Significance
The introduction of Wolbachia (an intracellular bacterium that does not infect higher organisms) into culicine mosquito populations from endemic areas is a promising strategy to prevent arboviral transmission. Anopheline mosquitoes were thought to be naturally refractory to Wolbachia, but a population of Anopheles gambiae from Burkina Faso infected with Wolbachia was recently reported. We identified a Wolbachia strain in A. gambiae mosquitoes from Mali (wAnga-Mali). wAnga-Mali infection was associated with reduced prevalence and intensity of sporozoite infection in field-collected females. Experimental infections indicate that wAnga-Mali infection reduces malaria transmission by a mechanism that affects sporozoites and opens the possibility of exploring the introduction of Wolbachia into natural populations of anophelines as a strategy to reduce disease transmission.
Keywords: Wolbachia, Plasmodium, mosquito, malaria, Anopheles
Abstract
A naturally occurring Wolbachia strain (wAnga-Mali) was identified in mosquitoes of the Anopheles gambiae complex collected in the Malian villages of Dangassa and Kenieroba. Phylogenetic analysis of the nucleotide sequence of two 16S rRNA regions showed that wAnga-Mali clusters with Wolbachia strains from supergroup A and has the highest homology to a Wolbachia strain isolated from cat fleas (Ctenocephalides). wAnga-Mali is different from two Wolbachia strains previously reported in A. gambiae from Burkina Faso (wAnga_VK5_STP and wAnga_VK5_3.1a). Quantitative analysis of Wolbachia and Plasmodium sporozoite infection in field-collected mosquitoes indicates that the prevalence and intensity of Plasmodium falciparum sporozoite infection is significantly lower in Wolbachia-infected females. The presence of Wolbachia in females from a laboratory Anopheles coluzzii (A. gambiae, M form) colony experimentally infected with P. falciparum (NF54 strain) gametocyte cultures slightly enhanced oocyst infection. However, Wolbachia infection significantly reduced the prevalence and intensity of sporozoite infection, as observed in the field. This indicates that wAnga-Mali infection does not limit early stages of Plasmodium infection in the mosquito, but it has a strong deleterious effect on sporozoites and reduces malaria transmission.
Despite recent strides in reducing the burden of malaria, this disease was still responsible for more than 400,000 deaths in 2015 (1). Most mortality is caused by Plasmodium falciparum infection in children from sub-Saharan Africa, where Anopheles gambiae and Anopheles coluzzii mosquitoes are the major disease vectors. These two anopheline species are traditionally known as the S and M molecular forms of A. gambiae, respectively (2, 3). They share limited genetic flow, occupy distinct ecological niches (4), and were only recently reclassified as different species (5). At present, vector control relies mostly on insecticide-based strategies, such as indoor spraying or long-lasting insecticide-treated nets. However, efficacy concerns and reports of evolving insecticide resistance highlight the need to develop alternative malaria control strategies (6).
Wolbachia is a genus of Gram-negative endosymbiotic proteobacteria that are vertically transmitted and commonly found in nematodes and arthropods (7). Several strains of Wolbachia are able to manipulate host reproduction by a mechanism known as cytoplasmic incompatibility (CI) (8, 9), which allows Wolbachia to reach high prevalence in natural populations (10, 11). Some strains of Wolbachia protect insect hosts from viral infections (12–14). For example, the presence of Wolbachia in Aedes aegypti mosquitoes prevented laboratory infections with dengue and other flaviviruses (15, 16). Based on these findings, a program to release Wolbachia-infected mosquitoes at several different test sites around the world was implemented, with the aim of spreading Wolbachia-mediated resistance to viruses in natural mosquito populations (17). Wolbachia also reduces mosquito susceptibility to other nonviral pathogens. For example, infection of Anopheles stephensi with the Aedes albopictus wAlbB Wolbachia strain reduced P. falciparum infection, but the effect was modest (18). Similar attempts to artificially infect A. gambiae with Wolbachia were limited to somatic tissues (19–21), suggesting that A. gambiae is less susceptible to Wolbachia infections. More recently, populations of Wolbachia-infected A. gambiae and A. coluzzii were identified in Burkina Faso. This strain was called wAnga (22), and mosquitoes positive for Wolbachia were reported to have a lower prevalence of Plasmodium infection (23). However, because the prevalence of Plasmodium infection in field-collected mosquitoes is relatively low (5.4% in this study), the analysis was based on a total of 12 P. falciparum-infected females (23).
In the present report, we identified A. gambiae s.l. mosquitoes in Mali that are naturally infected with wAnga-Mali, a Wolbachia strain that is different from the ones reported in Burkina Faso. wAnga-Mali was first detected in A. gambiae and A. coluzzii mosquitoes collected in 2010–2011, and its persistence was confirmed in recent collections in 2015–2016. We investigated the impact of Wolbachia on P. falciparum infection in the field, by analyzing a large number of naturally infected mosquitoes, and also under controlled laboratory conditions, by establishing a Wolbachia-infected A. coluzzii colony.
Results
Identification of Wolbachia in Natural Mosquito Populations from Mali.
