Abstract
The uplift and final connection of the Central American land bridge is considered the major event that allowed biotic exchange between vertebrate lineages of northern and southern origin in the New World. However, given the complex tectonics that shaped Middle America, there is still substantial controversy over details of this geographical reconnection, and its role in determining biogeographic patterns in the region. Here, we examine the phylogeography of Bothrops asper, a widely distributed pitviper in Middle America and northwestern South America, in an attempt to evaluate how the final Isthmian uplift and other biogeographical boundaries in the region influenced genealogical lineage divergence in this species. We examined sequence data from two mitochondrial genes (MT-CYB and MT-ND4) from 111 specimens of B. asper, representing 70 localities throughout the species’ distribution. We reconstructed phylogeographic patterns using maximum likelihood and Bayesian methods and estimated divergence time using the Bayesian relaxed clock method. Within the nominal species, an early split led to two divergent lineages of B. asper: one includes five phylogroups distributed in Caribbean Middle America and southwestern Ecuador, and the other comprises five other groups scattered in the Pacific slope of Isthmian Central America and northwestern South America. Our results provide evidence of a complex transition that involves at least two dispersal events into Middle America during the final closure of the Isthmus.
Introduction
Tropical Middle America, the region that extends from the Isthmus of Tehuantepec to the northwestern tip of South America, accounts for less than 0.7% of the Earth’s total land area, and yet, is one of the most biologically diverse territories on the planet [1–3]. The origin and maintenance of its tremendous diversity is attributed not only to Mesoamerica’s geographical location between two major continental masses but also to the intricate geological and climatological history that shaped the region since the late Cretaceous [4–6].
Among the myriad of events that resulted from these dynamics, perhaps the most significant was the rise and final connection of the Lower Central American Isthmus (LCA, but also known as the Isthmus of Panama). The uplift did not occur as a single incident but instead resulted from a series of geological and climatological events that started in the Oligocene and continued until the mid-Pliocene when the unbroken connection finally emerged [5–10, but see 11 for a different view]. This process allowed not only the evolution and redistribution of organisms in Mesoamerica [12, 13] but also was crucial in the exchange of species between the Northern and Southern continents, often referred as the Great American Biotic Exchange [14–18]. Despite its importance, there is still considerable debate about the timing of the final LCA connection, and whether pre-closure dispersal or vicariant events mediated the cladogenesis and biological diversification observed in the region [19, 20].
Several studies have shown the effectiveness of molecular phylogeographic approaches combined with robust estimations of divergence time to assess the role of putative geological events in the cladogenesis of species inhabiting Middle America [20–22]. This has allowed scientists to address questions about how historical divergence occurred across landscapes, especially in regions where there is little consensus on historical processes, as is the case of Middle America.
Pitvipers have been used to elucidate fine-scale historical biogeographical patterns, [19, 23–25] because most species have lower vagility than other vertebrates, which make them more prone to genetic isolation via vicariance. Moreover, the intrageneric relationships of pitvipers in Middle America are well known [26–30], and it seems that Mesoamerican pitviper lineages exhibit coincident temporal patterns of divergence that match major geological events that shaped the region. For instance, the tropical rattlesnake Crotalus durissus (sensu lato) has a northern origin [31], but its phylogeographic pattern is consistent with a gradual range expansion south that corresponded to the final uplift of the LCA isthmus, followed by a rapid dispersal into South America [32].
The effect of the LCA uplift and final closure on tropical snake lineages of South American origin is far less understood, although it is believed that most dispersed north only after the uplift of the Isthmus [29]. To assess the effect of the LCA uplift on South American taxa, we focus on the Central American lancehead pitviper, Bothrops asper. This species has close affinities with members of the B. atrox complex [sensu 27] a group mainly distributed east of the Andes [33], and is thus nested deeply within the genus Bothrops that is otherwise confined to South America [26, 27, 34]. However, B. asper extends its distribution from Colombia and northwestern Peru in South America to lowland Mexico and Central America. Given these affinities and its current distribution well into Middle America, it appears that Bothrops asper expanded its range rapidly northward once the LCA isthmus was finalized [34]. Thus, we predict that B. asper populations in Middle America will be more recent and exhibit lower divergence than those in South America.
To test this, we assess the phylogeographic pattern of B. asper using mtDNA markers across the species range and evaluate the time and mode in which the species colonized Mesoamerica.
Materials and methods
Specimens and laboratory methods
Bothrops asper individuals were collected from 70 localities throughout the species distribution (Fig 1 and S1 Table). Genomic DNA was extracted mostly from blood or shed skin following the procedure described in [35]. The use of biological material for this study was approved by the Research Committee of Instituto Clodomiro Picado (session No 06–2013) and by the Institutional Committee for Care and Use of Animals (CICUA) from Universidad de Costa Rica.
Fig 1. Distribution of Bothrops asper (red contour) in Middle and South America adapted from [33].
