We previously reported that Asp631 decreased the redox potential (Em) for heme 9 by 136 mV [1,362 mV according to Breuer et al. (1)] in native MtrF, whereas Asp631 protonation (corresponding to Asn631 mutation) did not significantly affect Em for heme 9, because deprotonation of other residues compensated for the Em shift (2). Breuer et al. (3) cannot observe the corresponding effect, since they fixed the protonation states of titratable residues (1).
According to Breuer et al. (1), residues make unusually large contributions to Em; for example, Asp228 decreases Em for heme 2 by −2,280 mV [−61 mV in our calculations (2)]. In their letter, Breuer et al. (3) state that “it is only meaningful to compare the sign but not the magnitude of the single-residue contributions”; however, in their original report, they focus on not only the sign but also the magnitude of the single-residue contributions, stating, for example, “we find that every charged residue in the environment of a cofactor contributes several tenths of volts” (1). The contribution of each residue to Em must be comparable to Em shifts experimentally measured upon mutations.
To verify Em obtained using their molecular dynamic (MD) simulation-based thermodynamic integration (TI) approach, we employed the slow-growth TI approach (4), as widely used for free energy calculations (e.g., ref. 5), wherein the redox states gradually transit from Fe3+ to Fe2+ (2). Breuer et al. (3) argue against the time scale of our TI simulations (10 ns), but they justify the time scale of their TI simulations (11 ns) assuming the structural “stiffness of the decaheme motif” (however, see below). We emphasize that any MD-based approaches are not appropriate in this case, irrespective of simulation time due to the following reasons.
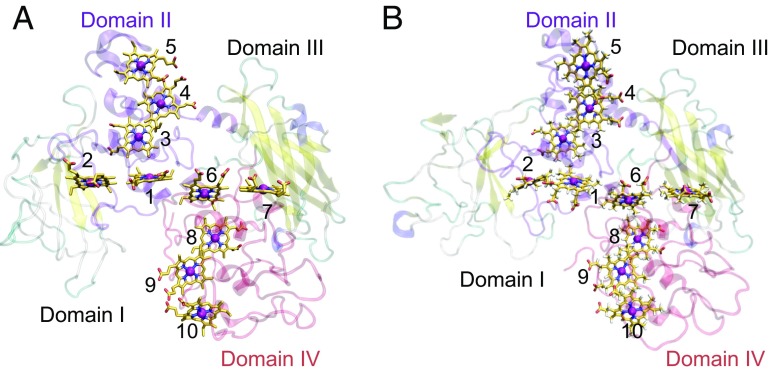
For the structural fluctuations (Fig. 1), we have pointed out that the side-chain orientations are incorrect in the β-barrel motif of domain I of the original MtrF crystal structure (e.g., hydrophobic residues are oriented toward the bulk solvent), resulting in remarkably high calculated B-factors (2). Nevertheless, Breuer et al. (1) have used the original side-chain orientations in the MtrF crystal structure. Heme 2 is closest to domain I (heme 7 to domain III). Notably, only Em values of the heme 2 and heme 7 pair differ significantly (130 mV) in their perfectly symmetrical Em profile (1). Moreover, even heme-binding domain IV is also unstable (Fig. 1). Since their simulations were still in the slow-decay process of domains I and IV, being far from equilibrium, Em profiles strongly depend on the MD-starting structure (e.g., obtained after equilibration for 0, 100, and 1,000 ns) as demonstrated in our test MD calculations (2). From the statement, “the considerable stiffness of the decaheme motif” in Breuer et al.’s letter (3), they have missed this point while calculating Em (1).
Fig. 1.
(A) Crystal structure of MtrF (9). Hemes 2 and 7 have been proposed to be located near the flavin-binding site (9, 10). (B) MtrF structure obtained after equilibration for 1.2 μs. Domains I and IV show remarkably large structural deviations from the original MtrF crystal structure. For comparison with their calculation, we also used the original MtrF crystal structure in MD-based TI calculations.
Finally, the Em for hemes 2 (−57 mV) and 7 (74 mV) reported by Breuer et al. (1) seem unlikely to support a role of bound flavin [−150 mV (6)] serving as the terminal electron acceptor (7, 8). Their MD simulation-based TI approach, using their geometry and fixing the protonation states of titratable residues and heme-propionic groups, is unlikely to provide functionally relevant values of Em in MtrF.
Supplementary Material
Acknowledgments
This research was supported by Japan Science and Technology Agency (JST) Core Research for Evolutional Science and Technology (Grant JPMJCR1656), JST Precursory Research for Embryonic Science and Technology, Japan Society for the Promotion of Science KAKENHI (Grant 17K15101 to H.C.W. and Grants JP16H06560 and JP26105012 to H.I.), the Japan Agency for Medical Research and Development, the Materials Integration for Engineering Polymers of Cross-Ministerial Strategic Innovation Promotion Program, the Interdisciplinary Computational Science Program in Center for Computational Sciences at University of Tsukuba, the TSUBAME Encouragement Program for Young/Female Users of Global Scientific Information and Computing Center at the Tokyo Institute of Technology, and the Joint Usage/Research Center for Interdisciplinary Large-Scale Information Infrastructures in Japan.
Footnotes
The authors declare no conflict of interest.
References
- 1.Breuer M, Zarzycki P, Blumberger J, Rosso KM. Thermodynamics of electron flow in the bacterial deca-heme cytochrome MtrF. J Am Chem Soc. 2012;134:9868–9871. doi: 10.1021/ja3027696. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe HC, Yamashita Y, Ishikita H. Electron transfer pathways in a multiheme cytochrome MtrF. Proc Natl Acad Sci USA. 2017;114:2916–2921. doi: 10.1073/pnas.1617615114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breuer M, Rosso KM, Blumberger J. Redox potentials in the decaheme cytochrome MtrF: Poisson–Boltzmann vs. molecular dynamics simulations. Proc Natl Acad Sci USA. 2017;114:E10028. doi: 10.1073/pnas.1716813114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Postma JPM, Berendsen HJC, Haak JR. Thermodynamics of cavity formation in water. A molecular dynamics study. Faraday Symp Chem Soc. 1982;17:55–67. [Google Scholar]
- 5.Bash PA, Singh UC, Langridge R, Kollman PA. Free energy calculations by computer simulation. Science. 1987;236:564–568. doi: 10.1126/science.3576184. [DOI] [PubMed] [Google Scholar]
- 6.Okamoto A, Hashimoto K, Nealson KH, Nakamura R. Rate enhancement of bacterial extracellular electron transport involves bound flavin semiquinones. Proc Natl Acad Sci USA. 2013;110:7856–7861. doi: 10.1073/pnas.1220823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards MJ, et al. Redox linked flavin sites in extracellular decaheme proteins involved in microbe-mineral electron transfer. Sci Rep. 2015;5:11677. doi: 10.1038/srep11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu S, Jangir Y, El-Naggar MY. Disentangling the roles of free and cytochrome-bound flavins in extracellular electron transport from Shewanella oneidensis MR-1. Electrochim Acta. 2016;198:49–55. [Google Scholar]
- 9.Clarke TA, et al. Structure of a bacterial cell surface decaheme electron conduit. Proc Natl Acad Sci USA. 2011;108:9384–9389. doi: 10.1073/pnas.1017200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brutinel ED, Gralnick JA. Shuttling happens: Soluble flavin mediators of extracellular electron transfer in Shewanella. Appl Microbiol Biotechnol. 2012;93:41–48. doi: 10.1007/s00253-011-3653-0. [DOI] [PubMed] [Google Scholar]