine recognition.
For BmPAPI, both the complete aromatic cage and the stabilizing glutamate residue (corresponding to Gly395 in human TDRD2) are present (Fig. S7C). Although the protruding loop is conserved, there are two insertions flanking the protruding loop of BmPAPI (Fig. S7C). These methylarginine-binding–specific structural features of BmPAPI may confer its preferential binding for methylarginine. However, the Tudor–SN interface also contributes to SIWI binding and piRNA trimming (Fig. 3 C and D).
Discussion
TDRD2 physically associates with PIWI proteins and plays a central role in piRNA biogenesis, spermatogenesis, and male fertility (5, 17, 20, 21). However, the molecular mechanism by which TDRD2 interacts and recruits PIWI proteins for piRNA biogenesis is poorly understood. In this study, we characterized the TDRD2–PIWIL1 interactions and verified the importance of the interactions for piRNA 3′-end trimming and maturation. The aromatic cage of TDRD2 appears to be less important than the Tudor–SN interface, as mutations of the aromatic cage had only minor effects on the PIWIL1–TDRD2 interaction (Fig. 2D). Although human TDRD2 shares sequence homology with BmPAPI, the binding studies unexpectedly reveal that they have different binding preferences against methylated arginine. A triple mutation could convert human TDRD2 into a BmPAPI-like binder of Rme2s (Table S3). However, even this TDRD2 mutant displayed weak selectivity for Rme2s. Thus, it is likely that additional structural differences contribute to the recognition of methylarginine PIWIL1.
Significance of Arginine Methylation-Independent TDRD2–PIWIL1 Binding for piRNA Trimming.
Although a dozen TDRD proteins with eTudor domains have been identified in humans, only TDRD2 exhibits nanomolar affinity for the GAR motif of PIWIL1 regardless of the presence or absence of methylation marks (Table S1). The affinity of other TDRD proteins for PIWIL1 peptides is at least one order of magnitude lower. Therefore, the high-affinity interactions may provide TDRD2 with advantages in competing effectively with other TDRD members for PIWI protein binding. Although the R10me2s was better tolerated than R4me2s, TDRD2 and even its triple mutant with an intact aromatic cage showed no detectable binding to a single Rme2s residue by ITC (Fig. S8B). In stark contrast, SND1 and other Rme2s-binding Tudor proteins can bind to the single Rme2s (8), further suggesting that TDRD2 is not an Rme2s binder. Notably, mutating the aromatic cage residue Tyr371 to aspartate increased the binding by approximately threefold, but mutating the aromatic cage residue Phe388 to aspartate had no effect on the binding affinity compared with the WT (Fig. S8C), strongly suggesting that the aromatic cage is not a prerequisite for PIWIL1 binding. TDRD2/BmPAPI plays conserved roles in piRNA maturation by facilitating the recruitment of PNLDC1/Trimmer to the pre-piRNA-loaded PIWI protein for 3′-end trimming. The interaction is mediated by the binding of the eTudor domain of TDRD2/BmPAPI to the arginine-rich N terminus of PIWIL1/SIWI. Our binding and structural results reveal that both human TDRD2 and silkworm BmPAPI are able to bind to the GAR motif of PIWI proteins with high binding affinity, while methylation of the arginine acts to fine-tune the binding affinity of the TDRD2-PIWIL1 (or BmPAPI-SIWI) interaction. It is conceivable that, to ensure the faithfulness of the important TDRD2–PIWIL1 interaction for piRNA trimming, constitutive protein–protein interaction is required. Therefore, a methylation-independent eTudor-binding mode should be favored to allow the constitutive TDRD2–PIWIL1 interaction. Here we show evolutionarily conserved arginine methylation-independent constitutive binding of TDRD2/BmPAPI to PIWIL1/SIWI, further highlighting the molecular basis for faithful trimming of piRNAs loaded on PIWI proteins.
A Common Recognition Mode for Unmethylated Arginine That Involves the Tudor–SN Interface.
The negatively charged surface of the Tudor–SN interface in TDRD2 is primarily responsible for binding PIWIL1 (Fig. 1D). It has previously been noted that, in addition to TDRD2, other eTudor proteins containing an incomplete aromatic cage are also involved in binding of PIWI proteins (2, 5, 14). We wondered whether these eTudor proteins could bind to PIWI proteins in a methylation-independent manner. Indeed, the first eTudor domain of TDRD7 (TDRD7-1) exhibited a higher binding affinity for Rme0-containing than for Rme2s-containing PIWIL1 peptide (Fig. S9A and Table S1). Furthermore, the key residues of TDRD2 essential for PIWIL1 binding are conserved in TDRD7-1 (Fig. S9B), because E596R mutation of TDRD7-1, which is equivalent to D440R mutation of TDRD2, nearly abolished the binding of PIWIL1 (Fig. S9A). Thus, it is likely that the Tudor–SN interface recognition mode is conserved among at least a subgroup of eTudor domain proteins. Notably, the interface of BAH–PHD or Ankyrin–Chromo has also been reported to recognize unmodified ligands (24, 25). For germline-specific TDRD family proteins, methylation-independent and methylation-dependent eTudor–PIWI interactions may act in a cooperative temporal–spatial fashion and together facilitate the formation of the piRNA machinery and optimal biogenesis of piRNAs.