Question marks in northwestern Venezuela and Peru indicate that part of the distribution which needs confirmation. Symbols represent localities of specimens included in analyses, showing their phylogeographical affinities: MY (Mexico-Yucatan), NCA (Caribbean Nuclear Central America), CICA (Caribbean Isthmian Central America), PICA (Pacific Isthmian Central America), WE (West Ecuador), TR (Tumbes region), CHOCO 1 (Darien-Colombian Chocó), CHOCO 2 (Ecuadorian Chocó), CCO (Caribbean Colombia), MV (Magdalena Valley). See text for elaboration.
The MT-ND4 and MT-CYB regions were amplified using the primer pairs described in [36, 37] respectively. For both genes, PCR reactions were set up to a final volume of 25 μl, using 2.0 μl genomic DNA, 0.4 μl of each primer (0.16 μM), 2.5 μl of 10 X PCR buffer (1X), 0.25 μl total dNTPs (100 μM), 1.0 μl of MgCl2 (2 mM), 0.2 μl of Taq polymerase (1 U), and 18.6 μl H2O). Typical amplification conditions involved initial denaturation at 94°C for 5 min, followed by 38 cycles of 94°C for 40 s, 54°C of annealing for 40 s, then 72°C for 1 min, followed by a final extension step of 72°C for 5 min. The amplified product was sequenced using the same primers by Macrogen (Seoul, S. Korea– http://dna.macrogen.com).
Alignment and data exploration
DNA sequences were edited using BioEdit version 7.0 [38], and then the alignment was verified by eye in GeneDoc [39]. Since we use coding genes in our analyses, all nucleotide sequences were translated into amino acids to evaluate the reading frame and ensure the absence of premature stop codons or other nonsense mutation [40]. We deposited novel sequences were deposited in GenBank, and the final nucleotide alignment matrix is available upon request. We calculated genetic distances (p-distance) from aligned sequences using Mega version 7 [41].
Phylogenetic analysis and divergence time estimation
For phylogenetic analyses we incorporate representatives of the following genera (# species) as outgroups: Lachesis (2), Ophryacus (2), Agkistrodon (2), Sistrurus (1), Crotalus (2), Bothriocophias (3), Rhinocerophis (4), Bothropoides (11), Bothriopsis (3), and Bothrops (12) (S1 Table).
We treated gaps in the alignment as missing data. We concatenated the different gene sequences (MT-ND4 and MT-CYB) for each specimen and analyzed the data jointly. Data for each gene was partitioned following [29], and Mega version 7 [41] was used to estimate the best-fitting models of nucleotide evolution for each partition independently. We select the best-fitting models according to Bayesian Information Criterion (BIC) [42].
We performed phylogenetic analyses of concatenated genes using maximum likelihood (ML), and Metropolis-Hastings coupled Markov chain Monte Carlo Bayesian methods (BMCMC). ML analyses were performed under different models of nucleotide evolution [43]. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log-likelihood value. We used a discrete Gamma distribution to model evolutionary rate differences among sites. These analyses were conducted in MEGA7 [41]. We use non-parametric bootstrap (10,000 replicates) to evaluate branch support in the phylogenetic reconstruction [44].
The BMCMC estimate of the phylogeny was inferred using MrBayes version 3.0B4 [45]. We executed three parallel MCMC runs simultaneously, each run for 20 x 106 generations with four Markov chains (one cold and three heated chains). Model parameters and the rate of evolution differ among each partition. We used Tracer 1.6 [46] for visualizing output parameters to ascertain stationarity and whether or not the duplicated runs had converged on the same mean likelihood. Runs appeared stationary before 106 generations, and we conservatively excluded the first 2.0 x 106 generations of each run as burn-in. All post-burn-in estimates (sampled every 1000 generations) were combined, and we summarized phylogeny and parameter estimates from this combined posterior distribution. Nodes were considered well supported if posterior probabilities > 0.95.
To evaluate phylogroup boundaries on our preferred phylogenetic tree, we performed Bayesian Poisson Tree Process (bPTP) implemented in http://www.exelixis-lab.org/software.html [47]. This method considers the number of substitutions between branching (speciation events) and assumes that each substitution has a small probability of generating speciation. Thus, if the number of changes is sufficiently large, the process follows a Poisson distribution. bPTP adds Bayesian support (BS) values to delimited species on the input tree, with higher node support indicates that all descendants of this node are more likely to be from one species [47].
We also implemented the Bayesian phylogeography approach (BPA, [48–49]) to evaluate the putative area of origin of B. asper-B. atrox lineages. BPA takes into account space-time domains when analyzing the evolutionary process, and uses localities as discrete states to allow inferences about the location of ancestral lineages. In order to reconstruct the ancestral state, we re-ran our Bayesian analysis without outgroups, using BEAST v.2.4.7 [50]. Three individual runs were performed for 10 x 106 generations with a sampling frequency of 10,000; under the same nucleotide substitution models used in our ML phylogenetic analysis (see Results). We apply the following parameters as priors: coalescence: constant size speciation process; clock rate: set at 0.005; and strict molecular clock. For each individual included in the analysis, we designate their country of origin as the state of locality. We are aware that this designation is broad and artificial, but we consider that it allows us to locate in present-day geography the spaces occupied by the ancestral lineages.
