Significance
This work used mass spectrometry imaging to visualize a key bacterial virulence factor, lipid A within tissues infected by Francisella novicida, a model organism for Francisella tularensis ssp. tularensis, the causative agent of tularemia. Lipid components of the host were mapped using the same technique, leading to identification of a lethal role for host lipid metabolism in Francisella infection. Combined with profiling of lipid inflammatory mediators, this work defined the dramatic polarity of the cyclooxygenase-2 axis in this infection and overproduction of the proinflammatory product prostaglandin E2. The host–pathogen lipid interface was reconstructed to demonstrate the spatial organization of the host lipid response in the spleen.
Keywords: host–pathogen interaction, lipid inflammation, cyclooxygenase pathway, mass spectrometry imaging, microbial pathogenesis
Abstract
Mass spectrometry imaging (MSI) was used to elucidate host lipids involved in the inflammatory signaling pathway generated at the host–pathogen interface during a septic bacterial infection. Using Francisella novicida as a model organism, a bacterial lipid virulence factor (endotoxin) was imaged and identified along with host phospholipids involved in the splenic response in murine tissues. Here, we demonstrate detection and distribution of endotoxin in a lethal murine F. novicida infection model, in addition to determining the temporally and spatially resolved innate lipid inflammatory response in both 2D and 3D renderings using MSI. Further, we show that the cyclooxygenase-2–dependent lipid inflammatory pathway is responsible for lethality in F. novicida infection due to overproduction of proinflammatory effectors including prostaglandin E2. The results of this study emphasize that spatial determination of the host lipid components of the immune response is crucial to identifying novel strategies to effectively address highly pathogenic and lethal infections stemming from bacterial, fungal, and viral origins.
Lipopolysaccharide (LPS) is a characteristic component of Gram-negative bacteria forming the majority of the outer leaflet of the outer membrane (1). Lipid A (endotoxin) is the lipophilic membrane anchor of LPS and when modified by addition of core and O-antigen to the diglucosamine backbone of lipid A forms full-length LPS (2). Endotoxin is the canonical ligand for Toll-like receptor 4 (TLR4) (3, 4) and endotoxin-like lipids are absent from the eukaryotic lipid repertoire. Individual bacterial lipid A structures are unique (5–7), although modifiable during growth (8, 9) to accommodate the diverse conditions encountered (10) during transmission from the environment to a warm-blooded host. Unlike the highly stimulatory endotoxin derived from enteric bacteria (10, 11), lipid A derived from Francisella species does not stimulate the canonical endotoxin pattern recognition receptor TLR4 (12) and is an important virulence factor. Mapping of the protein-based virulence determinant, Burkholderia mallei GroEL was recently demonstrated (13) using mass spectrometry imaging (MSI), highlighting the growing application of MSI to problems in microbial pathogenesis. Notably, direct mapping of an unlabeled, bacteria-borne lipid virulence factor within infected tissue has not yet been demonstrated.
MSI is a molecular imaging technique with rapidly expanding applications (14–16). MSI enables spatial resolution, or mapping of molecules within tissue by mass without the requirements for species-specific reagents, labels, or tags, and other indirect or direct tools typically associated with histological techniques. With respect to molecular origin (microbial, vector, host), MSI is also an unbiased discovery and validation tool because the output is observed exclusively as a mass/charge (m/z) ratio. Ignorance of species origin makes MSI an ideal tool for studying complex interactions in multispecies systems, such as host–pathogen (17) and vector–pathogen models. Matrix-assisted laser desorption/ionization (MALDI) is the most commonly used ionization method for MSI (18), although other ionization methods [such as desorption electrospray ionization (DESI) and liquid extraction surface analysis (LESA), and others] are also employed. MSI has been used in a wide array of applications ranging from fingerprint analysis to mapping drug distribution in tissue. MSI studies of cancer biology for biomarker discovery, therapeutic target identification, and fundamental pathology hold particular promise. In contrast, comparatively few systematic studies have leveraged MSI to describe the host–microbe relationship (19, 20).
MSI is well suited to mapping lipids (16, 21) and related metabolites (22), among many other classes of molecular targets. We have previously demonstrated methods to detect host and pathogen phospholipids (PLs) within mouse spleen (23) infected with the model organism Francisella novicida (F. novicida) alongside low-abundance detection of F. novicida lipid A extracted from infected tissues. The membrane of F. novicida contains both LPS and free lipid A, lacking core and O-antigen (24). F. novicida lipid A is readily soluble in solvent systems used for phospholipids, allowing direct detection of lipid A using MSI, free of radioactive labels, antibodies, or tags. In addition to targeted F. novicida lipid A mapping, the MSI datasets would also include the untargeted spectral information for both host and pathogen lipids.