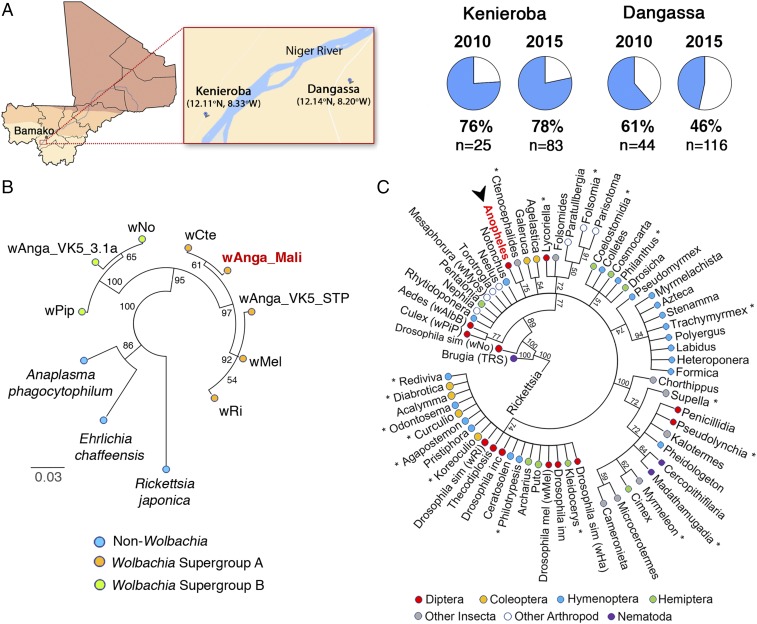
We first analyzed a collection of 13,321 A. gambiae s.l. mosquitoes collected in six Malian villages using the intradomiciliary spray-catch technique (24) (Table S1). The thorax and head region was dissected, homogenized, and part of the sample was used to detect the presence of P. falciparum sporozoites using an anticircumsporozoite protein (CSP) in ELISAs (24). A total of 205 females (1.53%) were positive for sporozoite infection. The rest of the homogenate from Plasmodium-positive samples was used to extract genomic DNA. A broad screen was carried out to analyze a large number of samples and try to identify a village(s) in which mosquitoes would be infected with Wolbachia. To this end, pools of 10 mosquitoes were analyzed for the presence of Wolbachia infection using the nested PCR-based assay previously used in Burkina Faso (23). One positive pool was detected in the village of Kenieroba and four in Dangassa. Analysis of individual mosquitoes in the positive pools confirmed that there were five Wolbachia-positive mosquitoes (one in each pool). Wolbachia was not detected in the pools of Plasmodium-infected females from the other four villages. We therefore decided to focus our studies in the villages of Kenieroba and Dangassa (Fig. 1A), where the presence of Wolbachia and Plasmodium coinfections had been confirmed.
Fig. 1.
A Wolbachia strain is present in A. gambiae and A. coluzzii mosquitoes from Mali. (A) Map of Mali and geographic localization of the villages of Dangassa and Kenieroba, where A. gambiae (S molecular form) and A. coluzzii (M molecular form) mosquitoes naturally infected with Wolbachia were identified (the geographic coordinates of the villages are indicated). The prevalence of Wolbachia in A. gambiae senso lato (s.l.) (wAnga-Mali) infection in samples collected in these two villages, in 2010 and 2015, is also indicated. (B) Phylogenetic analysis based on the alignment of a conserved region of the 16S rRNA gene using Wolbachia-specific primers. The sequence of wAnga-Mali (highlighted in red) clusters with Wolbachia strains from the supergroup A has the highest homology from Wolbachia isolated from cat fleas (Ctenocephalides) and is different from sequences previously reported for A. gambiae s.l. from Burkina Faso (Anga_VK5_STP and wAnga_VK5_3.1a). Sequences from other non-Wolbachia proteobacteria were also included, and the sequence from R. japonica was used as the reference outgroup. (C) Phylogenetic analysis based on the alignment of a variable region of the 16S rRNA gene from Wolbachia strains isolated from the nematode B. malayi, insects, and other arthropods. The sequence from R. japonica was used as the reference outgroup. Strains are identified by their host genus and color-coded according to their taxonomic order. The wAnga-Mali strain is shown red and is indicated by the arrowhead. Asterisks (*) denote when the consensus sequence for closely related sequences from the same genus was used. The numbers indicated the consensus support (%).
Development of a Quantitative Assay for Wolbachia Infection.
A quantitative PCR (qPCR)-based detection method was developed to establish both the prevalence and the intensity of Wolbachia infection in natural mosquito populations, and was compared with two other previously described PCR-based detection methods (23, 25). A set of 69 mosquitoes from Kenieroba and Dangassa that were not infected with Plasmodium were analyzed using three different methods to detect Wolbachia in the same sample. The 16S ribosomal RNA (rRNA) gene was amplified using regular PCR (W16S-Spec) (25), nested PCR (W16S-WE) (23), and a qPCR assay (W16S-qPCR) (Table S2). As expected, regular PCR was the least sensitive method and detected Wolbachia in 16% of the samples (11/69). Using nested PCR, 52% of samples (36/69) were positive in at least one of the technical duplicates. We found that with the regular and nested PCR assays, 22% and 19% of technical duplicates, respectively, were not concordant, suggesting that Wolbachia levels in mosquitoes from Mali are close to the limit of detection of these assays. As expected, the qPCR method was more sensitive. It detected Wolbachia (W+) in 67% of mosquitoes (46/69), and the correlation between technical replicates was high (R2 = 0.9978, P < 0.0001). Therefore, all other detections of Wolbachia in these studies were done using the qPCR method. All of the determinations of Wolbachia infection prevalence in these two villages (Fig. 1A) were done in females that were negative for P. falciparum infection, because they represent the majority of the population (97–99% of females were not infected with Plasmodium) and because a biological interaction between Wolbachia and P. falciparum could greatly bias the prevalence of Wolbachia in the Plasmodium-infected group. The prevalence of Wolbachia in female mosquitoes collected in 2010 from Dangassa (61%) was not significantly different from that in Kenieroba (76%) (Fig. 1A). However, in 2015, the prevalence (78%) and intensity of Wolbachia infection (Fig. S1) in Kenieroba were both significantly higher than in Dangassa (46%) (P < 0.00001, χ2 and P < 0.0001, Mann–Whitney, respectively) (Fig. 1A). A recent collection in 2016 confirmed that the prevalence of Wolbachia in Kenieroba was still very high (38/40 W+ females = 95%).
