Abstract
Coagulation factor XI (FXI) has become increasingly interesting for its role in pathogenesis of thrombosis. While elevated plasma levels of FXI have been associated with venous thromboembolism and ischemic stroke, its deficiency is associated with mild bleeding. We aimed to determine novel genetic and post-transcriptional plasma FXI regulators.
We performed a genome-wide association study (GWAS) for plasma FXI levels, using novel data imputed to the 1000 Genomes reference panel. Individual GWAS analyses, including a total of 16,169 European individuals from the ARIC, GHS, MARTHA and PROCARDIS studies, were meta-analysed and further replicated in 2,045 individuals from the F5L family, GAIT2 and MEGA studies. Additional association with activated partial thromboplastin time (aPTT) was tested for the top SNPs. In addition, a study on the effect of miRNA on FXI regulation was performed using in silico prediction tools and in vitro luciferase assays.
Three loci showed robust, replicating association with circulating FXI levels: KNG1 (rs710446, P-value = 2.07 × 10−302), F11 (rs4253417, P-value = 2.86 × 10−193), and a novel association in GCKR (rs780094, P-value = 3.56 ×10−09), here for the first time implicated in FXI regulation. The two first SNPs (rs710446 and rs4253417) also associated with aPTT. Conditional and haplotype analyses demonstrated a complex association signal, with additional novel SNPs modulating plasma FXI levels in both the F11 and KNG1 loci. Finally, eight miRNAs were predicted to bind F11 mRNA. Over-expression of either miR-145 or miR-181 significantly reduced the luciferase activity in cells transfected with a plasmid containing FXI-3’UTR.
These results should open the door to new therapeutic targets for thrombosis prevention.
Introduction
Coagulation factor XI (FXI) is a component of the contact pathway of coagulation with an important role in the propagation and stabilization of the thrombus and in the pathogenesis of thrombosis (1–5). Elevated plasma levels of FXI have been associated with increased risk of venous thromboembolism (VTE) and ischaemic stroke in most (1,6–8) but not all (9) studies. VTE and stroke constitute important, disabling and potentially fatal diseases with an incidence of 0.1–0.3% per year in Caucasians (10,11). Subjects with severe FXI deficiency have a reduced incidence of VTE and stroke (7,12) and only mild bleeding, and several genetic studies have revealed that genetic variants in or close to the F11 gene are associated with VTE (13–17) and ischemic stroke (18). Moreover, an association has been shown between the activated partial thromboplastin time (aPTT), which measures the efficacy of the intrinsic coagulation pathway and has been shown to be a good predictor for VTE (19), and single nucleotide polymorphisms (SNPs) in the F11 locus (20,21).
Substantial evidence from mice and non-human primates indicates that FXI deficiency/inhibition is protective against experimentally induced thrombosis or stroke (2,22) without excessive bleeding. Moreover, emerging studies using antisense oligonucleotides that specifically reduce plasma FXI levels have proved effective for prevention of postoperative VTE without bleeding risk (23), suggesting that the plasma level of FXI is an important determinant of thrombosis and that understanding its regulation can have important implications in the diagnosis and management of serious disease outcomes such as VTE and stroke. Because of its implication in VTE and stroke, and the minor bleeding risk, FXI—as other components of the intrinsic coagulation pathway—have become increasingly interesting as therapeutic targets for thrombosis.
While the implications of changes in FXI plasma levels for disease have been widely studied, the genetic regulation of FXI remains incompletely understood. In a previous genome-wide association study (GWAS) using data imputed to the HapMap reference panel, we identified KNG1 and F11 as the two main loci determining plasma FXI levels (24). Recent studies have demonstrated that use of 1000 Genomes (1000G)-imputed data can substantially improve the ability of GWAS to discover novel loci (25) and that the denser SNP coverage provided by imputation also allows us to study the genetic architecture of associated loci in greater detail, potentially revealing additional signals. Against this background, we have, in the present study, increased sample size 40-fold from the previous GWAS and have included novel data imputed to the 1000G reference panel in order to identify new loci that are important for FXI regulation and that could influence risk of venous and arterial thrombosis. In addition, we complement this genetic analysis with a study on the effect of microRNA (miRNA) on FXI regulation, with the overall aim to investigate novel regulatory mechanisms for FXI that could serve as therapeutic targets for treatment and prevention.
Results
Three loci, GCKR, KNG1 and F11, are robustly associated with circulating FXI levels
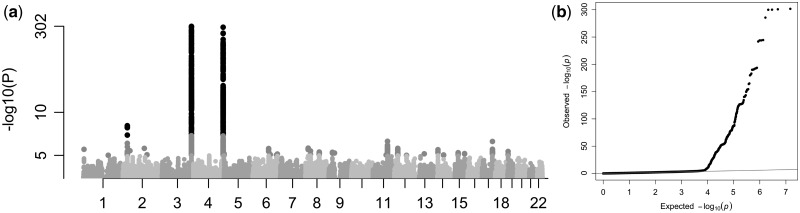
In the discovery analysis, five studies (ARIC, GHS-1, GHS-2, MARTHA, and PROCARDIS), including 16,169 European individuals, performed GWAS analyses against natural-logarithm-transformed FXI levels (lnFXI). After quality control (QC) and filtering for minor allele frequency (MAF > 0.01), 7,353,638 markers were included in a fixed-effect meta-analysis. Three loci contained associations with FXI plasma levels that were significant using the conventional genome-wide threshold of 5×10−8 (Fig. 1, Table 1, Supplementary Material, Table S2). The most significant association (rs710446, Beta = −0.0888, P-value = 2.07×10−302) was located on chromosome 3, and created a non-synonymous substitution (Ile581Thr) in the KNG1 gene; this confirms previous associations (24). A SNP on chromosome 4 (rs4253417, Beta = −0.0735, P-value = 2.86×10−193) was located in an intron on the structural gene coding for F11, which has previously been associated with FXI levels, albeit with different lead SNPs (18,24). Finally, a locus on chromosome 2 (rs780094, Beta = 0.0147, P-value = 3.56×10−09) was located in an intron of the glucokinase regulator (GCKR) gene, which has not previously been associated with FXI levels. The three lead SNPs were estimated, using GCTA (26,27), to explain 5.89% of the phenotypic variance in the PROCARDIS cohort. Summary results from the discovery meta-analysis have been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001002123.
Figure 1.
Results from the GWAS meta-analysis of natural-log-transformed plasma FXI levels in 16,169 European individuals (discovery cohort), adjusted for age, sex and population stratification; (a) Manhattan plot showing SNP P-values over chromosomes, (b) QQ-plot of expected and observed P-values (genomic control, Lambda = 1.006).
Table 1.
Associations with lnFXI in the discovery and replication studies, fixed-effect meta-analysis
| rs | Chr:pos | A1/A2 | FreqA1 | Betadisc (SE) | Pdisc (Phet) | Betarepl (SE) | Prepl (Phet) |
|---|---|---|---|---|---|---|---|
| Discovery | |||||||
| rs710446 | 3:186459927 | T/C | 0.590 | −0.0888 (0.00239) | 2.07 × 10−302 (1.76 × 10−05) | −0.125 (0.00944) | 9.74 × 10−40 (5.55 × 10−16) |
| rs4253417 | 4:187199005 | T/C | 0.592 | −0.0735 (0.00248) | 2.86 × 10−193 (2.23 × 10−07) | −0.0898 (0.00983) | 6.9 × 10−20 (1.11 × 10−15) |
| rs780094 | 2:27741237 | T/C | 0.407 | 0.0147 (0.00249) | 3.56 × 10−09 (0.0562) | 0.0238 (0.00965) | 0.0137 (0.262) |
| Conditional | |||||||
| rs4253421 | 4:187204937 | A/G | 0.118 | −0.0498 (0.00373) | 1.25 × 10−40 (5.45 × 10−4) | – | – |
| rs76438938 | 3:186461524 | C/T | 0.971 | −0.0765 (0.00711) | 6.07 × 10−27 (0.00211) | – | – |
Results from the GWAMA analysis, number of tested SNPs in the discovery analysis are 7,353,638 and for the conditional analyses 7,353,629. A1 = allele 1 A2 = allele 2. For SNPs displaying inflated heterogeneity (rs710446, rs4253417 andrs4253421) in an analysis, a random-effect version of the analysis was performed; in all cases, these analyses confirmed the discovered associations for the investigated SNPs (Supplementary Material, Table S2, S3, and S5).
The top SNP from each of the three genome-wide significant loci were tested for replication in 2,058 European individuals from the French-Canadian family study on Factor V Leiden (F5L) study, GAIT2 and MEGA studies (see further information in Supplementary Material, Table S1) and the results were combined in a fixed-effect meta-analysis (Table 1 and Supplementary Material, Table S3). All three loci were significantly associated with lnFXI in our replication studies, Table 1), at the Bonferroni-adjusted threshold α = 0.0167 (i.e. 0.05/3).
