Significance
H-NS is a nucleoid-associated protein that plays a major role in silencing pathogen genes. We discovered that the unstructured linker region connecting the N-terminal oligomerization and C-terminal DNA binding domains plays an important and surprising role in promoting DNA binding. Superresolution imaging identified H-NS foci that required DNA binding for their formation and were associated with the nucleoid. Removing the linker led to the disappearance of foci and a substantially lower affinity for DNA. It was proposed that H-NS compacts DNA, but decreasing DNA binding in cells did not lead to a relaxation of the nucleoid, suggesting H-NS does not play a major role in nucleoid compaction. Molecular dynamic simulations suggested that target acquisition by H-NS may involve sliding along the DNA.
Keywords: H-NS, nucleoid-associated proteins, superresolution microscopy, single-particle tracking, atomic force microscopy
Abstract
Nucleoid-associated proteins (NAPs) facilitate chromosome organization in bacteria, but the precise mechanism remains elusive. H-NS is a NAP that also plays a major role in silencing pathogen genes. We used genetics, single-particle tracking in live cells, superresolution microscopy, atomic force microscopy, and molecular dynamics simulations to examine H-NS/DNA interactions in single cells. We discovered a role for the unstructured linker region connecting the N-terminal oligomerization and C-terminal DNA binding domains. In the present work we demonstrate that linker amino acids promote engagement with DNA. In the absence of linker contacts, H-NS binding is significantly reduced, although no change in chromosome compaction is observed. H-NS is not localized to two distinct foci; rather, it is scattered all around the nucleoid. The linker makes DNA contacts that are required for gene silencing, while chromosome compaction does not appear to be an important H-NS function.
In bacteria, genetic information is highly organized in the cell, in a structure referred to as the nucleoid. Nucleoid-associated proteins (NAPs) are highly abundant proteins that organize and package DNA and also mediate gene regulation. One of these, H-NS, regulates ∼5% of the bacterial genome, mostly by gene silencing. Many of these silenced genes are involved in virulence in bacterial pathogens (1–3), conferring on H-NS the term “genome sentinel” (4, 5).
H-NS possesses two functional domains, an N-terminal oligomerization domain and a C-terminal DNA binding domain, separated by a flexible linker. The NMR structure of the isolated DNA binding domain is composed of a two-stranded β-sheet, an α-helix, and a 310-helix (6). Two conflicting NMR structures of the oligomerization domain exist as a parallel (7) or antiparallel (8) coiled-coil, and hence it is an open question as to how the C-terminal DNA binding domain is oriented on the DNA or whether H-NS is capable of adopting multiple orientations. The X-ray crystal structure of a longer N-terminal region (amino acids 1–83) is composed of two antiparallel dimerization sites of H-NS, and the second dimerization site is the basis for oligomerization (9).
Previous studies compared DNA binding of full-length H-NS protein with the isolated C-terminal DNA binding domain (10, 11). The C terminus displayed a substantially lower affinity for DNA (∼2,000-fold) compared with the full-length protein. This construct employed five residues of the flexible linker connecting the N-terminal and C-terminal DNA domains (10). As a result, we reasoned that the linker might be involved in promoting H-NS/DNA binding and perhaps we could separate gene regulatory functions from its proposed DNA compaction function (12–14). We therefore set out to examine the linker connecting H-NS domains. Our results demonstrate that charged residues in the linker play an important role in H-NS gene silencing, but H-NS does not play a major role in chromosomal compaction.
Single-molecule studies with magnetic tweezers and atomic force microscopy (AFM) identified two DNA binding modes of H-NS, bridging (12) and a stiffened filament (15), that were magnesium-dependent (16); see ref. 17 for a recent review. It was later established that gene silencing required formation of a stiffened protein filament, and relief of silencing resulted from proteins that bind and bend DNA, reorientating H-NS and allowing access for RNA polymerase (3, 18). Herein, we define a role for the unstructured linker that functions not as a passive tether but makes DNA contacts that promote engagement by the C-terminal DNA binding domain. In the absence of these initial contacts, a rigid filament required for gene silencing is not formed, and silencing is eliminated. Single-particle tracking experiments in live, single cells confirmed the alteration in H-NS binding to DNA; wild-type H-NS is almost exclusively bound to DNA (95%), whereas <21% of the linker deletion mutant is bound to DNA. Surprisingly, the nearly complete loss of H-NS binding to DNA did not lead to chromosome relaxation, as indicated by superresolution fluorescence microscopy, although cell elongation occurred.
Results
A Linker Deletion of H-NS Was Incapable of Gene Silencing.