Phylogenetic Analysis of Wolbachia in A. gambiae Mosquitoes from Mali.
A highly conserved region of the 16S rRNA gene was amplified using regular PCR with Wolbachia-specific primers (25) (Table S2). Sequencing of the PCR products (accession no. MF944114) confirmed that Wolbachia in A. gambiae mosquitoes from Mali (wAnga-Mali) clusters with Wolbachia strains of supergroup A (97–99.8% nucleotide identity) and has lower homology to Wolbachia strains of supergroup B (94–95%) and to other closely related bacterial species (Anaplasma phagocytophilum 93%; Ehrlichia chaffeensis 91%; Rickettsia japonica 88%) (Fig. 1B and Fig. S2). The sequences of this conserved region are also available for Wolbachia strains isolated from A. gambiae mosquitoes from Burkina Faso (we will refer to them as wAnga-BF) (22). Phylogenetic analysis indicates that one of the reported wAnga-BF sequences clusters with Wolbachia supergroup A (wAnga_VK5_STP) and shares 97% identity with wAnga-Mali, while the second strain (wAnga_VK5_3.1a) is more divergent, clusters with supergroup B, and has 94% identity with wAnga-Mali (Fig. 1B). Interestingly, wAnga-Mali has the highest homology (99.8%) to Wolbachia from cat fleas (Ctenocephalides).
We amplified a second region of 16S rRNA that is more variable, using primers that amplify both Wolbachia and closely related bacteria (25), which allows for a more detailed comparison between sequences from different Wolbachia strains. The wAnga-Mali sequence (accession no. MF944223) clustered with Wolbachia strains that infect other arthropods, and also had the highest homology to that from cat fleas (Ctenocephalides) (Fig. 1C). The phylogeny of the different Wolbachia strains does not match the phylogeny of their hosts. For example, wAnga-Mali is more closely related to Wolbachia from cat fleas than to those present in other mosquitoes, such as wAlbB (A. albopictus) and wPip (Culex pipiens), indicating that acquisition of this symbiont occurred by horizontal transfer. wAnga-Mali is evolutionarily very distant from Wolbachia that infects Brugia malayi (Fig. 1C), eliminating the possibility that wAnga-Mali could be a contamination from mosquitoes infected with this nematode.
The multilocus sequence typing (MLST) scheme is a universal genotyping tool for Wolbachia (26) that uses the sequence of specific regions from five conserved genes [gatB: aspartyl/glutamyl-tRNA(Gln) amidotransferase, subunit B; coxA: cytochrome c oxidase, subunit I; hcpA: conserved hypothetical protein; ftsZ: cell division protein; and fbpA: fructose-bisphosphate aldolase] to classify newly identified Wolbachia strains. We attempted to characterize wAnga-Mali using this system, but we were only able to amplify three of the five genes (accession nos. hcpA, MF946614; fbpA, MF946613; coxA, MF946612) (Figs. S3–S5). Multiple attempts to amplify the two remaining genes (gatB, ftsZ) were not successful, suggesting there may be some degree of sequence divergence in the primer region. The three amplified regions (from the hcpA, fbpA, coxA genes) all had higher homology to Wolbachia (91–93%) than to Ehrlichia (56–74%), Anaplasma (55–73%), or Rickettsia (46–70%) (Table S3). Taken together, these data indicate that Wolbachia is present in A. gambiae mosquitoes from Mali, and is not identical to wAnga-BF strains previously reported in Burkina Faso.
Correlation Between Wolbachia and Plasmodium Infections in the Field.