Conditional analysis identifies two independent signals at the KNG1 and F11 loci
The associations of previously discovered SNPs in the loci with related phenotypes are listed in Supplementary Material, Table S4. In addition to the top SNPs (rs710446, rs4253417, rs780094), there was, at each locus, a set of additional SNPs that displayed significant association with FXI levels (Table 2). Regional plots (Supplementary Materials, Figs S3–S5) suggested possible independent associations. To further investigate the association pattern at the three significantly associated loci, a secondary GWAS was performed in the discovery studies, conditioning on the three most significantly associated SNPs. The results of the fixed-effect meta-analysis of this conditional analysis are shown in Table 1 and Supplementary Material, Table S5. Additional, independently associated genome-wide significant associations were observed in the F11 locus at rs4253421 (Beta = −0.0498, P-value = 1.25×10−40) and in the KNG1 locus at rs76438938 (Beta = −0.0765, P-value = 6.07×10−27), to our knowledge previously not reported as associated with lnFXI. These SNPs were not in strong linkage disequilibrium (LD) with the lead F11 (rs4253417, r2 = 0.084, D' = 0.99) and KNG1 (rs710446, r2 = 0.033 and D' = 0.90) SNPs, respectively). Together with the three discovery SNPs, the two conditional SNPs were estimated, using GCTA (26,27), to explain 6.13% of the phenotypic variance in the PROCARDIS cohort. We used an approximate method, the stepwise selection procedure implemented in GCTA (27,28), to further investigate the signal pattern in the three loci. This analysis uses meta-analysis data combined with estimated pair-wise LD to identify the minimum subset of SNPs that jointly explains as much of the observed phenotype signal. For the GCKR and F11 loci, this approximate stepwise analysis found no other signals than the lead SNPs from our discovery and conditional analyses (Supplementary Material, Table S6). However, for the KNG1 locus, analyses suggested three additional signals, represented by rs5030095, rs9823431 and rs35575213, respectively, as lead SNPs (none of these is in strong LD with the lead SNPs from the discovery and conditional analyses, Supplementary Material, Table S7). We investigated these SNPs further in a haplotype analysis; however, the ensuing analyses focus on the five SNPs from our discovery and conditional analyses.
Table 2.
Summary of loci significantly associated with lnFXI in the discovery and conditional analyses.
| Start Chr:pos | Main gene | Discovery #GW-signif SNPs | Discovery top SNP | Conditional #GW-signif SNPs | Conditional top SNP |
|---|---|---|---|---|---|
| 3:186000000 | KNG1 | 470 | rs710446 | 219 | rs76438938 |
| 4:187000000 | F11 | 395 | rs4253417 | 84 | rs4253421 |
| 2:27000000 | GCKR | 5 | rs780094 | 0 | – |
For convenience, a locus is here considered to be 2 Mbp long. For the F11 locus, one additional SNP with genomic position 4:186990842 (i.e. just outside our conventional locus definition) was manually included.
Haplotype analyses reveal complex association structures at the F11 and KNG1 loci
To further resolve the LD patterns observed in the F11 and KNG1 loci, haplotype analyses were conducted in two of the discovery studies, PROCARDIS and MARTHA. For the F11 locus, we performed a haplotype association analysis of five SNPs with lnFXI, including our two lead F11 SNPs and three selected SNPs that have been associated with VTE in recent studies (rs3822057 and rs12186257 (29–31) and rs2036914 (30–32)). These five SNPs generated seven haplotypes with very similar distributions in PROCARDIS and MARTHA (Table 3). In both studies, all haplotypes carrying the T-allele at the lead rs4253417 were associated with decreased lnFXI levels. In addition, the single haplotype carrying the A-allele of the conditional rs4253421, and also carrying the rs4253417 T-allele, was associated with much lower lnFXI levels, supporting the result from our conditional analysis and suggesting that the rs4253417 and rs4253421 SNPs have additive independent effects on FXI levels.
Table 3.
Results from haplotype analysis of the F11 locus in the PROCARDIS and MARTHA studies
| rs4253417 | rs4253421 | rs3822057 | rs2036914 | rs12186257 | Haplotype frequencies | Additive effects on FXI levels Beta [CI] P-value |
|---|---|---|---|---|---|---|
| PROCARDIS | ||||||
| T | G | A | C | C | 0.041 | −0.017 [−0.059 to 0.025] p = 0.425 |
| T | G | A | T | C | 0.308 | −0.066 [−0.085 to 0.047] p = 1.18 10−11 |
| T | G | C | C | C | 0.122 | −0.066 [−0.091 to 0.041] p = 1.92 10−7 |
| T | A | A | T | G | 0.108 | −0.144 [-0.171 to 0.118] p = 5.93 10−27 |
| C | G | A | T | C | 0.019 | −0.013 [−0.077 to 0.050] p = 0.681 |
| C | G | A | T | G | 0.013 | −0.013 [−0.100 to 0.072] p = 0.757 |
| C | G | C | C | C | 0.355 | Intercept |
| MARTHA | ||||||
| T | G | A | C | C | 0.042 | −0.066 [−0.131 to 0.002] p = 0.044 |
| T | G | A | T | C | 0.308 | −0.093 [−0.122 to 0.065] p = 1.38 10−10 |
| T | G | C | C | C | 0.099 | −0.072 [−0.120 to 0.025] p = 0.0028 |
| T | A | A | T | G | 0.048 | −0.136 [−0.185 to 0.088] p = 3.32 10−8 |
| C | G | A | T | C | 0.028 | −0.061 [−0.140 to 0.018] p = 0.131 |
| C | G | A | T | G | 0.013 | −0.047 [−0.163 to 0.068] p = 0.421 |
| C | G | C | C | C | 0.428 | Intercept |
CI = confidence interval for Beta.
A similar haplotype analysis was performed on three SNPs from the KNG1 locus, our lead discovery and conditional SNPs (rs710446 and rs76438938, respectively) and one of the SNPs suggested by the GCTA analysis (rs5030095). Again, haplotype results for the PROCARDIS and MARTHA studies (Table 4) were similar. The three SNPs generated five common haplotypes. The three haplotypes carrying the rs710446 C-allele consistently displayed a higher effect size than those carrying the T-allele. Moreover, the rs76438938 T-allele and the rs5030095 G-allele both, apparently independently, further increased the effect size (Table 4).
Table 4.
Results from haplotype analysis of the KNG1 locus in the PROCARDIS and MARTHA studies
| rs710446 | rs76438938 | rs5030095 | Haplotype Frequencies | Additive effects on FXI levels Beta [CI] P-value |
|---|---|---|---|---|
| PROCARDIS | ||||
| T | C | G | 0.504 | Intercept |
| T | C | C | 0.087 | −0.011 [−0.046 to 0.019] P = 0.425 |
| C | C | G | 0.353 | +0.073 [0.055 to 0.092] P = 5.08 10−15 |
| C | C | C | 0.025 | +0.045 [−0.019 to 0.107] P = 0.162 |
| C | T | C | 0.029 | +0.147 [0.105 to 0.189] P = 5.20 10−12 |
| MARTHA | ||||
| T | C | G | 0.433 | Intercept |
| T | C | C | 0.101 | −0.039 [−0.090 to 0.013] P = 0.141 |
| C | C | G | 0.412 | +0.101 [0.073 to 0.128] P = 1.27 10−12 |
| C | C | C | 0.022 | +0.087 [−0.028 to 0.202] P = 0.137 |
| C | T | C | 0.031 | +0.138 [0.076 to 0.200] P = 1.13 10−5 |
CI = confidence interval for Beta.
Associations with expression level in human liver suggest possible effects on transcription of the GCKR gene
We investigated if any of the five lead SNPs from the discovery and conditional analyses could exert any cis-effect on expression of nearby genes (Table 5). As FXI is almost exclusively expressed in the liver, we limited our analysis to liver tissue. No eQTL showed significant association with mRNA expression after adjustment for multiple testing (11 genes tested for the KNG1 SNPs, 5 genes tested for the F11 SNPs and 21 genes tested for the GCKR SNPs). The strongest association was observed between SNP rs780094 and the GCKR gene (P-value = 0.00496, Bonferroni-adjusted significance threshold α = 0.05/21 = 0.00238).
Table 5.
