Significance
Cells cooperate to sense the direction of a chemical gradient by communicating with each other, which may be important when clumps of cancer cells metastasize or embryos develop. However, because each cell is distinct, we find these clumps are biased toward cells that are “loud”—sending inappropriately large signals. Cell clusters can reduce this bias by rearranging themselves so loud cells change their locations. This means the mechanical dynamics of the cluster matter—fluid, squishy clumps of cells are better at sensing than solid ones. If a single cell’s motion gets noisier, it will make the cluster more fluid—so adding noise can actually make a cluster of cells a better sensor.
Keywords: fundamental bounds, collective motion, chemotaxis, cell-to-cell variability, rheology
Abstract
When a single cell senses a chemical gradient and chemotaxes, stochastic receptor–ligand binding can be a fundamental limit to the cell’s accuracy. For clusters of cells responding to gradients, however, there is a critical difference: Even genetically identical cells have differing responses to chemical signals. With theory and simulation, we show collective chemotaxis is limited by cell-to-cell variation in signaling. We find that when different cells cooperate, the resulting bias can be much larger than the effects of ligand–receptor binding. Specifically, when a strongly responding cell is at one end of a cell cluster, cluster motion is biased toward that cell. These errors are mitigated if clusters average measurements over times long enough for cells to rearrange. In consequence, fluid clusters are better able to sense gradients: We derive a link between cluster accuracy, cell-to-cell variation, and the cluster rheology. Because of this connection, increasing the noisiness of individual cell motion can actually increase the collective accuracy of a cluster by improving fluidity.
Many cells follow signal gradients to survive or perform their functions, including white blood cells finding a wound, cells crossing a developing embryo, and cancerous cells migrating from tumors. Chemotaxis, sensing and responding to chemical gradients, is crucial in all of these examples (1, 2). Chemotaxis is traditionally studied by exposing single cells to gradients—but cells often travel in groups, not singly (3, 4). Collective cell migration is essential to development and metastasis (5) and can have remarkable effects on chemotaxis. Even when single cells cannot sense a gradient, a cluster of cells may cooperate to sense it. While collective chemotaxis is our primary focus, this “emergent” gradient sensing is found in response to many signals, including soluble chemical gradients (chemotaxis) (6–8), conditioned substrates (haptotaxis) (9), substrate stiffness gradients (durotaxis) (10), and electrical potential (galvanotaxis) (11, 12).
Cells can cooperate to sense gradients—but the physical principles limiting a cluster’s sensing accuracy are not settled. For single cells, the fundamental bounds on sensing chemical concentrations and gradients are well studied (13–23), showing unavoidable stochasticity in receptor–ligand binding limits chemotactic accuracy. Is this true for cell clusters? Is a cell cluster simply equivalent to a larger cell? The answer is no! There is an essential difference between many clustered cells and a single large cell: Even clonal populations of cells can have highly variable responses to signals, due to many factors, including intrinsic variations in regulatory protein concentrations (24–26). These cell-to-cell variations (CCVs) can be persistent over timescales much larger than the typical motility timescale of the cell (26). CCV has not been addressed in models of collective chemotaxis and it is not clear whether collective gradient sensing is limited by CCV or by stochastic receptor–ligand binding (7, 8, 27–33).
Using a combination of analytics and simulations, we show that unless CCV is tightly controlled, collective guidance of a cluster of cells is limited by these variations: Gradient sensing is biased toward cells with intrinsically strong responses. This bias swamps the effects of stochastic ligand–receptor binding. Cell clusters may reduce this error by time averaging their gradient measurements only if the cells rearrange their positions, creating an unavoidable link between the mechanics of the cell cluster and its gradient-sensing ability. As a result, surprising new tradeoffs arise: Clusters must balance using motility to follow a biased signal with using motility to reduce error and compromise between reducing noise and increasing cluster fluidity.
Gradient-Sensing Error Is Dominated by Cell-to-Cell Variation, Not Receptor Noise
We study a 2D model of gradient sensing with CCV and ligand–receptor dynamics where cells sense a chemoattractant with concentration gradient . Each cell at position measures local concentration, , via ligand–receptor binding, which is stochastic. This noise leads to unavoidable errors in the cluster’s estimate of . In addition, even if concentration is perfectly sensed, each cell responds differently to a fixed , which models known CCV in signal response (34, 35). As a result, when the cluster combines measurements from its cells, it may develop a drift in the direction of stronger-responding cells (Fig. 1). To combine these effects, we specify the “measured” signal in cell , , which is what the cluster believes the chemoattractant signal in cell to be, including ligand–receptor binding and CCV,
| [1] |
where is the mean concentration over the cluster, , are uncorrelated Gaussian noises with zero mean and unit variance, and are uncorrelated Gaussian noises with zero mean and variance . Stochastic fluctuations in ligand–receptor binding are taken into account in the term , where . This is the error in concentration sensing from a single snapshot of receptors with simple ligand–receptor kinetics and dissociation constant (ref. 14 and Review of Concentration Sensing Accuracy). Eq. 1 assumes that cell–cell variance additively corrupts the measurement of the concentration after an adaptation to the overall level of signal across the cluster . This is natural if the primary cell-to-cell variation is downstream of adaptation, as found to be a reasonable model in ref. 34. We expect similar results for CCV upstream of adaptation in certain limits (Cell-to-Cell Variation Upstream of Adaptation). We have also assumed that the CCV is uncorrelated between different cells; this is a useful initial model describing large variations in protein levels that remain localized within each cell. Extensions of the model could potentially address correlations arising from, e.g., extracellular vesicle transport or cell division, where daughter cells’ may be correlated.
Fig. 1.
