Significance
The majority of influenza vaccine antigens are prepared in chicken eggs. Human vaccine strains grown in eggs often possess adaptive mutations that increase viral attachment to chicken cells. Most of these adaptive mutations are in the hemagglutinin protein, which functions as a viral attachment factor. Here, we identify a hemagglutinin mutation in the current egg-adapted H3N2 vaccine strain that alters antigenicity. We show that ferrets and humans exposed to the current egg-adapted H3N2 vaccine strain produce antibodies that poorly neutralize H3N2 viruses that circulated during the 2016–2017 influenza season. These studies highlight the challenges associated with producing influenza vaccine antigens in eggs, while offering a potential explanation of why there was only moderate vaccine effectiveness during the 2016–2017 influenza season.
Keywords: influenza, hemagglutinin, antibody, vaccine
Abstract
H3N2 viruses continuously acquire mutations in the hemagglutinin (HA) glycoprotein that abrogate binding of human antibodies. During the 2014–2015 influenza season, clade 3C.2a H3N2 viruses possessing a new predicted glycosylation site in antigenic site B of HA emerged, and these viruses remain prevalent today. The 2016–2017 seasonal influenza vaccine was updated to include a clade 3C.2a H3N2 strain; however, the egg-adapted version of this viral strain lacks the new putative glycosylation site. Here, we biochemically demonstrate that the HA antigenic site B of circulating clade 3C.2a viruses is glycosylated. We show that antibodies elicited in ferrets and humans exposed to the egg-adapted 2016–2017 H3N2 vaccine strain poorly neutralize a glycosylated clade 3C.2a H3N2 virus. Importantly, antibodies elicited in ferrets infected with the current circulating H3N2 viral strain (that possesses the glycosylation site) and humans vaccinated with baculovirus-expressed H3 antigens (that possess the glycosylation site motif) were able to efficiently recognize a glycosylated clade 3C.2a H3N2 virus. We propose that differences in glycosylation between H3N2 egg-adapted vaccines and circulating strains likely contributed to reduced vaccine effectiveness during the 2016–2017 influenza season. Furthermore, our data suggest that influenza virus antigens prepared via systems not reliant on egg adaptations are more likely to elicit protective antibody responses that are not affected by glycosylation of antigenic site B of H3N2 HA.
Influenza viruses continuously acquire mutations in exposed epitopes of the hemagglutinin (HA) and neuraminidase (NA) proteins through a process called “antigenic drift” (1). Influenza vaccine effectiveness can be low when there is a mismatch between vaccine strains and circulating strains (2). Viral antigens included in influenza vaccines are routinely updated in an attempt to avoid antigenic mismatches (3, 4). Current seasonal influenza vaccines possess antigens from one H1N1 strain, one H3N2 strain, and one or two influenza B strains.
H3N2 viruses began circulating in humans in 1968. Most neutralizing antibodies (Abs) recognize antigenic sites on the globular head of H3 (designated sites A–E), while rare Abs bind to the more conserved HA stalk or sialic acid binding domains (5). Most H3N2 vaccine mismatches from 1968 to 2013 have been attributed to mutations in antigenic site B of HA (6). None of the site B mutations that emerged during this time period have led to new glycosylation sites on HA (6, 7). This is surprising, given that the addition of glycans on HA can dramatically affect Ab binding (8–13). Instead, new glycosylation sites have repeatedly emerged and fixed in other antigenically important regions of H3 (14).
Vaccine effectiveness was extremely low during the 2014–2015 influenza season (15). During that season, influenza vaccines possessed antigens from a 2012 H3N2 virus that belonged to the 3C.1 HA genetic clade, while the majority of circulating H3N2 strains belonged to the 3C.2a and 3C.3a genetic clades (16). Both the 3C.2a and 3C.3a viruses differed at residues in HA antigenic site B compared with the 2014–2015 H3N2 vaccine strain (17). Notably, 3C.2a viruses that circulated during the 2014–2015 season possessed a new predicted glycosylation site in HA antigenic site B (17), and the 2014–2015 influenza vaccine exhibited especially low effectiveness against this clade (18). In an effort to avoid an antigenic mismatch, the 2016–2017 influenza vaccine was updated to contain antigens from a 3C.2a H3N2 virus isolated in 2014 (19). The majority of H3N2 viruses that circulated in the Northern Hemisphere during the 2016–2017 influenza season were 3C.2a viruses; however, an interim estimate of vaccine effectiveness was only 43% against medically attended H3N2 infections (20). Vaccine effectiveness was especially low in individuals that were 18–49 y old (20). It is unclear why there was variable vaccine effectiveness during the 2016–2017 influenza season, given that the vaccine strain appeared to be well matched to most circulating strains.
The majority of antigens for influenza vaccines are prepared in fertilized chicken eggs, and the 2016–2017 egg-adapted 3C.2a vaccine strain lacks the site B glycosylation site that is present on circulating 3C.2a H3N2 strains (21). Here, we completed a series of studies to determine whether the difference in glycosylation of HA antigenic site B of H3N2 vaccine strains and circulating strains contributed to a previously unrecognized vaccine mismatch during the 2016–2017 influenza season.
Results
Recent H3N2 Viruses Possess a Glycosylation Site in Antigenic Site B of HA.