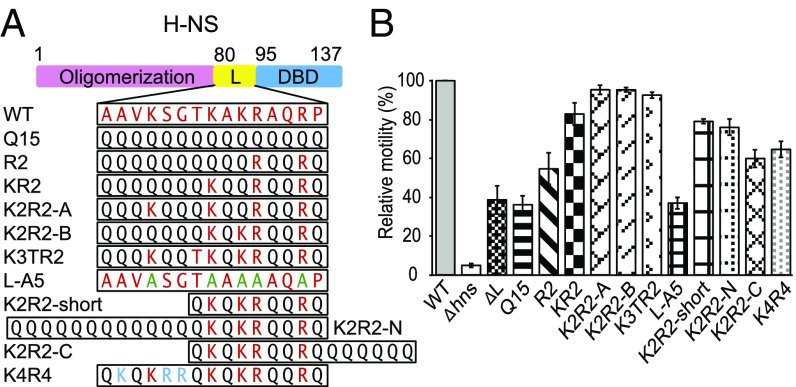
We constructed two chromosome mutants, one in which the N- and C-terminal domains were fused, completely lacking a linker (ΔL), and one in which the linker was replaced by a dummy linker (Q15) of 15 glutamine residues (19). We assayed H-NS function using motility, since H-NS represses hdfR, a repressor of flhDC, the master flagellar gene regulator (20). Thus, cell motility is positively correlated with H-NS function. An H-NS knock-out strain was constructed and the wild-type, null, ΔL, and Q15 linkers were compared on swarm plates (Fig. 1). The wild-type swarm diameter was 8 ± 0.1 cm, indicating the cells were motile and expressing flagella, whereas the hns null was nonmotile (0.3 cm, e.g., 4% compared with wild type). Strains expressing the ΔL or the Q15 linker exhibited significantly smaller halos (3.2 ± 0.3 and 3.1 ± 0.2 cm, respectively), ∼40% of the size of the wild-type swarm. We also examined the effect of the linker mutants at another locus, the csgD promoter, since we identified H-NS as a repressor of csgD in Salmonella (18). A promoter fusion of gfp to csgD (PcsgD-gfp) was constructed and inserted into the bacterial chromosome (SI Appendix, SI Material and Methods), and the fluorescence of the linker mutants was monitored in isolated, single Escherichia coli cells (SI Appendix, Fig. S1). As with the motility assay, linker mutants were nonfunctional and unable to repress PcsgD-gfp, leading to increased GFP fluorescence. The inability to repress was not a result of reduced expression of the mutants, as all of the mutant proteins were abundantly expressed and readily purified for AFM (discussed below). Thus, the H-NS linker does not function as a completely passive tether connecting two functional domains, and some amino acids in the linker contribute significantly to its gene regulation function.
Fig. 1.
Substitutions in the H-NS linker affect gene regulation. (A) L is the linker (amino acids 80–94) and DBD is the DNA binding domain. Original linker residues are in red, additional positive charged residues in blue, alanines in green, and glutamines in black. (B) Swarming motility assay of the linker substitution strains. The relative diameters of the swarm were normalized to the wild-type H-NS (100%). The mean was calculated from three independent experiments with at least two replicates.
Charged Residues in the Linker Were Essential for Function.
H-NS homologs are present in many species of Enterobacteriaceae. An alignment indicated a similarity of >92% and the linker region showed an identity ranging from 40 to 93% (SI Appendix, Fig. S2). The E. coli H-NS linker contains five charged residues that we suspected might contribute to function (21), based on a prediction of DNA-binding residues (DP-Bind). We reintroduced these residues (highlighted in red) into the Q15 linker beginning with two arginines at position 90 and 93 and assayed motility and PcsgD-gfp activity. In Fig. 1, solely adding arginine 90 and 93 (R2) to the Q15 linker had only a minor effect on activity (55% compared with wild type). Further addition of lysine 89, 87, and 83 and threonine 86 (predicted by DP-Bind) (K3TR2) restored full activity. Additional substitutions indicated that arginines 90 and 93 were essential, but a combination of two additional lysines was sufficient to restore activity (K2R2). Furthermore, the specific lysines that were present were not important (i.e., K2R2-A was identical to K2R2-B) (Fig. 1). These substitutions had similar effects at the csgD promoter (SI Appendix, Fig. S1). A net charge of +4 was a minimal requirement for full H-NS function. To further validate our finding we constructed L-A5, replacing the five positive charge residues with alanine in the native linker. L-A5 had a motility profile similar to that of ΔL and Q15 (Fig. 1). Thus, charged residues in the linker were essential for the ability of H-NS to regulate genes that it normally represses. Furthermore, altering the length of the linker or the positioning of charged residues with respect to the C terminus (76–79% of wild type) or adding additional charged residues (65% of wild type) had only slight effects on H-NS function (Fig. 1).
Linker Mutants Were Unable to Polymerize on DNA.
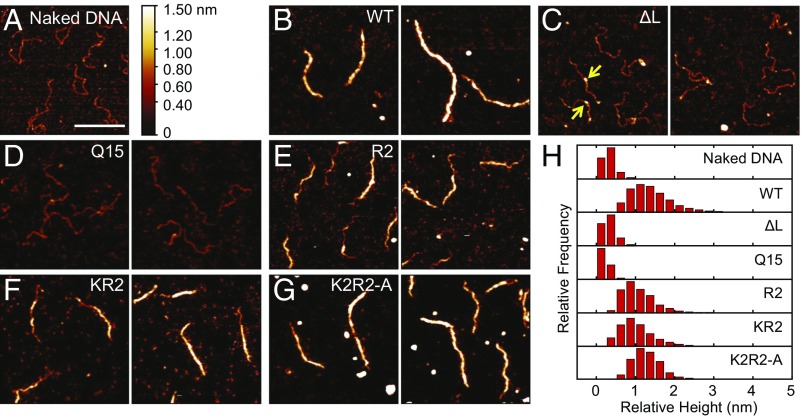
Our previous studies used magnetic tweezers and AFM to analyze H-NS binding functions (16, 17, 22), and identified gene silencing resulted from H-NS polymerization along DNA (22). It was therefore of interest to determine whether the linker mutants were capable of DNA binding and polymerization. The AFM image of a naked 755-bp PcsgD fragment (18) with no added protein is given in Fig. 2A. Addition of wild-type H-NS led to polymerization and complete coating of H-NS along the DNA (Fig. 2 B and H). In contrast, the ΔL and Q15 height histogram was similar to that of naked DNA (Fig. 2 C, D, and H). However, small foci were sometimes evident (yellow arrows Fig. 2C). Suspecting that the lack of binding was due to a reduced affinity of the linker mutants, we increased the protein concentration and the incubation time. The ΔL and Q15 mutant proteins formed small patches of filament in some places (SI Appendix, Fig. S3), indicating that they bound to DNA with lower affinity compared with the wild type. The AFM results verified the motility assay (Fig. 1), in that changes in the H-NS linker region had a large effect on DNA binding. These effects were further quantified using single-particle tracking photoactivatable localization microscopy (SptPALM) (discussed below). In contrast, the R2 mutant and the KR2 mutants were capable of polymerization, that is, the DNA was partially coated with some naked patches of DNA visible (Fig. 2 E and F). The K2R2 mutant was similar to wild-type H-NS (Fig. 2G). The inability of the ΔL and Q15 linker mutants to polymerize along PcsgD explained why they were incapable of gene silencing (Fig. 2 and SI Appendix, Fig. S3).