Results from eQTL associations of each of the three lead SNPs and the two conditional SNPs with the expression of neighboring genes (located within 500 Mbp) in human liver samples from the ASAP cohort.
| SNP | Gene | Fold change | Beta | SD | P-value |
|---|---|---|---|---|---|
| rs710446 | TBCCD1 | 0.9924 | −0.01104 | 0.03687 | 0.765 |
| ST6GAL1 | 1.027 | 0.03888 | 0.03559 | 0.276 | |
| RFC4 | 1.003 | 0.004982 | 0.03113 | 0.873 | |
| CRYGS | 0.9765 | −0.03428 | 0.03117 | 0.273 | |
| DNAJB11 | 1.006 | 0.009068 | 0.05579 | 0.871 | |
| FETUB | 1.035 | 0.04977 | 0.05485 | 0.365 | |
| KNG1 | 1.051 | 0.07149 | 0.03334 | 0.0332 | |
| HRG | 1.021 | 0.02942 | 0.02593 | 0.258 | |
| EIF4A2 | 1.023 | 0.03331 | 0.03678 | 0.366 | |
| ADIPOQ | 0.9744 | −0.0374 | 0.02193 | 0.0897 | |
| AHSG | 1.033 | 0.0466 | 0.03167 | 0.143 | |
| rs76438938 | TBCCD1 | 0.9392 | −0.09053 | 0.08882 | 0.309 |
| ST6GAL1 | 1.038 | 0.05356 | 0.08612 | 0.535 | |
| RFC4 | 1.038 | 0.0536 | 0.07433 | 0.472 | |
| CRYGS | 0.998 | −0.002872 | 0.07547 | 0.97 | |
| DNAJB11 | 1.004 | 0.005966 | 0.1338 | 0.964 | |
| FETUB | 0.9788 | −0.03097 | 0.1322 | 0.815 | |
| KNG1 | 0.9837 | −0.02375 | 0.08145 | 0.771 | |
| HRG | 0.9874 | −0.01829 | 0.06251 | 0.77 | |
| EIF4A2 | 0.9529 | −0.06959 | 0.08888 | 0.435 | |
| ADIPOQ | 1.092 | 0.1267 | 0.05254 | 0.0168 | |
| AHSG | 0.9706 | −0.04308 | 0.07623 | 0.573 | |
| rs4253417 | TLR3 | 1.008 | 0.01143 | 0.05101 | 0.823 |
| CYP4V2 | 0.964 | −0.05288 | 0.04612 | 0.253 | |
| F11 | 0.9862 | −0.02011 | 0.0353 | 0.57 | |
| KLKB1 | 1.015 | 0.02158 | 0.04275 | 0.614 | |
| rs4253421 | TLR3 | 0.9976 | −0.003492 | 0.07242 | 0.962 |
| CYP4V2 | 0.8927 | −0.1638 | 0.06467 | 0.0121 | |
| F11 | 1.018 | 0.02505 | 0.05012 | 0.618 | |
| MTNR1A | 1.033 | 0.04727 | 0.03058 | 0.124 | |
| KLKB1 | 1.046 | 0.06434 | 0.06055 | 0.289 | |
| rs780094 | SNX17 | 1.007 | 0.0104 | 0.02593 | 0.689 |
| PPM1G | 0.982 | −0.02613 | 0.02332 | 0.264 | |
| CCDC121 | 1.042 | 0.05874 | 0.03427 | 0.088 | |
| ZNF513 | 0.9945 | −0.007998 | 0.01743 | 0.647 | |
| FNDC4 | 0.9462 | −0.07973 | 0.03637 | 0.0295 | |
| IFT172 | 1.01 | 0.01428 | 0.02183 | 0.514 | |
| MPV17 | 0.9983 | −0.00252 | 0.03132 | 0.936 | |
| SUPT7L | 0.9863 | −0.01984 | 0.03583 | 0.58 | |
| DNAJC5G | 1.016 | 0.02329 | 0.02236 | 0.299 | |
| KRTCAP3 | 1.013 | 0.01855 | 0.02715 | 0.495 | |
| C2orf16 | 1.028 | 0.03919 | 0.0212 | 0.0659 | |
| GTF3C2 | 0.9973 | −0.003908 | 0.02108 | 0.853 | |
| GCKR | 0.926 | −0.1109 | 0.03904 | 0.00496 | |
| SLC4A1AP | 1.008 | 0.01146 | 0.03296 | 0.728 | |
| NRBP1 | 0.9907 | −0.01345 | 0.02053 | 0.513 | |
| GPN1 | 0.9959 | −0.005879 | 0.03988 | 0.883 | |
| UCN | 1.024 | 0.03448 | 0.03528 | 0.33 | |
| ZNF512 | 1.01 | 0.01398 | 0.0366 | 0.703 | |
| TRIM54 | 1.019 | 0.02703 | 0.02406 | 0.262 | |
| EIF2B4 | 0.9948 | −0.007518 | 0.0214 | 0.726 | |
| SLC30A3 | 1.002 | 0.002337 | 0.02607 | 0.929 |
Fold change is calculated from beta: fold change = 2beta.
Association results are most likely linked to the roles of KNG1 and F11 in the contact coagulation pathway
FXI has an important role in the propagation of the coagulation cascade by the contact pathway, which implicates the F11 gene as a plausible causal gene for VTE. To investigate how the discovered associations relate to VTE, the five FXI-associated SNPs were also tested for association with aPTT in 10,908 individuals from the ARIC, F5L, GAIT2, and MARTHA studies, using fixed-effect meta-analysis. The aPTT test reflects the activity of the intrinsic pathway of coagulation and has been inversely associated with VTE risk (33), To avoid bias due to technical differences in the measurements of aPTT across studies (see Material and Methods section), we performed a meta-analysis based on normalized effect sizes and standard errors. We additionally performed the meta-analysis based on sample-size-weighted P-values. We tested two models, model 1 was adjusted for age and sex covariates, and model 2 was additionally adjusted for lnFXI levels.
The two meta-analysis models yielded consistent results (Table 6, Supplementary Materials, S8 and S9). For model 1, the F11 and KNG1 SNPs all showed significant association in the expected directions with aPTT (P-value < 0.01, Bonferroni-correction for five tests), but not the GCKR SNP (rs780094, P-value = 0.125). With adjustment for lnFXI in model 2, the effect of all SNPs was strongly attenuated, and rs76438938 in the KNG1 locus was no longer significant.
Table 6.
Results of aPTT meta-analysis for models 1 and 2
| rs_locus | A1 | A2 | Freq1 | Beta | SE | P-value | Direction | P_het |
|---|---|---|---|---|---|---|---|---|
| Model 1 | ||||||||
| rs710446 | C | T | 0.412164 | −0.296423 | 0.009575 | 2.29 × 10−210 | — | 6.10 × 10−04 |
| rs4253417 | C | T | 0.419123 | −0.118784 | 0.009575 | 2.65 × 10−35 | — | 0.194202 |
| rs4253421 | G | A | 0.885623 | −0.083696 | 0.009575 | 2.44 × 10−18 | −+– | 0.130280 |
| rs76438938 | T | C | 0.028115 | −0.056257 | 0.009575 | 4.33 × 10−09 | — | 0.877173 |
| rs780094 | C | T | 0.592274 | 0.014700 | 0.009575 | 0.124693 | +++ | 0.770329 |
| Model 2 | ||||||||
| rs710446 | C | T | 0.411999 | −0.204035 | 0.009579 | 1.28 × 10−100 | — | 4.54 × 10−05 |
| rs4253417 | C | T | 0.418964 | −0.042991 | 0.009579 | 7.28 × 10−06 | −+– | 0.614175 |
| rs4253421 | G | A | 0.885574 | −0.031183 | 0.009579 | 0.001138 | − ++ | 0.255919 |
| rs76438938 | T | C | 0.028138 | −0.007322 | 0.009579 | 0.444662 | − +++ | 0.280294 |
| rs780094 | C | T | 0.592396 | −0.004186 | 0.009579 | 0.662094 | −+– | 0.700256 |
Results from the GWAMA fixed-effect meta-analysis based on normalized beta and standard errors. Because of the inflated heterogeneity for rs710446 in both model 1 and model 2 analyses, we performed a random-effect version of these analyses that confirmed the discovered association for this SNP (Supplementary Material, Table S9).
Functional annotation suggests effects at the protein sequence level only for rs710446 and rs76438938, but possible regulatory effects for all five FXI-associated SNPs.
We used the HaploReg web-service (34) to functionally annotate the five lead SNPs using information from publicly available genome annotation databases (Table 7). The two KNG1 SNPs are located in two distinct exons of the KNG1 gene. The strongest associated SNP, rs710446, is a missense mutation (I581T), while rs76438938 is a nonsense mutation (R376*). The remaining three SNPs are all intronic and located in the F11 and GCKR gene. There is substantial regulome information for the regions of interest. All SNPs, with the exception of rs4253417, were, based on liver reference epigenomes, reported to lie in predicted enhancer elements. These SNPs were also predicted to alter transcription factor binding motifs. However, only rs780094 has supporting ChIP-seq evidence for transcription factor and enhancer binding to this region in liver cell lines (Table 7).
Table 7.