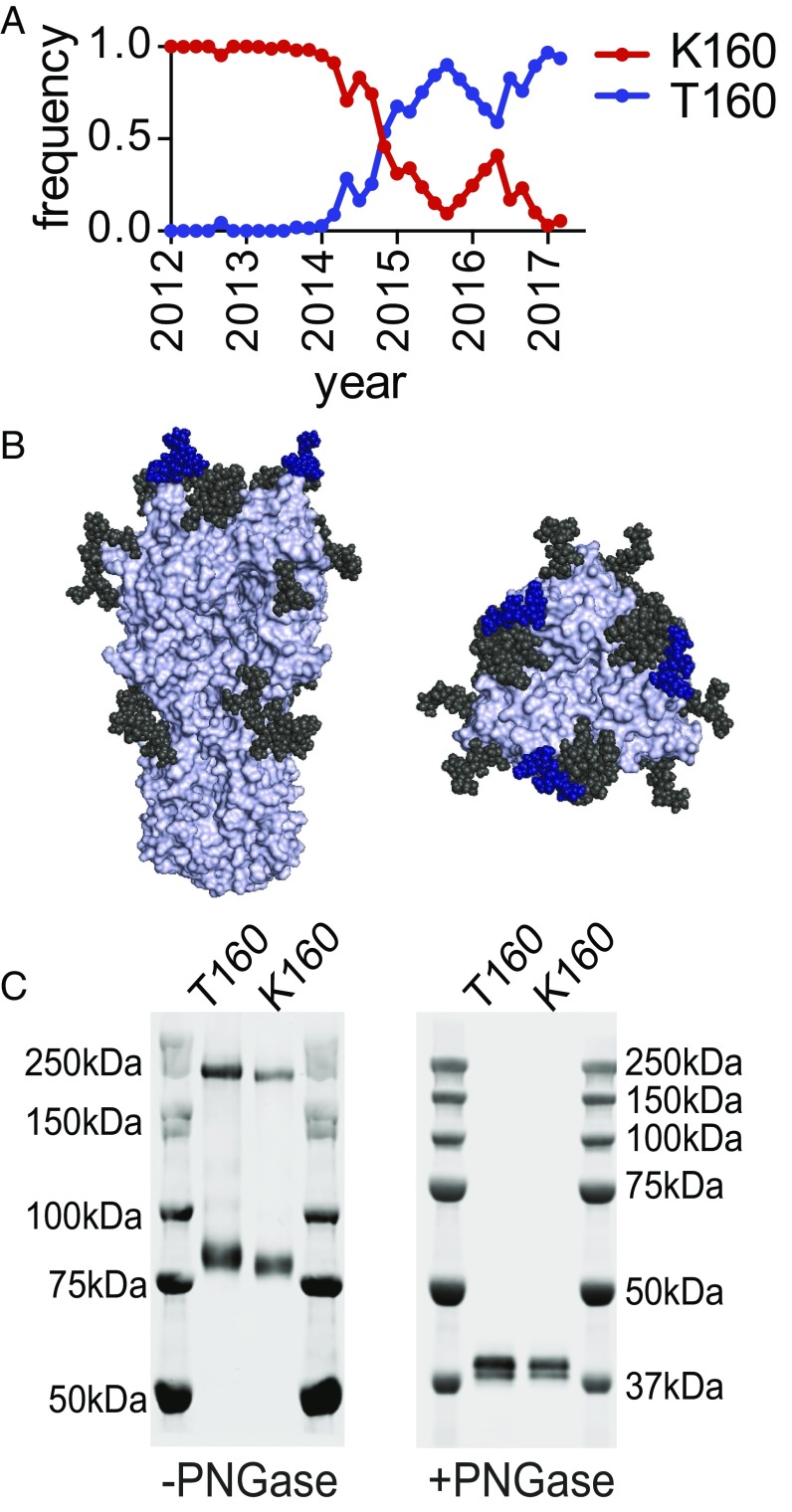
A K160T HA mutation rapidly rose to fixation during the 2014–2015 influenza season, and nearly all currently circulating H3N2 viruses possess threonine (T) at HA residue 160 (Fig. 1A). The K160T HA mutation is predicted to introduce an N-linked glycosylation site in antigenic site B of HA (Fig. 1B and Table S1). H3N2 viruses with the K160T HA mutation grow poorly in chicken eggs (21), and the 2016–2017 egg-adapted H3N2 vaccine strain possesses a T160K HA reversion mutation [reference: Global Initiative on Sharing All Influenza Data (GISAID) isolate ID EPI_ISL_189811]. We used reverse genetics to create H3N2 viruses possessing HAs with T160 and K160, and we completed Western blot analyses to determine whether the T160 and K160 HAs migrate differently in SDS/PAGE gels. HAs with T160 migrated with a higher molecular weight compared with HAs with K160 (Fig. 1C, Left). The HAs migrated similarly after PNGase treatment (Fig. 1C, Right), indicating that the difference in HA mobility in the absence of PNGase is due to differences in N-linked glycosylation.
Fig. 1.
Contemporary H3N2 viruses possess a new mutation that introduces a glycosylation site in antigenic site B of HA. (A) The K160T HA mutation rapidly rose to fixation during the 2014–2015 influenza season. Shown are frequencies estimated from viral samples (from GISAID database) collected from December 2011 to March 2017 and divided into 2-mo windows. (B) Putative glycosylated sites on contemporary HAs are shown on the A/Victoria/361/2011 HA trimer (PDB ID code 4O5I). The new putative site introduced by the K160T mutation is shown in blue, while the other putative sites are shown in black. (C) H3 viruses possessing either K160 HA or T160 HA were created by reverse genetics. The molecular weights of the HAs of these viruses were determined using Western blots with an anti-HA antibody, either with or without prior PNGase treatment. PNGase treatment was completed under reducing conditions. On the –PNGase gel, the upper bands correspond to HA trimers and the lower bands correspond to HA monomers.