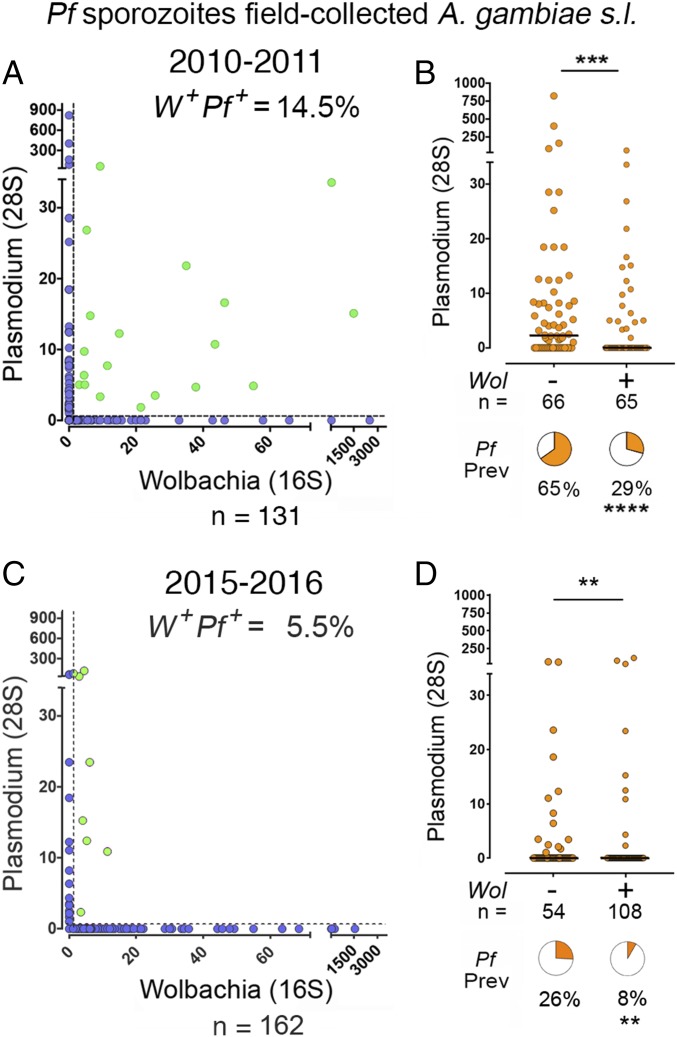
ELISAs were used to detect the presence of P. falciparum sporozoites in homogenates from the thorax and head region. Genomic DNA was extracted from 62 Plasmodium-positive and 69 Plasmodium-negative females collected in 2010–2011. The relative levels of Plasmodium and Wolbachia in each sample were determined by quantitative amplification of the 28S and 16S rRNA genes, respectively (Fig. 2A). We found that 65/131 (49.6%) of females were infected with Wolbachia. It is apparent from the distribution of the data that most values cluster either along the x or the y axis (Fig. 2A). The observed proportion of females coinfected with Wolbachia and Plasmodium (green dots, Fig. 2A) (19/131 = 14.5%) was lower than expected (23.4%) based on the prevalence of Plasmodium (47.3%) and Wolbachia (49.6%) infection (expected prevalence = 0.473 × 0.496 =0.234), indicating that females infected with one organism are less likely to be infected with the other. The prevalence of Plasmodium infection in Wolbachia positive (W+) females (29%) was significantly lower than that of Wolbachia negative (W−) females (65%, P < 0.0001, χ2) (Fig. 2B). The intensity of Plasmodium infection was also significantly reduced in W+ females (P < 0.001, Mann–Whitney). Genotyping revealed that 34 females (26%) were S form (A. gambiae), while 95 (72%) were M form (A. coluzzii). The prevalence of Wolbachia infection in A. gambiae (53.6%) was not significantly different from that of A. coluzzii (41.2%). Similar results were obtained when only A. coluzzii females (n = 95) were included in the analysis. The prevalence of Plasmodium infection in W+ A. coluzzii females (34.2%) was significantly lower than in W− females (66%) (P = 0.002, χ2), and the intensity of Plasmodium infection was also significantly lower (P = 0.005) in W+ females (Fig. S6). These findings from females collected in 2010–2011 were confirmed by analyzing mosquitoes collected in 2015–2016. ELISAs were done in 1,114 A. gambiae females, and 23 Plasmodium-positive mosquitoes were detected (2.0% infection prevalence). Genomic DNA was extracted from these 23 Plasmodium-positive and from 139 Plasmodium-negative females, and Wolbachia and Plasmodium infection levels were determined by qPCR (Fig. 2C). The prevalence of Plasmodium infection was also significantly lower (8%) in W+ than in W− females (26%, P < 0.01, χ2), and the intensity of Plasmodium infection was significantly reduced (P = 0.002, Mann–Whitney) (Fig. 2D). It is worth noting that the W+ females in the 2015–2016 collection that were infected with Plasmodium had low levels of Wolbachia infection (Fig. 2C). Taken together, these observations are in agreement with the previous report of reduced prevalence of Plasmodium infection in A. coluzzii females that carry wAnga-BF (23) and support the hypothesis that Wolbachia infection reduces Plasmodium infection in A. coluzzii. The sample size for A. gambiae S form (n = 34 in 2010–2011 and 13 in 2015–2016) was not large enough to carry out an independent analysis.
Fig. 2.
Effect of naturally occurring Wolbachia on Plasmodium sporozoite infection in field-collected mosquitoes. Wolbachia and Plasmodium levels in field-collected mosquitoes were determined by qRT-PCR. (A and C) Correlation between Wolbachia and Plasmodium levels. Coinfected mosquitoes (W+Pf+) are highlighted in green, and their relative abundance is indicated. (B and D) Levels of Plasmodium sporozoite infection in Wolbachia-infected and uninfected females. Each data point represents the level of Plasmodium infection in a single mosquito, and medians are indicated by the line. Value distributions were compared using the Mann–Whitney test. Pie charts represent the prevalence of Plasmodium sporozoite infection in Wolbachia-infected and uninfected samples. The prevalence was compared using the χ2 test (n = number of samples). **P < 0.01, ***P < 0.001, ****P < 0.0001.
Laboratory Adaptation of Wolbachia-Infected Mosquitoes and Effect of Wolbachia on Plasmodium Infections.
