Significance
Regulatory T cells (Tregs) play a critical role in maintaining immune tolerance to self-antigens and in suppressing excessive immune responses that may cause collateral damage to the host. Unlike other CD4+ T cells, Tregs have a distinct, yet-to-be-established metabolic machinery to produce energy for survival and function. Here we show that the metabolic sensor LKB1 is critical for the survival and function of Tregs through regulation of their cellular metabolism. Interestingly, AMP-activated protein kinase, the best-studied downstream kinase of LKB1, is largely dispensable for LKB1 function in Tregs; the MAP/microtubule affinity-regulating kinases and salt-inducible kinases may mediate its functions. We highlight LKB1 as metabolic regulator that links cellular metabolism to immune cell functions.
Keywords: Lkb1, Treg, Foxp3, cellular metabolism, autoimmune disease
Abstract
The metabolic programs of functionally distinct T cell subsets are tailored to their immunologic activities. While quiescent T cells use oxidative phosphorylation (OXPHOS) for energy production, and effector T cells (Teffs) rely on glycolysis for proliferation, the distinct metabolic features of regulatory T cells (Tregs) are less well established. Here we show that the metabolic sensor LKB1 is critical to maintain cellular metabolism and energy homeostasis in Tregs. Treg-specific deletion of Lkb1 in mice causes loss of Treg number and function, leading to a fatal, early-onset autoimmune disorder. Tregs lacking Lkb1 have defective mitochondria, compromised OXPHOS, depleted cellular ATP, and altered cellular metabolism pathways that compromise their survival and function. Furthermore, we demonstrate that the function of LKB1 in Tregs is largely independent of the AMP-activated protein kinase, but is mediated by the MAP/microtubule affinity-regulating kinases and salt-inducible kinases. Our results define a metabolic checkpoint in Tregs that couples metabolic regulation to immune homeostasis and tolerance.
The balance between protective immunity and excessive inflammation in the immune system requires tight regulation of the activation/differentiation of different types of immune cells (1). On recognition of foreign antigens and receiving proper costimulatory signals, naïve lymphocytes become activated and undergo rapid cell growth and proliferation, a process that requires sufficient energy generation and biosynthesis (2). The tailoring of cellular metabolism to discrete T cell subsets is central for proper T cell differentiation and function (2, 3).
T regulatory cells (Tregs) are a subset of CD4+ T cells that maintain immune system homeostasis through promoting self-tolerance and preventing autoimmune responses (4). Consistent with the idea that functionally distinct T cell subsets require distinct energetic and biosynthetic pathways to support their functional needs (5), Tregs are characterized by a unique metabolic signature that is different from conventional effector T cells (Teffs). For example, Th1, Th2, and Th17 express high levels of Glut1 and thus are highly glycolytic, while Tregs have low levels of Glut1 and high rates of lipid oxidation in vitro (3). This metabolic feature in Tregs led us to hypothesize that the LKB1–AMP-activated protein kinase (AMPK) axis, a central regulator that promotes mitochondrial oxidative metabolism rather than glycolysis (6), might play a critical role in Treg survival and function (3, 6–8). While AMPK has been identified as a key regulator of Teff immune response, the physiological relevance of AMPK in Tregs has not been clearly established. Moreover, whether LKB1, the upstream kinase of AMPK and a well-known metabolic sensor, plays a significant role in the cellular metabolism in Tregs, is unclear, although it is known to be involved in conventional T cell development and proliferation (9, 10).
Using genetic mouse models, here we found that LKB1, but not AMPK, is critical for the maintenance of cellular metabolism and energy homeostasis in Tregs. Tissue-specific deletion of Lkb1 in Tregs causes a severe autoimmune phenotype in mice. Tregs lacking Lkb1 have defective mitochondria, compromised oxidative phosphorylation (OXPHOS), depleted cellular ATP, and altered cellular metabolism pathways that impair their survival and function. We further show that the function of LKB1 in Tregs is mediated not by AMPK, but rather by MAP/microtubule affinity-regulating kinases (MARKs) and salt-inducible kinases (SIKs). Our findings highlight LKB1 as an essential metabolic regulator in Tregs that couples metabolic regulation to immune homeostasis and tolerance.