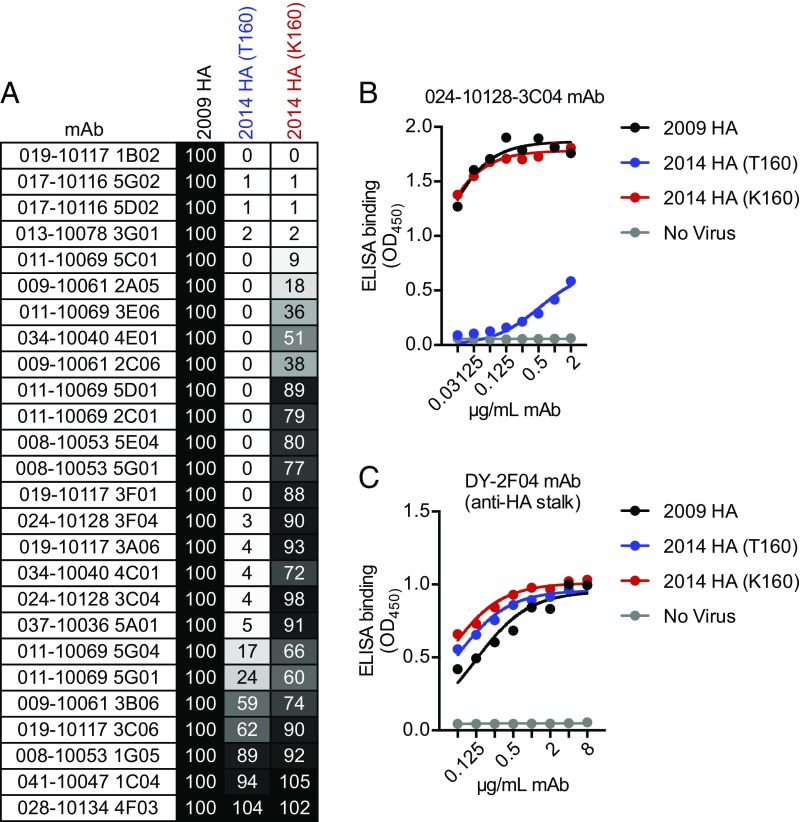
To determine whether the addition of this new antigenic site B glycan in HA affects antigenicity, we tested the binding of a panel of 26 human monoclonal antibodies that were elicited against an H3 virus from 2009, which lacks the new glycosylation site in antigenic site B of HA. All of these monoclonal antibodies bound to the HA of a 2009 virus (Fig. 2). The majority of these monoclonal antibodies (77%) bound to a 2014 virus with the egg-adaptive K160 HA (which lacks the glycosylation site), while only a few (23%) bound to a 2014 virus with T160 (which possesses the glycosylation site) (Fig. 2A). Raw binding data of a monoclonal antibody (024-10128-3C04) that binds efficiently to virus with K160 HA but not T160 HA is shown in Fig. 2B. As a control, we also measured binding of an HA stalk-reactive monoclonal antibody that efficiently recognizes virus with either T160 HA or K160 HA (Fig. 1C). Together, these data demonstrate that current circulating H3N2 influenza viruses possess a new glycosylation site in antigenic site B of HA that affects antigenicity, and that this glycosylation site is not present in current egg-adapted H3N2 vaccine strains.
Fig. 2.
Contemporary H3N2 viruses with T160 HA are antigenically distinct compared with H3N2 viruses with K160 HA. (A) ELISAs were completed to test the binding of 26 anti-H3 human monoclonal antibodies (mAbs) to a 2009 HA, a 2014 HA with T160, and a 2014 HA with K160. All antibodies in this panel were elicited by a 2009 HA following vaccination before the 2010–2011 season. Shown is percent binding of antibodies to the 2014 viruses relative to binding to the 2009 virus. (B) ELISA binding data for an antibody that binds efficiently to virus with K160 HA but not T160 HA is shown. (C) ELISA binding data for an antibody that recognizes a conserved epitope on the HA stalk is shown to verify that ELISA plates were coated with similar amounts of HA antigen.
Antibodies Elicited in Ferrets Exposed to the 2016–2017 Egg-Adapted H3N2 Vaccine Strain Poorly Neutralize a Circulating H3N2 Viral Strain.
We next completed a series of experiments to determine whether the current egg-adapted H3N2 vaccine strain (that possesses K160) elicits different types of antibodies compared with the current circulating H3N2 viral strain (that possesses T160). We infected ferrets with viruses possessing either K160 HA or T160 HA, and we completed foci reduction neutralization tests (FRNTs) using sera collected from these animals 28 d after infection. We completed FRNT assays rather than conventional hemagglutination–inhibition assays since 3C2.a H3N2 viruses inefficiently agglutinate red blood cells. Antibodies elicited by the egg-adapted vaccine strain possessing K160 HA recognized the egg-adapted vaccine strain fourfold to eightfold more efficiently than the currently circulating H3N2 strain that possesses T160 HA (Fig. 3A). This indicates that a large proportion of ferret antibodies elicited by the current egg-adapted H3N2 vaccine strain recognize the unglycosylated antigenic site B of HA. Interestingly, antibodies elicited by infection with the current circulating H3N2 viral strain possessing T160 HA recognized viruses with T160 HA and K160 HA equally (Fig. 3B). This is consistent with the hypothesis that the new HA glycosylation site effectively “shields” antigenic site B of HA, and that antibodies elicited by the viral strain possessing the new HA glycosylation site recognize epitopes not involving antigenic site B of HA.
Fig. 3.
Ferrets elicit different types of antibody responses when exposed to H3 viruses with K160 HA and T160 HA. Ferrets (n = 3 animals per group) were infected with viruses possessing (A) K160 HA or (B) T160 HA and sera were collected 28 d later. FRNTs were completed using viruses that possessed K160 HA or T160 HA. Neutralization titers are expressed as inverse dilution of sera that reduced foci by 90%. We completed three independent experiments with each sera. Shown are geometric means from the three independent experiments. Statistical significance was determined using a paired Student’s t test.