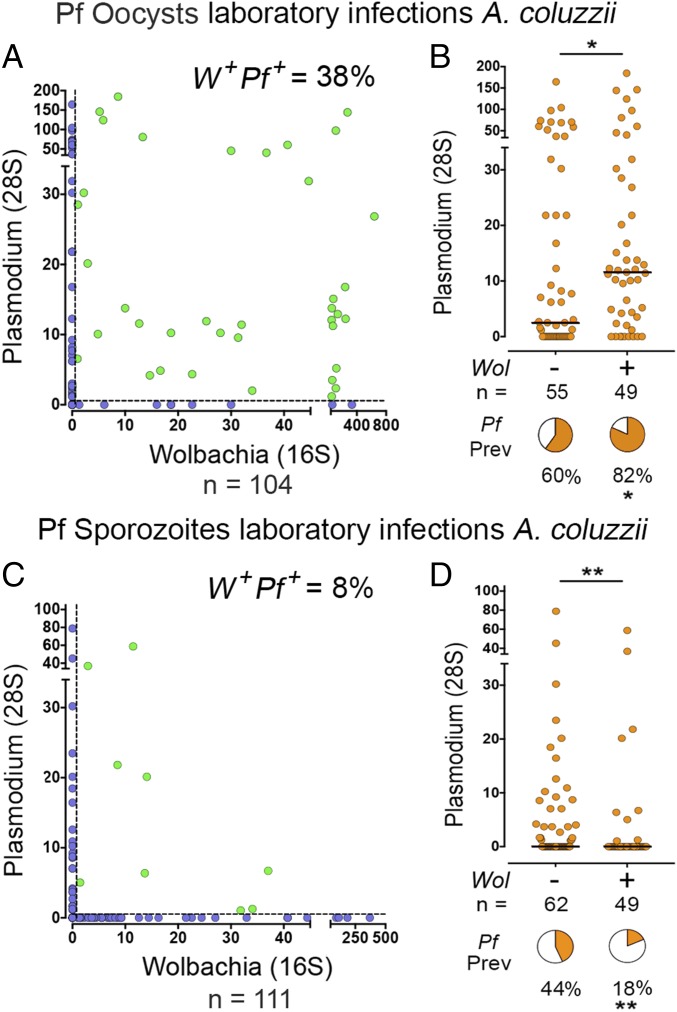
To further evaluate the effect of Wolbachia on Plasmodium transmission under controlled conditions, a colony of A. coluzzii mosquitoes was established from mosquito eggs brought from Mali (Dangassa) in 2015. The first three generations of field-collected mosquitoes were supplemented with male mosquitoes from an A. coluzzii colony established at NIH several years ago (A. gambiae M-form NIH) that were already adapted to mate in captivity. This colony was also established from eggs collected in Mali, and we confirmed that these mosquitoes were not infected with Wolbachia. The A. gambiae M-form NIH strain is a reference mosquito strain, and its genome has been sequenced (27). Mosquitoes were infected with P. falciparum (NF54 strain) by membrane feeding of gametocyte cultures, and the levels of both Plasmodium and Wolbachia infection were determined from genomic DNA extracted from midguts collected 8–10 d after infection. We were surprised to find that the observed prevalence of midgut coinfections (40/104 = 38%; Fig. 3A, green dots) was higher than expected, based on the prevalence of Wolbachia and Plasmodium infection (expected prevalence = 33%). Furthermore, opposite to what was observed in the field for sporozoite infections, the prevalence (P < 0.05, χ2) and the intensity of Plasmodium oocyst infection (P < 0.05, Mann–Whitney) were both moderately, but significantly, higher in females that carried Wolbachia infections. These data indicate that Wolbachia midgut infection does not negatively impact the early stages of Plasmodium development in the mosquito.
Fig. 3.
Effect of Wolbachia in laboratory-reared mosquitoes on Plasmodium infection. Wolbachia and Plasmodium levels in laboratory-reared A. coluzzii mosquitoes challenged with P. falciparum NF54 were determined by qRT-PCR. (A) Correlation between midgut Wolbachia and Plasmodium levels 10 d after infection. Coinfected mosquitoes (W+Pf+) are highlighted in green, and their relative abundance is indicated. (B) Midgut levels of Plasmodium infection in Wolbachia-infected and uninfected females. Each data point represents the level of Plasmodium infection in a single mosquito, and medians are indicated by the line. (C) Correlation between Wolbachia and Plasmodium levels 21 d after infection. Coinfected mosquitoes (W+Pf+) are highlighted in green, and their relative abundance is indicated. (D) Head-thorax levels of Plasmodium sporozoite infection in Wolbachia-infected and uninfected females. Each data point represents the level of Plasmodium infection in a single mosquito, and medians are indicated by the line. Value distributions were compared using the Mann–Whitney test. Pie charts represent Plasmodium prevalence in Wolbachia-infected and uninfected samples. Prevalence was compared using the χ2 test (n = number of samples). *P < 0.05, **P < 0.01.
Because the field studies were done by detecting sporozoite infection in the salivary glands, the effect of Wolbachia on this later stage of the parasite was also evaluated. Wolbachia and Plasmodium levels were analyzed in thorax and head samples collected 18–21 d after infection. Similar to what was observed in the field, the observed prevalence of coinfection was lower than expected (9/111 = 8%; Fig. 3C, green dots) based on the prevalence of Wolbachia and Plasmodium infections (expected prevalence = 14.3%). The prevalence of Plasmodium infection in W+ females (18%) was significantly lower (P < 0.004, χ2) than that of W− females (44%) (Fig. 3D). The intensity of Plasmodium infection was also significantly reduced (P < 0.01, Mann–Whitney). Taken together, these findings show that although Plasmodium oocyst infections were slightly higher in Wolbachia-infected females, the number of sporozoites was significantly reduced.