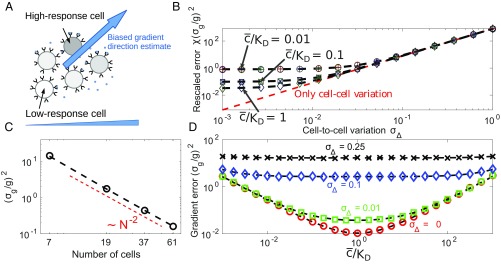
Cell-to-cell variation creates systematic biases that can be significantly larger than the effects of receptor–ligand binding. (A) Schematic of how cell-to-cell variation can create bias in gradient sensing toward high-signaling and away from low-signaling cells. (B) Gradient-sensing error , derived from numerical maximum likelihood (symbols), is well approximated by Eq. 2 (dashed lines) at low gradient strengths. Symbols are plotted for four cluster sizes: and cells (hexagonally packed clusters of unit spacing with layers, illustrated in Computation of χ for Cells in Hexagonally Packed Cluster). In A–D, we use and , in units where the cell–cell spacing is 1. (C) Gradient-sensing error decreases as cluster size increases as . In C, and . (D) Strong CCV can mask concentration dependence of accuracy. In absence of CCV, gradient-sensing accuracy is maximized when ; this effect is screened when CCV dominates gradient sensing. D is shown for cells.
To determine gradient-sensing accuracy, we perform maximum-likelihood estimation (MLE) of in Eq. 1, as in past approaches for single-cell gradient sensing (16). We obtain the MLE numerically (Materials and Methods and MLEs of Gradient Direction via Collective Guidance in the Presence of Cell–Cell Variation and Ligand–Receptor Noise) and thus the uncertainty (Fig. 1B, symbols), where is an average over CCV and ligand–receptor binding. For fixed and roughly circular (isotropic) cluster geometry, if the concentration change across the cluster is small, , can be approximated by assuming is constant across the cluster, resulting in
| [2] |
Here, is a shape parameter, and is cell position relative to cluster center of mass. Evaluating this expression reveals that it is an excellent approximation to the numerically obtained uncertainty (dashed lines, Fig. 1B).
The approximate expression for the uncertainty, Eq. 2, allows us to quantify the relative contribution of receptor–ligand fluctuations and CCV to the gradient-sensing error. For background concentrations near the receptor–ligand equilibrium constant and for typical receptor numbers in eukaryotic cells [ (36, 37)], can be smaller than 0.01. Protein concentrations, on the other hand, often vary between cells to 10–60% of their mean (25)—hence we estimate . Thus, we expect CCV to dominate gradient-sensing error and that the error from concentration sensing and receptor binding can be neglected completely if (Fig. 1B). Eq. 2 also reveals that CCV masks the impact of changing background concentration. When , gradient sensing is limited by ligand–receptor fluctuations and increases as moves away from (Fig. 1D)—accuracy decreases if either few receptors are bound or receptors are saturated. As CCV increases, no longer depends strongly on (Fig. 1D). Finally, Eq. 2 shows that gradient-sensing accuracy depends on the shape parameter and, therefore, on cluster size. For hexagonally packed clusters of cells with unit spacing (we measure in units of the cell diameter; Materials and Methods), a cluster with layers has cells and (Computation of χ for Cells in Hexagonally Packed Cluster); i.e., . Clusters of increasing size then have an error that decreases as (Fig. 1C); this scaling is similar to earlier results for single cells (Computation of χ for Cells in Hexagonally Packed Cluster).
Reducing Estimation Error by Time Averaging
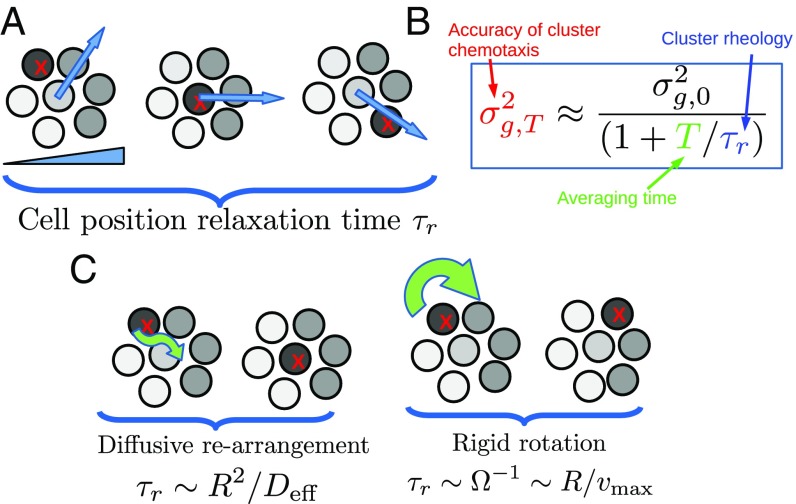
If a cluster made independent measurements, it could reduce by a factor of . In single-cell gradient sensing, independent measurements can be made by averaging over time—improving errors by a factor , where is the averaging time and the measurement correlation time. At first glimpse, time averaging seems unlikely to help with CCV, when correlation times for protein levels can be longer than cell division times, reaching 48 h in human cells (26). However, since gradient-sensing bias from CCV depends on the locations of strong- and weak-signaling cells within the cluster, time averaging can be successful if it is over a time long enough for the cluster to rearrange. This is true even if, as we initially assume, CCV biases are time independent. We expect gradient sensing error with time averaging, , will decrease by a factor of from , where is a correlation time related to cell positions (Fig. 2). Is this true, and how should we define ?
Fig. 2.
Time averaging links fluidity and accuracy. (A) Schematic drawing of how cell–cell rearrangement can change bias due to CCV. Shades of gray indicate measured signal ; a cell with strong response (marked with ) moves through the cluster, leading to biases in gradient estimate (blue arrow). The characteristic relaxation time for this bias is (main text). (B) This leads to a link between the timescale , which is a measure of the cluster’s rheology, and chemotactic accuracy (box). (C) Different rearrangement mechanisms will depend on cluster size in different ways (main text and Characteristic Timescales of Different Rearrangement Mechanisms).