Results
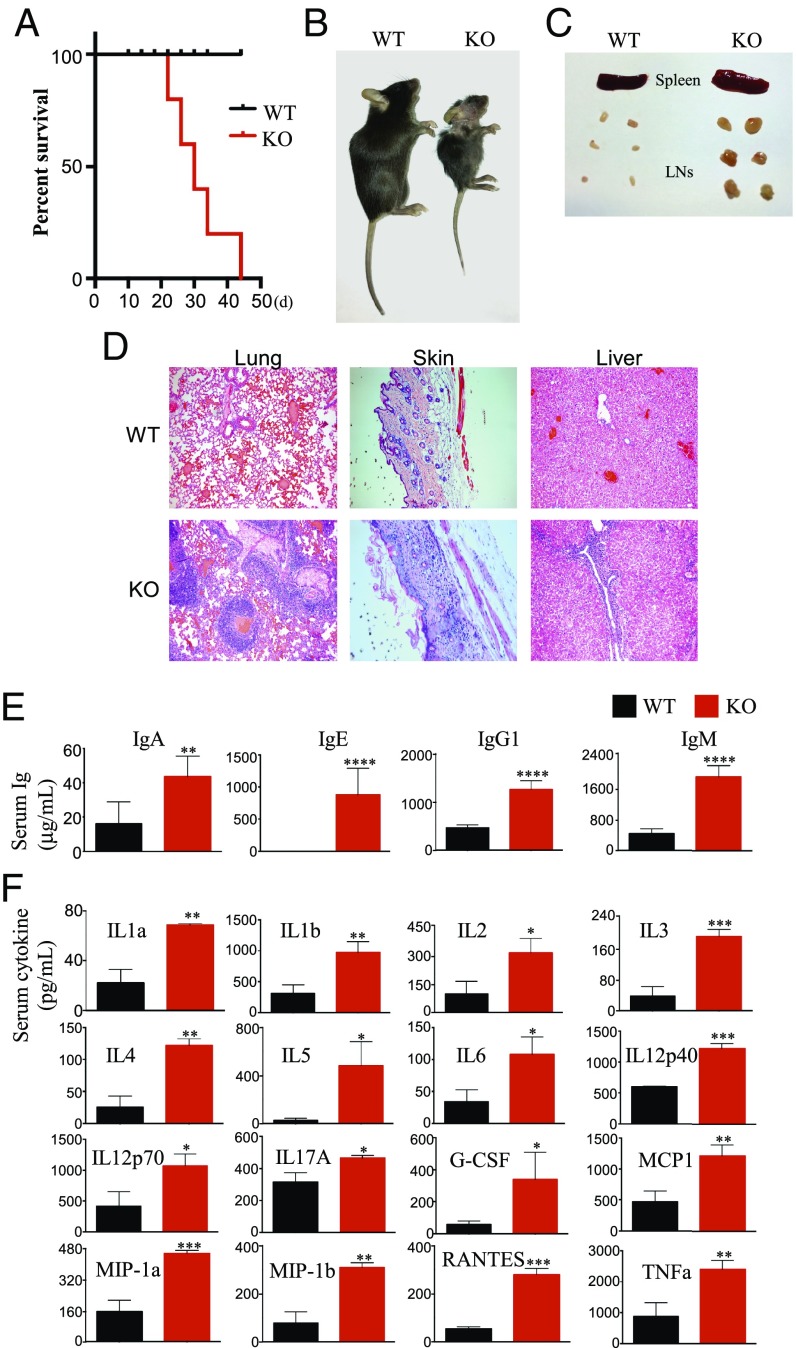
To explore the signaling pathways regulating Treg metabolism, we deleted Prkaa1, encoding the catalytic subunit of AMPK, and Lkb1 (also known as Stk11), encoding the upstream kinase regulating AMPK, selectively in Tregs (Prkaa1fl/fl and Lkb1fl/fl mice crossed to Foxp3YFPCre mice, respectively) (11). Surprisingly, the genetic deletion of AMPK in Tregs did not lead to any detectable abnormalities in mice under normal conditions. In marked contrast, mice lacking Lkb1 only in Tregs (Lkb1fl/flFoxp3YFPCre, designated KO) developed a profound inflammatory disorder, with all KO mice dying by 3–6 wk of age (Fig. 1A). At 3 wk, KO mice were markedly smaller than control mice (Lkb1+/+Foxp3YFPCre, designated WT), and displayed multiple symptoms suggestive of a profound immune response, including hunched posture, closed eyelids, crusty ears and tail, and skin ulcerations on the neck and upper back (Fig. 1B). Consistent with an unchecked inflammatory response, KO mice exhibited splenomegaly and lymphadenopathy (Fig. 1C), as well as extensive infiltration of leukocytes into multiple organs, including the lung, skin, and liver (Fig. 1D and Fig. S1A). Furthermore, the levels of serum Ig isotypes (Fig. 1E) and a wide array of inflammatory cytokines were significantly increased in the sera of KO mice (Fig. 1F). The phenotypic similarities of these mice with those harboring mutations in or deletion of Foxp3, the master regulator for Tregs (12–15), suggest an essential role of Lkb1 in Treg biology.
Fig. 1.