Evasion of the host innate immune system is one of the defining characteristics of a Francisella infection (25, 26). As a primary host evasion mechanism, Francisella goes undetected by TLR4 due to a combination of hypoacylation and acyl elongation of Francisella lipid A (12, 27). Secondary manipulations of the host response include ubiquitin-mediated degradation of MHCII, blocking of naïve T-cell maturation, and regulation of lipid-mediated inflammation (26). These host evasion mechanisms are a function of prostaglandin E2 (PGE2) release following infection in Francisella tularensis holarctica live vaccine strain (LVS) models (26). PGE2 is a downstream metabolic product of arachidonic acid (AA) initially in the form of a host membrane phospholipid acyl chain (28). AA is liberated from the host phospholipid via lipase enzymes and further processed to the potent lipid mediator PGE2 by a series of subsequent enzymatic conversions dependent on cyclooxygenase-2 (COX-2). LVS-infected macrophages release the lipid mediator PGE2, which blocks T cell proliferation, in a manner that uses not only cytosolic phospholipase A2 (cPLA2), but also phospholipase D (PLD) and diacylglycerol lipase (DAGL) (29–31). In addition to COX-2–dependent lipid mediators, the lipoxygenase (LOX) pathway is also up-regulated during LVS infection. Although these pathways have been investigated at the transcriptional level in Francisella infection, a thorough analysis of the resulting lipid mediators produced by COX and LOX has not been reported. The role of lipid metabolites in any infection is complex and can result in both pro- and antiinflammatory products (32), the division of which is largely temporal. The biochemical pathways controlling PGE2 release in directly infected cells have been well described, but to date, little is known about the systemic effects and dynamics of downstream COX-2 products resulting from Francisella infection. These findings directly couple the host evasion response of Francisella to the tightly regulated metabolism of membrane phospholipids.
Using MSI, we concurrently mapped both bacterial and host lipids and spatially resolved the depletion of AA-bearing phosholipids from the host during progression of a lethal infection in both 2D and 3D images. Further, using quantitative lipidomics, we evaluated the system-level production of lipid metabolites of the COX and LOX axes beyond the transcriptional regulation of the pathway. This work represents the mapping of unlabeled lipid A in tissue with the codetected host phospholipid response resulting in identification of a protective mechanism against lethal Francisella infection.
Results
Francisella Lipid A Accumulates in the Red Pulp.
Bacterial and host lipids are readily extracted in similar organic solvent systems (23, 33) used for total lipid isolation (34). We have previously established methods for observing F. novicida lipid A directly from extracts of infected murine tissues; however, this extract analysis lacked the ability to map the location of pathogen- and host-borne lipids to the infected tissues. To determine the distribution of F. novicida lipid A in an active infection, we adapted our previously reported conditions to visualize F. novicida lipid A on thin tissue sections using MSI. The major host-adapted structure (35) of F. novicida lipid A is detected as a deprotonated negative ion (m/z 1,665, Fig. 1 A and B) by MALDI mass spectrometry. F. novicida lipid A is exclusive to the pathogen and was used as a marker for bacterial location and to determine lipid A abundance within infected tissue using MALDI MSI while simultaneously mapping the host phospholipid response.
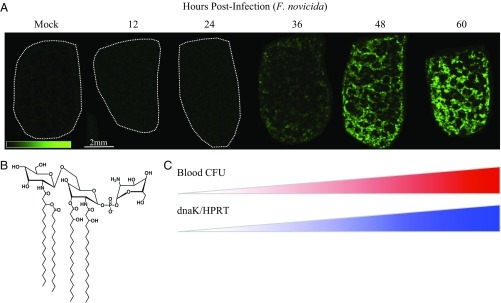
Fig. 1.
Endotoxin mapping in splenic red pulp by mass spectrometry imaging (MSI) corresponds to septic transition in mouse infection with F. novicida. (A) Ion image of F. novicida lipid A in bisected mouse spleen sections, 10-μm thickness, bisection face oriented up, 75-μm spatial resolution by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF). False color ion intensity is given in green color scale, 0–45 arbitrary units (a.u.) normalized, total ion current (TIC). Dotted white outlines are given for negative tissues. Data are representative of three parallel replicates. (B) Predominant structure of F. novicida lipid A given as the negative ion [M-H]− observed at m/z 1,665, supported by fragmentation (SI Appendix, Table S1). (C) Red heatbar: bacterial dissemination (F. novicida) to blood following subcutaneous infection (derived from SI Appendix, Fig. S3A), colony forming units (CFU) per milliliter (mL) cardiac blood, postmortem, n = 3 per time point. Blue heatbar: confirmation of bacterial transcript in the opposite halves of bisected spleens from A, qRT-PCR. F. novicida dnaK transcript is normalized to murine Hprt. n = 3 per time point (derived from SI Appendix, Fig. S3B).
Using a lethal murine model for F. tularensis, we detected F. novicida lipid A in the spleen at 36 hours postinfection (h.p.i.) (Fig. 1A). At 36 h.p.i., F. novicida lipid A mapped exclusively to the splenic red pulp at 36 h.p.i. (SI Appendix, Fig. S1) at low signal intensity followed by an increase at 48 and 60 h.p.i. (Fig. 1A and SI Appendix, Fig. S2). Detection of lipid A in the spleen corresponded to bacterial dissemination with viable organisms detected in the blood at 24 h.p.i. with a septic bacterial load (>103 cfu/mL) at 36 h.p.i. (SI Appendix, Fig. S3A). By 48 h.p.i., lipid A was also detected in the central area of white pulp nodules corresponding to a previous observation (36) that necrotic regions of the splenic white pulp contain replicating organisms late in infection. Bacterial burden counts in F. novicida-infected spleens showed 108 cfu/spleen at 36 h.p.i., concurrent with detection of F. novicida lipid A (SI Appendix, Fig. S3B). Bacterial transcript (F. novicida dnaK) was present at 24 h.p.i. and increased by 5 logs (fold change) at 36 h.p.i. (SI Appendix, Fig. S3C) normalized to murine control transcript (Mus musculus Hprt), confirmed by the presence of organisms (SI Appendix, Fig. S3 D and E) in the splenic red pulp of parallel infected mice. To characterize the major lipid A structure present in the infected spleens, we prepared total lipid extracts and performed fragmentations on the MALDI-produced m/z 1,665.2 ion (SI Appendix, Fig. S4 and Table S1). Thirteen major ions (35) were identified, allowing structural characterization (Fig. 1B). These results represent the direct in situ observation, mapping, and molecular characterization of a bacterial lipid A using an MSI approach, suggesting MS-coupled methods can be developed for early detection and mapping of sepsis without the requirement for molecular tags, specific detection reagents, or other indirect methods (37, 38).