Our earlier results suggest that CCV dominates the gradient-sensing error. Ligand–receptor noise will also be even less relevant in the presence of time averaging, as the receptor relaxation time [seconds to minutes (38)] is much faster than that for cluster rearrangement (tens of minutes or longer). We therefore completely neglect ligand–receptor binding fluctuations, allowing an analytical solution for the MLE (Materials and Methods and MLEs of Gradient Direction via Collective Guidance in the Presence of Cell–Cell Variation and Ligand–Receptor Noise).
How much does time averaging reduce error? If we average the MLE over a time by applying a kernel , i.e., we define and , we can derive (Detailed Derivation of Time-Averaged Gradient-Sensing Error)
| [3] |
where is the normalized cell position–position correlation function, is its Fourier transform, and is the error in the absence of time averaging. To derive Eq. 3, we make two approximations: (i) The cluster has a constant and isotropic shape and (ii) rearrangement of cell positions relative to the center of mass is independent of the particular values of . The first approximation is not necessary, but is a useful simplification; a generalized result is given in Detailed Derivation of Time-Averaged Gradient-Sensing Error. The second approximation assumes that averaging over CCV and averaging over cell positions are independent. This decoupling approximation is necessary to characterize cluster fluidity and mechanics separately from the details of signaling. It excludes, e.g., models where cells with larger-than-average sort out from the cluster. We discuss potential errors due to this approximation later in this paper.
For exponential position–position correlation functions and averaging, and , where is the Heaviside step function, Eq. 3 is simple:
| [4] |
In other words, gradient-sensing accuracy can be improved by taking independent measurements in a time . [The assumption of exponential averaging here is not crucial, and many averaging functions can be used in Eq. 3. We choose this average because it can be computed easily by a combination of accumulation and decay; i.e., .]
Crucial in this reduction is the position–position correlation time which depends on the cluster rearrangement mechanism. Two natural mechanisms are persistent cluster rotation and neighbor rearrangements within the cluster (Fig. 2C). These mechanisms may coexist, as when cells slide past one another during cluster rotation (39). can depend on cluster size; for diffusive rearrangements, we expect , and for persistently rotating clusters, (Characteristic Timescales of Different Rearrangement Mechanisms).
We have assumed that the CCV is time independent over our scale of interest—consistent with the long memory found in ref. 26. If changes faster than the cluster rearranges, our results can be straightforwardly modified. Generalizations of Eqs. 3 and 4 to this case are provided in Detailed Derivation of Time-Averaged Gradient-Sensing Error.
Tradeoffs in Collective Accuracy and Motility: Cluster Rotation
Our central result (Eq. 3) shows that clusters can improve their chemotactic accuracy by changing cell positions. The simplest mechanism to do this is cluster rotation, which occurs in border cell clusters (40) and transiently in leukocyte clusters (7). When should a cluster actively rotate to increase its accuracy? Rotation creates an important tradeoff: More work must be put into rotating and therefore less into crawling up the gradient. [We note that this is most relevant if motility is a large portion of the cluster’s energy budget, a complex question that may be cell-type dependent (41–43).]
For constant work of motility, the maximum speed of a cluster of radius that rotates with angular speed is (Materials and Methods)
| [5] |
where and the maximum cluster speed absent rotation. The cluster cannot rotate faster than .
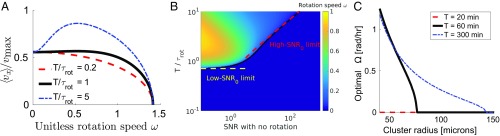
If a cluster follows its best estimate with speed given by Eq. 5, it can improve its velocity in the gradient direction by rotating when the averaging time is long compared with (Fig. 3A). (The cluster’s directionality is always improved by rotating, so there is no tradeoff unless speed of motion matters.) We find that the optimal rotation speed that maximizes the up-gradient speed depends only on the signal-to-noise ratio (SNR) without rotation, and (Fig. 3B and Materials and Methods). Mammalian cells have speeds in the range of micrometers per minute and radii of tens of micrometers, so to benefit from averaging (), must be longer than tens of minutes. The timescale and both depend on cluster size—larger clusters with more cells are both better gradient sensors and more difficult to drive to large angular speeds. As a consequence, the optimal is highly cluster-size dependent: As cluster size decreases, there is a continuous transition to nonzero optimal if is sufficiently long (Fig. 3C).
Fig. 3.
Cluster rotation can improve directed cluster motility. (A) As the averaging time is increased above the characteristic rotational timescale , the mean cluster velocity in the gradient direction is maximized for nonzero rotational speed. in A. is the unitless rotational speed. (B) Rotation improves chemotaxis at long averaging times and low (bad gradient sensing in the absence of rotation). Color map shows the value of that maximizes , found by numerical evaluation; black line shows the contour. (C) Cluster rotation is preferred at small cluster radii. In this graph, is estimated by using , where is the number of cells per unit area in the cluster (Materials and Methods).
Linking Chemotactic Accuracy and Fluidity
Clusters with more cell rearrangement are more accurate by Eq. 4. To further quantify the consequences of cell rearrangements, we model a cluster of cells as self-propelled particles that follow the cluster estimate with a noise characterized by angular diffusion and with cell–cell connections modeled as springs of strength between Delaunay neighbors (Materials and Methods). We emphasize that the angular diffusion parameterized by is an additional source of noise: As increases, cells are less accurate in following the cluster’s estimate of the gradient. These two parameters are systematically varied to study the effects of cluster fluidity on chemotactic accuracy.