Fig. 2.
DNA binding is impaired in H-NS linker mutants. (A) The csgD promoter in the absence of H-NS protein. (B) Wild-type H-NS binds and polymerizes along the DNA, forming a stiffened filament. Linker mutants lacking the linker (C) or with the polyQ linker (D) form small foci (yellow arrows) due to reduced binding affinity (SI Appendix, Fig. S3) and fail to polymerize. Addition of positive charges improves binding, leading to DNA polymerization in E and F. The K2R2 mutant (G) binds similarly to the wild-type protein. Two panels show different representative images. All proteins were present at 600 nM, and binding was performed in 50 mM KCl, 2 mM MgCl2, 10 mM Tris⋅HCl (pH 7.4) buffer. (H) Relative height distribution histograms were obtained from DNA contours of each experiment. (Scale bar: 200 nm.)
DNA Binding Impairment Does Not Affect Oligomerization.
The N-terminal oligomerization domain of H-NS was important for H-NS/DNA nucleoprotein filament formation and also affected gene repression (9, 11, 20). To test whether the linker mutants were impaired in their ability to polymerize along DNA because of a defect in oligomerization, we examined whether the ΔL mutant existed as an oligomer in solution. His-tagged wild-type and ΔL mutant proteins were analyzed using gel filtration liquid chromatography (SI Appendix, Fig. S4). The chromatography profile of ΔL was similar to that of wild-type H-NS. Wild-type and ΔL protein peaks were shifted to a lower retention volume, indicating oligomer formation. Hence, the reduced binding affinity of the H-NS mutants and an inability to form nucleoprotein filaments resulted from a direct effect of linker substitutions or its deletion and was not due to a failure to oligomerize. Furthermore, incorporating a dimerization mutant L30P, which eliminates H-NS repression (23), into the ΔL mutant had no additional effect on DNA binding (SI Appendix, Fig. S5).
Foci Formation Required DNA Binding and Was Absent in ΔL.
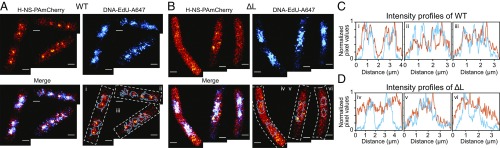
Since the ΔL mutant had reduced affinity for DNA and was unable to polymerize along DNA (Fig. 2) we performed two-color sequential single-molecule localization microscopy (SMLM) to investigate the localization of H-NS and the nucleoid. We compared wild-type H-NS and ΔL, which exhibited the most extreme differences in both AFM and motility assays. Wild-type H-NS and ΔL were linked to photoactivatable mCherry (PAmCherry) using a 12-aa linker. The activity of the wild-type fusion was equivalent to the wild-type native protein, based on the motility assay (SI Appendix, Fig. S6). We first imaged wild-type H-NS-PAmCherry and ΔL-PAmCherry using PALM (Fig. 3 A and B), followed by direct stochastic optical reconstruction microscopy (dSTORM) imaging on the nucleoid labeled with Alexa 647 coupled to 5′-ethynyl-2-deoxyuridne (EdU) which was incorporated into the genome (Fig. 3 A and B). H-NS and nucleoid images were merged to compare their localization. In the wild type, dense regions of H-NS were evident, as reported previously (24). However, in addition to the dense foci, many molecules of H-NS were evident throughout the cell outside the foci. Dense foci were not observed in the ΔL mutant (compare Fig. 3 A and B). Occasionally, there were cells with smaller clusters on the sides (second cell in Fig. 3B), but they were morphologically distinct from the wild-type clusters, which were aligned along the cell length. Next, we used density-based spatial clustering of applications with noise (DBSCAN) to determine the wild-type cluster size (25). The average cluster diameter was 273 ± 108 nm (SI Appendix, Fig. S7). The percentage of molecules that were inside the clusters was 26% (SI Appendix, SI Material and Methods). A previous study reported a cluster size of ∼360 nm and 60 ± 25% of the total localizations were in the cluster (24). The differences in cluster size reported previously were most likely due to imaging during different stages of the cell cycle (26). We imaged cells during midexponential growth (OD600 = 0.6), whereas the previous study imaged cells in lag phase (OD600 = 0.01).
Fig. 3.
Two-color sequential SMLM of H-NS-PAmCherry and DNA-EdU-Alexa Fluor 647 (A647). Wild-type H-NS (A) and ΔL (B). Top left in A and B are images of H-NS localization, and right panels in A and B are nucleoid localization. Merged images of H-NS and the nucleoid are in the bottom left. Bottom right panels in A and B include the outline of the nucleoids in cyan. Boxes outlined with dotted white lines indicate a 0.9-µm-thick line drawn along the long axis of the cell integrating pixel values across the width of the line creating the intensity profiles in C and D. The indices i to vi correspond to intensity profile plots in C and D. C and D are intensity profiles of H-NS (orange) and DNA (cyan) localizations for wild-type H-NS (C) and ΔL (D). (Scale bars: 0.5 µm.)