For all analyses performed on BEAST, each run was analyzed in Tracer [46] to confirm that effective sample sizes (ESS) were sufficient for all parameters (posterior ESS values > 300). LogCombiner® and TreeAnnotator® (both available in the BEAST package) were used to infer the ultrametric tree after discarding 10% of the samples from each run.
We estimated divergence times using the Bayesian relaxed clock method with uncorrelated lognormal rates among branches across the B. asper phylogeny, assuming a birth-death process for the speciation model implemented in BEAST v.2 [50]. This method incorporates heterogeneity rates based on Bayesian inference and allows the simultaneous use of different evolutionary parameters for each dataset [51, 52].
Posterior distributions of parameters were approximated using three independent MCMC analyses of 20 x 106 generations each, with samples retained every 1,000 generations. Samples from the two runs, which yielded similar results, were combined and convergence of the chains was checked using the program Tracer 1.6 [46].
We used three calibration points: (1) the minimum age of Sistrurus estimated from the fossil evidence of the most recent ancestor (TMRCA) of Sistrurus + Crotalus [53]. Using a log-normal prior with zero offset (hard upper bound) of 8 million years ago (Mya), a mean of 0.01, and standard deviation (SD) of 0.76, this estimation produced a median age centered at 9 Mya and a 95% prior credible interval (PCI) extending to 11.5 Mya [54] (2) The basal divergence within the crown Agkistrodon clade (A. piscivorous-A. contortrix divergence) in the Late Miocene [54]. Using a log-normal prior with zero offset of 6 Mya, a mean of 0.01, and SD of 0.42, this estimation produced a median age of 7 Mya and a 95% PCI extending to 8 Mya [54]. Although these are very narrow constraints based on a patchy fossil record, true divergence dates will probably be older than the oldest known fossil, thus making these lognormal distributions with hard lower bounds a conservative assumption [25, 55]. (3) The estimated age of divergence between C. ruber and C. atrox according to [56] due to the Pliocene marine incursion of the Sea of Cortés [57], using a lognormal mean of 1.1 and SD of 0.37, no zero offset. This produced a median age centered at 3 Mya and a 95% PCI extending to 5.5 Mya that coincides with time estimations for the development of Sea of Cortés [57]. There is still some debate on the time and level of isolation of the California peninsula during those marine incursions [58], so we perform our analyses including and excluding this calibration point. No differences in the estimation of divergence time retrieved by both approaches were noticed.
Testing for range expansion
We analyze the distribution of pairwise nucleotide differences (mismatch distribution) to infer the existence of demographic growth, following [59] method implemented in ARLEQUIN 3.5 [60]. Distribution curves were analyzed assuming constant and non-constant sizes for the observed empirical distribution. We use rg as a measure of "statistical raggedness" [61] and R2 statistic [62] to determine the correspondence between the observed and theoretical curves. Also, we employed Fu and Li [63], and Tajima ɵ statistics to detect possible changes in population sizes [62, 64].
Results
Sequence analysis
We generated new sequences for B. asper (MT-CYB: 101; MT-ND4: 90), B. atrox (MT-CYB: 5; MT-ND4: 1), B. colombiensis (MT-CYB: 9; MT-ND4: 9), B. isabelae (MT-CYB: 5; MT-ND4: 5) and B. venezuelensis (MT-CYB: 2; MT-ND4: 1). From 174 specimens, including 111 in the ingroup, we obtained a concatenated matrix of 1442 bp for the two genes (758bp for MT-CYB and 684bp for MT-ND4).
The alignment was unambiguous, and the inferred amino acid sequence contained no stop codons, which suggest a mitochondrial origin sequence rather than nuclear insertion. Including outgroups, we recover 545 polymorphic sites from all sequences, 387 of them are informative. For B. asper, we recover 296 polymorphic sites, 237 informative.
Phylogenetic analysis and genetic distances
Selected models of nucleotide evolution differ among all partitions, the model chosen was GTR+Г for the first and third codon position of MT-CYB and all MT-ND4 partitions, and HKY+ I for the second MT-CYB position. Under ML, the tree with the highest log likelihood (-9078.04) is congruent with the phylogenetic relationships recovered under the BMCMC method, with only minor differences among the outgroups. Thus, we only show the results of the Bayesian analysis here (Fig 2).
Fig 2. Bayesian phylogeny of relationships among members of B. asper from different physiographic regions (names as in Fig 1).
Two distinct B. asper lineages are depicted in red (clade A) and green (clade B) branches. Red bars indicate phylogroups supported by the bPTP analysis. Support for each node is shown as posterior probability (PP, Bayesian inference) or Bootstrap value (BS, Maximum likelihood).
Our analysis recovered a partial phylogeny of Bothrops that is consistent with previous reconstructions of lancehead phylogenies based on morphology and/or mtDNA [30, 34, 65–68].
We recovered a deep split of B. asper into two well-supported lineages (clades A and B, Fig 2), differing by average p-distances of 5.7% (MT-CYB) and 4.3% (MT-ND4), and paraphyly of B. asper concerning the B. atrox group. However, the node placing the atrox group as sister to B. asper clade B (Fig 2) is weakly supported, and our analyses using BEAST recovered B. asper as monophyletic (see Figs 3 and 4).