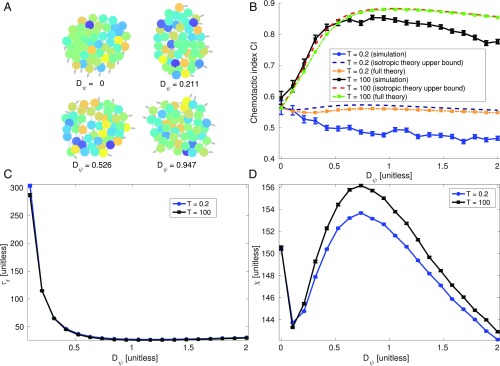
Cluster Fluidity Improves Cluster Chemotaxis.
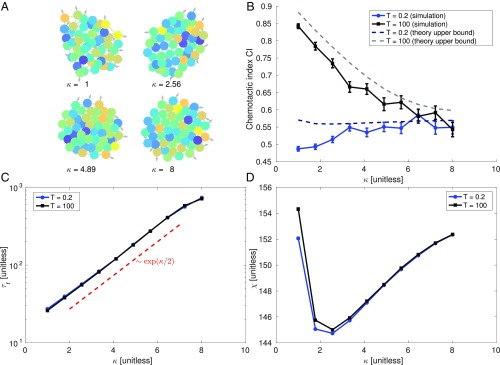
Within our model, increasing cell–cell adhesion makes clusters more ordered, moving between fluid-like and crystalline states (Fig. 4A). As a consequence, rearrangement slows significantly (Fig. 4C) with [ is single exponential].
Fig. 4.
Cluster fluidity increases cluster accuracy. (A) Typical configurations of cell clusters (plotted from simulations with ). Color indicates measured signal , lines connect neighboring cells, and the arrows indicate the polarity . (B) When time averaging is significant, chemotactic index of clusters decreases as cell–cell adhesion stiffness is increased; this behavior is captured by the upper bound computed using Eq. 3. (C) Positional relaxation time increases with stiffness roughly as ; does not strongly depend on averaging time . (D) Cluster size parameter is not strongly dependent on in this range of . All plots are computed by averaging over 600 simulations of cells, each composed of time steps with . , , , , and . The first time units of the simulation are discarded, to allow the system to reach a steady state. Error bars indicate 68% CIs.
Cluster structure and size change when clusters fluidize (Fig. 4A), which may in principle affect the shape parameter , which also strongly affects the chemotactic accuracy (Eq. 2). However, in our simulations is not significantly dependent on , changing by under 10% (Fig. 4D). Averaging time also has only a weak effect on cluster shape and dynamics—changes in and when the averaging time is increased by orders of magnitude are small (Fig. 4). This is consistent with our assumption decoupling the gradient estimate and cell rearrangements, suggesting clusters should obey the bound [3].
We can, using the results in Reducing Estimation Error by Time Averaging, predict the cluster chemotactic index, , where is the cluster velocity. Assuming , we can compute the CI from given by Eq. 4 (Materials and Methods). This requires parameters and (measured from simulations) and , , and (known). We note that our approach, which extracts and from cell trajectories, could also be applied to experimental data; in that case, would still be known, but the extent of time averaging () and the error due to CCV () would have to be determined by fitting to the data. This prediction should be an upper bound to the measured CI, because our model includes additional noise beyond the assumptions of Eq. 4, via . As expected, the cluster CI decreases significantly as clusters solidify and the relaxation time increases. The simulation data qualitatively follow the predicted upper bound (Fig. 4B). When the averaging time is reduced below typical relaxation times, the CI significantly decreases. In addition, for this short time averaging, changing cluster stiffness no longer strongly affects the CI.
Our model describes cell–cell adhesion by controlling the stiffness of springs connecting circular cells. Within this scheme, increasing adhesion reduces cluster fluidity; other models and experiments have suggested adhesion increases jamming and decreases rearrangement (44). However, descriptions of epithelial sheets using models that resolve cell shape, including vertex and cellular Potts models, suggest that increasing adhesion actually promotes a more fluid state (45–48). With this in mind, whether adhesion increases or decreases cluster fluidity may be an open question. We use adhesion in our model as a simple control knob to adjust cluster fluidity; our results in Eq. 4 do not depend on the origin of the tissue’s fluidity, only on its characteristic relaxation time .
Increasing the Stochasticity of Single-Cell Motility Can Increase Cluster Accuracy.
Any mechanism that fluidizes the cluster can decrease the correlation time . Because of this, increasing motility noise can improve cluster chemotactic accuracy (Fig. 5). We increase single-cell angular noise and see an initial sharp increase in cluster CI as (Fig. 5B). At larger values of , cluster CI decreases below the bound set by Eq. 4, as the additional noise added degrades the gradient-following behavior. Without significant time averaging (), additional noise primarily impedes chemotactic accuracy.
Fig. 5.
Finite levels of stochasticity in single-cell motility increase accuracy. (A) Typical configurations of cell clusters (plotted from simulations with ). Color indicates measured signal , lines connect neighboring cells, and the arrows indicate the polarity . (B) When time averaging is significant, the CI of clusters first increases as increasing single-cell noise fluidizes the cluster and then falls below the upper bound computed using Eq. 3. We also plot an extended theory not assuming cluster isotropy, derived in Detailed Derivation of Time-Averaged Gradient-Sensing Error. Simulations for small may slightly exceed the upper bound (main text). (C) Positional relaxation time first decreases and then increases as is increased; the point , which has effectively infinite, is not shown. does not strongly depend on averaging time . (D) Cluster size parameter weakly depends on fluidization by . All plots are computed by averaging over 600 simulations of cells, each composed of time steps with . , , , , and . The first time units of the simulation are discarded, to allow the system to reach a steady state. Error bars indicate 68% CIs.