In the merged image it was evident that wild-type H-NS foci were colocalized with the nucleoid (Fig. 3C). This image is significant, because it indicates that the foci observed in superresolution images of H-NS (ref. 24 and Fig. 3A) resulted from H-NS binding to DNA (absent in the ΔL strain) and were not the result of PAmCherry-driven oligomerization, as previously suggested (27). Furthermore, when we grew cells in rich media (LB) the foci were no longer evident in the wild type, probably due to rapid growth (SI Appendix, Fig. S8). We also imaged the R2 and K2R2-A mutants. Both mutants have nucleoid-associated foci (SI Appendix, Fig. S9). Thus, positive charges of the linker were essential for association of H-NS with the nucleoid in single cells.
Elimination of H-NS Binding Does Not Affect Nucleoid Compaction.
H-NS was proposed to perform two distinct functions: nucleoid compaction and gene regulation. Clearly, the absence of DNA binding of the ΔL mutant drastically affected gene regulation (Fig. 1 and SI Appendix, Fig. S1). To examine whether elimination of H-NS binding to DNA affected nucleoid compaction, the size of the nucleoid in the wild type and ΔL mutant were quantified in cells stained with DAPI, using structured-illumination microscopy (SIM) (SI Appendix, SI Material and Methods). For this experiment, SIM was favored over SMLM, because we could rapidly image more cells (SI Appendix, Fig. S10). The average nucleoid area of wild-type H-NS cells was 0.43 ± 0.11 μm2 (n = 107). In contrast, the nucleoid of the ΔL mutant was smaller (0.38 ± 0.10 μm2, n = 112). The cell length of the ΔL mutant was longer (2.83 ± 0.46 μm) compared with the wild type (2.41 ± 0.41 μm) (Figs. 3 and 4). As a positive control, we added novobiocin to the wild-type culture. Novobiocin inhibits DNA gyrase ATPase activity, relaxing DNA supercoils (28). Novobiocin-treated wild-type cells had larger nucleoids (0.51 ± 0.15 μm2, n = 150) compared with the untreated control, indicating that DNA relaxation/compaction can be determined using this method.
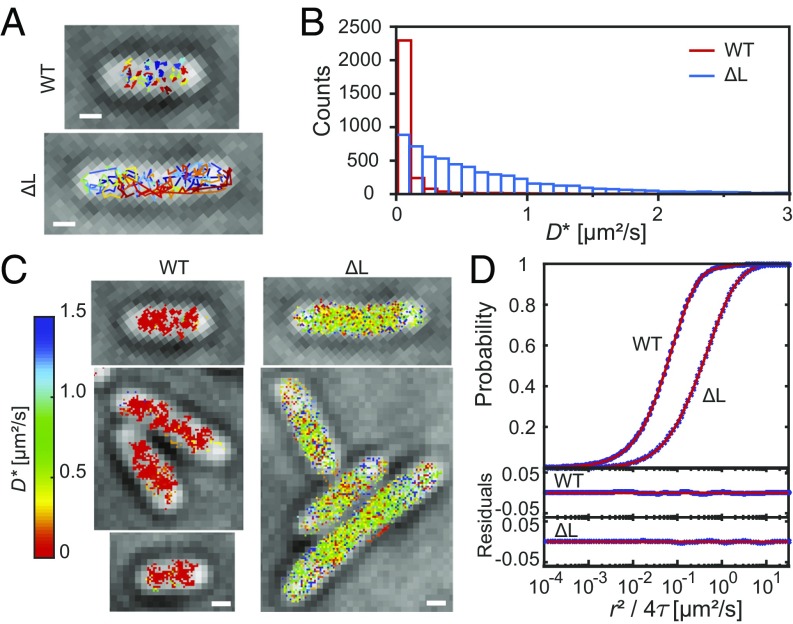
Fig. 4.
SptPALM of wild-type H-NS and ΔL. (A) Representative cells and tracks of wild-type and ΔL strains. (B) Apparent diffusion coefficient D* histogram obtained from the slope of the first three points of the MSD plot calculated from individual tracks. (C) Diffusion map constructed from the D* obtained from B by converting the x–y vector coordinates of all of the spots in the tracks into pixel coordinates (50-nm size). The color map ranges from D* = 0 (red) to 1.5 µm2/s (dark blue). Values >1.5 µm2/s were also dark blue pixels. (D) CPD plot of both wild-type and ΔL strains. Blue circles represent the experimental data, and red lines represent the fit of the data to SI Appendix, Eq. 3. (Scale bars: 0.5 µm.)
SptPALM Identified a Reduction in DNA Binding by ΔL.
The localization of ΔL was very different from wild-type H-NS (Fig. 3), due to its low affinity for DNA, evident in the AFM images (Fig. 2 and SI Appendix, Fig. S3). We therefore used SptPALM (29) in live bacteria to measure the apparent diffusion coefficients D* of wild-type H-NS and ΔL (Fig. 4 A and B and SI Appendix, Fig. S11). The D* values of wild-type H-NS were <0.2 µm2/s [i.e., similar to values obtained for the NAP HU (30) and RNA polymerase (31)]. In contrast, D* values of ΔL were distributed over a broad range, clearly indicating that a substantial fraction of ΔL molecules was either not bound to the nucleoid or was transiently bound. LacI proteins that made nonspecific interactions with DNA had a D* value of about 0.4 µm2/s (32), and transient, nonspecific RNA polymerase interactions with DNA during promoter searching had a D* value of 0.36 µm2/s (31). Given these values, we sorted the H-NS tracks into three categories based on D* values: (i) <0.2 µm2/s, (ii) 0.2–0.6 µm2/s, and (iii) >0.6 µm2/s (SI Appendix, Fig. S12). The tracks were also converted into a pixelated D* heat map displaying the spatial distribution of D* (Fig. 4C and SI Appendix, Fig. S13). From the heat map, the tracks of wild-type H-NS, which has low D* values (indicated by the red/orange pixels), were localized as clusters in the cell. In contrast, the ΔL tracks were distributed throughout the cell with a broad range of D* values.