Antibodies Elicited in Humans Vaccinated with the Egg-Adapted H3N2 Strain Poorly Neutralize a Circulating H3N2 Viral Strain.
We completed additional antigenic tests using sera isolated from humans (18–49 y old) before and after vaccination during the 2016–2017 influenza season. Although the majority of influenza vaccine antigens are prepared in fertilized chicken eggs, a small fraction of vaccine antigens are produced in insect cells or canine kidney cells. To account for this, we measured human antibody responses elicited by vaccine antigens prepared in eggs (Fluzone; n = 22 donors), MDCK cells (Flucelvax; n = 26 donors), and insect cells (Flublok; n = 22 donors). Importantly, the H3 antigens in the Fluzone and Flucelvax vaccines possess the egg-adapted K160 HA, while the recombinant H3 antigen in the Flublok vaccine possesses T160 HA.
Vaccine effectiveness was lower in younger adults compared with older adults during the 2016–2017 season (20). Interestingly, we found that younger adults had higher prevaccination titers to the egg-adapted K160 HA compared with older individuals (Spearman’s rho = 0.3, P < 0.01; raw data from three independent experiments are shown in Table S2). Following vaccination, K160 HA titers were also higher in younger adults compared with older individuals (Spearman’s rho = 0.4), even after adjusting for higher prevaccination titers to K160 HA (Table S3). We speculate that the observed age-related differences in antibody titers to K160 HA might be due to birth year-related differences in H3N2 exposure history. Antibody titers to T160 HA were lower than antibody titers to K160 HA both before and after vaccination (P < 10−10 for each, Wilcox rank sum test), but there were no birth year-related effects on antibody titers to T160 HA (P = 0.23 prevaccination, P = 0.33 postvaccination; Table S3).
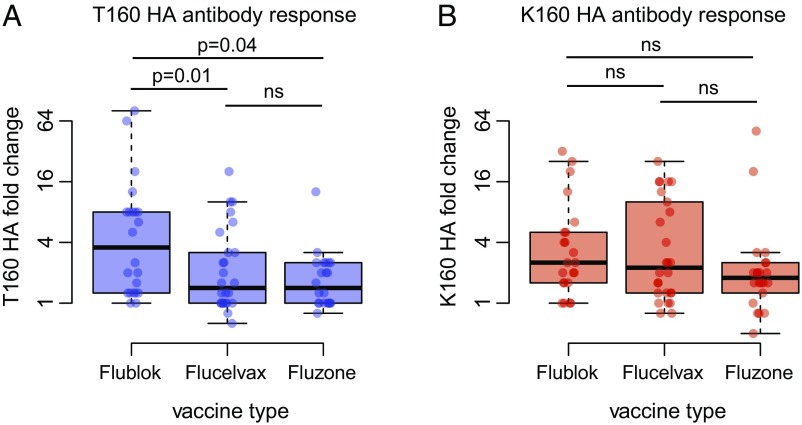
Some individuals in our study mounted strong antibody responses against T160 HA following vaccination (Table S2). Importantly, the majority of these individuals received the Flublok vaccine that possesses T160 HA (Fig. 4A). Antibody titers against T160 HA increased approximately fourfold following vaccination with Flublok, whereas there were only minimal increases in T160 HA antibody reactivity in most individuals following vaccination with Fluzone and Flucelvax (Fig. 4A). Conversely, antibody increases against the K160 HA were similar following vaccination with the three different vaccine antigens (Fig. 4B). These differential effects of the vaccines remained statistically significant after adjusting for age, prevaccination titer, and vaccination history (Table S3). These data indicate that H3N2 vaccine antigens produced during the 2016–2017 influenza season in eggs and MDCK cells (that possess K160 HA) elicit antibody responses that react poorly to current circulating H3N2 virus strains, whereas H3N2 vaccine antigens produced in insect cells (that possess T160 HA) elicit antibody responses that react efficiently to current circulating H3N2 viral strains.
Fig. 4.
Vaccine antigens possessing K160 HA and T160 HA elicit different responses in humans. Donors were vaccinated with seasonal influenza vaccines, and sera were collected before and 28 d after vaccination. FRNTs were completed using viruses that possessed T160 HA or K160 HA. (A) Flublok induced higher fold changes to T160 HA than did Flucelvax and Fluzone (P = 0.01 and P = 0.04 in adjusted analysis, respectively; Table S3; ns, nonsignificant). (B) The vaccine types did not differ in their ability to induce responses to K160 HA (P > 0.1 in adjusted analysis; Table S3). Thick horizontal lines show the median fold changes of the geometric mean titers. Colored rectangles indicate the interquartile range, and whiskers indicate the 150% interquartile ranges. Individual data points are superimposed. See Table S2 for raw titer data.
The Effect of Repeat Vaccination on Anti-H3 Antibody Responses During the 2016–2017 Influenza Season.
Many individuals receive influenza vaccines every year; however, recent data suggest that repetitive vaccinations may be associated with reduced antibody responses (22–24) and reduced vaccine effectiveness (25–27) during some influenza seasons. To determine whether prior vaccinations impacted the development of H3 antibodies following vaccination of donors in our study, we examined neutralizing antibody data in relation to vaccine history. Donors in our study self-reported vaccination history in the previous two seasons (2012–2013 and 2013–2014). We excluded individuals who reported having received the live attenuated vaccine in previous seasons (n = 1) and individuals who did not remember their vaccination status in either season (n = 7), and then we grouped individuals into three categories: unvaccinated both seasons, vaccinated in only one season, and vaccinated in both seasons. Interestingly, vaccination history was uncorrelated with prevaccine titers to T160 HA and K160 HA, but it was the strongest independent predictor of final antibody titers and fold changes to both viral strains following vaccination (Table S3). Individuals that had been vaccinated in the previous 2 y exhibited overall lower antibody boosts (Fig. S1), after adjusting for age and prevaccination antibody titers (Table S3).