Why can extra noise help sensing? For , all cells follow the best estimate precisely, leading to an ordered cluster (Fig. 5A) with effectively infinite. As is increased, the cluster fluidizes, and the relaxation time decreases strongly (Fig. 5C), resulting in more independent measurements. As in Fig. 4, this fluidization is relevant only if the averaging time exceeds the relaxation time, so when , the effect of increasing is solely detrimental to chemotaxis.
In deriving our bound, we made two key approximations: cluster isotropy and decoupling. These approximations are exact for the rigid cluster rotation in Tradeoffs in Collective Accuracy and Motility: Cluster Rotation, but only approximate for this self-propelled particle model. As a consequence, at small , simulated clusters have chemotactic indexes slightly exceeding our predictions (Fig. 5B). This error likely arises from emergent couplings between cluster shape and —clusters may spread perpendicular to , weakening the decoupling approximation (Fig. 5A). The approximation of cluster isotropy can be removed (Detailed Derivation of Time-Averaged Gradient-Sensing Error) and does not resolve the violation of the bound (Fig. 5B). Despite this potential error source, the model captures CI variation over a broad range of parameters (Bounds Capture Variation of CI over a Large Range of Parameters).
Discussion
Our study results in several predictions and suggestions for experiments that investigate collective chemotaxis. For example, we predict that, when CCV is large, gradient-sensing error is insensitive to background concentration (Fig. 1C). This is consistent with recent measurements on developing organoids that show that the up-gradient bias is not strongly dependent on mean concentration (8), although in contrast with results on lymphocyte clusters (7). Furthermore, if CCV limits collective chemotaxis, clusters that gradient sense in vivo should have tightly regulated expression of proteins relevant to the signal response. Interestingly, measurements of zebrafish posterior lateral line primordium (49) show tightly controlled Sdf1 signaling, measured by Cxcr4b internalization, suggesting that CCV may be small enough to allow for accurate collective gradient sensing.
We also show that there is a direct link between fluidity and chemotaxis as illustrated by Eq. 3. Verifying this expression in experiments requires simultaneous measurement of several quantities, including cluster size, cluster rearrangement, and signal gradient. Therefore, care has to be taken when modifying experimental conditions as these might change many key quantities simultaneously. Altering adhesion, for example, changes both cluster fluidity and spreading as shown in a recent study using neural crest clusters (50), creating a confounding factor. Nevertheless, these types of experiments may be successful in setting bounds on possible time averaging and the link between fluidity and chemotaxis. In principle, given detailed cell position tracks, our analysis could be performed on experimental data. However, there are significant technical details to be worked out. In comparing theory and simulation, we have full tracks of every cell in the cluster over every time point, allowing simple computations; experimental data, however, may have only transient or partial tracks or show clusters that fragment or merge. Critical in this comparison will be assessing potential sources of bias from missing data or including more complex cluster dynamics.
Our results suggest that many recent experiments may need reinterpretation. Measured chemotactic accuracies can depend on cluster size (6, 7, 31); these results have been modeled without time averaging or CCV (7, 27, 28, 31). Our results show that rearrangement times also influence chemotaxis—and that depends on cluster size. Cluster relaxation dynamics are therefore an unexplored potential issue for interpreting collective gradient-sensing experiments.
Essential in the reduction of gradient-sensing errors due to CCV is the existence of a biochemical or mechanical memory that can perform a time average over tens of minutes. There are several possibilities. First, memory could be external to the cluster—e.g., stored in extracellular matrix structure or a long-lived trail (51). Second, supracellular structures like actin cables influence cell protrusion and leader cell formation (52), suggesting that collective directional memory could be kept by regulating actin cable formation and maintenance. Third, memory may be kept at the individual cell level by cells attempting to estimate their own bias level and compensating for it. This contrasts with our straightforward average of the collective estimate , but could be an important alternative mechanism.
Our model states that the cluster can take its information about signal levels, modulated only by CCV and ligand–receptor noise, and make the optimal computation of the gradient estimator . We do not explicitly identify a biochemical or mechanical mechanism by which the cluster can compute this estimate. However, at least in the limit where receptor noise is negligible and , the estimate depends simply on the cluster geometry. To compute , the cluster would have to be able to estimate only the cell location relative to the cluster center of mass ; this could potentially be done by secreting and sensing a signal. Other possibilities could include generalizing the tug-of-war scheme proposed in ref. 27 so that cells farther away from the cluster center exert larger forces. We also note that, even if cell clusters use a different estimator than the optimal MLE we have described here [e.g., estimators derived from local excitation, global inhibition (LEGI), or tug-of-war models (7, 27, 28, 30)], similar qualitative results will hold: Relaxation of any position-dependent estimator will be faster if the cluster is more fluid.
We have so far addressed gradient sensing in only two dimensions. In higher dimensions, we expect some aspects of our results to be relatively unchanged; for instance, Eq. 2 will change only by a prefactor, and the results of Eq. 3 straightforwardly generalize to three dimensions. However, the scaling laws that determine how the shape factor changes with cluster size and the relaxation times change with cluster size will naturally differ in three dimensions. For instance, we would expect .
Our results are critical for understanding the ubiquitous phenomenon of collective gradient sensing. The importance of CCV provides a valuable design principle: CCV must either be tightly controlled or be mitigated by time averaging. We also established a surprising link between a central mechanical property of a cluster—its rheology—and its sensing ability. This connects mechanical transitions like unjamming (47) to sensing, opening up more areas of study. In addition, our results show cluster accuracy depends strongly on cluster rearrangement mechanism. Finally, our results show that noise in cell motility can be beneficial for collective sensing.
Materials and Methods
MLE of Gradient Direction in the Presence of Cell–Cell Variation and Ligand–Receptor Noise.