To quantify the fraction of ΔL molecules that were not bound to the nucleoid, the cumulative probability distribution (CPD) method was employed. The CPD plot of ΔL was shifted to the right of wild-type H-NS, indicating increased mobility (Fig. 4D). A three-diffusion component model best fit the data (SI Appendix, Eq. 3). D* values obtained for wild-type H-NS were D1 = 0.040 ± 0.002 µm2/s (35%), D2 = 0.12 ± 0.01 µm2/s (60%), and D3 = 1.05 ± 0.15 µm2/s (5%). Proteins that bind to the nucleoid have D* values <0.2 µm2/s (30, 31). Hence, D1 and D2 values of 0.040 and 0.12 µm2/s represent H-NS bound to DNA. The presence of two bound D* components could be due to the DNA binding modes of H-NS (16, 17). Only 5% of wild-type H-NS molecules had a fast D3, indicating that most of the H-NS in the cell was bound to DNA.
ΔL apparent diffusion coefficients were D1 = 0.12 ± 0.01 µm2/s (21%), D2 = 0.54 ± 0.03 µm2/s (55%), and D3 = 1.96 ± 0.11 µm2/s (24%). The appearance of a 0.54 µm2/s D2 in the mutant was indicative of transient DNA binding. The very slow D1 component (0.040 µm2/s) in the wild type was no longer detected and D2 decreased from 60 to 21% (D1 was 0.12 µm2/s in ΔL). In addition, the D3 fast fraction (1.05 µm2/s in the wild type) increased from 5 to 24% and was almost twofold faster (1.96 µm2/s) in ΔL. We also determined diffusion coefficients of the R2 and K2R2-A mutants (SI Appendix, Table S3). K2R2-A and the wild type had similar levels of the unbound fraction of H-NS, while the R2 mutant was between ΔL and the wild type, in keeping with its motility phenotype and AFM profile (Figs. 1 and 2). These observations identify the linker as being essential for tight binding of H-NS to DNA and for the appearance of foci in superresolution images (Fig. 3). In the absence of the linker, ΔL can barely bind to DNA, as we observed in the AFM (Fig. 2). This led to the appearance of D2 (0.54 µm2/s, 55%), which could arise from the C-terminal DNA binding domain making transient contact with DNA but being unable to switch into the tighter binding mode due to the loss of the linker.
Linker Residues Initiated DNA Contact by Electrostatic Interaction.
Next we employed all-atom molecular dynamics (MD) simulations to explore the role of the charged linker residues in promoting DNA binding. The interaction between the csgD promoter high-affinity sequence ATTTTAATATAACGAGTTAC and H-NS (amino acids 80–137) was investigated. DNA and H-NS were initially placed ∼3.5 nm apart, and production runs of 500 ns were performed to investigate the interaction between freely diffusing DNA and H-NS molecules. In nine independent simulations with different initial orientations of DNA and H-NS, the linker bound to DNA within 10 ns after simulations commenced, and H-NS remained associated with DNA throughout its duration. Two snapshots of DNA-H-NS complexes at 500 ns of the combined nine simulations are shown in Fig. 5 (the remaining are in Movie S1), revealing that the linker can bind both major and minor grooves. Initial interaction led to a decrease in electrostatic potential between the linker and DNA (SI Appendix, Fig. S14), that is, identical to the electrostatic potential of the sole five lysine/arginine residues and DNA. This result indicated that the electrostatic attraction between the linker and DNA critically depends on the positively charged residues (Figs. 1, 2, and 5).
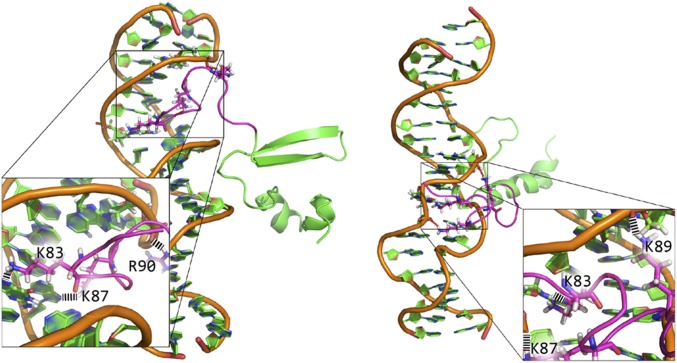
Fig. 5.
Two of nine snapshots from independent trajectories of MD simulations with different initial conformations. The C terminus of H-NS (amino acids 95–137) is indicated in green and the linker (amino acids 80–94) indicated in magenta. All trajectories feature a stable attraction between DNA and the H-NS linker. Key residues forming H-bonds are shown in stick representation and H-bonds are shown in black dashed lines. See SI Appendix, Fig. S14 and Movie S1 for remaining snapshots.