Discussion
Wolbachia is widely prevalent among arthropods and is thought to be naturally present in as many as 65% of insect species (28). Other species can be infected under laboratory conditions (29–31). Interestingly, anophelines display a remarkable degree of refractoriness to Wolbachia. A stable infection of A. stephensi with wAlbB has been established (18), but all other laboratory infections of anophelines have been limited to somatic tissues, and failed to infect the germ line and did not propagate to the offspring (19–21). Field populations of anophelines were thought to be resistant to Wolbachia infections. However, A. gambiae populations from Burkina Faso naturally infected with Wolbachia were recently reported (22). It is not clear whether some Wolbachia strains evolved specific adaptations that allowed them to colonize mosquitoes of the A. gambiae complex in West Africa, or whether genetic differences already present in West African anophelines favored Wolbachia invasion. The observation that two independent PCR amplifications with standard primers from the MLST universal genotyping tool (gatB, ftsZ) failed in wAnga-Mali–infected mosquitoes suggests that wAnga-Mali is more divergent in these regions than in other Wolbachia strains. The prevalence and distribution, as well as seasonal fluctuations of Wolbachia infections in natural anopheline mosquito populations in Africa and other continents, remain to be established.
We investigated whether the presence of Wolbachia was a rare local event, limited to Burkina Faso, or a common occurrence in West Africa. We first identified natural populations of A. gambiae and A. coluzzii infected with Wolbachia in the Malian villages of Kenieroba and Dangassa during the wet season of 2010; recent collections from the same villages, in 2015–2016, confirmed that Wolbachia is still circulating. The levels of wAnga-Mali we detected in anopheline mosquitoes are very low and close to the detection limit of PCR-based assays. For comparison, the copies of the 16S rRNA gene from wAnga-Mali that we detect are usually less than 1% of total mosquito genome copies of S7 rRNA. This is a remarkable difference from what has been reported in field-released wMel-infected A. aegypti (32), or in wAlbB infections of A. albopictus (33) or A. stephensi (18), where Wolbachia genome copies are close to a 1:1 ratio with mosquito genome copies. Phylogenetic analysis of two different regions of 16S rRNA indicates that wAnga-Mali is more similar to a Wolbachia strain isolated from cat fleas (Ctenocephalides) (Fig. 1 B and C) (99.8% identity) than to the two strains isolated from A. gambiae complex mosquitoes in Burkina Faso (94 and 97% identity) (Fig. 1B), suggesting that the acquisition of Wolbachia in Burkina Faso and Mali were independent events. Interestingly, it was reported that amplification of fbpA from the wAnga-BF strain failed (22), while we successfully amplified this gene in wAnga-Mali using the same primers, further suggesting some sequence divergence between wAnga-Mali and wAnga-BF. A detailed comparison of the variable region of the 16S rRNA with the isolates from Burkina Faso, as well as whole-genome sequencing, would be necessary to obtain a clear picture of the origin and spread of Wolbachia in natural mosquito populations in Africa.
Wolbachia infections of culicine mosquitoes and other dipterans were shown to be protective against several viral pathogens and to reduce Plasmodium gallinaceum infection in A. aegypti (12–14, 16). Somatic Wolbachia infections of anophelines have further suggested that Wolbachia could also confer partial protection of mosquitoes against Plasmodium (19). Furthermore, a lower Plasmodium prevalence was observed in mosquitoes carrying PCR-detectable levels of Wolbachia in Burkina Faso. A total of 221 blood-fed A. coluzzii obtained from homes were analyzed 5 d after collection. There were 12 infected females, one in the group infected with Wolbachia (1/116 = 0.8%) and 11 in those mosquitoes that did not carry Wolbachia (11/105 = 10.4%). The detection of the parasite was done using a PCR assay, so it is not clear whether early oocysts or sporozoites from a previous infection were detected. In the present study, we increased the number of infected mosquitoes by prescreening a large sample set using a CSP-based ELISA in homogenates from the thorax and head region to detect sporozoites. We also found a strong negative association between Wolbachia infection and the prevalence of P. falciparum sporozoite infection (P < 0.0001, χ2). The effect on A. coluzzii is clear, but the sample size of A. gambiae (S form) was too small to be analyzed independently. Although Wolbachia infections have a similar prevalence in A. coluzzii and A. gambiae, it is not clear whether the same negative correlation with Plasmodium infection will also be observed in A. gambiae (S form).
Environmental factors and host age affect both Wolbachia levels and its effects on host immunity (34–37), and neither factor can be controlled in field-collected mosquitoes. To control such variables, we established a colony of A. coluzzii mosquitoes from eggs brought from Dangassa, Mali, in 2015. Infections with the African NF54 strain of P. falciparum confirmed that Wolbachia negatively impacts sporozoite development. Interestingly, this negative effect was not observed up to the oocyst stage. Furthermore, the effect was opposite, with a modest but significant increase in midgut infection, suggesting that parasites are protected from the effect of Wolbachia while inside the oocysts. Perhaps Wolbachia infection depletes some nutrients, such as membrane lipids, required by sporozoites, or may activate a stronger immune response that targets sporozoites when they are released into the hemolymph. Alternatively, sporozoites may be damaged in the salivary gland, either during the traversal, as they come in direct contact with the cell cytoplasm, or as they accumulate in the secretory cavity where they are constantly bathed by salivary gland secretions. Stable wAlbB infection in A. stephensi significantly reduced the number of oocysts, but had a much stronger effect on salivary gland sporozoites (18). The mechanism by which Wolbachia infection affects sporozoites remains to be elucidated.