We compute the MLE of gradient direction given the measured signal at cell , , given by Eq. 1. If the cluster of cells is in a shallow linear gradient, with concentration at the cluster’s center of mass , then and thus . is then with and ; i.e., .
are uncorrelated between cells, with a Gaussian distribution of zero mean and SD ; i.e., with the Kronecker delta function. As and are both Gaussian, the sum of these variables is also Gaussian, and the likelihood of observing a configuration of measured signals is , where is the probability density function of observing the configuration given parameters . The likelihood is
| [6] |
where is the mean value of and is its variance. We want to apply the method of maximum likelihood by finding the gradient parameters that maximize this likelihood; i.e.,
| [7] |
However, because of the complex dependence of on the gradient , this is not possible analytically. We perform this optimization numerically, using a Nelder–Mead method (Matlab’s fminsearch), with an initial guess set by the maximum for (i.e., neglecting concentration-sensing noise), which can be found exactly. For numerical convenience, we maximize the log-likelihood , up to an additive constant.
In the limit of (neglecting concentration noise), our model becomes a simple linear regression, and the log likelihood can be maximized analytically by finding such that (MLEs of Gradient Direction via Collective Guidance in the Presence of Cell–Cell Variation and Ligand–Receptor Noise). The result is , where . This estimator is simplest in the limit of roughly circular (isotropic) clusters, where . In this case, , where .
Cluster Rotation Dynamics.
How much speed does a cluster lose by rotating? One possibility is to assume the power expended in generating motility is constant. Consider a circular cluster propelling itself over a surface, with the cells having velocity ; we expect that the frictional force per unit area between the cluster and substrate will be , where is a friction coefficient with the substrate. If all of the power available for motility is going into driving the cluster over the substrate, then we can write . If the cluster is traveling as a rigid, circular cluster with its maximum possible velocity, , then , where is the translational drag coefficient of the cluster. If, instead, the cluster puts its entire power into rigid-body rotation with (in polar coordinates), then , where is the rotational drag coefficient of the cluster. In general, the power dissipated if the cluster is moving rigidly with velocity and angular speed is and hence we find that the speed ) that a cluster rotating with angular velocity is able to travel to obtain is
| [8] |
This quantifies one reasonable tradeoff between speed and angular velocity for a cluster. If the power available for cell motility is a small amount of the cell’s energy budget (41–43), other tradeoffs may be more important and additional modeling will be necessary.
We consider a circular cluster traveling toward its best estimate of the gradient with speed given by Eq. 8 and traveling in the direction of the estimator . We can then determine when the cluster maximizes its mean velocity in the direction of the increasing gradient, which we choose to be , , as a function of . This average is
| [9] |
We know from our results above and in Detailed Derivation of Time-Averaged Gradient-Sensing Error that, for a fixed configuration, has a Gaussian distribution with mean and variance given by Eq. 3. The average of depends only on , with (Computing Chemotactic Indexes, below).
Given the angular velocity , we can work out the distribution of by Eq. 3. We know , and hence and its Fourier transform , and thus .
By rescaling to unitless parameters, we then find that
| [10] |
where is the unitless angular velocity, is the usual SNR with no averaging, is the ratio of the averaging time to the characteristic rotational time , and is the function given by Eq. 18. When is sufficiently small and sufficiently large, has a maximum at finite (Fig. 3).
In the limit of low SNR, , and we find is maximized by when and otherwise. For the large SNR limit, and rotation will increase the mean velocity in the direction of the gradient when . More generally, it is possible to find the value of that maximizes numerically. We show the complete phase diagram in Fig. 3B.
Particle-Based Model of Collective Cell Migration.
We use a minimal model of collective cell migration, describing cells as self-propelled particles connected by springs,
| [11] |
| [12] |
| [13] |
| [14] |
where is a Gaussian Langevin noise with zero mean and , with the Kronecker delta. In this model, the orientation of an individual cell is the cluster’s best estimate of the gradient direction, , plus a noise which varies from cell to cell. here controls the persistence of this noise and its amplitude; when is increased, each individual cell is worse at following the estimate . The cell–cell forces are
| [15] |
where and . The forces are only between neighboring cells , where we define neighboring cells as any cells connected by the Delaunay triangulation of the cell centers (Fig. 4A); this approach resembles that of ref. 53. We use the Euler–Maruyama method to integrate Eqs. 11–14.
Simulation Units.
We have chosen our parameters in the simulation and throughout the paper to be measured in units where the equilibrium cell–cell separation (i.e., the cell diameter is unity), and the velocity of a single cell in the absence of cell–cell forces has unit magnitude. For, e.g., neural crest cells, the cell diameters are of order 20 m and the cell speeds on the order of micrometers per minute—so a unitless time of corresponds to roughly in real time. However, cell size and speed vary strongly from cell type to cell type, so we prefer to present these results in their unitless form so that they can be more easily converted.
Computing Chemotactic Indexes.
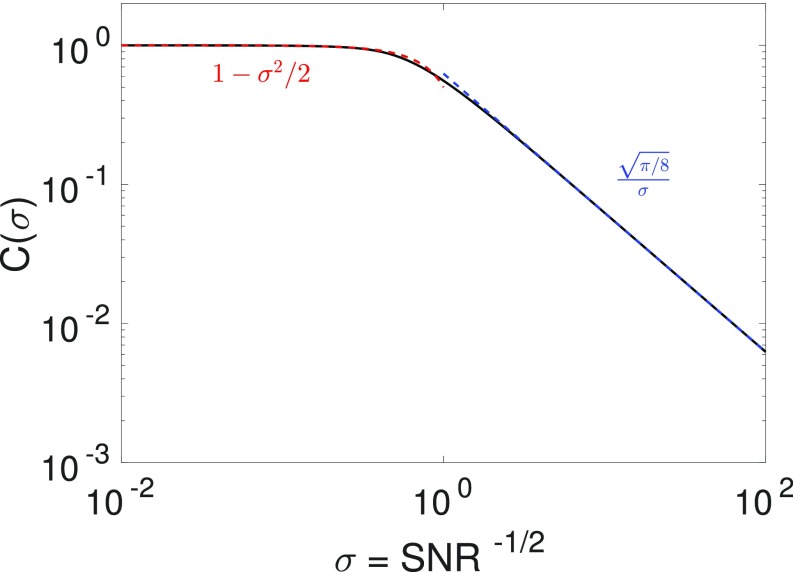
If we use the maximum-likelihood method to make an estimate for the direction in which the cell moves, how do we translate between the uncertainty and the distribution of velocities? We found that the MLE for the gradient is , with a Gaussian random variable with zero mean and variance —and similar results for the time-average . One measure of this estimate’s accuracy is the instantaneous chemotactic index—or the cosine of the angle between the estimate and the gradient direction. To compute this, if without loss of generality, we find
| [16] |
| [17] |
| [18] |
where , with (Fig. 6). These results carry over naturally to the time-averaged case if remains Gaussian—we find .