Summary of HaploReg results for the three discovery SNPs and the two conditional SNPs
| SNP | Gene | Functional effect | Chromatin state in liver tissue | Proteins bound in liver cell lines | Transcription factor binding motifs changed |
|---|---|---|---|---|---|
| rs780094 | GCKR | intron | Enhancers | FOXA2; MAFK; RXRA | AP-1; DBP; Foxi1; HNF4; Maf; Myf; RP58; TCF12; p300 |
| rs710446 | KNG1 | missense; intron | Enhancers; Genic enhancers | – | AIRE; AP-1; CCNT2 |
| rs76438938 | KNG1 | nonsense | Enhancers; Flanking Active TSS | – | GLI; LF-A1 |
| rs4253417 | F11 | intron | – | – | Gfi1 |
| rs4253421 | F11 | intron | Enhancers | – | CHOP::CEBPalpha; NF-kappaB; RREB-1; Smad |
Investigation of potential post-transcriptional regulation
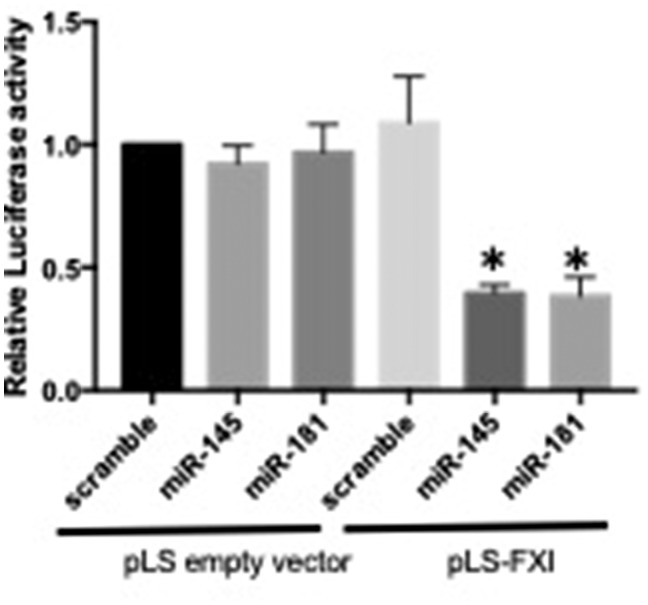
Previous results have indicated that post-transcriptional regulation of F11 RNA expression in the liver can affect circulating FXI levels (35). We therefore investigated a potential post-transcriptional mechanism for the regulation of FXI levels. Specifically, using an in silico consensus approach, we first predicted binding of miRNAs to the F11 3’ UTR, 5’ UTR or promoter regions (the top hits are shown in Table 8). Eight miRNAs were predicted to target F11 mRNA. Three of these have previously been experimentally tested (hsa-miR-181a-5p6, hsa-miR-16-5p6 and hsa-miR-23a-3p6; only the first was confirmed to bind to F11) (36), while the remaining five (hsa-miR-15a-5p, hsa-miR-15b-5p, hsa-miR-145-5p, hsa-miR-150-5p and hsa-miR-424-5p) remain to be experimentally validated. Of these latter five, miR-145 had the highest expression in liver (36) and was predicted to bind to the 3’ UTR region of F11, 383 bases from the F11 stop codon. For this reason, we selected miR-145 for further in vitro validation. We used an in vitro luciferase assay in HEK293 cells to validate that miR-145 directly targets F11 mRNA at its 3’-UTR; HEK293 cells, as the established cell source for luciferase reporter assays, were chosen for providing the highest possible transfection efficiency (26,27,29,30,36,37). The previously validated miR-181 (26,27,29–31,36) was used as a positive miRNA control. Overexpression of either miR-145 or miR-181 significantly reduced the luciferase activity in cells transfected with pLS-FXI-3’UTR, but not the control pLS vector, suggesting that miR-145 (and miR-181) indeed targets F11 mRNA through its 3’-UTR (Fig. 2) and regulate FXI levels. Finally, to connect this result to our genetic analysis, we investigated our significant F11-locus SNPs close to the miR-145 and miR-181binding sites. Three SNPs (rs1062547, rs4253430, and rs4253429) and a small deletion (4:187210318TA/A) were located within the reference 3’UTR for F11 (see Supplementary Section 2); since all of them were significantly associated with FXI levels (P-values = 2.7×10−41, 1.7×10−41, 7.7×10−27, and 5.1×10−9, respectively), we first hypothesized that a possible effect of these SNPs could be to affect the binding of miR-145/miR-181 and thereby affect the regulation of FXI. To investigate if SNPs in the F11 3’UTR affect binding of miR-145 or miR-181 to the F11 3’ UTR, by changing the binding site sequence or through changes in secondary structure of the 3’UTR, we estimated haplotypes for these four SNPs in one of our cohorts (PROCARDIS, including 3525 individuals) and found four haplotypes. We investigated in silico the effect of miR-145 and miR-181a-5p binding in F11 3’UTR sequences corresponding to these four haplotypes using several public prediction tools that estimate the thermodynamic stability of the secondary structure of the putative miRNA-target pair, miRNA accessibility to binding site, and binding energy for the different sequences (See Supplementary Material, Section 2). We did not find evidence that any of the SNPs had a functional effect on miRNA binding to the F11 3’ UTR, none of the investigated SNPs changed the binding target sequence, and there were no significant changes in miRNA binding energy between the different UTR haplotypes. Our results could not show any evidence of an effect of SNPs in 3’UTR on miRNA binding, suggesting that other mechanisms, probably the effect of LD with SNPs located somewhere else in the gene, might explain the association between these SNPs and FXI levels.
Table 8.
Prediction of miRNAs binding to the F11 mRNA.
| miRNA | MirWalk | TargetScan | miRanda | mirSVR | Tarbase | Predicted Binding | Si score |
|---|---|---|---|---|---|---|---|
| hsa-miR-15a-5p | 6,0,3 | −0.114 | 145 | −0.1365 | 0.46 | 3' and promoter | 4 |
| hsa-miR-15b-5p | 5,0,3 | −0.114 | 149 | −0.1365 | 0.464 | 3' and promoter | 4 |
| hsa-miR-145-5p | 7,0,2 | −0.287 | 155 | −1.1535 | 0.751 | 3' and promoter | 5 |
| hsa-miR-150-5p | 6,0,4 | −0.111 | 140 | −0.2513 | 0.637 | 3' and promoter | 4 |
| hsa-miR-424-5p | 7,1,2 | −0.133 | 157 | −0.1338 | 0.555 | 3' and promoter | 5 |
| hsa-miR-181a-5p | 6,3,2 | −0.047 | 145 | −0.3645 | 0.515 | 3', 5' and promoter | 5 |
| hsa-miR-16-5p | 6,0,3 | −0.114 | 146 | −0.129 | 0.464 | 3’ and promoter | 5 |
| hsa-miR-23a-3p | 6,0,3 | −0.073 | 140 | −0.6707 | 0.664 | 3’ and promoter | 4 |
Figure 2.
Results from the luciferase assay of miRNA binding to F11 mRNA. The plot shows luciferase activity after 24h of cells co-transfected with either an empty luciferase reporter plasmid (pLS), or a plasmid containing the FXI 3’UTR (pLS-FXI-3’UTR), together with scramble control, miR-145 or miR-181.
Discussion
Coagulation factor XI is produced in the liver and circulates in the blood as a homodimer in complex with high molecular weight kininogen (HK or sometimes HMWK), also produced in the liver. HK also works as a co-factor in contact-induced activation of coagulation factor XII, which activates FXI; in this process, HK is cleaved by kallikrein to form bradykinin. Activated FXI, in turn, activates coagulation factor IX, which will start the common pathway of the coagulation cascade, leading to clot-formation.
In this study, we have made a thorough investigation of genetic and potential post-transcriptional regulators of plasma FXI levels and their connection to activation of the contact coagulation pathway. In a previous study (24,26,27,29,31,32,36), we showed that the KNG1 and F11 loci are important determinants of plasma FXI levels based on a GWAS of a mere 398 individuals. The present study, representing a 40–fold increase in sample size (of note, the current sample is non-overlapping with that in the previous study), confirmed these associations, and identified additional independent signals in both loci. Moreover, we identified a novel third associated locus in the GCKR gene region, a gene that has been associated with multiple other traits, including coagulation factor VII, and Protein C (PC) levels (1–5,26,27,29,30,32,36,38–40). The KNG1 and F11 SNPs were also associated with aPTT, which support previously published associations of these loci with VTE (1–5,13,15,27,29–31,36,41,42). Finally, we confirmed the effect of miR-181 and demonstrated the effect of a novel miRNA, miR-145, in the regulation of plasma FXI concentration.
Consistent with what was reported in the previous GWAS (24,26,27,29–32,36), the most robustly associated locus contained the KNG1 gene. Several genes are located within the KNG1 locus, including other biologically plausible genes such as those coding for histidine-rich glycoprotein (HRG) or adiponectin (ADIPOQ). Our results for the main discovery SNP, rs710446, as well as the main conditional SNP, rs76438938, point to KNG1 as the main candidate gene in the region (Supplementary Material, Table S10). The C-allele of rs710446 causes an exonic, nonsynonymous mutation in KNG1, thus affecting the amino acid sequence of the protein, and therefore potentially also its function, while rs76438938 is a nonsense mutation. KNG1 codes for HK, which has a central role in the transport and activation of FXI. FXI circulates as a homodimer in complex with HK. A probable mechanism for the effect of one or both SNPs on FXI levels could be by influence on the availability (or functionality) of HK for complex binding, thereby indirectly affecting the levels of circulating FXI. Our results show that the genetic structure underlying the KNG1 association with FXI is more complex than previously thought. Our conditional and haplotype analyses, indeed, revealed that the T-allele of the nonsense rs76438938 was also associated with markedly increased FXI levels and that a third KNG1 polymorphism could also be involved in the regulation of FXI levels. Further deep genetic investigations at the KNG1 locus are required to better understand the KNG1-driven genetic regulation of FXI levels.