In all bound conformations lysine and arginine residues interacted with the phosphate backbone and bases through approximately five H-bonds, with approximately three contributed by the two arginines and approximately two by the three lysines (SI Appendix, Fig. S15). In the bound state the conformation of the linker was highly dynamic, consistent with the nature of nonspecific electrostatic interactions between positively charged residues and DNA. This implies a potential capability of the linker to slide diffusively along DNA without dissociation, since such diffusion does not involve significant free energy cost. Within the simulation time scale the C terminus of H-NS was trapped in the vicinity of DNA by the linker, but did not stably bind the DNA (Movie S1). Binding of the C terminus to the minor groove (33) likely involves deformation of DNA that does not occur within our MD-accessible time scale.
Discussion
H-NS Does Not Appear to Play a Role in Chromosome Compaction.
Previous optical tweezers and AFM measurements of H-NS bridging of two distinct DNA molecules suggested that H-NS was involved in compaction of the nucleoid (12–14). The results of these in vitro measurements were not compatible with our direct image analysis of the nucleoid (SI Appendix, Fig. S10). If H-NS were to play a significant role in chromosome compaction, reducing its DNA binding ability from 95 to 21% should lead to relaxation of the chromosome, which was not observed. Although the cells in the ΔL mutant were noticeably longer (17% > wild type), the increase in length was likely due to the many cell envelope genes regulated by H-NS (34); compaction functions are most likely provided by redundant NAPs such as FIS (35) or HU (36–38). Our observation that H-NS does not play a major role in nucleoid compaction also explains why hns deletions are tolerated in E. coli, since nucleoid relaxation would be expected to be deleterious. In previous reports, the number of origins in an hns null was less than the wild type (39–41), with fewer DNA replication forks and less DNA content. This could explain why ΔL has a more compact nucleoid (12% smaller than wild type). Nevertheless, complete relaxation of the chromosome, in which the nucleoid expands significantly, was not observed, indicating that H-NS does not play a major role in nucleoid compaction.
Polymerization Is a Function of DNA Binding Affinity.
The N terminus of H-NS is involved in oligomerization, and two distinct regions contribute to its oligomerization ability (9). Oligomerization was not affected by linker substitutions, because the ΔL mutant still retained its ability to form higher-order complexes (SI Appendix, Fig. S4). One would expect that oligomerization propensity might somehow be coupled to the ability of H-NS to polymerize along DNA. However, the linker mutants were unable to polymerize (Fig. 2), and this property was related to their reduced binding affinity (SI Appendix, Fig. S3). At higher concentrations polymerization was evident, but it was not nearly as robust as the wild-type H-NS.
A Role for Linkers in Proteins.
Multidomain proteins are joined by linker peptides that connect protein domains and provide functions such as cooperative interdomain interactions, allowing flexibility or preserving biological activity. A survey of 1,280 linkers in native proteins grouped the average length of linkers in multidomain proteins into small (4.5 residues), medium (9.1 residues), and large linkers (21 residues) (42). Natural linkers adopt various conformations in secondary structure to exert their functions. Q-linkers were observed at the boundaries of functionally distinct domains in a variety of bacterial regulatory and sensory transduction proteins (43). A flexible, disordered linker separates the two distinct functional domains of H-NS. Disordered linkers have been overlooked in biology, because the conventional structure–function paradigm requires a fixed, 3D structure for function. One demonstration of linker conformation effects on transcription is the AraC bacterial transcriptional regulator. In one conformation (suspected to be a random coil) AraC forms a repressive loop between distant molecules bound to DNA, whereas a change in linker conformation (proposed to be an increase in helicity) reorients the AraC dimer, and it binds to adjacent, activating sites on DNA (44).
Recent studies have discovered that disordered proteins are abundant in proteomes across all domains of life and functionally significant. About 39% of proteins in the Eukaryota, 10% in Archaea, and 9% in Bacteria have long disordered regions (>30 disordered residues) (45). Intrinsically disordered proteins and protein regions are highly enriched in eukaryotic DNA-binding proteins (∼68%), in contrast to ∼8% of bacterial DNA-binding proteins (45). These disordered regions are rich in positively charged residues that tend to form clusters in the tails of DNA binding domains. The role of disordered linkers in DNA-binding has been mainly studied in eukaryotic transcriptional factors. For example, the cocrystal structure of Drosophila Scr-DNA (a homeodomain transcription factor) shows positive charges arginine and histidine from the extended disordered region inserted into the minor groove that are required for DNA binding (46). MD simulations of homeodomain proteins Antp, NK2, HoxD9, and p53 identified electrostatic interactions between the disordered region and DNA that facilitate specific DNA searching by one-dimensional sliding or hopping (47). With the exception of HoxD9, the other homeodomain proteins possess four to five net positive charges (as does H-NS). The efficiency of the DNA search was dependent on the length and net charge (47).
The H-NS linker has five positively charged residues in the linker; among these, K87, K89, and R90 are clustered in the center, while K83 and K93 are flanking them. The net charge is +5, similar to the net charge of NK2 and p53. Our MD simulation suggested that binding to DNA by H-NS was initiated by the insertion of the disordered linker, driven by nonspecific electrostatic forces, followed by a subsequent one-dimensional sliding mode, very similar to homeodomain proteins (47). Since the number of charged residues, their distribution, and the length of the disordered linker are critical for the sliding search mode (47), we examined the effect of linker length and spacing between the charged clusters to the DNA binding domain (Fig. 1, K2R2-N, K2R2-C, and K2R2 short). Only minor effects on the position and length (60–80% of wild type) of the disordered linker were observed. Reducing the number of positive charges to +0 eliminated function, and a +4 positive charge was sufficient for maximal activity. Increasing the number of positive charges to +8 reduced activity to 60% of wild type. Taken together, our results reveal an exquisite sensitivity to overall charge in the disordered linker of H-NS to +4–5.