Discussion
It is unclear why there was only moderate vaccine effectiveness during the 2016–2017 influenza season, given that the 2016–2017 vaccine strains appeared to be well matched to most circulating viral strains (20). Our data suggest that a mismatch in antigenic site B of H3N2 viruses, caused by the propagation of the vaccine strain in eggs, likely contributed to this low vaccine effectiveness. When influenza viruses are passaged in new hosts, antigenic properties of hemagglutinin can change since many residues of this protein are involved with both receptor binding and antibody recognition (28–31). This is not the first example of antigenic properties of influenza viruses being altered by egg adaptations. In 1978, Kilbourne (32) showed that there were two antigenically distinct viruses in the 1976 X-53 swine influenza vaccine and that these viruses had different growth properties in chicken embryos and canine kidney cells. In 1983, Webster and coworkers (33) demonstrated that influenza B viruses acquire mutations that alter antigenicity when propagated in chicken eggs. Egg adaptations resulting in antigenic changes have also been reported for H3N2 viruses (34–37). It has been proposed that H3N2 egg adaptations contributed to low vaccine effectiveness during the 2012–2013 influenza season (38), although this remains controversial (39).
Nonetheless, the majority of influenza vaccine antigens continue to be prepared in eggs (4). One solution to the problem of egg adaptations is to simply produce influenza antigens via a baculovirus system or in cell culture. The baculovirus system seems particularly well suited to avoid adaptive mutations; however, there are also potential problems with this approach. HA antigens prepared in insect cells are glycosylated with less complex sugars compared with mammalian cells (40), and it is possible that this affects antigenicity. In our study, some individuals vaccinated with baculovirus-prepared Flublok (that possessed T160 HA) mounted strong antibody responses that effectively neutralized a virus possessing an HA with a glycosylated antigenic site B; however, some Flublok-vaccinated individuals mounted antibody responses that reacted poorly to viruses with T160 HA.
It is important to note that there is more HA antigen in Flublok vaccine formulations compared with conventional egg-based vaccine formulations. It is possible that the increased amount of antigen in Flublok contributed to higher antibody responses against viruses that possess T160 HA; however, this is likely not the case since compared with the other vaccines tested, Flublok did not induce a significantly greater response to K160 HA. Therefore, Flublok did not generate an overall higher antibody response in our study, but rather an antibody response that was better able to recognize viruses possessing T160 HA. This is interesting in light of a recent study demonstrating that the Flublok vaccine elicited more protective antibody responses in older adults compared with egg-based vaccines during the 2014–2015 season (41). This warrants further investigation because there was a large H3 antigenic mismatch during this season (17). It is possible that the Flublok vaccine elicited antibodies of different specificities compared with egg-based vaccines during the 2014–2015 influenza season, perhaps to epitopes involving conserved residues of the HA receptor binding pocket.
Cell culture-expressed HA antigen might also avoid problems associated with egg adaptations. The 2017–2018 recommended H3N2 component is the same as the 2016–2017 recommended H3N2 component, but, starting this year, vaccine manufacturers that prepare antigens via cell culture systems will be allowed to use viral strains that have not been previously adapted to grow in eggs (https://www.cdc.gov/flu/protect/vaccine/cell-based.htm). H3N2 vaccine strains grown in this new system may possess HAs with a glycosylated antigenic site B, although mutations that abrogate this glycosylation site have been previously reported upon serial passage of clade 3C.2a H3N2 viruses in MDCK cells (21).
Interim reports indicate that the 2016–2017 vaccine protected younger individuals from H3N2 infection less effectively compared with older individuals (20). In our study, we found that younger adults were more likely to produce antibodies that efficiently neutralized K160 HA but weakly neutralized T160 HA after vaccination. These age-related differences may be due to differences in prior exposure histories. It is clear that prior influenza exposures can affect how an individual responds to new antigenically distinct viral strains (42). For example, individuals exposed to the 2009 pandemic H1N1 strain produce antibodies that target epitopes that are conserved in previously circulating seasonal H1N1 viral strains (43–46), particularly viral strains that they were likely exposed to in childhood. Early childhood infections also likely impact susceptibility to pandemic influenza virus strains. Gostic et al. (47) recently demonstrated that early childhood infections with either H1N1 or H3N2 influenza viruses are associated with protection from H5N1 and H7N9 viruses later in life, presumably through induction of cross-reactive antibodies against epitopes that are conserved between these different viruses. Further studies should explore if early childhood antigenic “imprinting” with different H3N2 viral strains affects the specificity of antibodies elicited by seasonal influenza vaccination.
Vaccine effectiveness against H3N2 is often low (2), especially among repeat vaccinees (25–27). Antigenic mismatch due to incorrect strain selection is an established cause of low vaccine effectiveness (2). Future studies should address whether egg-adapted mutations constitute another form of antigenic mismatch that alters vaccine effectiveness in other influenza virus seasons. Antigens with egg-adapted mismatches might recall preexisting immune responses, including responses to previous vaccines. A major effort should be made to develop and utilize new systems that produce influenza antigens that are not dependent on egg or cell culture-adaptive mutations. Antigens that do not possess adaptive mutations will likely offer better protection against influenza virus strains that circulate in the human population.
Materials and Methods
Viruses.