The second strongly FXI-associated locus was the F11 locus with two intronic SNPs, rs4253417 and rs4253421, that independently influenced FXI levels. The involvement of F11, the gene encoding FXI, could be expected. Neither rs4253417 nor rs4253421 (or SNPs in LD with these SNPs, Supplementary Material, Table S11) appears to have an effect on protein sequence, but functional annotation suggests potential regulatory functions; however, neither of the SNPs appears to have any effect on the mRNA expression of F11, nor on any surrounding gene. Consequently, we investigated if a post-transcriptional mechanism, specifically regulation by miRNA, could be operating. MiRNAs can have a regulating effect both on the transcriptional and post-transcriptional level. An increasing number of miRNAs have recently been involved in disease regulation, but the understanding of the role of miRNAs in the regulation of the coagulation pathway and their relevance to VTE remains in its infancy. A previous study (26,27,30–32,36) implicated a role for miR-181a-5p in FXI regulation. Our results confirm the effect of miR-181a-5 and combine bioinformatics-based prediction of miRNA binding to the 3’UTR region of F11 with validation using a luciferase assay to demonstrate a regulatory effect on FXI by the miRNA hsa-mir-145-5p. However, we could find no evidence supporting that the FXI associations in our study are mediated by SNPs affecting the binding of these two miRNAs.
GCKR codes for a glucokinase regulatory protein that inhibits glucokinase in liver and pancreatic-islet cells. This protein regulates the uptake and storage of dietary glucose and is important in the switch between feeding and fasting periods, and as such has been associated with several key metabolic pathways, especially for lipid and glucose metabolism. GCKR appears to be highly pleiotropic and genetic variations, specifically the SNP rs780094 and the nonsense SNP rs1260326, which are in LD with rs780094 (r2 = 0.92; Supplementary Materials, Tables S4 and S12), have been associated with numerous metabolic traits, including type 2 diabetes (T2D) (1–5,26,27,29–32,36), maturity-onset diabetes of the young, nonalcoholic fatty liver disease, platelet count, and blood levels of glucose, fasting insulin, plasma-triglyceride, and total cholesterol (1–5,26,27,29–32,36), and also metabolites such as uric acid, creatinine and albumin (27,29–32). This is the first study associating GCKR with FXI levels, and the mechanism behind this association is therefore unclear. However, GCKR has also been associated with plasma viscosity (24,26,30–32) and coagulation proteins such as FVII (1–5,29–32,36,43), and PC (20,29–31), and also with the inflammatory marker c-reactive protein (CRP) (30–32,44,45), which in turn has been shown to be associated with VTE (33,46). The GCKR gene is mainly expressed in liver and catalyzes the initial step in the utilization of glucose, providing glucose-6-phosphate. As suggested for PC and FVII (39), GCKR could affect the function of several glycosylated proteins synthesized in the liver by altering the glucose required for their glycosylation. This could suggest a possible mechanism for the effect on FXI function—as FXI also has several N-glycosylation sites positioned at key areas of the glycoprotein that binds ligands involved in the coagulation cascade (47)—but not its levels. However, a feedback regulatory loop on FXI could be mediated by FVII or PC within the coagulation cascade, both of which are regulated by GCKR. In our results, rs780094 displays no eQTL effects on either F11 or KNG1 expression. This strengthens the hypothesis of post-transcriptional or feedback regulatory mechanisms at a protein level or, alternatively, it might simply reflect that our study was underpowered to detect trans-effects.
This might also be the reason why we could not see an association between rs780094 and aPTT. Plasma FXI has previously been robustly associated with aPTT and VTE. If FXI is causal in this association, we would therefore expect, from the Mendelian Randomization (MR) principle (48), that genetic variants that regulate FXI levels are also associated with aPTT and VTE. We verified this for all SNPs in KNG1 and F11, which had clear associations with aPTT, while the effect of the GCKR SNP rs780084 was much weaker and non-significant. When adjusting aPTT for FXI levels, all associations were markedly attenuated, indicating that, for all SNPs, the effect on aPTT is indeed mediated through their effect on FXI levels. As aPTT is a more complex phenotype, depending also on other factors than FXI, the power of the aPTT analysis would be expected to be lower than that of the lnFXI analysis (notice also that the two analyses are based on different sets of cohorts). The association P-values for aPTT are overall weaker than those for lnFXI and, in particular, the absence of a strong effect on FXI plasma levels for rs780084 resulted in an even weaker, insignificant, effect on aPTT. In addition, one of the criteria for MR to be applicable is the absence of pleiotropy, a condition that does not apply to GCKR (31). Most likely, the effects of GCKR on other related phenotypes like FVII and PC levels act as confounders for the effects on FXI, and contribute to the loss of power to detect an association. Clearly, further studies are needed to understand the biological mechanism by which this regulatory protein influences plasma FXI.
The use of 1000 genomes-imputed data has clear benefits compared with our previously published GWAS based on HapMap-imputed data: by allowing inclusion of more SNPs in the meta-analysis, it also enables discovery of additional signals with greater statistical significance—our top SNP in the F11 locus rs4253417 has a P-value = 2.86×10−193 in our discovery analysis, to be compared with the P-value =1.02×10−165 for rs4253399 (in the present analysis), which was the top SNP in the analysis based on genotyped data in (24), cf. Supplementary Material, Table S4. Moreover, the greater number of queried SNPs allowed a more comprehensive picture of the SNP structure of the locus (395 SNPs for the KNG1 locus, 470 SNPs for the F11 locus and five for the GCKR locus), and allowed the discovery of multiple signals in the locus.
To conclude, our results confirm KNG1 and F11 as important loci for FXI plasma level regulation, and discover GCKR, which has been reported to have pleiotropic effects on other coagulation proteins, as a novel interesting locus determining plasma FXI levels. Finally, we also demonstrate post-transcriptional regulation of FXI protein expression by a novel miRNA (miR-145-5p) binding to F11-UTR region. Moreover, our results suggest a potential mediating role of HK, the gene product of KNG1, and a future direction could therefore include studies on circulating HK levels. Given the role of FXI in determining risk of thrombosis, and the accumulating evidence suggesting that molecular determinants of FXI levels could be potential targets for anti-coagulation therapies that would carry a lesser bleeding risk compared with current anti-coagulant therapies, the novel genetic and post-transcriptional regulators of FXI in plasma found in the present study should open the door to new therapeutic targets for thrombosis prevention.
Materials and methods
Study sample
The study consists of a discovery sample of 16,169 individuals of European ancestry from five studies (ARIC, PROCARDIS, GHS-1, GHS-2, and MARTHA) and a replication sample of 2,045 individuals from three studies (F5Leiden Cohort, MEGA, and GAIT2); for details, see Supplementary Material, Table S1. All studies were approved by the appropriate research ethics committees, and all respondents signed informed consent prior to participation.
Phenotype determinations
Plasma FXI concentration was determined using immunoassay (ELISA or electrochemiluminescence), coagulometry, a functional assay or a clotting assay. Results were expressed in U/ml, 1 unit being defined as the amount of activity of FXI in 1 ml of plasma. For details, please refer to Supplementary Material, Table S1.
Activated partial thromboplastin time (aPTT) was measured in plasma, using automated coagulometry, either as the raw clotting time in seconds (three studies) or as a normalized ratio between the clotting time of the sample and the clotting of a laboratory reference standard (one cohort); for details see Supplementary Material, Table S1.
Genotyping and imputations and QC
Genome-wide genotyping chips from Illumina or Affymetrix were used by all studies (Supplementary Material, Table S1). All studies imputed variant dosages using the phase I version 3 or version 2 reference panels from the 1000 Genomes Project using MACH or IMPUTE (see further Supplementary Material, Table S1) (49–52).
Association analysis result files from the different studies were quality-controlled using the R package EasyQC (53) to harmonize any discrepancy in marker names between studies and to perform basic exclusions (incomplete and invalid data, monomorphic SNPs and SNPs with a sample size < 30, a minor allele count < 6, failed or low quality imputation, i.e. rsq < 0.3 or allele mismatches). Duplicate SNPs (i.e. SNPs mapped to the same genomic position due to differences in the version of 1000G Phase I reference panel used for imputation) were excluded from analysis. The complete EasyQC summary output can be found in Supplementary Material, Table S13. A total of 9,966,893 SNPs were included in the meta-analysis. Only SNPs in autosomes were interrogated.
Association analyses
Detailed information about statistical software and cohort-specific analysis parameters for individual studies is given in Supplementary Material, Table S1. All meta-analyses were performed by two different analysts using two different softwares; fixed-effect meta-analysis was performed using METAL (54) and GWAMA (55), while random-effect analysis was performed with GWAMA only. Heterogeneity was assessed Cochran’s heterogeneity test as implemented in GWAMA. In all meta-analyses, adjustment for multiple testing was performed using the Bonferroni correction.