Fig 3. Maximum clade credibility phylogeny for Bothrops asper, retrieved from mtDNA sequences.
Branches are colored according to the most probable “location state” of their descendant nodes. Values in branches indicate the location set probability of the ancestral state, in this case, the probability that the origin of the branch occurred in the region that is now Colombia. Bayesian support for clades as in Fig 2.
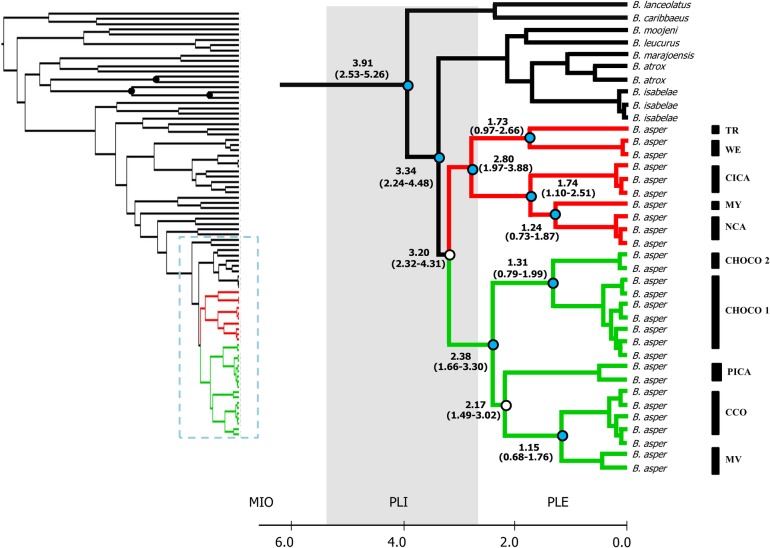
Fig 4. Bayesian estimates of divergence time (Mya) for the lancehead phylogeny.
Left: Overall tree showing the calibration points (black dots) for time divergence estimations. Right: B. asper phylogeny showing the mean and 95% confidence intervals (in parenthesis) for divergence time estimates at each node. Clades A and B are depicted as in Fig 2. Grey bar indicates the extension of the Pliocene. Well supported nodes (PP > 0.95) for divergence estimations are shown in light blue, whereas weakly supported nodes are shown in white. B. asper phylogroup names as in Fig 1.
Clade A includes B. asper specimens from the Caribbean coast of Middle America and southwestern Ecuador. In this clade, five well supported (PP > 0.9) groups were retrieved in our bPTP analysis (Fig 2): (1) specimens from localities along the Gulf of Mexico coast and Yucatán Peninsula, including Petén (MY); (2) Nuclear Central America (NCA), including specimens from the Caribbean slopes of Honduras, Guatemala, and Belize; (3) Caribbean Isthmian Central America (CICA), from the Caribbean lowlands of Nicaragua and Costa Rica; (4) Western Ecuador (WE), from Guayas, Manabí, Los Ríos and Chimborazo provinces in Ecuador; (5) Tumbes biogeographic region (TR, that includes specimens from Loja in southern Ecuador). MY and NCA form a well-supported clade that is distributed within the same biogeographical unit (Veracruzan Province sensu Morrone [69]).
Clade B also includes five distinct B. asper groups (Fig 2): (1) Pacific Isthmian Central America (PICA) from Pacific localities of Costa Rica and Panama; (2) Northern Choco (CHOCO 1) from central and eastern Panama, and Pacific coast of Colombia; (3) Chocoan Ecuador (CHOCO 2), from Esmeralda and Pichincha, Ecuador; (4) Caribbean Colombia (CCO) localities from the Caribbean lowlands of Colombia; and (5) Rio Magdalena valley (MV), from localities along the upper Magdalena basin. CHOCO 1 and CHOCO 2 form a well-supported clade that is distributed within the same biogeographical unit (Choco-Darien Province [69]). The groups spread west of the Cordillera Occidental (CCO and MV) also form a supported clade (Fig 2) and are included in Morrone’s Magdalena Province [69].
Average uncorrected p-distances for MT-ND4 were lower than those for MT-CYB in most groups (Table 1). The pairwise divergence between the B. atrox group and B. asper lineages ranged between 3.1% and 6.1% for MT-CYB, and 4.3% and 5.9% for MT-ND4. The separation between B. asper clades A and B ranged between 3.6% and 8.2% for MT-CYB and 3.3% and 5.1% for MT-ND4: the greatest pairwise divergence observed between individuals from MY and those from CCO in Colombia for (MT-CYB p distance = 8.2%, Table 1). Even in Costa Rica, the mean pairwise divergence between snakes from the Caribbean region (CICA) and those in the Pacific lowlands (PICA) was relatively high, 6.5% (MT-CYB) and 4.6% (MT-ND4), despite that these populations occur only a few kilometers apart. The lowest mean divergence was noticed between snakes distributed in the Magdalena Valley and those in the Caribbean region of Colombia (Table 1).