The HA gene of a representative clade 3C.2a H3N2 virus (A/Colorado/15/2014) was cloned into the vector pHW2000, and the T160K mutation was introduced to remove the predicted glycosylation motif. Viruses containing H3 and N2 genes with A/Puerto Rico/34/1934 internal genes were rescued using the influenza reverse-genetics system by transfecting a coculture of 293T and MDCK-SIAT1 cells. Transfection supernatants were collected 3 d after transfection and stored at −80 °C for use in neutralization assays. HA and NA genes of virus expansions grown on MDCK-SIAT1 cells were sequenced to confirm that no additional mutations arose during transfection.
Glycosylation Western Blotting and Glycan Modeling.
Viruses possessing T160 and K160 HA were expanded on MDCK-SIAT1 cells and then concentrated at 20,000 rpm (72,128 × g) for 1 h at 4 °C using an SW-28 rotor (Beckman Coulter). The amount of HA in each sample was normalized by ELISA with a human anti-HA monoclonal antibody for subsequent SDS/PAGE and Western blotting. To cleave N-linked glycans, samples were treated with PNGase-F (New England Biolabs) under reducing conditions. PNGase-treated and untreated samples were run on SDS/PAGE gels (Thermo Fisher Scientific) and transferred to a nitrocellulose membrane (Thermo Fisher Scientific). Blots were probed using anti-HA tag primary antibody clone HA-7 (product number 59658; Sigma-Aldrich) and an anti-mouse secondary antibody (product number 926-32212; Licor). To structurally model the N158 glycosylation introduced by T160, a K160T mutation was introduced into the crystal structure (PDB ID code 4O5I) using the program PyMol. Basic N-linked glycans were added to predicted N-linked glycosylation sites in the crystal structure using the GlyProt web server: www.glycosciences.de/modeling/glyprot/php/main.php.
Human Monoclonal Antibody Binding.
Human monoclonal antibodies were previously isolated from donor peripheral blood mononuclear cells following vaccination with the 2010–2011 influenza vaccine (48). ELISA plates were coated overnight at 4 °C with a 2009 virus (A/Victoria/210/2009; the H3N2 component of the 2010–2011 influenza vaccine), a glycosylated 2014 clade 3C2.a virus with T160 HA, or an unglycosylated 2014 clade 3C2.a virus with K160 HA. ELISA plates were blocked with a 3% (wt/vol) BSA solution in PBS for 2 h. Plates were washed three times with PBS containing 0.1% Tween 20, and serial dilutions of each monoclonal antibody in ELISA buffer [1% BSA (wt/vol) in PBS] were added to plates in 1% BSA in PBS. After 2 h of incubation, plates were again washed and a peroxidase-conjugated anti-human secondary antibody (product number 109-036-098; Jackson ImmunoResearch) diluted in ELISA buffer was added. After a 1-h incubation, plates were washed, and a 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (product number 50–00-03; Seracare) was added. The TMB reaction was stopped by adding HCl, and absorbance was measured using a plate reader. One-site specific binding curves were fitted in Prism software and the maximal binding against the 2009 vaccine strain was calculated for each monoclonal antibody. After background subtraction (from ELISA plates that did not have coated virus), binding of each monoclonal antibody relative to the vaccine strain was determined using the absorbance values at the lowest concentration of antibody that gave greater than 90% of maximal binding against the vaccine strain. Equivalent coating of each virus was checked using the stalk-reactive DY-2F04 monoclonal antibody.
Ferret Sera.
Ferret antisera were prepared at the Association for Assessment and Accreditation of Laboratory Animal Care-accredited company Noble Life Sciences using protocols approved by the Noble Life Sciences Institutional Animal Care and Use Committee. Naive Fitch ferrets were intranasally infected with 2 × 105 PFU of virus expressing either K160 HA or T160 HA. Serum samples were collected before infection and 28 d following infection. Sera used in antigenic assays were treated with receptor-destroying enzyme (Denka Seiken) for 2 h at 37 °C, and the enzyme was then heat-inactivated at 55 °C for 30 min.
Human Sera.
Experiments using human sera were conducted with the approval of the University of Rochester and University of Pennsylvania Institutional Review Boards. Informed consent was obtained for all individuals enrolled. Serum samples were collected at the University of Rochester before and 28 d following vaccination. Serological experiments were completed at the University of Pennsylvania using deidentified samples. Before assays, sera samples were treated with receptor-destroying enzyme (Denka Seiken) for 2 h at 37 °C, and the enzyme was then heat-inactivated at 55 °C for 30 min.
FRNT Assays.
Serum samples were serially diluted in 96-well round-bottom plates containing serum-free media. Approximately 200 focus-forming units of reverse-genetics transfection supernatant of each virus were added to each diluted sera sample and the virus–sera mixtures were incubated at room temperature for 1 h. The virus–sera mixtures were then added to confluent monolayers of MDCK-SIAT1 cells and incubated at 37 °C for 1 h. After incubation, cells were washed with serum-free media and an overlay medium of serum-free media containing 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) (Thermo Fisher Scientific), gentamycin (Thermo Fisher Scientific), and 0.5% (wt/vol) methylcellulose (Sigma-Aldrich) was added to cells. Infected monolayers were incubated for 18 h, after which the overlay medium was aspirated and the cells were fixed and permeabilized with ice-cold methanol–acetone [1:1 (vol/vol)]. Infected monolayers were stained with anti-NP monoclonal antibody IC5-1B7 (product number NR-43899; BEI Reagent Resources) and an anti-mouse peroxidase-conjugated secondary antibody (product number 855563; MP Biomedicals). A TMB substrate (product number 5510-0030; Seracare) was added to visualize foci. Following staining, plates were imaged and foci were quantified using an ELISPOT reader (Cellular Technologies Limited). FRNT90 titers were reported as the reciprocal of the highest dilution of sera that reduced the number of foci by at least 90%, relative to control wells that had no serum or monoclonal antibody added. Undetectable titers were assigned a value of 10. All FRNT assays were repeated three times on separate days.