Discovery GWAS analysis
For the discovery analysis, the ARIC, GHS1 and 2, GAIT2, Martha and PROCARDIS studies performed linear regression analyses using an additive model of inheritance between 1000 Genome-imputed SNPs and natural-logarithm-transformed FXI levels, with adjustments for age, sex, and accounting for population stratification and cryptic relatedness (adjustment for MDS or PCA components). PROCARDIS further used adjustment for coronary artery disease status and familial clustering to account for family structure. A total number of 16,169 European individuals were meta-analysed using an inverse-standard error model with fixed-effects. For significantly associated SNPs displaying significant heterogeneity among studies, a supplementary meta-analysis, using an inverse-standard error model with random-effects, was performed to control the discovered association. For convenience of analysis, we initially divided each chromosome into several 2 Mbp-sized ‘loci’ (while arbitrary, this preliminary locus definition worked well in practice for this study); each of these loci was represented by a lead SNP, defined as the most significantly associated SNP that fulfilled the following additional criteria of MAF > 0.01 and consistent direction of effect size across discovery studies. We later refined these locus definitions using conditional analysis (see below).
Lead SNPs with P-values below the pre-defined genome-wide threshold of 5×10−8 (in total three SNPs) were chosen for replication. Replication was performed in a total of 2,045 individuals from the F5Leiden, GAIT2 and MEGA studies using, in each cohort, linear regression with natural-logarithm-transformed FXI, and adjusting for age, sex, and accounting for population stratification and cryptic relatedness (adjustment for MDS or PCA components). The cohort analysis results were combined using a fixed-effect meta-analysis; SNPs for which significant heterogeneity was indicated were confirmed using a random-effect meta-analysis.
Conditional analysis
In order to investigate the existence of additional functional SNPs in the associated loci, all discovery studies (ARIC, GHS1 and 2, GAIT2, MARTHA and PROCARDIS) repeated the association analyses, using the same number of individuals and covariates, but now additionally conditioning on the three lead SNPs from the main analysis (rs710446, rs4253417 and rs780094). Meta-analysis and correction for multiple testing were performed as in the discovery analysis.
To investigate the existence of further independent signals present at the discovered loci, we used the approximate conditional joint selection (–cojo-slct) analysis implemented in the GCTA package (28). This analysis uses summary statistics from association analyses and an estimate of the LD structure of relevant regions in an approximate stepwise model selection of a set of independently associated SNPs for a pre-defined significance level (here P-value < 5×10−8). We used the same summary results from the ARIC, GHS1 and 2, GAIT2, MARTHA and PROCARDIS studies as in discovery, and used the hard-called WTCCC data, described below, for a robust estimate of the LD-structure.
Haplotype analysis
Haplotype analyses for the relevant associated SNPs were performed in the PROCARDIS and MARTHA studies using the THESIAS software (56), and association with lnFXI was adjusted for age, sex and population structure (principal components). Haplotype analyses were performed on best-guessed genotypes from imputed SNPs with imputation quality score greater than 0.8. Five SNPs were included in the haplotype analysis for the F11 locus; the lead SNPs from the present study, rs4253417 and rs4253421, and three SNPs previously associated with VTE (rs3822057 and rs12186257 (17) and rs2036914 (13)). These SNPs were all very well imputed in both studies. The SNPs were in strong LD and generated seven haplotypes with a frequency greater than 0.01 (Table 3). For the KNG1 locus, the aim was to include all SNPs identified in the discovery, conditional and the GCTA approximate conditional joint selection analyses; however, two of the SNPs from the GCTA analysis failed the imputation quality filter, so the final analysis comprised SNPs rs710446, rs76438938 and rs5030095, which generated five haplotypes.
Association with APTT
The five candidate SNPs (i.e. the three lead SNPs from the main analysis plus the two additional SNPs from the conditional analyses, rs780094, rs710446, rs4253417, rs4253421 and rs76438938) were tested in the F5Leiden, GAIT2, MARTHA studies (a total of 10,908 subjects) for association with aPTT, adjusting for age, sex, and, if applicable, principal components (model 1), and also with additional adjustment for FXI levels (model 2). As aPTT was reported as the raw aPTT clotting time in three studies, but as a normalized aPTT ratio in one cohort, meta-analysis weighted by standard errors () was not applicable. We instead used two other meta-analysis approaches: In the first of these, we followed a previous approach to the same problem (20) and performed a sample-size-weighted meta-analysis on P-values weighted by the sample size (N); in the second, we created normalized aPTT measures () by their standard deviation (), that is, .
eQTL analyses
The five candidate SNPs derived from the discovery and conditional analyses (rs710446, rs4253417, rs780094, rs76438938 and rs4253421) were subsequently tested for association with expression levels of nearby genes (defined as located within 500 Mbp in both directions) in a total of 211 human liver samples obtained from the Advanced Study of Aortic Pathology (ASAP) study (57). The ASAP study is a prospective, single-centre, observational cohort study of patients with aortic valve and ascending aortic disease, undergoing elective open-heart surgery at the Cardiothoracic Surgery Unit, Karolinska University Hospital in Stockholm, Sweden. The inclusion criteria were patients aged 18 or above with aortic valve disease (i.e. aortic stenosis or regurgitation) and/or ascending aorta dilatation (aneurysm or ectasia of the ascending aorta including the aortic root) but devoid of coronary artery disease (defined as lacking significant stenosis on coronary angiogram) and primarily not planned for other concomitant valve surgery. Global gene expression in the ASAP cohort was measured using the Affymetrix GeneChip® Human Exon 1.0 ST array and the core subset of Affymetrix meta probe sets. For specific details on quality control procedures, see (58). SNPs rs710446 and rs780094 were genotyped in ASAP on the Illumina 1M chip, while rs4253417, rs76438938 and rs4253421 were imputed to 1000G. Genotypes of the five candidate SNPs were tested for association with gene expression levels using additive linear models in R, with each genotype being coded as 0, 1 or 2.
Functional analysis and LD estimation
We used HaploReg v4 (34) to explore and summarize the functional annotation of associated SNPs from our analyses.
To obtain robust estimates of the LD structure, we used the controls from the Wellcome Trust Case Control Consortium (WTCCC). We imputed the WTCCC genotyped data for 5667 subjects into the phase I version 3 reference panels from the 1000 Genomes Project using IMPUTE. We then hard-called this data using an in-house C ++ program and a cutoff genotype probability > 0.8 and converted the result to plink binary format. For estimation of pairwise LD between specified SNPs, we used the plink v1.90b2m (59,60).
miRNA analysis
Prediction of miRNA binding
To find miRNA predicted to bind to the 3’ UTR, the 5’ UTR or the promoter region of the F11 mRNA, we devised a consensus algorithm combining the results from several public methods and databases for prediction of miRNA binding. In brief, an initial score was assigned to each miRNA; this score was then incremented by 1 for each of the following criteria that was true: i) TargetScan v 6.2 (61–64) context+ score < 0.045; ii) miRanda v 3.3a (65) score > 145; iii) mirSVR (66) score ≤ 0.1; iv) positive prediction from miRWalk 2.0 (67) 5’ UTR algorithms ≥ 50%; v) positive prediction from miRWalk 2.0 3’ UTR algorithms >50%; vi) positive prediction from miRWalk 2.0 promoter ≥ 50%. TargetScan and miRanda was set to use the mirBase release 21 database (68,69) and a cutoff was required for a miRNA to be selected. We limited our search to miRNA that were expressed in the liver, which was defined by any of the following criteria were fulfilled: 1) Clone count > 1 in SmirnaDB version 2009-05-08. 2) Normalized clone count > 0.00072 in microRNA.org version 2010-11-01 (70). 3) Reported as expressed in the liver in (36): supporting Supplementary Material, Table S1. Lastly, Tarbase (71) was used to investigate if miRNAs found were already experimentally validated.
Luciferase reporter assay
To validate if the miRNA of interest changed the expression of FXI, a luciferase report assay was performed as described (72). In brief, HEK293 cells were seeded on 24-well plates (1 × 105 cells/well). At 50–60% confluence, cells were transfected with luciferase reporter plasmid pLS, pLS-FXI-3’UTR (100 ng/well, Active Motif, Switchgear Genomics), together with scramble control, miR-145 or miR-181 (10 nM final concentration) using DharmaFECT Duo Transfection Reagent (Switchgear) according to the manufacturer’s protocol. After a 24-hour transfection period, luciferase activity was quantified using the LightSwitch™ Luciferase Assay Kit (Switchgear) according to the manufacturer’s protocol.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful to Mary Cushman for advice in the project design and selection of samples and to Mattias Frånberg for valuable discussions on analysis methods. The authors thank the staff and participants of the ARIC study for their important contributions.
Conflict of Interest statement. None declared.