Release of Wolbachia-infected A. aegypti is one of the most promising strategies to prevent transmission of dengue and other flaviviruses in endemic areas. The rapid success of this intervention is dependent on CI triggered by Wolbachia infection, a natural mechanism that drives this bacterium into natural insect populations. In brief, CI is defined as offspring mortality when Wolbachia-infected males mate with females that are not infected. Mortality does not occur when infected males mate with females that are also infected. CI gives Wolbachia-infected females a reproductive advantage that becomes more effective as the prevalence of Wolbachia infection increases in the population, allowing these bacteria to reach very high frequencies within a few generations. However, CI is not found in every Wolbachia strain. The wAnga-BF strain did not induce CI under laboratory conditions (23). We did not directly evaluate the presence of CI in our colony, but the prevalence of Wolbachia infection did not increase during the time of this study (5–15 generations), as would be expected if CI was taking place. CI-associated genes were recently identified in Drosophila (38, 39), but nongenetic factors also seem to affect the manifestation of CI (37, 40, 41). In general, the levels of infection present in mosquitoes from Mali are very low, and the mechanisms or interactions with the mosquito immune system or with other symbionts that may limit Wolbachia infection are unknown. Adaptation to tissue culture and genetic analysis of wAnga strains will be paramount to analyze whether the genes responsible for CI are present in Wolbachia strains circulating in mosquitoes from West Africa.
Collectively, our data support the hypothesis that Wolbachia infection, even at low levels, negatively impacts Plasmodium sporozoite infection in A. coluzzii. In theory, the release of Wolbachia-infected mosquitoes could be a promising strategy to reduce transmission, but the lack of clear CI could be an important limitation. It might be feasible, however, to develop a genetically modified Wolbachia to induce CI, or to select Wolbachia strains that can spread efficiently in natural A. gambiae populations. The high prevalence (78%) and increased intensity of Wolbachia infection in the village of Kenieroba during the 2015 and 2016 collections, that was not observed in Dangassa, is very interesting and warrants further investigation. This observation suggests that there may be local differences in the adaptation of Wolbachia to mosquitoes between these two villages, and/or differences in the time when these bacteria were introduced. It also indicates that, under some conditions, Wolbachia infection can reach a high prevalence and that infection levels in the population can increase substantially over time, to levels that could disrupt P. falciparum malaria transmission.
Materials and Methods
Intradomiciliary collections A. gambiae females were done in several Malian Villages using the spray-catch technique, and mosquitoes were screened for the presence of P. falciparum sporozoites using ELISAs and for Wolbachia infections using nested PCR. Genomic DNA was extracted from a similar number of females that were either positive or negative for Plasmodium infection, and the prevalence and intensity of Wolbachia and P. falciparum sporozoite infections were determined by qPCR. A laboratory colony of A. gambiae M-form (A. coluzzi) was established, and the effect of Wolbachia infection on the prevalence and intensity of midgut oocysts and salivary gland P. falciparum infection was evaluated. Detailed information on the methodology used can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Andre Laughinghouse and Kevin Lee for insectary support and Ana Beatriz Barletta Ferreira, Julio Castillo, Nitin Kamath, and Rebecca Greene for experimental assistance. This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in GenBank, https://www.ncbi.nlm.nih.gov/genbank/ (accession nos. MF946612–MF946614, MF944223, and MF944114).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716181114/-/DCSupplemental.
References
- 1.World Health Organization . World Malaria Report 2015. WHO; Geneva: 2016. [Google Scholar]
- 2.Esnault C, et al. High genetic differentiation between the M and S molecular forms of Anopheles gambiae in Africa. PLoS One. 2008;3:e1968. doi: 10.1371/journal.pone.0001968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.della Torre A, et al. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol Biol. 2001;10:9–18. doi: 10.1046/j.1365-2583.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- 4.Lehmann T, Diabate A. The molecular forms of Anopheles gambiae: A phenotypic perspective. Infect Genet Evol. 2008;8:737–746. doi: 10.1016/j.meegid.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coetzee M, et al. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619:246–274. [PubMed] [Google Scholar]
- 6.Ranson H, et al. Pyrethroid resistance in African anopheline mosquitoes: What are the implications for malaria control? Trends Parasitol. 2011;27:91–98. doi: 10.1016/j.pt.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Werren JH, Baldo L, Clark ME. Wolbachia: Master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 8.Breeuwer JA, Werren JH. Microorganisms associated with chromosome destruction and reproductive isolation between two insect species. Nature. 1990;346:558–560. doi: 10.1038/346558a0. [DOI] [PubMed] [Google Scholar]
- 9.O’Neill SL, Karr TL. Bidirectional incompatibility between conspecific populations of Drosophila simulans. Nature. 1990;348:178–180. doi: 10.1038/348178a0. [DOI] [PubMed] [Google Scholar]
- 10.Turelli M, Hoffmann AA. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature. 1991;353:440–442. doi: 10.1038/353440a0. [DOI] [PubMed] [Google Scholar]