Table 1. Net divergences (uncorrected p-distances) for MT-CYB (below diagonal) and MT-ND4 (above diagonal) within B. asper groups, and among them and related clades.
For group names see text.
| Groups | WGMD | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Antillean Bothrops | 3.9\4.5 | 5.0 | 4.9 | 4.0 | 3.7 | 4.8 | 4.9 | 5.2 | 4.6 | 4.5 | 4.4 | 4.3 | |
| 2 | B. atrox group | 2.6\2.9 | 4.1 | 5.0 | 4.5 | 4.3 | 4.4 | 5.2 | 5.9 | 5.0 | 5.0 | 4.5 | 5.1 | |
| 3 | MY | 0.1\0.2 | 5.3 | 6.0 | 3.4 | 2.0 | 4.4 | 4.8 | 5.1 | 4.5 | 4.6 | 4.7 | 4.7 | |
| 4 | NCA | 0.0\01 | 4.5 | 5.7 | 2.7 | 1.3 | 3.7 | 3.6 | 4.9 | 3.5 | 3.7 | 3.7 | 3.8 | |
| 5 | CICA | 0.1\0.2 | 4.6 | 4.6 | 4.0 | 3.9 | 3.4 | 3.8 | 4.6 | 3.3 | 3.3 | 3.4 | 3.4 | |
| 6 | TR | 0.0\0.0 | 3.7 | 3.1 | 4.9 | 4.3 | 3.1 | 3.1 | 4.8 | 4.4 | 4.3 | 4.3 | 4.8 | |
| 7 | WE | 0.0\0.1 | 4.5 | 5.0 | 6.0 | 4.3 | 3.9 | 2.6 | 4.8 | 4.4 | 4.8 | 4.3 | 4.4 | |
| 8 | PICA | 0.4\0.6 | 5.4 | 5.4 | 7.5 | 7.5 | 6.5 | 5.9 | 6.1 | 4.0 | 4.3 | 3.8 | 3.9 | |
| 9 | CHOCO1 | 0.2\0.4 | 4.5 | 3.4 | 5.3 | 6.0 | 4.9 | 3.6 | 5.1 | 4.1 | 1.5 | 2.5 | 3.0 | |
| 10 | CHOCO2 | 0.3\0.2 | 4.7 | 3.9 | 6.7 | 6.6 | 5.1 | 4.0 | 4.9 | 3.0 | 2.4 | 2.9 | 3.4 | |
| 11 | CCO | 0.6\0.3 | 6.3 | 6.1 | 8.2 | 7.4 | 6.2 | 5.7 | 5.7 | 4.8 | 4.8 | 5.4 | 2.4 | |
| 12 | MV | 0.1\0.3 | 5.2 | 5.2 | 6.5 | 6.5 | 5.3 | 5.6 | 5.7 | 3.9 | 3.0 | 4.5 | 1.8 |
WGMD, Within Group Mean Distance.
Within B. asper groups, we observed the highest divergence among individuals in PICA (mean p-distances = 0.4% and 0.6%, for MT-CYB and MT-ND4 gene, respectively); whereas we recorded the lowest value within the MV group (mean p-distances = 0.1 and 0.3%, for both genes).
We observed valuable sample sizes for all parameters in all BEAST analyses and our estimation of convergence statistics in Tracer indicated that all analyses had converged (ESS > 384). The root state posterior probabilities for all locations range between <0.01 and 0.78, with Colombia receiving the highest probability (Fig 3). Thus, the ancestral branch that led to the B. atrox complex exhibited high probability for that locality (P > 0.78), although it is somewhat lower in the branch leading to B. asper (P > 0.5, Fig 3). Similarly, the probability that Colombia was the state at the root of Clade B is high (P = 0.63), despite the low probability observed in the branch leading to PICA. On the other hand, neither Colombia nor any other locality dominates the probabilities associated with the root of Clade A, and therefore the location state of its ancestor remains unclear.
Divergence times
According to our Bayesian estimations of divergence time (Fig 4) the origin of the B. asper-B. atrox group occurred approximately 3.91 Mya (CI95% = 2.53 to 5.26 Mya) when this clade diverged from the Antillean lanceheads (B. caribbaeus and B. lanceolatus). The divergence between the B. atrox species group and the B. asper lineages is estimated to have occurred soon after, about 3.34 (CI95% = 2.44 to 4.48 Mya). The separation between B. asper clades A and B is dated at mid-Pliocene, approximately 3.20 Mya (CI95% = 2.32 to 4.31 Mya). Within clade A, the Caribbean Middle American groups diverged from those in Southwestern Ecuador almost simultaneously, as the estimation time is 2.80 Mya (CI95% = 1.97 to 3.88 Mya).
The divergence between sister groups (MY-NCA/CICA) and (WE/TR) occurred more recently approximately 1.74 Mya (CI95% = 1.10 to 2.51 Mya), at the Pliocene-Pleistocene boundary (Fig 4).