Visualization of Splenic Lipid Inflammation.
We next sought to spatially resolve alterations in the host lipidome following progression to lethal sepsis resulting from F. novicida infection and evaluate their contribution to pathogenesis. Using MSI, we mapped splenic lipids across the infection time course (Fig. 2 and SI Appendix, Fig. S5 B–G).
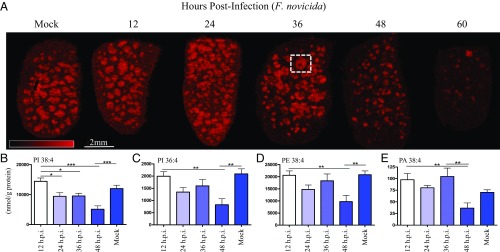
Fig. 2.
Arachidonic acid (AA)-containing phospholipids of the splenic white pulp are depleted during F. novicida infection. (A) Image of m/z 885.6 ion channel identified at 1-stearoyl, 2-arachidonyl-phosphatidylinositol (SAPI), [M-H]−, 75-μm spatial resolution, MALDI-TOF. Serial sections of spleens are as in Fig. 1A. False color ion intensity is given in red color scale, 0–325 a.u., normalized (TIC). White box outlines white pulp nodule with an active germinal center (identified in black box, SI Appendix, Fig. S1). (B–E) Quantitative LC/MS of parent phospholipid containing arachidonyl group normalized to total input splenic protein content (in grams) evaluated every 12 hours postinfection (h.p.i.), lethal dose F. novicida, SQ. (B) SAPI, m/z 885.6. (C) Palmitoyl, arachidonyl phosphatidylinositol (PI) 16:0/20:4 (PI 36:4), m/z 857.6. (D) Stearoyl, arachidonyl phosphatidylethanolamine (PE) 18:0/20:4 (PE 38:4), m/z 766.5. (E) Stearoyl, arachidonyl phosphatidic acid (PA) 18:0/20:4 (PA 38:4), m/z 723.5. Lipid levels were analyzed using one-way ANOVA for injection time points, *P < 0.05, **P < 0.01, ***P < 0.001 (n = 10). Error bars denote SEM.
This analysis revealed a high abundance ion (m/z 885.6), increasing through 36 h.p.i. (Fig. 2A), in the white pulp of naïve spleens. This ion is organized toward the outer regions of the white pulp with several white pulp centers (site of germinal center formation) depleted of signal. At 48 h.p.i., we observed a marked decreased in abundance in the white pulp with nearly complete loss at 60 h.p.i. (Fig. 2A). On-tissue MALDI-TOF/TOF (SI Appendix, Fig. S6) confirmed the identity of this ion to be 1-stearoyl, 2-arachidonyl-phoshphatidylinositol (SAPI) [also phoshphatidylinositol (PI) 18:0/20:4, positions assigned based on relative abundance of fragments]. Using zymosan-stimulated murine peritoneal macrophages, Gil-de-Gómez et al. (39) demonstrated the use of SAPI as a reservoir for AA released from membrane phospholipids for production of eicosanoids, host signaling molecules made by enzymatic processing or oxidation of AA (29, 40). The progressive loss of SAPI abundance was consistent with the onset of sepsis (Fig. 1). F. novicida-infected spleens showed destruction of the white pulp at 48 h.p.i. and hemorrhagic necrosis at 60 h.p.i. (SI Appendix, Fig. S7 E and F), characteristics of rapid progression to severe sepsis, suggesting that SAPI depletion is involved with the process of end-stage sepsis. Average spleen weight peaked at 36 h.p.i. (SI Appendix, Fig. S7H) and was consistent with cellular migration including polymorphonuclear cells (PMNs), hemosiderin-laden macrophages, and immature lymphoid cells, as well as significant and sustained increases in IL-1β and TNF-α (SI Appendix, Fig. S8). To determine whether the loss of SAPI was related to the lethal infected or to the presence of an activated immune response, we used an avirulent mutant of F. novicida. F. novicida ΔlpxD1 is a virulence-attenuated mutant (9) of lipid A biosynthesis; during infection with F. novicida ΔlpxD1, the bacteria disseminate and ultimately elicit a protective immune response. At 48 h.p.i., F. novicida ΔlpxD1 was disseminated to the spleen, but no loss of SAPI signal was observed (SI Appendix, Fig. S9). A subcutaneous dose of 5 × 106 cfu F. novicida ΔlpxD1 in mice loads the spleen between 105 and 106 cfu through 48 h.p.i., and decreases are observed on day 3 and beyond (9). The loss of SAPI abundance in the wild-type (WT) F. novicida infection is a result of the tissue destruction leading to multiorgan dysfunction syndrome and mortality rather than a result of an activated splenic immune response.