Fig. 6.
Relationship between instantaneous CI and SNR. is plotted numerically from definition in Eq. 18.
The integral for cannot be solved analytically, but we can find asymptotic forms for or evaluate it numerically. For , we find , and for .
Supplementary Material
Acknowledgments
We thank Albert Bae and Monica Skoge for useful discussions and many scientists from the Gordon Research Conference on Directed Cell Motility for interesting questions and reference suggestions. B.A.C. also thanks Kristen Flowers for several useful suggestions. This work was supported by NIH Grant P01 GM078586.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: MATLAB simulation code for the MLE method, self-propelled particle simulations, and subsequent analysis as well as simulation output sufficient to reproduce the relevant figures is available online via the github repository: https://github.com/bcamley/celltocellreproduce.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712309114/-/DCSupplemental.
References
- 1.Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: A network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine H, Rappel WJ. The physics of eukaryotic chemotaxis. Phys Today. 2013;66:24–30. doi: 10.1063/PT.3.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hakim V, Silberzan P. Collective cell migration: A physics perspective. Rep Prog Phys. 2017;80:076601. doi: 10.1088/1361-6633/aa65ef. [DOI] [PubMed] [Google Scholar]
- 4.Camley BA, Rappel WJ. Physical models of collective cell motility: From cell to tissue. J Phys D Appl Phys. 2017;50:113002. doi: 10.1088/1361-6463/aa56fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 6.Theveneau E, et al. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malet-Engra G, et al. Collective cell motility promotes chemotactic prowess and resistance to chemorepulsion. Curr Biol. 2015;25:242–250. doi: 10.1016/j.cub.2014.11.030. [DOI] [PubMed] [Google Scholar]
- 8.Ellison D, et al. Cell–cell communication enhances the capacity of cell ensembles to sense shallow gradients during morphogenesis. Proc Natl Acad Sci USA. 2016;113:E679–E688. doi: 10.1073/pnas.1516503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winklbauer R, Selchow A, Nagel M, Angres B. Cell interaction and its role in mesoderm cell migration during Xenopus gastrulation. Dev Dyn. 1992;195:290–302. doi: 10.1002/aja.1001950407. [DOI] [PubMed] [Google Scholar]
- 10.Sunyer R, et al. Collective cell durotaxis emerges from long-range intercellular force transmission. Science. 2016;353:1157–1161. doi: 10.1126/science.aaf7119. [DOI] [PubMed] [Google Scholar]
- 11.Li L, et al. E-cadherin plays an essential role in collective directional migration of large epithelial sheets. Cell Mol Life Sci. 2012;69:2779–2789. doi: 10.1007/s00018-012-0951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lalli ML, Asthagiri AR. Collective migration exhibits greater sensitivity but slower dynamics of alignment to applied electric fields. Cell Mol Bioeng. 2015;8:247–257. doi: 10.1007/s12195-015-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg HC, Purcell EM. Physics of chemoreception. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaizu K, et al. The Berg-Purcell limit revisited. Biophys J. 2014;106:976–985. doi: 10.1016/j.bpj.2013.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu B, Chen W, Rappel WJ, Levine H. Physical limits on cellular sensing of spatial gradients. Phys Rev Lett. 2010;105:048104. doi: 10.1103/PhysRevLett.105.048104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu B, Chen W, Rappel W-J, Levine H. How geometry and internal bias affect the accuracy of eukaryotic gradient sensing. Phys Rev E. 2011;83:021917. doi: 10.1103/PhysRevE.83.021917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endres RG, Wingreen NS. Maximum likelihood and the single receptor. Phys Rev Lett. 2009;103:158101. doi: 10.1103/PhysRevLett.103.158101. [DOI] [PubMed] [Google Scholar]
- 18.Endres RG, Wingreen NS. Accuracy of direct gradient sensing by single cells. Proc Natl Acad Sci USA. 2008;105:15749–15754. doi: 10.1073/pnas.0804688105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuller D, et al. External and internal constraints on eukaryotic chemotaxis. Proc Natl Acad Sci USA. 2010;107:9656–9659. doi: 10.1073/pnas.0911178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueda M, Shibata T. Stochastic signal processing and transduction in chemotactic response of eukaryotic cells. Biophys J. 2007;93:11–20. doi: 10.1529/biophysj.106.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews BW, Iglesias PA. An information-theoretic characterization of the optimal gradient sensing response of cells. Plos Comput Biol. 2007;3:e153. doi: 10.1371/journal.pcbi.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bialek W, Setayeshgar S. Physical limits to biochemical signaling. Proc Natl Acad Sci USA. 2005;102:10040–10045. doi: 10.1073/pnas.0504321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segota I, et al. High fidelity information processing in folic acid chemotaxis of dictyostelium amoebae. J R Soc Interface. 2013;10:20130606. doi: 10.1098/rsif.2013.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niepel M, Spencer SL, Sorger PK. Non-genetic cell-to-cell variability and the consequences for pharmacology. Curr Opin Chem Biol. 2009;13:556–561. doi: 10.1016/j.cbpa.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sigal A, et al. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 27.Camley BA, Zimmermann J, Levine H, Rappel W-J. Emergent collective chemotaxis without single-cell gradient sensing. Phys Rev Lett. 2016;116:098101. doi: 10.1103/PhysRevLett.116.098101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camley BA, Zimmermann J, Levine H, Rappel WJ. Collective signal processing in cluster chemotaxis: Roles of adaptation, amplification, and co-attraction in collective guidance. PLoS Comput Biol. 2016;12:e1005008. doi: 10.1371/journal.pcbi.1005008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varennes J, Han B, Mugler A. Collective chemotaxis through noisy multicellular gradient sensing. Biophys J. 2016;111:640–649. doi: 10.1016/j.bpj.2016.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mugler A, Levchenko A, Nemenman I. Limits to the precision of gradient sensing with spatial communication and temporal integration. Proc Natl Acad Sci USA. 2016;113:E689–E695. doi: 10.1073/pnas.1509597112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai D, et al. Modeling and analysis of collective cell migration in an in vivo three-dimensional environment. Proc Natl Acad Sci USA. 2016;113:E2134–E2141. doi: 10.1073/pnas.1522656113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Copenhagen K, et al. 2017. Frustration induced phases in migrating cell clusters. arXiv:1705.00025.