Lkb1 deletion in Tregs leads to a fatal early-onset autoimmune disorder. (A) Percent survival of WT (Lkb1+/+,Foxp3YFPCre) and KO (Lkb1fl/fl,Foxp3YFPCre) mice (n = 30–40). (B) Representative images of 35-d-old WT and KO mice. (C) Representative images of spleen and peripheral LNs from 21-d-old WT and KO mice. (D) H&E staining of lung, skin, and liver sections of WT and KO mice. (Magnification: 40×.) (E) Concentrations of IgA, IgE, IgG1, and IgM in sera of WT and KO mice (n = 8–10) determined by ELISA. (F) Concentrations of cytokines in sera of WT and KO mice (n = 8–10) determined by the Bio-Plex Pro Mouse Cytokine 23-Plex Immunoassay. Statistical significance was determined by Student's unpaired t test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
In line with the severe immunopathology, the T cell population was expanded in KO mice, with the total T cell numbers increasing by ∼50% in the spleen and by ∼450% in the lymph nodes (LNs) (Fig. S1B). In addition, an increase in memory/effector cells (CD44hiCD62Llo) was accompanied by a decrease in the CD4+/CD8+ ratio in KO mice (Fig. 2A and Fig. S1C). Consequently, KO mice had significant increases in IFN-γ–, IL-17–, and IL-4–producing CD4+ T cells (Fig. 2B). This marked imbalance in the immune system of KO mice suggests that LKB1 is crucial for the development and/or function of Tregs. Indeed, the percentage of Tregs in CD4+ T cells was significantly reduced in both the spleen and LNs of KO mice (Fig. 2C). Notably, this reduction was not due to developmental defects, because at 12 d, when the inflammatory symptoms are barely detectable, the percentage of thymic Tregs was similar in WT and KO mice (Fig. S2). However, the absolute number of Tregs was higher in KO mice compared with WT mice, suggesting that loss of LKB1 compromises the suppressive function of Tregs. Consistent with this, Lkb1-deficient Tregs were less competent than WT Tregs at suppressing naïve CD4+ T responder (Tresp) cell proliferation in vitro (Fig. 2D). Furthermore, we also tested the suppressive function of KO Tregs in vivo in a Teff transfer-induced colitis model (CD4+CD45RBhiCD25− cells injected into Rag1−/− mice; Fig. 2E). While WT Tregs could stall the development of colitis in recipient mice, Lkb1-deficient Tregs were unable to prevent disease progression (Fig. 2 F and G and Fig. S3).
Fig. 2.
Ablation of Lkb1 abrogates Treg function and results in uncontrolled immune activation. (A) Flow cytometry analysis of CD44 and CD62L expression on CD4+ T cells in spleen and peripheral LNs of WT and KO mice (n = 3–5). (B) Flow cytometry analysis of cytokine production by splenic CD4+ T cells from WT and KO mice (n = 3–5). (C) Treg percentage in spleen and LNs of 3- to 4-wk-old WT and KO mice (n = 3–5). (D) In vitro suppression of CFSE-labeled WT naïve CD4+ T responder cells (Tresp) by WT and KO Tregs. (E–G) WT or KO Tregs purified from CD45.1−CD45.2+ mice and Teffs (CD4+CD45RBhiCD25−) isolated from CD45.1+CD45.2− mice were mixed and transferred in to Rag1−/− recipient mice (n = 5) through retroorbital injection. Changes in body weight (F) and H&E staining of colon sections (magnification: 40×) (G) of each group are shown. Data are shown as mean ± SD and are representative of three experiments. Statistical significance was determined by Student's unpaired t test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Because LKB1 is required for maintaining mitochondrial mass, cellular metabolism, and energy homeostasis in liver and muscle (16, 17), we posited that the loss of LKB1 compromises the cellular metabolism of Tregs. Indeed, decreased mitochondrial mass and membrane potential were seen in KO Tregs, leading to reductions in both mitochondrial oxygen consumption rate (OCR) and maximal mitochondrial oxidative capacity, along with a significantly lower basal ATP level (Fig. 3 A–C). Notably, the reduced mitochondrial respiration in Lkb1-deficient Tregs was accompanied by significantly compromised glycolysis, as measured by the extracellular acidification rate (ECAR) (Fig. 3B). The decreased overall metabolic activity in KO Tregs was in agreement with a reduced cellular level of reactive oxygen species (ROS) (Fig. 3D).
Fig. 3.
Lkb1 is required for Tregs to maintain cellular metabolism and energy homeostasis. (A) Mitochondrial mass and membrane potential of freshly purified WT and KO Tregs assayed by Mitotracker and DilC1(5) staining, respectively. (B) OCR and ECAR in freshly purified WT and KO Tregs under basal conditions and in response to sequential addition of oligomycin, fluorocarbonyl cyanide phenylhydrazone (FCCP), rotenone (Rtn), and antimycin A (AA). (C and D) ATP concentrations and ROS levels in freshly purified WT and KO Tregs, respectively. All data are representative of three experiments and are expressed as mean ± SD. (E and F) KEGG pathway analysis and functional annotation of down-regulated gene categories in KO Tregs, respectively. (G) Representative down-regulated gene lists in KO Tregs. Statistical significance was determined by Student's unpaired t test (**P < 0.01; ***P < 0.001).