Polyunsaturated fatty acids (PUFAs), such as AAs are liberated from the membrane phospholipids of the host cell membrane (29) by phosphoactivated phospholipases, such as cPLA2 (41) and are subsequently converted (42) to potent inflammatory mediators. AA is available to multiple pathways (43) (COX, LOX, cytochrome P450) to yield both proinflammatory and proresolving metabolites (44); the fate of free AA in the cytosol depends on the available and phosphoactivated enzyme pool. Components of both the COX (45) and LOX (46) pathways are up-regulated in Francisella infection, although the specific membrane phospholipid source of AA and the activities of the downstream products have not been defined.
To quantify the AA-bearing phospholipids, as well as free AA and relevant downstream metabolites of the COX and LOX pathways, we used a liquid chromatography/mass spectrometry (LC/MS) multiple reaction monitoring approach. This analysis identified a decrease in four AA-containing phospholipids that also showed corresponding loss of abundance in splenic white pulp (SI Appendix, Fig. S5); SAPI (PI 18:0/20:4), PI 16:0/20:4, phosphatidylethanolamine (PE) 18:0/20:4, and phosphatidic acid (PA) 18:0/20:4 at 48 h.p.i. (Fig. 2 B–E). Both PI 16:0/20:4 and SAPI (PI 18:0/20:4) shared similar distributions (SI Appendix, Fig. S5 B and C), were depleted in late-stage white pulp, and shared a strong correlation in pixel intensities (value = 0.928, SI Appendix, Fig. S10C), suggesting these PIs may serve as an AA pool for splenic eicosanoid production and suggest that these decreases in parent phospholipids were due to enzymatic loss of AA secondary to cPLA2 liberation. Quantitation of cPLA2 (cPla2) and COX-2 (Ptgs2) transcripts showed a significant induction by 36 h.p.i. (SI Appendix, Fig. S11) with COX-2 induced 10-fold (log2) at 36 and 48 h.p.i. that was sustained throughout the course of infection. To further characterize COX-2 induction, we investigated the eicosanoid amplification mechanism of high-mobility group box-1 (HMGB1) protein synergism (47) with IL-1β, which plays a central role in the regulation of immune and inflammatory responses to infection. Together, HMGB1 and decreased levels of IL-1β result in the production of inflammatory mediators, including IL-6, COX-2, and PGE2, and eliciting PGE2 at levels comparable to high IL-1β alone, and thus may contribute to the sharp induction of COX-2 at 36 h.p.i. (SI Appendix, Fig. S11B). Splenic IL-1β transcript was induced over 3-fold at 24 h.p.i. (SI Appendix, Fig. S8) and HMGB1 protein was present at all early time points (SI Appendix, Fig. S11C), likely contributing to the up-regulation of the COX-2 pathway. The fates of PI, PE, and PA lysolipids were not evaluated, but reports of mammalian membrane-bound O-acyltransferase (MBOAT) proteins suggest they are subsequently involved in arachidonate recycling (48) by neutrophils.
COX-2 Metabolites Contribute to Lethality in Francisella Infection.
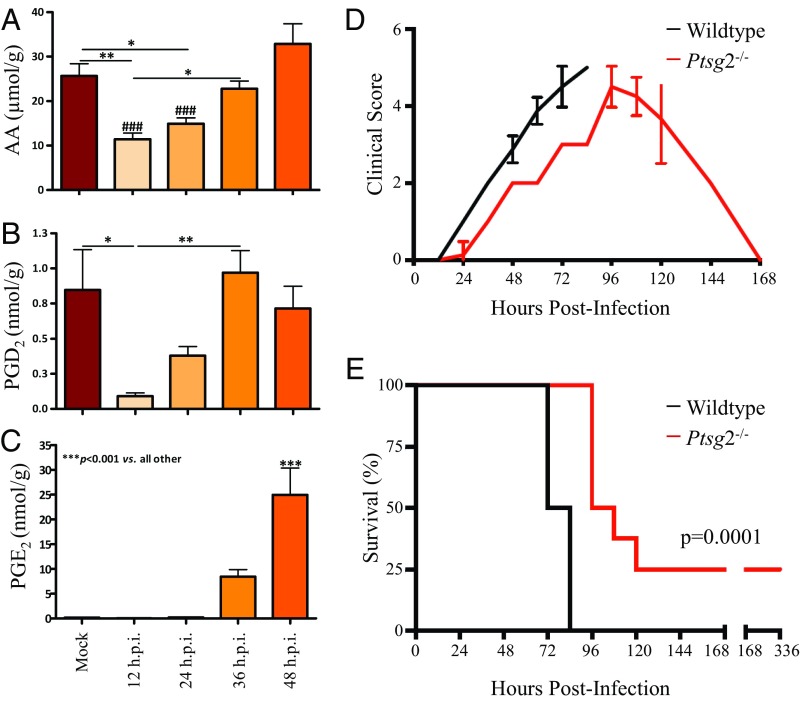
Having identified depletion of four AA-bearing phospholipids, we next determined the extent of PGE2 induction, as well as production of a variety of other inflammatory lipid mediators derived from AA (SI Appendix, Fig. S12) in the F. novicida-infected spleens. In naïve spleens, the initial AA pool was depleted by two-thirds at 12 h.p.i. and did not recover to baseline level until 36–48 h.p.i. (Fig. 3A). These results are in agreement with the depletion timeframe of SAPI (and other AA-bearing phospholipids) observed at 24–48 h.p.i. by extraction/quantitation and 36–60 h.p.i. by MSI, suggesting that SAPI is a major reservoir for early phase donation of AA for lipid inflammation. Pro- and antiinflammatory products of both the COX and LOX pathways were detected by LC/MS analysis. Prostaglandin D2 (PGD2, an antiinflammatory product) was initially depleted, but returned to baseline levels at 48 h.p.i. (Fig. 3B), whereas PGE2 (a highly proinflammatory product) was first detected at 36 h.p.i. and induced >50-fold by 48 h.p.i. (Fig. 3C). The LOX-dependent pathway showed a less than fivefold induction of proinflammatory 12(S)- and 15(S)-hydroxyeicosatetraenoic acids (HETEs) and a twofold reduction in the proresolving lipoxygenase A4 (LXA4) (SI Appendix, Fig. S12). These results suggested a central and potent role for COX-2–dependent products in propagating early phase inflammation, as an eicosanoid storm launching rapid lethality in F. novicida infection. We therefore evaluated the progression of F. novicida infection (SQ) in COX-2 knockout (KO) mice (Ptgs2−/−) compared with WT (C57BL/6) mice. COX-2 KO mice experienced a delay in clinical onset of disease and were significantly protected from lethal infection with F. novicida compared with wild-type controls (Fig. 3 D and E). At 72 h.p.i., the spleens of COX-2 KO mice maintained intact white pulp (SI Appendix, Fig. S13). COX-2–dependent metabolic products aggravate the early phase of F. novicida infection and contribute to rapid lethality.