- 33.Varennes J, Fancher S, Han B, Mugler A. 2017. Emergent versus individual-based multicellular chemotaxis. Phys Rev Lett 119:188101. [DOI] [PubMed]
- 34.Wang CJ, Bergmann A, Lin B, Kim K, Levchenko A. Diverse sensitivity thresholds in dynamic signaling responses by social amoebae. Sci Signal. 2012;5:ra17. doi: 10.1126/scisignal.2002449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samadani A, Mettetal J, van Oudenaarden A. Cellular asymmetry and individuality in directional sensing. Proc Natl Acad Sci USA. 2006;103:11549–11554. doi: 10.1073/pnas.0601909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hesselgesser J, et al. Identification and characterization of the CXCR4 chemokine receptor in human T cell lines: Ligand binding, biological activity, and HIV-1 infectivity. J Immunol. 1998;160:877–883. [PubMed] [Google Scholar]
- 37.Macdonald JL, Pike LJ. Heterogeneity in EGF-binding affinities arises from negative cooperativity in an aggregating system. Proc Natl Acad Sci USA. 2008;105:112–117. doi: 10.1073/pnas.0707080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, Rappel WJ, Kerr R, Levine H. Quantifying noise levels of intercellular signals. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;75:061905. doi: 10.1103/PhysRevE.75.061905. [DOI] [PubMed] [Google Scholar]
- 39.Rappel WJ, Nicol A, Sarkissian A, Levine H, Loomis WF. Self-organized vortex state in two-dimensional Dictyostelium dynamics. Phys Rev Lett. 1999;83:1247. [Google Scholar]
- 40.Combedazou A, et al. Myosin II governs collective cell migration behaviour downstream of guidance receptor signalling. J Cell Sci. 2016;130:97–103. doi: 10.1242/jcs.179952. [DOI] [PubMed] [Google Scholar]
- 41.Purcell EM. Life at low Reynolds number. Am J Phys. 1977;45:3–11. [Google Scholar]
- 42.Flamholz A, Phillips R, Milo R. The quantified cell. Mol Biol Cell. 2014;25:3497–3500. doi: 10.1091/mbc.E14-09-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katsu-Kimura Y, Nakaya F, Baba SA, Mogami Y. Substantial energy expenditure for locomotion in ciliates verified by means of simultaneous measurement of oxygen consumption rate and swimming speed. J Exp Biol. 2009;212:1819–1824. doi: 10.1242/jeb.028894. [DOI] [PubMed] [Google Scholar]
- 44.Garcia S, et al. Physics of active jamming during collective cellular motion in a monolayer. Proc Natl Acad Sci USA. 2015;112:15314–15319. doi: 10.1073/pnas.1510973112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiang M, Marenduzzo D. Glass transitions in the cellular Potts model. Europhys Lett. 2016;116:28009. [Google Scholar]
- 46.Bi D, Lopez J, Schwarz J, Manning ML. A density-independent rigidity transition in biological tissues. Nat Phys. 2015;11:1074–1079. [Google Scholar]
- 47.Bi D, Yang X, Marchetti MC, Manning ML. Motility-driven glass and jamming transitions in biological tissues. Phys Rev X. 2016;6:021011. doi: 10.1103/PhysRevX.6.021011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park JA, et al. Unjamming and cell shape in the asthmatic airway epithelium. Nat Mater. 2015;14:1040–1048. doi: 10.1038/nmat4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venkiteswaran G, et al. Generation and dynamics of an endogenous, self-generated signaling gradient across a migrating tissue. Cell. 2013;155:674–687. doi: 10.1016/j.cell.2013.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuriyama S, et al. In vivo collective cell migration requires an lpar2-dependent increase in tissue fluidity. J Cell Biol. 2014;206:113–127. doi: 10.1083/jcb.201402093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim K, et al. Neutrophil trails guide influenza-specific CD8+ T cells in the airways. Science. 2015;349:aaa4352. doi: 10.1126/science.aaa4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reffay M, et al. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat Cell Biol. 2014;16:217–223. doi: 10.1038/ncb2917. [DOI] [PubMed] [Google Scholar]
- 53.Meineke FA, Potten CS, Loeffler M. Cell migration and organization in the intestinal crypt using a lattice-free model. Cell Prolif. 2001;34:253–266. doi: 10.1046/j.0960-7722.2001.00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.