Models of Initial and Final Antibody Titers and Fold Responses.
Three replicate measurements of prevaccination and postvaccination antibody titers from each serum sample (Table S2) were geometrically averaged, and the geometric mean titers were used for model fitting. Linear models that included age and vaccination history (receipt of trivalent inactivated vaccine in neither, only one, or both of the last 2 y) were fitted to log2 prevaccination titers as continuous and factor variables, respectively. Models were similarly fitted to log2 postvaccination antibody titers, also including log2 prevaccination titers and vaccine group (Flublok, Flucelvax, and Fluzone) as continuous and factor variables, respectively. Likelihood maximization was performed with the lm package in R, version 3.3.1. Code for the analysis is available at https://cobeylab.github.io/H3N2_glycosylation/.
Supplementary Material
Acknowledgments
We acknowledge the authors and the originating and submitting laboratories of the sequences from GISAID’s EpiFlu Database on which Fig. 1A was based (see Supporting Information for list of laboratories that contributed sequence data to GISAID’s EpiFlu Database). This work was supported by the National Institute of Allergy and Infectious Diseases [1R01AI113047 (to S.E.H.); 1R01AI108686 (to S.E.H.); DP2AI117921 (to S.C.); CEIRS HHSN272201400005C (to P.C.W., A.J.S., J.J.T., S.C., and S.E.H.)]. S.E.H. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Conflict of interest statement: S.J.Z., K.P., M.E.G., K.K., S.D.P., P.C.W., A.J.S., S.C., and S.E.H. have no conflicts of interest. J.J.T. is an advisor (nonpaid) for Protein Sciences and is on the scientific advisory board or received consulting payments for Sequiris, Medicago, Takeda, and Flugen. J.J.T.’s laboratory has also received support from Sanofi.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712377114/-/DCSupplemental.
References
- 1.Yewdell JW. Viva la revolución: Rethinking influenza a virus antigenic drift. Curr Opin Virol. 2011;1:177–183. doi: 10.1016/j.coviro.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belongia EA, et al. Variable influenza vaccine effectiveness by subtype: A systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis. 2016;16:942–951. doi: 10.1016/S1473-3099(16)00129-8. [DOI] [PubMed] [Google Scholar]
- 3.Carrat F, Flahault A. Influenza vaccine: The challenge of antigenic drift. Vaccine. 2007;25:6852–6862. doi: 10.1016/j.vaccine.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 4.Schultz-Cherry S, Jones JC. Influenza vaccines: The good, the bad, and the eggs. Adv Virus Res. 2010;77:63–84. doi: 10.1016/B978-0-12-385034-8.00003-X. [DOI] [PubMed] [Google Scholar]
- 5.Krammer F, Palese P, Steel J. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr Top Microbiol Immunol. 2015;386:301–321. doi: 10.1007/82_2014_408. [DOI] [PubMed] [Google Scholar]
- 6.Koel BF, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 7.Smith DJ, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 8.Skehel JJ, et al. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci USA. 1984;81:1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wrigley NG, et al. Electron microscopy of influenza haemagglutinin-monoclonal antibody complexes. Virology. 1983;131:308–314. doi: 10.1016/0042-6822(83)90499-3. [DOI] [PubMed] [Google Scholar]
- 10.Abe Y, et al. Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J Virol. 2004;78:9605–9611. doi: 10.1128/JVI.78.18.9605-9611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das SR, et al. Fitness costs limit influenza A virus hemagglutinin glycosylation as an immune evasion strategy. Proc Natl Acad Sci USA. 2011;108:E1417–E1422. doi: 10.1073/pnas.1108754108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei CJ, et al. Cross-neutralization of 1918 and 2009 influenza viruses: Role of glycans in viral evolution and vaccine design. Sci Transl Med. 2010;2:24ra21. doi: 10.1126/scitranslmed.3000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medina RA, et al. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci Transl Med. 2013;5:187ra70. doi: 10.1126/scitranslmed.3005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherry JL, Lipman DJ, Nikolskaya A, Wolf YI. Evolutionary dynamics of N-glycosylation sites of influenza virus hemagglutinin. PLoS Curr. 2009;1:RRN1001. doi: 10.1371/currents.RRN1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmerman RK, et al. US Flu VE Investigators 2014–2015 Influenza vaccine effectiveness in the United States by vaccine type. Clin Infect Dis. 2016;63:1564–1573. doi: 10.1093/cid/ciw635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Mello T, et al. Centers for Disease Control and Prevention (CDC) Update: Influenza activity—United States, September 28, 2014–February 21, 2015. MMWR Morb Mortal Wkly Rep. 2015;64:206–212. [PMC free article] [PubMed] [Google Scholar]
- 17.Chambers BS, Parkhouse K, Ross TM, Alby K, Hensley SE. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014–2015 influenza season. Cell Rep. 2015;12:1–6. doi: 10.1016/j.celrep.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flannery B, et al. Enhanced genetic characterization of influenza A(H3N2) viruses and vaccine effectiveness by genetic group, 2014–2015. J Infect Dis. 2016;214:1010–1019. doi: 10.1093/infdis/jiw181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anonymous Recommended composition of influenza virus vaccines for use in the 2016–2017 northern hemisphere influenza season. Wkly Epidemiol Rec. 2016;91:121–132. [PubMed] [Google Scholar]