To gain insight into the molecular mechanisms underlying the defects in Lkb1-deficient Tregs, we compared the transcriptomes of WT and KO Tregs. To exclude potentially secondary effects caused by systemic inflammation, we performed RNA-Seq with KO Tregs from phenotypically normal female Foxp3YFPCre/WT heterozygous mice. The X chromosome location of the Foxp3-driven Cre, combined with random X inactivation in females (18), allowed us to sort KO Tregs (Lkb1fl/flFoxp3YFPCre) from female Lkb1fl/flFoxp3YFPCre/WT mice, in which coexisting WT Tregs (Lkb1fl/flFoxp3WT) prevent the marked inflammatory response seen in male mice. Remarkably, loss of Lkb1 led to extensive transcriptional changes, with up-regulated expression of 1,363 genes and down-regulated expression of 1,306 genes (nominal P < 0.01). Pathway analysis indicated that the down-regulated genes were enriched in metabolism-related pathways, including OXPHOS, nucleotide metabolism, amino acid synthesis and metabolism, glycolysis, and the tricarboxylic acid (TCA) cycle (Fig. 3E). Functional annotation analysis indicated that many of the down-regulated genes in KO Tregs fell into the functional categories that maintain mitochondrial integrity and energy homeostasis (Fig. 3F). Notably, genes encoding subunits of the electron transport chain (complexes I–IV) and ATP synthases (complex V) were specifically down-regulated on Lkb1 deletion in Tregs (Fig. 3G, Left), consistent with defective integrity and function of mitochondria (Fig. 3 A–C). Other representative down-regulated genes include those involved in pyrimidine/purine metabolism (Fig. 3G, Middle), glycolysis, biosynthesis of amino acids, and cysteine and methionine metabolism (Fig. 3G, Right). The altered expression levels of representative genes were confirmed by quantitative RT-PCR (qRT-PCR) (Fig. S4). In addition, an array of Treg signature suppressive molecules (19–22) were also down-regulated in KO Tregs (Fig. S5), in agreement with their defective in vitro and in vivo suppression functions (Fig. 2 E and F).
Given the overall defects in cellular metabolism, we questioned whether Lkb1 deficiency would compromise Treg survival. Indeed, a significantly lower proportion of KO Tregs were able to survive when cultured in vitro (TCR and IL-2 stimulation for 3 d; Fig. 4A). Furthermore, few surviving KO Tregs were found at 5 wk posttransplantation in the Teff-induced colitis model, while WT Tregs survived and proliferated (Fig. 4B). Interestingly, although Foxp3 was still maintained at high levels in surviving WT Tregs, a small percentage of surviving KO Tregs lost Foxp3 expression (Fig. 4B), consistent with a recent observation that LKB1 can also maintain the stability of Foxp3 expression (23). In addition, the level of cleaved caspase-3 in KO Tregs was more than double that found in WT Tregs at 12 h after in vitro stimulation with TCR and IL-2 (Fig. 4C), consistent with increased apoptosis and decreased survival (Fig. 4 A and B). Furthermore, KO Tregs had elevated levels of the DNA damage response marker phospho(Ser139)-histone H2AX (phospho-H2AX) (24, 25) (Fig. 4D). Taken together, these results indicate that increased cellular stress in Lkb1-deficient Tregs led to their failure in survival and expansion both in vitro and in vivo.
Fig. 4.
Lkb1 deletion compromises Treg survival, and its downstream kinases MARKS and SIKS mediate Lkb1 function in Tregs. (A) Flow cytometry analysis of survival of WT and KO Tregs over time in vitro. (B) WT or KO Tregs purified from CD45.1−CD45.2+ mice and Teffs (CD4+CD45RBhiCD25−) isolated from CD45.1+CD45.2− mice were mixed and retroorbitally injected in Rag1−/− recipient mice (n = 5). Cellularity was analyzed at 5 wk after initial transfer. (C) Apoptosis in WT and KO Tregs assayed by cleaved caspase-3 staining after in vitro culturing for 12 h. (D) DNA damage response in WT and KO Tregs assayed by phosphor-H2A.x staining after in vitro culturing for 12 h. (E) Expression profile of Lkb1 downstream kinases in Tregs. (F) Flow cytometry analysis of the survival of WT Tregs in response to different combination of inhibitors targeting AMPK, MARKS, SIKS, and NUAKS. All data are representative of three experiments. Statistical significance was determined by Student's unpaired t test [not significant (n.s.): P > 0.05; ***P < 0.001; ****P < 0.0001].