Fig. 3.
Cyclooxygenase (COX)-2 inflammatory pathway contributes to rapid lethality in F. novicida infection. (A–C) Quantitative LC/MS of substrate and metabolites of COX-2 enzyme normalized to total input splenic protein content (in grams) evaluated every 12 h.p.i., lethal dose F. novicida, SQ. (A) Free AA. (B) Prostaglandin D2 (PGD2). (C) Prostaglandin E2 (PGE2). Lipid levels were analyzed using one-way ANOVA for injection time points, *P < 0.05, **P < 0.01, ***P < 0.001 as given by bars, ###P < 0.001 versus 48 h.p.i. (n = 10). Error bars denote SEM. (D and E) Infection in wild-type versus COX-2 knockout (Ptgs2−/−) mice (n = 8) with F. novicida, SQ. (D) Clinical scores evaluated every 12 h.p.i. using a 5-point scale (SI Appendix, SI Methods). (E) Survival percentages, survival of COX-2 knockout mice evaluated using log-rank test, *P = 0.0001. Representative of two replicate survival studies, both significant at P < 0.05.
Three-Dimensional Reconstruction of the Host–Pathogen Interaction.
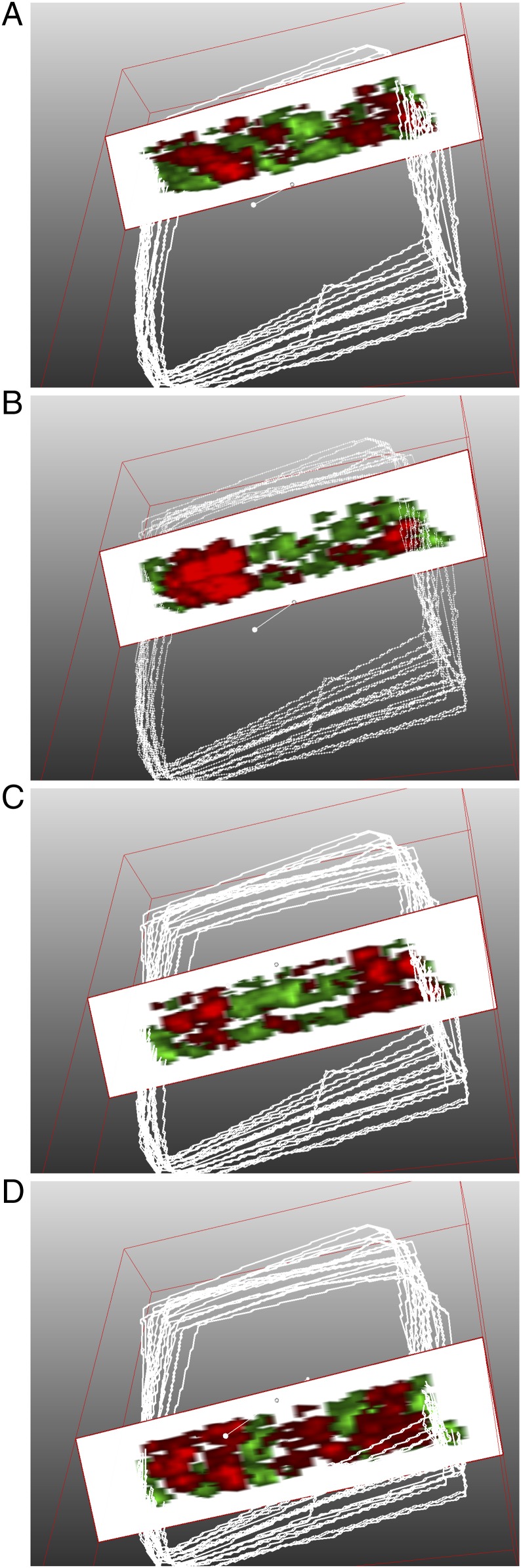
To illustrate the complex interface between host and pathogen lipids, we reconstructed the host–pathogen interface using 3D MSI. Recent advances in high-speed (49) and high-spatial resolution MALDI-MSI facilitated 3D imaging and subsequent volume renderings of infected spleens. Using the white pulp-excluded lipid A signature and the white pulp-biased SAPI signature, we reconstructed the host–pathogen molecular interface. Spleens from F. novicida-infected animals (48 h.p.i.) were serially sectioned, SAPI/lipid A were codetected in negative ion mode using an ultra-high speed MALDI-TOF, and the images were rendered into 3D volumes. Cutaway views of the 3D volume rendering (Movie S1) demonstrate the extent of lipid A exclusion to the red pulp (green, Fig. 4) that extends the length and width of the tissue. SAPI signal (red, Fig. 4B) is present throughout multiple, reconstructed white pulp nodules. In an organized white pulp nodule (36 h.p.i., Fig. 2A) reduction of SAPI was observed in the center of several nodules corresponding to germinal centers, suggesting lower SAPI abundance in lymphoid cells. At 48 h.p.i., the distribution of lipid A completely encircles the white pulp (Fig. 1A); therefore, the edge of lipid A signatures clearly outlines the edges of the white pulp. Using this marker, there are several lipid A-annotated white pulp nodules with low peripheral (marginal zone) SAPI abundance. SAPI depletion in the marginal zone, an area dedicated to phagocytosis and transition from red-to-white pulp, suggests stimulation and AA release in the resident antigen-presenting cells.