Funding
The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. PROCARDIS was supported by the European Community Sixth Framework Program (LSHM-CT- 2007-037273), AstraZeneca, the British Heart Foundation, the Wellcome Trust (Contract No. 075491/Z/04), the Swedish Research Council, the Knut and Alice Wallenberg Foundation, the Swedish Heart-Lung Foundation, the Torsten and Ragnar Söderberg Foundation, the Strategic Cardiovascular and Diabetes Programs of Karolinska Institutet and Stockholm County Council, the Foundation for Strategic Research and the Stockholm County Council. Bengt Sennblad acknowledges funding from the Magnus Bergvall foundation and the foundation for old servants. Maria Sabater-Lleal is recipient of the European Hematology Association and the International Society of Thrombosis and Hemostasis (EHA-ISTH) fellowship and acknowledges funding from Swedish Heart-Lung Foundation (20130399), Åke Wiberg (400546009), Lars Hiertas Minne (F02014-0357), and Magnus Bergvall's foundations (2014-00242). The Gutenberg Health Study is funded through the government of Rhineland-Palatinate (‘Stiftung Rheinland-Pfalz für Innovation’, contract AZ 961-386261/733), the research programs ‘Wissenschafft Zukunft’ and ‘Center for Translational Vascular Biology (CTVB)’ of the Johannes Gutenberg-University of Mainz, and its contract with Boehringer Ingelheim and PHILIPS Medical Systems, including an unrestricted grant for the Gutenberg Health Study. Philipp S. Wild is funded by the Federal Ministry of Education and Research (BMBF 01EO1503) and he is PI of the German Center for Cardiovascular Research (DZHK). The MARTHA project was supported by grants from the Program Hospitalier de Recherche Clinique. Statistical data analysis in MARTHA was performed on the C2BIG high-performance computing system funded by the Region Ile de France, the Université Pierre et Marie Curie and the ICAN Institute for Cardiometabolism and Nutrition (ANR-10-IAHU-05). GAIT2 was supported partially by grants PI-11/0184, Pi-14/0582, RD12/0042/0032 and UIN2013-50833 from the Instituto Carlos III (Fondo de Investigación Sanitaria—FIS). The French-Canadian family study on Factor V Leiden was partially supported by the Canadian Institutes of Health Research and the Heart and Stroke Foundation of Canada; France Gagnon is supported by a Canada Research Chair in Genetic Epidemiology.
References
- 1. Yang D.T., Flanders M.M., Kim H., Rodgers G.M. (2006) Elevated factor XI activity levels are associated with an increased odds ratio for cerebrovascular events. Am. J. Clin. Pathol., 126, 411–415. [DOI] [PubMed] [Google Scholar]
- 2. Tucker E.I., Marzec U.M., White T.C., Hurst S., Rugonyi S., McCarty O.J.T., Gailani D., Gruber A., Hanson S.R. (2009) Prevention of vascular graft occlusion and thrombus-associated thrombin generation by inhibition of factor XI. Blood, 113, 936–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meijers J.C., Tekelenburg W.L., Bouma B.N., Bertina R.M., Rosendaal F.R. (2000) High levels of coagulation factor XI as a risk factor for venous thrombosis. N. Engl. J. Med., 342, 696–701. [DOI] [PubMed] [Google Scholar]
- 4. Gailani D., Smith S.B. (2009) Structural and functional features of factor XI. J. Thromb. Haemost., 7 Suppl 1, 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doggen C.J.M., Rosendaal F.R., Meijers J.C.M. (2006) Levels of intrinsic coagulation factors and the risk of myocardial infarction among men: Opposite and synergistic effects of factors XI and XII. Blood, 108, 4045–4051. [DOI] [PubMed] [Google Scholar]
- 6. Siegerink B., Rosendaal F.R., Algra A. (2014) Antigen levels of coagulation factor XII, coagulation factor XI and prekallikrein, and the risk of myocardial infarction and ischemic stroke in young women. J. Thromb. Haemost., 12, 606–613. [DOI] [PubMed] [Google Scholar]
- 7. Salomon O., Steinberg D.M., Koren-Morag N., Tanne D., Seligsohn U. (2008) Reduced incidence of ischemic stroke in patients with severe factor XI deficiency. Blood, 111, 4113–4117. [DOI] [PubMed] [Google Scholar]
- 8. Folsom A.R., Tang W., Roetker N.S., Heckbert S.R., Cushman M., Pankow J.S. (2015) Prospective study of circulating factor XI and incident venous thromboembolism: The Longitudinal Investigation of Thromboembolism Etiology (LITE). Am. J. Hematol., 10.1002/ajh.24168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Folsom A.R., George K.M., Appiah D. (2015) Lack of association of plasma factor XI with incident stroke and coronary heart disease: The Atherosclerosis Risk in Communities (ARIC) Study. Atherosclerosis, 243, 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holst A.G., Jensen G., Prescott E. (2010) Risk factors for venous thromboembolism. Circulation, 4;121:1896–1903. [DOI] [PubMed] [Google Scholar]
- 11. Naess I.A., Christiansen S.C., Romundstad P., Cannegieter S.C., Rosendaal F.R., Hammerstrøm J. (2007) Incidence and mortality of venous thrombosis: a population-based study. J. Thromb. Haemost., 5, 692–699. [DOI] [PubMed] [Google Scholar]
- 12. Salomon O., Steinberg D.M., Zucker M., Varon D., Zivelin A., Seligsohn U. (2011) Patients with severe factor XI deficiency have a reduced incidence of deep-vein thrombosis. Thromb. Haemost., 105, 269–273. [DOI] [PubMed] [Google Scholar]
- 13. Bezemer I.D., Bare L.A., Doggen C.J.M., Arellano A.R., Tong C., Rowland C.M., Catanese J., Young B.A., Reitsma P.H., Devlin J.J., et al. (2008) Gene variants associated with deep vein thrombosis. JAMA-J. Am. Med. Assoc., 299, 1306–1314. [DOI] [PubMed] [Google Scholar]
- 14. Li Y., Bezemer I.D., Rowland C.M., Tong C.H., Arellano A.R., Catanese J.J., Devlin J.J., Reitsma P.H., Bare L.A., Rosendaal F.R. (2009) Genetic variants associated with deep vein thrombosis: the F11 locus. J. Thromb. Haemost., 7, 1802–1808. [DOI] [PubMed] [Google Scholar]
- 15. Austin H., De Staercke C., Lally C., Bezemer I.D., Rosendaal F.R., Hooper W.C. (2011) New gene variants associated with venous thrombosis: a replication study in White and Black Americans. J. Thromb. Haemost., 9, 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rovite V., Maurins U., Megnis K., Vaivade I., Pečulis R., Rits J., Prave S., Klovins J. (2014) Association of F11 polymorphism rs2289252 with deep vein thrombosis and related phenotypes in population of Latvia. Thromb. Res., 134, 659–663. [DOI] [PubMed] [Google Scholar]
- 17. Germain M., Chasman D.I., de Haan H., Tang W., Lindström S., Weng L.C., de Andrade M., de Visser M.C.H., Wiggins K.L., Suchon P., et al. (2015) Meta-analysis of 65,734 individuals identifies TSPAN15 and SLC44A2 as two susceptibility loci for venous thromboembolism. Am. J. Hum. Genet., 96, 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hanson E., Nilsson S., Jood K., Norrving B., Engström G., Blomstrand C., Lindgren A., Melander O., Jern C. (2013) Genetic variants of coagulation factor XI show association with ischemic stroke up to 70 years of age. PLoS One, 8, e75286.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zakai N.A., Ohira T., White R., Folsom A.R., Cushman M. (2008) Activated partial thromboplastin time and risk of future venous thromboembolism. Am. J. Med., 121, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang W., Schwienbacher C., Lopez L.M., Ben-Shlomo Y., Oudot-Mellakh T., Johnson A.D., Samani N.J., Basu S., Gögele M., Davies G., et al. (2012) Genetic Associations for Activated Partial Thromboplastin Time and Prothrombin Time, their Gene Expression Profiles, and Risk of Coronary Artery Disease. Am. J. Hum. Genet., 91, 152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gaunt T.R., Lowe G.D.O., Lawlor D.A., Casas J.P., Day I.N.M. (2013) A gene-centric analysis of activated partial thromboplastin time and activated protein C resistance using the HumanCVD focused genotyping array. Eur. J. Hum. Genet., 21, 779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosen E.D., Gailani D., Castellino F.J. (2002) FXI is essential for thrombus formation following FeCl3-induced injury of the carotid artery in the mouse. Thromb. Haemost., 87, 774–776. [PubMed] [Google Scholar]
- 23. Büller H.R., Bethune C., Bhanot S., Gailani D., Monia B.P., Raskob G.E., Segers A., Verhamme P., Weitz J.I.. FXI-ASO TKA Investigators (2015) Factor XI antisense oligonucleotide for prevention of venous thrombosis. N. Engl. J. Med., 372, 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sabater-Lleal M., Martinez-Perez A., Buil A., Folkersen L., Souto J.C., Bruzelius M., Borrell M., Odeberg J., Silveira A., Eriksson P., et al. (2012) A genome-wide association study identifies KNG1 as a genetic determinant of plasma factor XI Level and activated partial thromboplastin time. Arterioscl. Throm. Vas., 32, 2008–2016. [DOI] [PubMed] [Google Scholar]
- 25. Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C., et al. (2015) A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet., 47, 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., et al. (2010) Common SNPs explain a large proportion of the heritability for human height. Nat. Genet., 42, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang J., Lee S.H., Goddard M.E., Visscher P.M. (2011) GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet., 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A.F., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J., et al. (2012) Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet., 44, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., et al. (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet., 42, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ng M.C.Y., Saxena R., Li J., Palmer N.D., Dimitrov L., Xu J., Rasmussen-Torvik L.J., Zmuda J.M., Siscovick D.S., Patel S.R., et al. (2013) Transferability and fine mapping of type 2 diabetes loci in African Americans: the Candidate Gene Association Resource Plus Study. Diabetes, 62, 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brouwers M.C.G.J., Jacobs C., Bast A., Stehouwer C.D.A., Schaper N.C. (2015) Modulation of Glucokinase Regulatory Protein: A Double-Edged Sword?. Trends Mol. Med., 21, 583–594. [DOI] [PubMed] [Google Scholar]