Within clade B, lineages from the Choco biogeographic region diverged from all other groups about 2.38 Mya (CI95% = 1.66 to 3.30 Mya) coinciding with the separation between PICA and the groups located east of the Colombian Andes (CCO and MV, Fig 3). Finally, the divergence estimates between MV and CCO and between the Choco-Ecuador and Choco Colombia-Panama groups occurred well into the Pleistocene (Fig 4).
Testing for demographic expansion
A total of 41 and 47 unique haplotypes were recovered from MT-CYB and MT-ND4 sequences respectively. MY, CICA, and CHOCO exhibit a relatively higher number of distinct haplotypes (5, 7, and 9, respectively), whereas we noticed only four in MV, despite this last group has the most extensive sample size (Table 2).
Table 2. Mistmatch distribution statistics (MT-CYB and MT-ND4) for B. asper phylogroups.
Group names as in Fig 1. No data available for TR and CHOCO 2 due to small sample sizes.
| Groups | N CYB/ND4 |
Mismatch distribution |
Rg | R2 | D* |
Tajima's D (θT) |
|---|---|---|---|---|---|---|
| MY | 7/4 | Unimodal | 0.33/0.25 | 0.14/0.27 | 0.23/0.59 | -0.88/0.59 |
| NCA | 9/9 | Unimodal | 0.07/0.18 | 0.18/0.22 | 0.23/-1.68 | 0.20/-1.51 |
| CICA | 8/6 | Unimodal | 0.14/0.06* | 0.12/0.25* | -0.72/-1.26 | -0.70/-1.23 |
| WE | 5/5 | Bimodal | 0.68/0.05 | 0.40/0.27 | 0.23/-1.05 | -0.97/-1.05 |
| PICA | 13/11 | Bimodal | 0.14/0.06 | 0.11/0.15 | -0.55/0.49 | -0.56/0.01 |
| CHOCO1 | 16/11 | Unimodal | 0.02/0.04 | 0.13/0.09* | -0.59 /-1.71 | -0.56/-1.44 |
| CCO | 12/11 | Unimodal | 0.04/0.09 | 0.11/0.14 | -0.79/-0.44 | -0.64/-0.80 |
| MV | 5/30 | Unimodal | 0.50/0.07 | 0.13/0.08 | -3.32*/-3.46* | -2.09*/-2.07* |
Except for WE and PICA, all other studied groups showed unimodal patterns of mismatch distribution curves (Table 2). Also, most groups exhibited low values for the neutrality test statistics applied here (Table 2). These combined results are often interpreted as an indication of selective sweep or population expansion [63–64]. However, in our analysis only MV show significant values for the Fu and Li’s D* and Tajima’s ɵ, therefore supporting signs of recent demographic growth in that group (Table 2).
Discussion
Bothrops asper exhibits robust genetic partitioning that accounts for at least ten distinct mitochondrial phylogroups, included in two separate lineages. The groups occupy different geographic regions and show private haplotypes, indicating a clear division among them. We found evidence of transition zones only in Peten (Guatemala) and in central Panama, but in both cases, contact occurs between sibling groups within a single B. asper lineage (MY/NCA and PICA/CHOCO1, respectively). Conversely, we did no observe mitochondrial admixture between B. asper lineages, nor even in Isthmian Central America where they converge. Additional sampling in this region, especially in western Panama, could help to establish the extent to which these lineages are introgressing and the precise geographical boundaries in this apparent admixture zone.
Phylogeographic history
The observed molecular variation might, in part, result from the complicated geological and climatic history that shaped the distribution of the species during the Pliocene-Pleistocene, in addition to the species own ability to spread into the new empty niches that arose during that period.
As stated before, the sister relationship between B. asper and the B. atrox group has been established previously [27, 34, 65]; and a monophyletic B. asper was recovered in our BEAST estimations of divergence time and the putative ancestral distributions of our clades. Our estimates of divergence time suggest that the B. asper-B. atrox group ancestor was distributed in South America no older than 4.48 Mya (Fig 4). Also, BPA analysis resulted in Colombia as that the most probable 'state of locality' for the deep branches in the phylogeny of B. asper (Fig 3). We interpret these combined results as evidence that the diversification of B. asper occurred in the northwestern region of South America by the early Pliocene.
The allopatric distribution of B. atrox group and B. asper lineages on each side of the Eastern Andes Cordillera, suggests that the final uplift of this mountain range played a significant role in the cladogenesis of these lanceheads. Thus, this Andean Cordillera reached no more than 40% of its present elevation during much of the Neogene, but intense mountain building followed in the late Miocene and, especially in the Pliocene when elevations increased rapidly [70–71]. As a result, we hypothesize that a B. asper stock diverged to the west of the Eastern Andes, likely within the foothills along the Pacific coast of northern South America, as supported by the relatively high number of haplotypes observed in that region.
During that time, this ancient stock was further isolated by several marine incursions in northern South America, especially at the Magdalena Valley and the Maracaibo basin to the west, and the presence of three sea corridors in lower Central America to the north: the Atrato seaway, the Panama Portal, and the San Carlos Basin [72]. These marine transgressions resulted from changes in sea level that occurred during the warm climate periods of the Pliocene [73–75], but also during the Quaternary interglacial periods when warmer and wetter climates dominated again. Isolation during the cycles of sea incursions allowed rapid species evolution in the trans-Andean region, as has been recognized for several organisms by Nores [76], and is feasible that similar forces managed the divergence within B. asper.