As an evolutionarily conserved regulator of cellular metabolism, LKB1 functions upstream of the AMPK-related kinases (26–28). To identify potential downstream mediators in Tregs, we profiled the expression of its known substrates AMPK, SIK1/2/3, MARK1/2/3/4, BRSK1/2, NUAK1/2, and SNRK. Eight of these kinases were expressed at detectable levels, with Mark2 and Snrk having the highest relative expression (Fig. 4E). When assaying the survival of WT Tregs in response to TCR/IL-2 stimulation in vitro (Fig. 4 A and F, Top), we screened chemical inhibitors for those that could replicate the compromised survival seen in KO Tregs. Interestingly, inhibitors targeting AMPK, MARK, SIK, and NUAK had little effect as single agents on WT Treg survival, suggesting redundancy in the downstream pathways (Fig. 4F, Top); however, the combination of the four inhibitors reduced the ability of WT Tregs to survive the TCR/IL-2 treatment to a level similar to that of KO Tregs. Combinations of three of the four inhibitors indicated that the AMPK and NUAK pathways were unlikely critical effectors of LKB1, while MARK and SIK inhibitors were required to mimic the KO phenotype (Fig. 4F, Middle). Furthermore, the dual combination of the MARK and SIK inhibitors was sufficient to down-regulate WT Treg survival, while inhibitors targeting AMPK and NUAK had no effect (Fig. 4F, Bottom), implicating the MARK and SIK kinases as critical effectors of LKB1 in maintaining energy homeostasis in Tregs.
Discussion
In recent years, immunometabolism has emerged as a rapidly advancing field at the interface of immunology and metabolism (29–31). Consistent with the notion that cellular metabolism underpins T cell function, we identify an essential role for the metabolic sensor LKB1 in Treg survival and function, as Treg-specific deletion of Lkb1 in mice leads to an uncontrolled, systemic autoimmune disorder with reduced Treg number and compromised Treg function. We further demonstrate that Lkb1-deficient Tregs have defective mitochondria and thus compromised OXPHOS with depleted cellular ATP levels. Gene expression analyses indicate that glycolysis is also compromised in KO Tregs, raising the possibility that the observed reduction in ECAR may be a consequence of reduced glucose uptake (Fig. 3 E and G). In addition, many other cellular metabolism pathways, such as pyrimidine/purine metabolism, biosynthesis of amino acids, and cysteine and methionine metabolism, are also dysregulated in Lkb1-deficient Tregs. We conclude that Lkb1 deletion leads to major defects in cellular metabolism and energy homeostasis that have profound negative impacts on Treg survival and function, similar to those reported for hematopoietic cells (32–34).
To our surprise, LKB1 regulates multiple metabolic pathways in Tregs independently of AMPK, relying instead on MARK and SIK kinases as downstream metabolic effectors. Thus, while LKB1 is the essential upstream kinase of AMPK, it uses variant downstream effectors in a cell type-specific manner (16, 33, 35). MARK2 has been implicated in immune system homeostasis (36) and metabolism (37), but the role of SIKs in metabolism is not well characterized (38, 39). Further dissecting the roles of LKB1 downstream kinases, such as MARKs and SIKs, would help elucidate how LKB1 links metabolic regulation to Treg fitness and immune homeostasis.
It has been increasingly recognized that cellular metabolism exerts a major impact on Treg function and even fate determination, although very few regulators/signaling pathways have been identified so far. The hypoxia-inducible factor 1 (HIF-1)-mediated glycolytic program suppresses inducible Treg (iTreg) generation but promotes Th17 differentiation (40). In addition, Treg-specific deletion of Raptor, encoding the defining subunit of mTORC1, leads to loss of Treg function (but not survival) associated with impaired lipid biosynthesis, particularly in the mevalonate pathway (10). Through governing energy homeostasis and a wide array of cellular metabolic pathways, our study has demonstrated that LKB1 controls not only Treg function but also survival, lending further strength to the idea that cellular metabolism fundamentally influences Treg biology.
LKB1 is known to play indispensable roles in T cells in general. Tamás et al. (9) and MacIver et al. (41) have shown that LKB1 is important for the survival and proliferation of thymocytes and peripheral T cells, consistent with our finding that LKB1 governs Treg survival. Interestingly, although Lkb1-deficient T cells have diminished proliferative capacity, MacIver et al. (41) reported an activated T cell phenotype in mutant CD4+ and CD8+ T cells with enhanced cytokine production. Because the in vivo function of Tregs absolutely requires an intact Lkb1, T cell-specific deletion of Lkb1 would similarly destroy Treg function completely and thus shift the immune balance toward uncontrolled immune activation. Our results might explain why Lkb1-deficient CD4+ and CD8+ T cells have reduced survival/proliferation but at the same time are highly activated.