Materials and Methods
All proteins used in this study were produced using a previously established method (11). Crystals were obtained by the sitting-drop vapor-diffusion method at 18 °C. Protein expression, purification, crystallization, structure determination, and biochemical assays are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Wolfram Tempel and Aiping Dong for their help with structure determination; Dr. Atsushi Miyawaki for providing the CS-RfA-EVBsd shRNA expression vector; Guillermo Senisterra for help with the thermo-shift assay; and the staff at beamline 08ID of the Canadian Light Source. The Structural Genomics Consortium is a registered charity (no. 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through the Ontario Genomics Institute, Innovative Medicines Initiative (European Union/European Federation of Pharmaceutical Industries and Associations) Unrestricted Leveraging of Targets for Research Advancement and Drug Discovery Grant 115766, Janssen, Merck & Co., Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation, Takeda, and the Wellcome Trust. This work was in part supported by Ministry of Education, Culture, Sports, Science and Technology KAKENHI Grant JP26113007 (to Y.T.), Japan Society for the Promotion of Science KAKENHI Grant JP17K17673 (to N.I.), and NIH Grant R01HD084494 (to C.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID codes 5J39 and 6B57). The TDRD2 low-resolution crystal structure used for molecular replacement has been deposited in Zenodo (https://doi.org/10.5281/zenodo.1021859).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711486114/-/DCSupplemental.
References
- 1.Maurer-Stroh S, et al. The Tudor domain ‘royal family’: Tudor, plant agenet, chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 2.Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol. 2011;12:629–642. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- 3.Lu R, Wang GG. Tudor: A versatile family of histone methylation ‘readers’. Trends Biochem Sci. 2013;38:546–555. doi: 10.1016/j.tibs.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel DJ, Wang Z. Readout of epigenetic modifications. Annu Rev Biochem. 2013;82:81–118. doi: 10.1146/annurev-biochem-072711-165700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, et al. Mouse PIWI interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci USA. 2009;106:20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Côté J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 7.Liu K, et al. Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PLoS One. 2012;7:e30375. doi: 10.1371/journal.pone.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tripsianes K, et al. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol. 2011;18:1414–1420. doi: 10.1038/nsmb.2185. [DOI] [PubMed] [Google Scholar]
- 9.Liu H, et al. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 2010;24:1876–1881. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathioudakis N, et al. The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse PIWI proteins and piRNA biogenesis factors. RNA. 2012;18:2056–2072. doi: 10.1261/rna.034181.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu K, et al. Structural basis for recognition of arginine methylated PIWI proteins by the extended Tudor domain. Proc Natl Acad Sci USA. 2010;107:18398–18403. doi: 10.1073/pnas.1013106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ku H-Y, Lin H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl Sci Rev. 2014;1:205–218. doi: 10.1093/nsr/nwu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010;24:636–646. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vagin VV, et al. Proteomic analysis of murine PIWI proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawkins SM, Buchold GM, Matzuk MM. Minireview: The roles of small RNA pathways in reproductive medicine. Mol Endocrinol. 2011;25:1257–1279. doi: 10.1210/me.2011-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handler D, et al. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011;30:3977–3993. doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxe JP, Chen M, Zhao H, Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32:1869–1885. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reuter M, et al. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 19.Wasik KA, et al. RNF17 blocks promiscuous activity of PIWI proteins in mouse testes. Genes Dev. 2015;29:1403–1415. doi: 10.1101/gad.265215.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izumi N, et al. Identification and functional analysis of the pre-piRNA 3′ trimmer in silkworms. Cell. 2016;164:962–973. doi: 10.1016/j.cell.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda S, et al. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA. 2013;19:1405–1418. doi: 10.1261/rna.040428.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiol J, et al. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell. 2012;47:970–979. doi: 10.1016/j.molcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 23.Ren R, et al. Structure and domain organization of Drosophila Tudor. Cell Res. 2014;24:1146–1149. doi: 10.1038/cr.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, et al. Structural basis for the unique multivalent readout of unmodified H3 tail by Arabidopsis ORC1b BAH-PHD cassette. Structure. 2016;24:486–494. doi: 10.1016/j.str.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holdermann I, et al. Chromodomains read the arginine code of post-translational targeting. Nat Struct Mol Biol. 2012;19:260–263. doi: 10.1038/nsmb.2196. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bricogne G, et al. 2016. BUSTER (Global Phasing Ltd., Cambridge, UK), Version X.Y.Z.
- 30.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H, Sidhu SS. Studying binding specificities of peptide recognition modules by high-throughput phage display selections. Methods Mol Biol. 2011;781:87–97. doi: 10.1007/978-1-61779-276-2_6. [DOI] [PubMed] [Google Scholar]
- 36.Zhong N, et al. Optimizing production of antigens and Fabs in the context of generating recombinant antibodies to human proteins. PLoS One. 2015;10:e0139695. doi: 10.1371/journal.pone.0139695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.