Considering its South American origin, and given the low posterior probabilities for the ancestral locality that we retrieved for the root of Clade A (Fig 3), we postulated that the early separation between lineages in B. asper was driven by a dispersal event north into Caribbean Mesoamerica. In addition, the divergence between lineages in mid-Pliocene coincides with estimations of the final closure of Isthmian Central America, a dynamic event that, as previously mentioned, has shaped the region’s biogeography.
There is still considerable debate regarding the dynamics and timing of the final closure of Isthmian Central America. Some authors argue that the Isthmus arose as a series of islands in a shallow sea with a concluding land connection established in the late Pliocene [9, 10]; whereas others suggest that it emerged as a continuous land bridge since the Miocene [11, 72, 77], only broken by the three previously mentioned sea gates that eventually followed a north-to-south closure at the end of the Neogene [78]. Regardless of the paleogeographic model of emergence, colonization of Caribbean Mesoamerica by B. asper may have entailed crossing water corridors, a challenge that lancehead pitvipers seem suitable to carry out [65]. In support of this view, present-day surface currents in the southern Caribbean are mostly directed toward the northwest [79], and passive transportation from the north Caribbean coast of South America to northern Mesoamerica has been demonstrated experimentally [80].
According to our divergence time estimations, the separation within the South American clade B started in late Pliocene, when a group reached the Pacific coast of Isthmian Central America (PICA). The route involved in this expansion to Mesoamerica is unclear, but most likely it also involved passive transportation through the Atrato seaway when global climate was warm [81]. By then, the Talamanca mountain range in Isthmian Central America isolated this population from the Caribbean lowlands [82, 83]. This hypothesis explains why Caribbean and Pacific B. asper populations in Mesoamerica are not sister taxa. Close affinities between lineages along the Pacific slope of Isthmian Central America and those in the Choco region in Colombia have also been reported in other taxa: frogs [84, 85], birds [86] and snakes [23, 87], suggesting a comparable history of colonization.
Hence, we postulate that B. asper invaded Mesoamerica in at least two, chronologically separated independent events. This trend coincides with the two-step dispersal-pulse hypothesis proposed by Savage [88] to explain the unequal distribution of amphibians and reptiles of South American origin inhabiting Middle America. According to this author, the first and earlier episode took place some 3.4 MYA when the sea level lowered [89], and a handful of South American taxa were able to invade Mesoamerica before the final closure of the isthmus. Consequently, few South American taxa have distributions that extend as far north as Mexico. Savage’s [88] second dispersion episode presumably occurred almost a million years later, after the final closure of the Isthmian bridge. As an outcome of this late invasion, the majority of genera of South American origin have distributions that do not reach beyond southern Nicaragua or Costa Rica. Chronologically, Savage’s first pulse corresponds to the split of B. asper lineages, whereas his second pulse matches the invasion of B. asper from South America to PICA.
Alternatively, a single dispersal event into Middle America is also possible, especially if incomplete lineage sorting [90] or secondary (interspecific) gene flow is contemplated. Both are distinct phenomena but produce very similar patterns of shared genetic diversity that in turn could affect species integrity.
During range expansion, some alleles could increase their frequencies due to gene drift, thus, “surfing” into fixed spatial sectors at the expanding front. Lineages that arise as a consequence of demographic expansion are temporary but tend to last longer in species with limited dispersal abilities [91]. Genetic surfing has been shown to explain patterns of spatial assorting of gene variation in loci with weak effective dispersal, such as mitochondrial DNA. Thus, genetic surfing should be more frequent in species that exhibit male-biased dispersal, and that have relatively low vagility, as it has been recently reported in the coral snake Micrurus tener [92]. Although the dispersal patterns of B. asper are unknown, male-biased dispersal is expected in this species, as it has been described in several other species of vipers, comprising both crotalines [93] and viperines [94].
Our mismatch distribution analyses for demographic expansion provide evidence that MV and CCO groups represent recent populations that diverged in the early Pleistocene. Thus, it is possible that the sorting of these haplotype groups resulted from gene surfing and not due to divergence in allopatry. However, no further evidence for demographic expansion was recovered in other populations, not even in those located at the northern extreme of the species distribution. Furthermore, we did not observe divergent mitochondrial haplotypes in sympatry in the putative ancestral populations, which is a crucial component to evidence of gene surfing, therefore limiting our interpretation. Further sampling of these populations and the inclusion of genome-wide markers in the analyses could resolve the question of whether the spatial sorting of haplotype groups in B. asper resulted from allopatric divergence or genetic surfing during demographic expansion.