- 20.Flannery B, et al. Interim estimates of 2016–17 seasonal influenza vaccine effectiveness—United States, February 2017. MMWR Morb Mortal Wkly Rep. 2017;66:167–171. doi: 10.15585/mmwr.mm6606a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y, et al. The characteristics and antigenic properties of recently emerged subclade 3C.3a and 3C.2a human influenza A(H3N2) viruses passaged in MDCK cells. Influenza Other Respir Viruses. 2017;11:263–274. doi: 10.1111/irv.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson MG, et al. Effects of repeated annual inactivated influenza vaccination among healthcare personnel on serum hemagglutinin inhibition antibody response to A/Perth/16/2009 (H3N2)-like virus during 2010–11. Vaccine. 2016;34:981–988. doi: 10.1016/j.vaccine.2015.10.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung VKY, et al. Influenza vaccination responses: Evaluating impact of repeat vaccination among health care workers. Vaccine. 2017;35:2558–2568. doi: 10.1016/j.vaccine.2017.03.063. [DOI] [PubMed] [Google Scholar]
- 24.Huang KA, Chang SC, Huang YC, Chiu CH, Lin TY. Antibody responses to trivalent inactivated influenza vaccine in health care personnel previously vaccinated and vaccinated for the first time. Sci Rep. 2017;7:40027. doi: 10.1038/srep40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLean HQ, et al. Impact of repeated vaccination on vaccine effectiveness against influenza A(H3N2) and B during 8 seasons. Clin Infect Dis. 2014;59:1375–1385. doi: 10.1093/cid/ciu680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohmit SE, et al. Influenza vaccine effectiveness in the community and the household. Clin Infect Dis. 2013;56:1363–1369. doi: 10.1093/cid/cit060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skowronski DM, et al. A perfect storm: Impact of genomic variation and serial vaccination on low influenza vaccine effectiveness during the 2014–2015 season. Clin Infect Dis. 2016;63:21–32. doi: 10.1093/cid/ciw176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hensley SE, et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326:734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, et al. Single hemagglutinin mutations that alter both antigenicity and receptor binding avidity influence influenza virus antigenic clustering. J Virol. 2013;87:9904–9910. doi: 10.1128/JVI.01023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gambaryan AS, Robertson JS, Matrosovich MN. Effects of egg-adaptation on the receptor-binding properties of human influenza A and B viruses. Virology. 1999;258:232–239. doi: 10.1006/viro.1999.9732. [DOI] [PubMed] [Google Scholar]
- 31.Gambaryan AS, et al. Effects of host-dependent glycosylation of hemagglutinin on receptor-binding properties on H1N1 human influenza A virus grown in MDCK cells and in embryonated eggs. Virology. 1998;247:170–177. doi: 10.1006/viro.1998.9224. [DOI] [PubMed] [Google Scholar]
- 32.Kilbourne ED. Genetic dimorphism in influenza viruses: Characterization of stably associated hemagglutinin mutants differing in antigenicity and biological properties. Proc Natl Acad Sci USA. 1978;75:6258–6262. doi: 10.1073/pnas.75.12.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schild GC, Oxford JS, de Jong JC, Webster RG. Evidence for host-cell selection of influenza virus antigenic variants. Nature. 1983;303:706–709. doi: 10.1038/303706a0. [DOI] [PubMed] [Google Scholar]
- 34.Katz JM, Webster RG. Efficacy of inactivated influenza A virus (H3N2) vaccines grown in mammalian cells or embryonated eggs. J Infect Dis. 1989;160:191–198. doi: 10.1093/infdis/160.2.191. [DOI] [PubMed] [Google Scholar]
- 35.Kilbourne ED, et al. Influenza A virus haemagglutinin polymorphism: Pleiotropic antigenic variants of A/Shanghai/11/87 (H3N2) virus selected as high yield reassortants. J Gen Virol. 1993;74:1311–1316. doi: 10.1099/0022-1317-74-7-1311. [DOI] [PubMed] [Google Scholar]
- 36.Meyer WJ, et al. Influence of host cell-mediated variation on the international surveillance of influenza A (H3N2) viruses. Virology. 1993;196:130–137. doi: 10.1006/viro.1993.1461. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, Zhou H, Jin H. The impact of key amino acid substitutions in the hemagglutinin of influenza A (H3N2) viruses on vaccine production and antibody response. Vaccine. 2010;28:4079–4085. doi: 10.1016/j.vaccine.2010.03.078. [DOI] [PubMed] [Google Scholar]
- 38.Skowronski DM, et al. Low 2012–13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS One. 2014;9:e92153. doi: 10.1371/journal.pone.0092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cobey S, et al. 2017. Despite egg-adaptive mutations, the 2012–13 H3N2 influenza vaccine induced comparable antibody titers to the intended strain. bioRxiv:10.1101/158550.
- 40.An Y, et al. Comparative glycomics analysis of influenza hemagglutinin (H5N1) produced in vaccine relevant cell platforms. J Proteome Res. 2013;12:3707–3720. doi: 10.1021/pr400329k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunkle LM, et al. PSC12 Study Team Efficacy of recombinant influenza vaccine in adults 50 years of age or older. N Engl J Med. 2017;376:2427–2436. doi: 10.1056/NEJMoa1608862. [DOI] [PubMed] [Google Scholar]
- 42.Cobey S, Hensley SE. Immune history and influenza virus susceptibility. Curr Opin Virol. 2017;22:105–111. doi: 10.1016/j.coviro.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li GM, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci USA. 2012;109:9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linderman SL, et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013-2014 influenza season. Proc Natl Acad Sci USA. 2014;111:15798–15803. doi: 10.1073/pnas.1409171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J Exp Med. 2013;210:1493–1500. doi: 10.1084/jem.20130212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gostic KM, Ambrose M, Worobey M, Lloyd-Smith JO. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science. 2016;354:722–726. doi: 10.1126/science.aag1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henry Dunand CJ, et al. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J Clin Invest. 2015;125:1255–1268. doi: 10.1172/JCI74374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.