During the preparation of this paper, two studies describing the importance of LKB1 in metabolic and functional fitness, as well as in maintaining Treg cell lineage identity, have been published (23, 42). Consistent with our findings, both studies described similar autoimmune phenotypes in mice harboring a Treg-specific deletion of Lkb1 that lead to early fatality. In addition, defective mitochondrial metabolism and compromised suppressor function are commonly reported for Tregs lacking LKB1 (Fig. S5). Interestingly, lineage tracing experiments have found that LKB1 stabilizes Foxp3 expression (18), consistent with the low Foxp3 expression in the few surviving KO Tregs in our Teff transfer-induced colitis model (Fig. 4B). Notably, our study identified severely compromised in vivo and in vitro survival of KO Tregs (Fig. 4 A and B), due largely to dysregulated cellular metabolism, in agreement with the concept of function exhaustion proposed by Yang et al. (42). Deletion of LKB1 in Tregs manifested as defective mitochondria, compromised OXPHOS, depleted cellular ATP, and altered cellular metabolism pathways (Fig. 3). Surprisingly, the striking global metabolic changes in KO Tregs observed by us and Yang et al. (42) were not captured in the study by Wu et al. (23). Nonetheless, together these studies establish a critical role of LKB1 in Treg biology (Fig. S6). By stabilizing Foxp3 expression and maintaining metabolic fitness, LKB1 fundamentally underpins Treg survival and function to preserve immune homeostasis and tolerance.
Materials and Methods
Mice.
Lkb1fl/fl, Foxp3YFPCre, CD45.1+, Rag1−/−, C57BL/6 mice were purchased from Jackson Laboratory. Lkb1fl/flFoxp3YFPCre mice were used at age 3–4-wk unless indicated otherwise, with age- and sex-matched WT mice containing the Foxp3YFPCre allele serving as controls. All mice were maintained at the Salk Institute of Biological Studies specific pathogen-free animal facility in accordance with institutional regulations. All mouse experiments were performed with the approval of the institutional animal care and use committee (IACUC) of the Salk Institute.
Flow Cytometry.
Cell surface markers were stained in 1×PBS containing 2% FBS with indicated antibodies. Anti-CD4 (RM4-5), anti-CD8 (53-6.7), anti-CD25 (PC61), anti-CD44 (IM7), anti-CD62L (MEL-14), anti-CD45.1 (A20), and anti-CD45.2 (104) were purchased from eBioscience. Intracellular staining was performed using the Foxp3/Transcription Factor Staining Kit (eBioscience) with indicated antibodies. For intracellular cytokine staining, cells were stimulated for 4 h with phorbol 12-myristate 13-acetate (50 ng/mL; Sigma-Aldrich) and ionomycin (1 μg/mL; Sigma-Aldrich), and then treated for another 1 h with Golgi-Plug (BD Biosciences). Anti–IFN-γ (XMG1.2), anti-IL17A (eBio17B7), and anti–IL-4 (eBio 11B11) were purchased from eBioscience, and anti–cleaved caspase-3(Asp175) (D3E9), anti–phospho-H2A.x(Ser139) (20E3), and anti-LC3b (D11) were purchased from Cell Signaling Technology. All data collection was performed with a FACSAria II cell sorter (BD Biosciences), and data analysis was done with FlowJo software (Tree Star).
Cell Isolation and FACS Sorting.
Total CD25+ cells were isolated from spleen and LNs of 3- to 4-wk-old male (unless indicated otherwise) WT (Lkb1+/+,Foxp3YFPCre) and KO (Lkb1fl/fl,Foxp3YFPCre) mice with anti-CD25-PE (7D4) and anti-PE Microbeads (Miltenyi Biotech). For RNA-sequencing (RNA-Seq) samples, total CD25+ cells were isolated from 4-wk-old female WT (Lkb1+/+,Foxp3YFPCre/Cre) and heterozygous (Lkb1fl/fl,Foxp3YFPCre/+) mice. Foxp3YFP+CD4+CD25+ cells were further purified from total CD25+ cells by sorting on a FACSAria II cell sorter.
Serum Ig and Cytokine Measurement.
Serum IgM, IgG1, and IgA concentrations were measured using the SBA Clonotyping System (SouthernBiotech). For IgE, ELISA was performed using biotinylated anti-IgE detection antibody (BD Pharmingen) and streptavidin-conjugated HRP. Serum cytokine concentrations were measured using the Bio-Plex Pro Mouse Cytokine 23-Plex Immunoassay (Bio-Rad) according to the manufacturer’s instructions.
In Vitro Suppression Assay.
In this assay, CD62L+CD44–CD4+ T cells labeled with CellTrace carboxyfluorescein diacetate succinimidyl ester (CFSE; Life Technologies) served as responder T cells. Antigen-presenting cells were prepared by depleting T cells from WT B6 splenocytes using Thy1-specific MACS beads. Responder cells (5 × 104) were cultured with irradiated antigen-presenting cells (2 × 105) and anti-CD3 (2C11; 0.3 μg/mL) in the absence or presence of various numbers of Tregs for 72 h. The division of responder T cells was assessed by dilution of CellTrace CFSE, and the division index was calculated using FlowJo software.