Fig. 4.
Three-dimensional molecular reconstruction of the host–pathogen lipid profile visualizes the lethal septic interface. (A–D) Slice cutaway views of 3D volume reconstruction from two-channel image. MALDI-TOF, 20 μm x–y spatial resolution, 12-μm section thickness, negative ion mode, formalin-fixed tissue (center third of F. novicida-infected spleen, 48 h.p.i.) prepared for endotoxin and host lipid simultaneous coimaging. White outlines are the tissue margins from MSI setup and define the outer scan area, not necessarily the tissue perimeter. Green: m/z 1,665, identified as lipid A (Fig. 1 and SI Appendix, Fig. S4). Red: m/z 885.6, identified as SAPI (Fig. 2 and SI Appendix, Fig. S6). Intensities are optimized for 3D view and for fidelity to 2D images (Movie S1).
Discussion/Conclusion
We have directly mapped a known bacterial virulence factor by MSI, in vivo. Before this work, MSI studies in infection models have focused on molecules involved in the host response. These results presented here underscore the role of lipid-mediated inflammation in propagating severe pathology in a bacterial infection. F. novicida infection results in the sharp induction of the proinflammatory lipid mediator PGE2, resulting in the free and replenished cytosolic AA pool. SAPI and other AA-containing phospholipids serve as an important cellular reservoir for the upstream COX-2–mediated precursors of PGE2, a result of F. novicida infection that can be visualized at the first step using MSI (Fig. 2A). Using 3D molecular imaging (SAPI/endotoxin), we defined the host–pathogen interface, used it to illuminate an organized immune response, and defined a role for SAPI depletion as a biomarker for lipid-involved inflammation in tissues. These studies simultaneously map an established bacterial virulence factor (F. novicida lipid A) with a host lipid inflammatory precursor (SAPI) for use in identifying and defining molecular mechanisms of bacterial pathogenesis.
Materials and Methods
Expanded methods are available in SI Appendix.
Bacterial Strains and Growth Conditions.
F. novicida strain U112 wild type was cultured on tryptic soy broth or agar containing 0.1% l-cysteine supplementation at 37 °C and with 225 rpm orbital shaking, where applicable. The mutant strain F. novicida ΔlpxD1 (9) was cultured under the same conditions in the presence of kanamycin at 20 μg/mL.
Mouse Infections and Tissue Collection.
Adult female C57BL/6 and Ptgs2−/− (COX-2 KO) mice received a subcutaneous dose of 300 cfu (wild-type F. novicida) in 50 μL sterile PBS. Infections with F. novicida ΔlpxD1 were performed as above with a dose of 5.5 × 106 cfu. Following carbon dioxide narcosis and thoracotomy, spleen and blood were collected. Bisected spleens were either snap frozen on liquid nitrogen or formalin fixed for 18 h, then snap frozen and stored at −80 °C for imaging experiments and RNA extraction. Blood was collected in tubes containing 10% (by volume) citrate buffer as an anticoagulant and serially plated for cfu enumeration. Whole spleens were homogenized in 1 mL PBS and serially plated for cfu enumeration. All studies were performed under humane guidelines approved by the Institutional Animal Care and Use Committee at the University of Maryland Baltimore.
Mass Spectrometry Imaging.
For 2D MALDI-TOF images, fresh frozen spleen was sectioned at 10-μm thickness and at 13 μm for both 2D MALDI-Fourier transform ion cyclotron resonance (FTICR) and 3D MALDI-TOF images. All sections for MSI were heat-mounted to indium-tin oxide (ITO) slides, desiccated, and spray coated with norharmane (NRM) matrix using a Bruker ImagePrep (for 2D MALDI-TOF) or a SunCollect Matrix Sprayer (for 2D MALDI-FTICR and 3D MALDI-TOF images). Matrix mixtures consisted of 12 mg/mL NRM in 2:1:0.5 parts chloroform:methanol:water for lipid A imaging (23) or 10 mg/mL NRM in 1:2:0.5 parts chloroform:methanol:water for phospholipid imaging. Two-dimensional MALDI-TOF and FTICR images were captured at 75 μm rastering, whereas the 3D reconstruction was performed at 20 μm rastering. All images were normalized to total ion current (TIC).
Quantitation of Eicosanoid and Phospholipids.