Implications of the MD Simulations.
MD simulations suggest a fast initial binding of the H-NS linker to DNA. Initial binding features a nonspecific electrostatic interaction between positively charged residues in the linker domain and DNA, which possibly allows diffusive sliding of H-NS along DNA. In three of nine 500-ns simulations the bound molecule diffused over a length scale of half of a helical repeat of DNA; the 1D diffusion coefficient was roughly estimated to be on the order of nanometers squared per microsecond, the same order of magnitude of the diffusion coefficient of transcription factors sliding on nonspecific DNA sequences (48). A one-dimensional “sliding” motion of protein along nonspecific DNA can drastically increase the rate of specific binding targeting (49, 50). Therefore, the simulation suggests that target site searching of H-NS on DNA involves an initial nonspecific electrostatic attraction by the linker, followed by 1D sliding along the DNA and final localization at a high-affinity, AT-rich site. This 1D sliding may also facilitate incorporation of a newly bound H-NS into an H-NS filament, which would then be unable to slide further. Such sliding-assisted site searching was previously observed for LacI searching for its specific binding sites (32).
Implications of H-NS DNA Binding Affinity and Specificity.
In the present work we determined that deletion of the linker domain or deletion of positive charged residues in the linker drastically reduced the binding of H-NS to DNA, indicating that the H-NS linker was the predominant factor that contributed to the DNA binding affinity of H-NS. This nonspecific electrostatic interaction between the linker and DNA is not in conflict with reports of H-NS binding to specific AT-rich nucleation sites (51). Specificity can be achieved as long as the specific site provides a conformational feature that can further increase the attractive interaction between H-NS and DNA. Recent studies reported that recognition of a specific H-NS binding site depended on indirect readouts based on DNA shape and electrostatics, rather than base-specific contacts (33, 52). The most preferred bound conformation involved narrower minor grooves, compared with B-form DNA, which is characteristic of AT tracts (33). Binding involves Arg90 of H-NS as a key residue, inserting its side chain deep into the minor groove. This binding conformation was also observed in our MD simulations (Movie S1). Therefore, this additional affinity between H-NS and AT-rich DNA can result in preferential formation of an H-NS filament at AT-rich regions.
Materials and Methods
Details of materials and methods are given in SI Appendix. A list of strains and plasmids are in SI Appendix, Table S1, and primers are given in SI Appendix, Table S2. In brief, AFM imaging was performed with 600 nM H-NS and 755 bp E. coli csgD. For SMLM imaging, 10 µM EdU was added to the cells, grown in M9 medium, to OD600 ∼0.2. Cells were harvested at OD600 ∼ 0.6 and fixed with 1.5% paraformaldehyde. PALM imaging was performed, followed by click-chemistry, incorporating Alexa647 for dSTORM imaging. SptPALM was performed at 17 ms per frame, and only tracks with five or more spots were analyzed. MD simulations were performed using Gromacs 5.1.1 with the ff99sb-ildn-NMR force field.
Supplementary Material
Acknowledgments
We thank Mike Heilemann and Christoph Spahn (Goethe University) for invaluable comments. This work was supported by Grants NIHAI-123640 and VAIOBX-000372 (to L.J.K.) and a Research Center of Excellence in Mechanobiology from the Ministry of Education, Singapore.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716721114/-/DCSupplemental.
References
- 1.Prajapat MK, Saini S. Interplay between Fur and HNS in controlling virulence gene expression in Salmonella typhimurium. Comput Biol Med. 2012;42:1133–1140. doi: 10.1016/j.compbiomed.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Sun F, et al. H-NS is a repressor of major virulence gene loci in Vibrio parahaemolyticus. Front Microbiol. 2014;5:675. doi: 10.3389/fmicb.2014.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walthers D, et al. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J Biol Chem. 2011;286:1895–1902. doi: 10.1074/jbc.M110.164962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucchini S, et al. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navarre WW, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 6.Shindo H, et al. Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli. FEBS Lett. 1995;360:125–131. doi: 10.1016/0014-5793(95)00079-o. [DOI] [PubMed] [Google Scholar]
- 7.Esposito D, et al. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J Mol Biol. 2002;324:841–850. doi: 10.1016/s0022-2836(02)01141-5. [DOI] [PubMed] [Google Scholar]
- 8.Bloch V, et al. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat Struct Biol. 2003;10:212–218. doi: 10.1038/nsb904. [DOI] [PubMed] [Google Scholar]
- 9.Arold ST, Leonard PG, Parkinson GN, Ladbury JE. H-NS forms a superhelical protein scaffold for DNA condensation. Proc Natl Acad Sci USA. 2010;107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shindo H, et al. Identification of the DNA binding surface of H-NS protein from Escherichia coli by heteronuclear NMR spectroscopy. FEBS Lett. 1999;455:63–69. doi: 10.1016/s0014-5793(99)00862-5. [DOI] [PubMed] [Google Scholar]
- 11.Sette M, et al. Sequence-specific recognition of DNA by the C-terminal domain of nucleoid-associated protein H-NS. J Biol Chem. 2009;284:30453–30462. doi: 10.1074/jbc.M109.044313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dame RT, et al. DNA bridging: A property shared among H-NS-like proteins. J Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 14.Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winardhi RS, Yan J, Kenney LJ. H-NS regulates gene expression and compacts the nucleoid: Insights from single-molecule experiments. Biophys J. 2015;109:1321–1329. doi: 10.1016/j.bpj.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai SK, et al. The horizontally-acquired response regulator SsrB drives a Salmonella lifestyle switch by relieving biofilm silencing. Elife. 2016;5:e10747. doi: 10.7554/eLife.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattison K, Oropeza R, Kenney LJ. The linker region plays an important role in the interdomain communication of the response regulator OmpR. J Biol Chem. 2002;277:32714–32721. doi: 10.1074/jbc.M204122200. [DOI] [PubMed] [Google Scholar]