In Vivo Colitis Model.
Rag1−/− mice were injected i.v. with 0.4 × 106 Teffs (CD4+CD45RBhiCD25−), either alone or with 0.2 × 106 WT or KO Tregs. Mice were weighed and examined every week for signs of disease and then euthanized for tissue harvest at 7–8 wk (or as indicated otherwise). Cells from mesenteric LNs were subjected to flow cytometry analysis with the indicated antibodies. Colon tissue were fixed in 10% neutral buffered formalin (Thermo Fisher Scientific) and stained with H&E.
In Vitro Treg Culture.
WT (Lkb1+/+,Foxp3YFPCre) and KO (Lkb1fl/fl,Foxp3YFPCre) Tregs were FACS-sorted from total CD25+ cells isolated from spleen and LNs of 3- to 4-wk-old male mice. Tregs were generally cultured for 72 h with 500–1,000 U/mL human IL-2 (Peprotech) and Mouse T-Activator CD3/CD28 Dynabeads (Life Technologies) with a bead-to-cell ratio of 3:1 unless specified otherwise. For the experiments testing the effects of inhibitors, 0.5 μM AMPK inhibitor (dorsomorphin dihydrochloride; Tocris), 50 μM MARK inhibitor (MARK/Par-1 Activity Inhibitor, 39621; Calbiochem), 1 μM NUAK inhibitor (WZ 4003; Tocris), and 50 nM SIK inhibitor (HG-9–91-01; Cayman Chemical) were added simultaneously with IL-2 and CD3/CD28 Dynabeads.
RNA Sequencing.
Total RNA was extracted with TRIzol (Life Technologies) and the RNeasy Mini Kit (Qiagen) from Tregs that had been FACS-purified from 4-wk-old female WT mice (Lkb1+/+,Foxp3YFPCre/Cre) and mice heterozygous for Foxp3-Cre (Lkb1fl/fl,Foxp3YFPCre/WT). Sequencing libraries were prepared from 100 ng of total RNA using the TruSeq RNA Sample Preparation Kit v2 (Illumina), and then validated using the Agilent 2100 Bioanalyzer system, normalized, and pooled for sequencing. Sequencing was carried out on the Illumina HiSeq 2500 sequencing system using bar-coded multiplexing and a 100-bp read length. Reads were aligned to the mouse genome (mm10) using STAR (43). RNA-Seq data were normalized and visualized using HOMER (44) (homer.ucsd.edu/homer/) to generate custom tracks for the UCSC Genome Browser (genome.ucsc.edu/). Heat maps were generated using the open source software GENE-E (https://software.broadinstitute.org/GENE-E/), and pathway analysis and functional annotation of significantly regulated genes were performed using DAVID (david.ncifcrf.gov).
qPCR.
Total RNA was extracted with TRIzol reagent (Life Technologies) and the RNeasy Mini Kit (Qiagen), and cDNA was prepared using iScript reagent (Bio-Rad). qRT-PCR was subsequently performed using SsoAdvanced SYBR Green Supermix reagent on the CFX384 PCR detection system (Bio-Rad). Relative expression values were determined using the standard curve method, and abundance was normalized to cyclophilin. Values represent averages from three biological samples. Primer sequences are provided in Table S1.
Metabolic Assays.
For ROS, mitochondrial mass, and membrane potential measurements, freshly isolated Tregs were incubated with 2.5 uM CellROX Deep Red, 20 nM MitoTracker Deep Red FM, and 50 nM DilC1(5) (Life Technologies), respectively, at 37 °C for 15–30 min and then analyzed by flow cytometry. ATP was measured from lysates of 500,000 freshly isolated Tregs prepared using the boiling water method (45) with an ATP Determination Kit (Life Technologies). For the Seahorse assay, 500,000 freshly isolated Tregs were plated using Cell-Tak (Corning), and OCR and ECAR were measured on a Seahorse XF96 analyzer (Agilent) in the presence of the mitochondrial inhibitor oligomycin (2 μM), mitochondrial uncoupler FCCP (1 μM), and respiratory chain inhibitor antimycin A/rotenone (2 μM).
Supplementary Material
Acknowledgments
We thank L. Ong and C. Brondos for administrative assistance and H. Juguilon, Y. Dai, L. Chong, E. Banayo, M. He, B. Collins, and Y. Liang for technical assistance. R.M.E. is an investigator of the Howard Hughes Medical Institute and the March of Dimes Chair in Molecular and Developmental Biology at the Salk Institute. R.M.E. was funded by grants from the National Institutes of Health (NIH) (DK057978, HL105278, HL088093, and CA014195), the Leona M. and Harry B. Helmsley Charitable Trust (2017-PG-MED001), the Fondation Leducq, and Ipsen/Biomeasure. Y.Z. was supported by NIH Grant 105278. This research was supported by the National Institute of Environmental Health Sciences under Award P42ES010337. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. N.H. was supported by T32 Institutional Research Training Grant 5T32CA009370.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. SRP098763).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715363114/-/DCSupplemental.