Extraction and quantitation of PLs and eicosanoids were performed according to previously published methods (50). Results were normalized to protein content in the aqueous fraction of the lipid extraction using the bicinchoninic acid (BCA) assay according to manufacturer’s instructions (Thermo Scientific Pierce).
RNA Extraction and Quantitative RT-PCR.
RNA was extracted from spleens using the TriZol extraction method according to manufacturer’s protocol (LifeTech). Comparative fold changes were determined using the ΔΔCt method and bacterial transcript abundance was evaluated using the copy count method. Primers corresponsing to F. novicida genes were designed using the RealTime PCR Design Tool (IDT).
Supplementary Material
Acknowledgments
We thank Drs. Harold R. Neely and Martin F. Flajnik for their contributions to the early development of this work; Dr. Stefanie Vogel for generous donation of the Ptgs2−/− knockout mice and guidance; Dr. Dennis Treade of SCiLS for assistance with 3D MSI video rendering; and Dr. Sung Hwan Yoon for review and critique of the fragmentation data. A.J.S. was supported by NIH Grants AI007540 and AI095190. This work was supported in part by NIH Grants AI123820 and GM111066 (to R.K.E.) and the Dutch Province of Limburg (S.R.E. and R.M.A.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712887114/-/DCSupplemental.
References
- 1.Raetz CRH. Biochemistry of endotoxins. Annu Rev Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 2.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 4.Beutler B, Rietschel ET. Innate immune sensing and its roots: The story of endotoxin. Nat Rev Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 5.Trent MS, Stead CM, Tran AX, Hankins JV. Diversity of endotoxin and its impact on pathogenesis. J Endotoxin Res. 2006;12:205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- 6.Gunn JS. Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotoxin Res. 2001;7:57–62. [PubMed] [Google Scholar]
- 7.Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 8.Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in lipid A structure in the pathogenic yersiniae. Mol Microbiol. 2004;52:1363–1373. doi: 10.1111/j.1365-2958.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, et al. LPS remodeling is an evolved survival strategy for bacteria. Proc Natl Acad Sci USA. 2012;109:8716–8721. doi: 10.1073/pnas.1202908109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott AJ, Oyler BL, Goodlett DR, Ernst RK. Lipid A structural modifications in extreme conditions and identification of unique modifying enzymes to define the Toll-like receptor 4 structure-activity relationship. Biochimi Biophys Acta. 2017;1862:1439–1450. doi: 10.1016/j.bbalip.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raetz CRH. Bacterial endotoxins: Extraordinary lipids that activate eucaryotic signal transduction. J Bacteriol. 1993;175:5745–5753. doi: 10.1128/jb.175.18.5745-5753.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunn JS, Ernst RK. The structure and function of Francisella lipopolysaccharide. Ann N Y Acad Sci. 2007;1105:202–218. doi: 10.1196/annals.1409.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaros TG, Blancett CD, Bell TM, Natesan M, Ulrich RG. Serum biomarkers of Burkholderia mallei infection elucidated by proteomic imaging of skin and lung abscesses. Clin Proteomics. 2015;12:7. doi: 10.1186/s12014-015-9079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry–Looking beyond classical histology. Nat Rev Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 15.Amstalden van Hove ER, Smith DF, Heeren RMA. A concise review of mass spectrometry imaging. J Chromatogr A. 2010;1217:3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 16.Gode D, Volmer DA. Lipid imaging by mass spectrometry:–A review. Analyst (Lond) 2013;138:1289–1315. doi: 10.1039/c2an36337b. [DOI] [PubMed] [Google Scholar]
- 17.Moore JL, Caprioli RM, Skaar EP. Advanced mass spectrometry technologies for the study of microbial pathogenesis. Curr Opin Microbiol. 2014;19:45–51. doi: 10.1016/j.mib.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heeren RMA. Getting the picture: The coming of age of imaging MS. Int J Mass Spectrom. 2015;377:672–680. [Google Scholar]
- 19.Attia AS, et al. Monitoring the inflammatory response to infection through the integration of MALDI IMS and MRI. Cell Host Microbe. 2012;11:664–673. doi: 10.1016/j.chom.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore JL, et al. Imaging mass spectrometry for assessing temporal proteomics: Analysis of calprotectin in Acinetobacter baumannii pulmonary infection. Proteomics. 2014;14:820–828. doi: 10.1002/pmic.201300046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy RC, Hankin JA, Barkley RM. Imaging of lipid species by MALDI mass spectrometry. J Lipid Res. 2009;50(Suppl):S317–S322. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miura D, Fujimura Y, Wariishi H. In situ metabolomic mass spectrometry imaging: Recent advances and difficulties. J Proteomics. 2012;75:5052–5060. doi: 10.1016/j.jprot.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Scott AJ, et al. Norharmane matrix enhances detection of endotoxin by MALDI-MS for simultaneous profiling of pathogen, host and vector systems. Pathog Dis. 2016;74:ftw097. doi: 10.1093/femspd/ftw097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, et al. Structure and biosynthesis of free lipid A molecules that replace lipopolysaccharide in Francisella tularensis subsp. novicida. Biochemistry. 2006;45:14427–14440. doi: 10.1021/bi061767s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steiner DJ, Furuya Y, Metzger DW. Host-pathogen interactions and immune evasion strategies in Francisella tularensis pathogenicity. Infect Drug Resist. 2014;7:239–251. doi: 10.2147/IDR.S53700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones CL, et al. Subversion of host recognition and defense systems by Francisella spp. Microbiol Mol Biol Rev. 2012;76:383–404. doi: 10.1128/MMBR.05027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cowley SC, Elkins KL. Immunity to Francisella. Front Microbiol. 2011;2:26. doi: 10.3389/fmicb.2011.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 29.Humes JL, et al. Macrophages synthesis and release prostaglandins in response to inflammatory stimuli. Nature. 1977;269:149–151. doi: 10.1038/269149a0. [DOI] [PubMed] [Google Scholar]