- 32. Hu Z., Liu J., Song Z., Hou Q., Fan X., Hou D. (2015) Variants in the Atherogenic ALOX5AP, THBD, and KNG1 Genes Potentiate the Risk of Ischemic Stroke via a Genetic Main Effect and Epistatic Interactions in a Chinese Population. J. Stroke Cerebrovasc. Dis., 24, 2060–2068. [DOI] [PubMed] [Google Scholar]
- 33. Fabian M.R., Sonenberg N. (2012) The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol., 19, 586–593. [DOI] [PubMed] [Google Scholar]
- 34. Ward L.D., Kellis M. (2011) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Safdar H., Cleuren A.C.A., Cheung K.L., Gonzalez F.J., Vos H.L., Inoue Y., Reitsma P.H., van Vlijmen B.J.M. (2013) Regulation of the F11, Klkb1, Cyp4v3 gene cluster in livers of metabolically challenged mice. PLoS One, 8, e74637.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salloum-Asfar S., Teruel-Montoya R., Arroyo A.B., García-Barberá N., Chaudhry A., Schuetz E., Luengo-Gil G., Vicente V., González-Conejero R., Martínez C. (2014) Regulation of Coagulation Factor XI Expression by MicroRNAs in the Human Liver. PLoS One, 9, e111713.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Remy I., Michnick S.W. (2006) A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods, 3, 977–979. [DOI] [PubMed] [Google Scholar]
- 38. Smith N.L., Chen M.H., Dehghan A., Strachan D.P., Basu S., Soranzo N., Hayward C., Rudan I., Sabater-Lleal M., Bis J.C., et al. (2010) Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation, 121, 1382–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang W., Basu S., Kong X., Pankow J.S., Aleksic N., Tan A., Cushman M., Boerwinkle E., Folsom A.R. (2010) Genome-wide association study identifies novel loci for plasma levels of protein C: the ARIC study. Blood, 116, 5032–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Taylor K.C., Lange L.A., Zabaneh D., Lange E., Keating B.J., Tang W., Smith N.L., Delaney J.A., Kumari M., Hingorani A., et al. (2011) A gene-centric association scan for Coagulation Factor VII levels in European and African Americans: the Candidate Gene Association Resource (CARe) Consortium. Hum. Mol. Genet., 20, 3525–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bruzelius M., Bottai M., Sabater-Lleal M., Strawbridge R.J., Bergendal A., Silveira A., Sundström A., Kieler H., Hamsten A., Odeberg J. (2015) Predicting venous thrombosis in women using a combination of genetic markers and clinical risk factors. J. Thromb. Haemost., 13, 219–227. [DOI] [PubMed] [Google Scholar]
- 42. Germain M., Saut N., Greliche N., Dina C., Lambert J.C., Perret C., Cohen W., Oudot-Mellakh T., Antoni G., Alessi M.C., et al. (2011) Genetics of venous thrombosis: insights from a new genome wide association study. PLoS One, 6, e25581.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang W., Cushman M., Green D., Rich S.S., Lange L.A., Yang Q., Tracy R.P., Tofler G.H., Basu S., Wilson J.G., et al. (2015) Gene-centric approach identifies new and known loci for FVIII activity and VWF antigen levels in European Americans and African Americans. Am. J. Hematol., 90, 534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ellis J., Lange E.M., Li J., Dupuis J., Baumert J., Walston J.D., Keating B.J., Durda P., Fox E.R., Palmer C.D., et al. (2014) Large multiethnic Candidate Gene Study for C-reactive protein levels: identification of a novel association at CD36 in African Americans. Hum. Genet., 133, 985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ridker P.M., Paré G., Parker A., Zee R.Y.L., Danik J.S., Buring J.E., Kwiatkowski D., Cook N.R., Miletich J.P., Chasman D.I. (2008) Loci related to metabolic-syndrome pathways including LEPR, HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am. J. Hum. Genet., 82, 1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Folsom A.R., Lutsey P.L., Astor B.C., Cushman M. (2009) C-reactive protein and venous thromboembolism. A prospective investigation in the ARIC cohort. Thromb. Haemost., 102, 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Faid V., Denguir N., Chapuis V., Bihoreau N., Chevreux G. (2014) Site-specific N-glycosylation analysis of human factor XI: Identification of a noncanonical NXC glycosite. Proteomics, 14, 2460–2470. [DOI] [PubMed] [Google Scholar]
- 48. Smit R.A.J., Trompet S., De Craen A.J.M., Jukema J.W. (2014) Using genetic variation for establishing causality of cardiovascular risk factors: overcoming confounding and reverse causality. Neth. Heart J., 22, 186–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li Y., Willer C., Sanna S., Abecasis G. (2009) Genotype imputation. Annu. Rev. Genomics Hum. Genet., 10, 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Howie B.N., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genetics, 5, e1000529.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Consortium, T.1.G.P., The 1000 Genomes Consortium Participants are arranged by project role, T.B.I.A.A.F.A.W.I.E.F.P.I.A.P.L.A.I., author, C., committee, S., Medicine, P.G.B.C.O., BGI-Shenzhen, Broad Institute of MIT and Harvard, European Bioinformatics Institute, Illumina, Max Planck Institute for Molecular Genetics., et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 490, 56–65. [Google Scholar]
- 52. Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol., 34, 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Winkler T.W., Day F.R., Croteau-Chonka D.C., Wood A.R., Locke A.E., Mägi R., Ferreira T., Fall T., Graff M., Justice A.E., et al. (2014) Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc., 9, 1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mägi R., Morris A.P. (2010) GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics, 11, 288.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tregouet D.A., Garelle V. (2007) A new JAVA interface implementation of THESIAS: testing haplotype effects in association studies. Bioinformatics, 23, 1038–1039. [DOI] [PubMed] [Google Scholar]
- 57. Jackson V., Petrini J., Caidahl K., Eriksson M.J., Liska J., Eriksson P., Franco-Cereceda A. (2011) Bicuspid aortic valve leaflet morphology in relation to aortic root morphology: a study of 300 patients undergoing open-heart surgery. Eur. J. Cardiothorac. Surg., 40, e118–e124. [DOI] [PubMed] [Google Scholar]
- 58. Folkersen L., van't Hooft F., Chernogubova E., Agardh H.E., Hansson G.K., Hedin U., Liska J., Syvänen A.C., Paulsson-Berne G., Paulssson-Berne G., et al. (2010) Association of genetic risk variants with expression of proximal genes identifies novel susceptibility genes for cardiovascular disease. Circ. Cardiovasc. Genet., 3, 365–373. [DOI] [PubMed] [Google Scholar]
- 59. Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience, 4, 559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gaunt T.R., Rodríguez S., Day I.N. (2007) Cubic exact solutions for the estimation of pairwise haplotype frequencies: implications for linkage disequilibrium analyses and a web tool ‘CubeX’. BMC Bioinformatics, 8, 428.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lewis B.P., Burge C.B., Bartel D.P. (2005) Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell, 120, 15–20. [DOI] [PubMed] [Google Scholar]
- 62. Grimson A., Farh K.K.H., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. (2007) MicroRNA Targeting Specificity in Mammals: Determinants beyond Seed Pairing. Molecular Cell, 27, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. (2011) Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol., 18, 1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Friedman R.C., Farh K.K.H., Burge C.B., Bartel D.P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res., 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. (2004) Human MicroRNA targets. PLoS Biol., 2, e363.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Betel D., Koppal A., Agius P., Sander C. (2010) Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol., 11:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dweep H., Sticht C., Pandey P., Gretz N. (2011) miRWalk–database: prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J. Biomed. Inform., 44, 839–847. [DOI] [PubMed] [Google Scholar]
- 68. Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. (2007) miRBase: tools for microRNA genomics. Nucleic Acids Res., 36, D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kozomara A., Griffiths-Jones S. (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res., 39, D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell, 129, 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vlachos I.S., Paraskevopoulou M.D., Karagkouni D., Georgakilas G., Vergoulis T., Kanellos I., Anastasopoulos I.L., Maniou S., Karathanou K., Kalfakakou D., et al. (2015) DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res., 43, D153–D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li Y., Challagundla K.B., Sun X.X., Zhang Q., Dai M.S. (2015) MicroRNA-130a associates with ribosomal protein L11 to suppress c-Myc expression in response to UV irradiation. Oncotarget., 6, 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Consortium R.E., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature, 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Consortium E.P. (2011) A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol., 9, e1001046.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kheradpour P., Kellis M. (2014) Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res., 42, 2976–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.