References
- 1.Kronenberg M, Rudensky A. Regulation of immunity by self-reactive T cells. Nature. 2005;435:598–604. doi: 10.1038/nature03725. [DOI] [PubMed] [Google Scholar]
- 2.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michalek RD, et al. Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Zheng Y. Regulatory T cell identity: Formation and maintenance. Trends Immunol. 2015;36:344–353. doi: 10.1016/j.it.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coe DJ, Kishore M, Marelli-Berg F. Metabolic regulation of regulatory T cell development and function. Front Immunol. 2014;5:590. doi: 10.3389/fimmu.2014.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardie DG. AMPK: A key regulator of energy balance in the single cell and the whole organism. Int J Obes. 2008;32:S7–S12. doi: 10.1038/ijo.2008.116. [DOI] [PubMed] [Google Scholar]
- 7.Blagih J, et al. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity. 2015;42:41–54. doi: 10.1016/j.immuni.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Zhang BB, Zhou G, Li C. AMPK: An emerging drug target for diabetes and the metabolic syndrome. Cell Metab. 2009;9:407–416. doi: 10.1016/j.cmet.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Tamás P, et al. LKB1 is essential for the proliferation of T-cell progenitors and mature peripheral T cells. Eur J Immunol. 2010;40:242–253. doi: 10.1002/eji.200939677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng H, et al. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 13.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 14.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 15.Wildin RS, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 16.Koh HJ, et al. Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3. Mol Cell Biol. 2006;26:8217–8227. doi: 10.1128/MCB.00979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw RJ, et al. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 19.Beyersdorf N, Ding X, Tietze JK, Hanke T. Characterization of mouse CD4 T cell subsets defined by expression of KLRG1. Eur J Immunol. 2007;37:3445–3454. doi: 10.1002/eji.200737126. [DOI] [PubMed] [Google Scholar]
- 20.Huang CT, et al. Role of LAG-3 in regulatory T cells. Immunity. 2004;21:503–513. doi: 10.1016/j.immuni.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Joller N, et al. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity. 2014;40:569–581. doi: 10.1016/j.immuni.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu D, et al. Lkb1 maintains Treg cell lineage identity. Nat Commun. 2017;8:15876. doi: 10.1038/ncomms15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 25.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shorning BY, Clarke AR. Energy sensing and cancer: LKB1 function and lessons learnt from Peutz-Jeghers syndrome. Semin Cell Dev Biol. 2016;52:21–29. doi: 10.1016/j.semcdb.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 29.Loftus RM, Finlay DK. Immunometabolism: Cellular metabolism turns immune regulator. J Biol Chem. 2016;291:1–10. doi: 10.1074/jbc.R115.693903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: Insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rathmell JC. Metabolism and autophagy in the immune system: Immunometabolism comes of age. Immunol Rev. 2012;249:5–13. doi: 10.1111/j.1600-065X.2012.01158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan B, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurumurthy S, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeppesen J, et al. LKB1 regulates lipid oxidation during exercise independently of AMPK. Diabetes. 2013;62:1490–1499. doi: 10.2337/db12-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurov JB, et al. Immune system dysfunction and autoimmune disease in mice lacking Emk (Par-1) protein kinase. Mol Cell Biol. 2001;21:3206–3219. doi: 10.1128/MCB.21.9.3206-3219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurov JB, et al. Loss of the Par-1b/MARK2 polarity kinase leads to increased metabolic rate, decreased adiposity, and insulin hypersensitivity in vivo. Proc Natl Acad Sci USA. 2007;104:5680–5685. doi: 10.1073/pnas.0701179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundberg TB, et al. Small-molecule screening identifies inhibition of salt-inducible kinases as a therapeutic strategy to enhance immunoregulatory functions of dendritic cells. Proc Natl Acad Sci USA. 2014;111:12468–12473. doi: 10.1073/pnas.1412308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takemori H, Okamoto M. Regulation of CREB-mediated gene expression by salt inducible kinase. J Steroid Biochem Mol Biol. 2008;108:287–291. doi: 10.1016/j.jsbmb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacIver NJ, et al. The liver kinase B1 is a central regulator of T cell development, activation, and metabolism. J Immunol. 2011;187:4187–4198. doi: 10.4049/jimmunol.1100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang K, et al. Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature. 2017;548:602–606. doi: 10.1038/nature23665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang NC, Ho WM, Chen YH, Hu ML. A convenient one-step extraction of cellular ATP using boiling water for the luciferin-luciferase assay of ATP. Anal Biochem. 2002;306:323–327. doi: 10.1006/abio.2002.5698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.