- 20.Ko M, Park C. H-NS-Dependent regulation of flagellar synthesis is mediated by a LysR family protein. J Bacteriol. 2000;182:4670–4672. doi: 10.1128/jb.182.16.4670-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giangrossi M, Wintraecken K, Spurio R, de Vries R. Probing the relation between protein-protein interactions and DNA binding for a linker mutant of the bacterial nucleoid protein H-NS. Biochim Biophys Acta. 2014;1844:339–345. doi: 10.1016/j.bbapap.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Lim CJ, Lee SY, Kenney LJ, Yan J. Nucleoprotein filament formation is the structural basis for bacterial protein H-NS gene silencing. Sci Rep. 2012;2:509. doi: 10.1038/srep00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueguchi C, Seto C, Suzuki T, Mizuno T. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J Mol Biol. 1997;274:145–151. doi: 10.1006/jmbi.1997.1381. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Li GW, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ester M, Kriegel H-P, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. In: Simoudis E, Han J, Fayyad UM, editors. Proceedings of the Second International Conference on Knowledge Discovery and Data Mining (KDD-96) AAAI; Palo Alto, CA: 1996. pp. 226–231. [Google Scholar]
- 26.Liu Y, et al. A model for chromosome organization during the cell cycle in live E. coli. Sci Rep. 2015;5:17133. doi: 10.1038/srep17133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc Natl Acad Sci USA. 2014;111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cameron AD, Stoebel DM, Dorman CJ. DNA supercoiling is differentially regulated by environmental factors and FIS in Escherichia coli and Salmonella enterica. Mol Microbiol. 2011;80:85–101. doi: 10.1111/j.1365-2958.2011.07560.x. [DOI] [PubMed] [Google Scholar]
- 29.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 30.Sanamrad A, et al. Single-particle tracking reveals that free ribosomal subunits are not excluded from the Escherichia coli nucleoid. Proc Natl Acad Sci USA. 2014;111:11413–11418. doi: 10.1073/pnas.1411558111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stracy M, et al. Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. Proc Natl Acad Sci USA. 2015;112:E4390–E4399. doi: 10.1073/pnas.1507592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon BR, et al. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc Natl Acad Sci USA. 2011;108:10690–10695. doi: 10.1073/pnas.1102544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hommais F, et al. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 35.Skoko D, et al. Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J Mol Biol. 2006;364:777–798. doi: 10.1016/j.jmb.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: Formation of flexible hinges and rigid filaments. Proc Natl Acad Sci USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnurr B, Vorgias C, Stavans J. Compaction and supercoiling of single, long DNA molecules by HU protein. Biophys Rev Lett. 2006;01:29–44. [Google Scholar]
- 38.Xiao B, Johnson RC, Marko JF. Modulation of HU-DNA interactions by salt concentration and applied force. Nucleic Acids Res. 2010;38:6176–6185. doi: 10.1093/nar/gkq435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atlung T, Hansen FG. Effect of different concentrations of H-NS protein on chromosome replication and the cell cycle in Escherichia coli. J Bacteriol. 2002;184:1843–1850. doi: 10.1128/JB.184.7.1843-1850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morigen, Odsbu I, Skarstad K. Growth rate dependent numbers of SeqA structures organize the multiple replication forks in rapidly growing Escherichia coli. Genes Cells. 2009;14:643–657. doi: 10.1111/j.1365-2443.2009.01298.x. [DOI] [PubMed] [Google Scholar]
- 41.Helgesen E, Fossum-Raunehaug S, Skarstad K. Lack of the H-NS protein results in extended and aberrantly positioned DNA during chromosome replication and segregation in Escherichia coli. J Bacteriol. 2016;198:1305–1316. doi: 10.1128/JB.00919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.George RA, Heringa J. An analysis of protein domain linkers: Their classification and role in protein folding. Protein Eng. 2002;15:871–879. doi: 10.1093/protein/15.11.871. [DOI] [PubMed] [Google Scholar]
- 43.Wootton JC, Drummond MH. The Q-linker: A class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989;2:535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]
- 44.Malaga F, et al. A genetic and physical study of the interdomain linker of E. Coli AraC protein–A trans-subunit communication pathway. Proteins. 2016;84:448–460. doi: 10.1002/prot.24990. [DOI] [PubMed] [Google Scholar]
- 45.Wang C, Uversky VN, Kurgan L. Disordered nucleiome: Abundance of intrinsic disorder in the DNA- and RNA-binding proteins in 1121 species from eukaryota, bacteria and archaea. Proteomics. 2016;16:1486–1498. doi: 10.1002/pmic.201500177. [DOI] [PubMed] [Google Scholar]
- 46.Joshi R, et al. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vuzman D, Levy Y. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol Biosyst. 2012;8:47–57. doi: 10.1039/c1mb05273j. [DOI] [PubMed] [Google Scholar]
- 48.Blainey PC, et al. Nonspecifically bound proteins spin while diffusing along DNA. Nat Struct Mol Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 50.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat Struct Mol Biol. 2007;14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 52.Cordeiro TN, et al. Indirect DNA readout by an H-NS related protein: Structure of the DNA complex of the C-terminal domain of Ler. PLoS Pathog. 2011;7:e1002380. doi: 10.1371/journal.ppat.1002380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.