- 11.Turelli M, Hoffmann AA. Cytoplasmic incompatibility in Drosophila simulans: Dynamics and parameter estimates from natural populations. Genetics. 1995;140:1319–1338. doi: 10.1093/genetics/140.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chrostek E, et al. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: A phenotypic and phylogenomic analysis. PLoS Genet. 2013;9:e1003896. doi: 10.1371/journal.pgen.1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedges LM, Brownlie JC, O’Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322:702. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- 14.Glaser RL, Meola MA. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One. 2010;5:e11977. doi: 10.1371/journal.pone.0011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker T, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–453. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- 16.Moreira LA, et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139:1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann AA, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476:454–457. doi: 10.1038/nature10356. [DOI] [PubMed] [Google Scholar]
- 18.Bian G, et al. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340:748–751. doi: 10.1126/science.1236192. [DOI] [PubMed] [Google Scholar]
- 19.Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 2011;7:e1002043. doi: 10.1371/journal.ppat.1002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kambris Z, et al. Wolbachia stimulates immune gene expression and inhibits plasmodium development in Anopheles gambiae. PLoS Pathog. 2010;6:e1001143. doi: 10.1371/journal.ppat.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes GL, Vega-Rodriguez J, Xue P, Rasgon JL. Wolbachia strain wAlbB enhances infection by the rodent malaria parasite Plasmodium berghei in Anopheles gambiae mosquitoes. Appl Environ Microbiol. 2012;78:1491–1495. doi: 10.1128/AEM.06751-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldini F, et al. Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nat Commun. 2014;5:3985. doi: 10.1038/ncomms4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw WR, et al. Wolbachia infections in natural Anopheles populations affect egg laying and negatively correlate with Plasmodium development. Nat Commun. 2016;7:11772. doi: 10.1038/ncomms11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolo G, et al. Malaria transmission in relation to rice cultivation in the irrigated Sahel of Mali. Acta Trop. 2004;89:147–159. doi: 10.1016/j.actatropica.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Werren JH, Windsor DM. Wolbachia infection frequencies in insects: Evidence of a global equilibrium? Proc Biol Sci. 2000;267:1277–1285. doi: 10.1098/rspb.2000.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baldo L, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawniczak MKN, et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330:512–514. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia?–A statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grenier S, et al. Successful horizontal transfer of Wolbachia symbionts between Trichogramma wasps. Proc Biol Sci. 1998;265:1441–1445. [Google Scholar]
- 30.Riegler M, Charlat S, Stauffer C, Merçot H. Wolbachia transfer from Rhagoletis cerasi to Drosophila simulans: Investigating the outcomes of host-symbiont coevolution. Appl Environ Microbiol. 2004;70:273–279. doi: 10.1128/AEM.70.1.273-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki T, Kubo T, Ishikawa H. Interspecific transfer of Wolbachia between two lepidopteran insects expressing cytoplasmic incompatibility: A Wolbachia variant naturally infecting Cadra cautella causes male killing in Ephestia kuehniella. Genetics. 2002;162:1313–1319. doi: 10.1093/genetics/162.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frentiu FD, et al. Limited dengue virus replication in field-collected Aedes aegypti mosquitoes infected with Wolbachia. PLoS Negl Trop Dis. 2014;8:e2688. doi: 10.1371/journal.pntd.0002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mousson L, et al. The native Wolbachia symbionts limit transmission of dengue virus in Aedes albopictus. PLoS Negl Trop Dis. 2012;6:e1989. doi: 10.1371/journal.pntd.0001989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sumi T, Miura K, Miyatake T. Wolbachia density changes seasonally amongst populations of the pale grass blue butterfly, Zizeeria maha (Lepidoptera: Lycaenidae) PLoS One. 2017;12:e0175373. doi: 10.1371/journal.pone.0175373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds KT, Thomson LJ, Hoffmann AA. The effects of host age, host nuclear background and temperature on phenotypic effects of the virulent Wolbachia strain popcorn in Drosophila melanogaster. Genetics. 2003;164:1027–1034. doi: 10.1093/genetics/164.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murdock CC, Blanford S, Hughes GL, Rasgon JL, Thomas MB. Temperature alters Plasmodium blocking by Wolbachia. Sci Rep. 2014;4:3932. doi: 10.1038/srep03932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mouton L, Henri H, Bouletreau M, Vavre F. Effect of temperature on Wolbachia density and impact on cytoplasmic incompatibility. Parasitology. 2006;132:49–56. doi: 10.1017/S0031182005008723. [DOI] [PubMed] [Google Scholar]
- 38.LePage DP, et al. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature. 2017;543:243–247. doi: 10.1038/nature21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beckmann JF, Ronau JA, Hochstrasser M. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol. 2017;2:17007. doi: 10.1038/nmicrobiol.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynolds KT, Hoffmann AA. Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet Res. 2002;80:79–87. doi: 10.1017/s0016672302005827. [DOI] [PubMed] [Google Scholar]
- 41.Duron O, Fort P, Weill M. Influence of aging on cytoplasmic incompatibility, sperm modification and Wolbachia density in Culex pipiens mosquitoes. Heredity (Edinb) 2007;98:368–374. doi: 10.1038/sj.hdy.6800948. [DOI] [PubMed] [Google Scholar]
- 42.Beier JC. Vector incrimination and entomological inoculation rates. In: Doolan DL, editor. Malaria Methods and Protocols. Springer; Berlin: 2002. pp. 1–12. [DOI] [PubMed] [Google Scholar]
- 43.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 44.Hughes GL, et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc Natl Acad Sci USA. 2014;111:12498–12503. doi: 10.1073/pnas.1408888111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ifediba T, Vanderberg JP. Complete in vitro maturation of Plasmodium falciparum gametocytes. Nature. 1981;294:364–366. doi: 10.1038/294364a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.