- 30.Woolard MD, et al. Francisella tularensis-infected macrophages release prostaglandin E2 that blocks T cell proliferation and promotes a Th2-like response. J Immunol. 2007;178:2065–2074. doi: 10.4049/jimmunol.178.4.2065. [DOI] [PubMed] [Google Scholar]
- 31.Navratil AR, Brummett AM, Bryan JD, Woolard MD. Francisella tularensis LVS induction of prostaglandin biosynthesis by infected macrophages requires specific host phospholipases and lipid phosphatases. Infect Immun. 2014;82:3299–3311. doi: 10.1128/IAI.02060-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serhan CN, Ward PA, Gilroy DW. Fundamentals of Inflammation. Cambridge Univ Press; Cambridge, UK: 2010. [Google Scholar]
- 33.Li Y, Wang X, Ernst RK. A rapid one-step method for the characterization of membrane lipid remodeling in Francisella using matrix-assisted laser desorption ionization time-of-flight tandem mass spectrometry. Rapid Commun Mass Spectrom. 2011;25:2641–2648. doi: 10.1002/rcm.5168. [DOI] [PubMed] [Google Scholar]
- 34.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 35.Shaffer SA, Harvey MD, Goodlett DR, Ernst RK. Structural heterogeneity and environmentally regulated remodeling of Francisella tularensis subspecies novicida lipid A characterized by tandem mass spectrometry. J Am Soc Mass Spectrom. 2007;18:1080–1092. doi: 10.1016/j.jasms.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasmussen JW, et al. Phenotypic, morphological, and functional heterogeneity of splenic immature myeloid cells in the host response to tularemia. Infect Immun. 2012;80:2371–2381. doi: 10.1128/IAI.00365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaurand P. Imaging mass spectrometry of thin tissue sections: A decade of collective efforts. J Proteomics. 2012;75:4883–4892. doi: 10.1016/j.jprot.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Scott AJ, et al. Mass spectrometry imaging enriches biomarker discovery approaches with candidate mapping. Health Phys. 2014;106:120–128. doi: 10.1097/HP.0b013e3182a4ec2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gil-de-Gómez L, et al. A phosphatidylinositol species acutely generated by activated macrophages regulates innate immune responses. J Immunol. 2013;190:5169–5177. doi: 10.4049/jimmunol.1203494. [DOI] [PubMed] [Google Scholar]
- 40.Scott WA, Zrike JM, Hamill AL, Kempe J, Cohn ZA. Regulation of arachidonic acid metabolites in macrophages. J Exp Med. 1980;152:324–335. doi: 10.1084/jem.152.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu ZH, de Carvalho MS, Leslie CC. Regulation of phospholipase A2 activation by phosphorylation in mouse peritoneal macrophages. J Biol Chem. 1993;268:24506–24513. [PubMed] [Google Scholar]
- 42.Dennis EA. Regulation of eicosanoid production: Role of phospholipases and inhibitors. Nat Biotechnol. 1987;5:1294–1300. [Google Scholar]
- 43.Serhan CN, Haeggström JZ, Ayoub SS. Lipid mediators in acute inflammation and resolution: Eicosanoids, PAF, resolvins, and protectins. In: Serhan CN, Ward PA, Gilroy DW, editors. Fundamentals of Inflammation. Cambridge Univ Press; Cambridge, UK: 2009. pp. 153–174. [Google Scholar]
- 44.Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: Dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol. 2008;8:349–361. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole LE, et al. Macrophage proinflammatory response to Francisella tularensis live vaccine strain requires coordination of multiple signaling pathways. J Immunol. 2008;180:6885–6891. doi: 10.4049/jimmunol.180.10.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh A, et al. Lipoxin A4, a 5-lipoxygenase pathway metabolite, modulates immune response during acute respiratory tularemia. J Leukoc Biol. 2017;101:531–542. doi: 10.1189/jlb.4A0815-365RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leclerc P, et al. IL-1β/HMGB1 complexes promote the PGE2 biosynthesis pathway in synovial fibroblasts. Scand J Immunol. 2013;77:350–360. doi: 10.1111/sji.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gijón MA, Riekhof WR, Zarini S, Murphy RC, Voelker DR. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J Biol Chem. 2008;283:30235–30245. doi: 10.1074/jbc.M806194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogrinc Potočnik N, Porta T, Becker M, Heeren RMA, Ellis SR. Use of advantageous, volatile matrices enabled by next-generation high-speed matrix-assisted laser desorption/ionization time-of-flight imaging employing a scanning laser beam. Rapid Commun Mass Spectrom. 2015;29:2195–2203. doi: 10.1002/rcm.7379. [DOI] [PubMed] [Google Scholar]
- 50.Lerner R, Post J, Loch S, Lutz B, Bindila L. Targeting brain and peripheral plasticity of the lipodome in acute kainic acid-induced epileptic seizures in mice via quantitative mass spectometry. Biochim Biophys Acta. 1862;1862:255–267. doi: 10.1016/j.bbalip.2016.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.