Another possible scenario is that mtDNA introgression from B. atrox into B. asper occurred in the past, as this could explain the paraphyletic relation of this last species retrieved in our analyses (Fig 2). Introgression between sibling species is more widespread than previously thought and is reported in a great variety of plant and animal taxa [95, 96] including snakes [97, 98]. Ancient introgression could occur even if no evidence of shared haplotypes is available, as has been recently reported by Ruane et al. [98] for milk snakes genus Lampropeltis. For species that exhibit male-biased dispersal, as expected in B. asper, rates of introgression in mtDNA markers are often higher compared with those from biparentally inhered nuclear DNA markers [95]. Unfortunately, we did not include nuclear markers in our analysis, and none of our mitochondrial haplotypes were shared among our phylogroups, thus precluding further comparisons to evaluate introgression.
Relation between Caribbean Middle American and Ecuadorian groups
The observed sister relationship between MNCA/CICA and the WE/TS groups depicted in clade A (Fig 2) seems incongruous, as no biogeographical connection has ever been suggested between the mesic forests of Caribbean Middle American and the seasonally dry environments of southwestern Ecuador [99]. Biogeographically, the northern portion of Caribbean Middle America is dominated by Mesoamerican influences whereas southwestern Ecuador has influences from the Pacific dominion [100]. Therefore, biotic similarities of this last region are expected to be mainly with the geographically proximal Chocoan biogeographic region, as has been described for bats [101] and woody plants [102]. It is possible that observed affinities resulted from retention of ancestral polymorphisms followed by lineage sorting, but the sample size of our Ecuadorian populations prevent further analysis at this point.
Phenotypic variation in B. asper
Across Mesoamerica, B. asper populations show remarkable differences in natural history traits [103], morphological variation [104, 105], venom composition [106, 107], and biological effects of venoms [108–110]. The extent of intraspecific variation in B. asper venoms could affect the capacity of antivenoms to neutralize toxins from snakes of geographically separated populations [111]; thus in Costa Rica polyvalent antivenom is produced from a mixture that includes B. asper venoms from the Caribbean and Pacific regions of that country [112, 113]. All these features suggest independence in the evolutionary history of at least the two B. asper lineages revealed in this study. In fact, Aragón and Gubensek [114] have suggested that given that the differences in morphology, toxin composition, and biological effect of venoms are so notable, populations in the Central American Isthmus warrant taxonomic recognition. Further, sequence divergences observed among B. asper Caribbean Mesoamerican populations and those in Pacific/South American (5.8 ± 0.2%) are on par to those reported among different species in other snake groups: Lachesis stenophrys and L. melanocephala with a 5.3% [115] the Naja nigricollis species complex between 4.7% to 8.3% [116], and the genus Agkistrodon between 4.0% to 6.4% [28].
Nevertheless, we refrain from proposing any taxonomic changes for these B. asper lineages at present, as their phylogenetic relationships with the B. atrox group remains unclear. Further phylogenetic resolution of our tree could be attained using other sequence markers, such as nuclear genes, as studies in birds [117], mammals [118, 119] and reptiles [120] suggest. These characters exhibit low homoplasy levels and are known to be useful in recovering phylogenetic information from taxonomic groups that have experienced adaptive radiation in a short period [119], therefore resulting in more robust phylogenies. Also, a more thorough sampling that includes other localities from Panama and especially from Venezuela and Ecuador might be necessary to further resolve the phylogenetic affinities within the group.
Concluding remarks
Divergence within B. asper lineages appears to result from the tectonic events and sea transgressions that have shaped Mesoamerica and northern South America since the early Pliocene. Our findings support the view that B. asper invaded Mesoamerica multiple times during the complex history of the final closure of Isthmian portal, and that Savage’s [88] view of two invasion pulses to explain differential distribution of taxa of South American origin in Mesoamerica might hold for this species. However, our data suggest that Bothrops asper, as currently understood, probably consists of a complex of related lineages that follow their evolutionary trajectories. A more robust revision of these lineages and their taxonomic status is presently underway.
Supporting information
Accession Numbers With Asterisks Are Sequences Obtained Of GeneBank.
(DOCX)
Acknowledgments
Twenty years ago, J.A. Campbell asked one of us how to interpret the current distribution pattern of B. asper in Mesoamerica. In part, this study was inspired by that question. J.A. Campbell, F. Joyce, V. Nuñez, and U. Kuch contributed with tissues and information. V. Martinez from Universidad de Panama kindly contributed with tissues and important information from that country. M.E. Barragán, C. Barrio-Amorós, J.P. Diasparra, A. Freire-Lascano, T. Garel, A. Mijares-Urrutia, J.A. Quijada-Mascareñas, D.C. Taphorn, R.D.G Theakston, L. Thomas, J.-M. Touzet, G.L. Underwood, D.A. Warrell, and J.L. Yrausquin helped with additional sampling and information. C. Bravo, J. Serrano and D. Zuñiga kindly assisted with figures. M.A. Méndez allowed the use of his molecular facilities at Universidad de Chile and contributed with early developments of this project. W. Lamar, D. Wasko, and S. Vargas substantially improved the manuscript with their comments and suggestions. W. Lamar, F. Bonilla, and A. Solórzano are acknowledged for spending many nights looking for the cryptic B. asper in the rainforest of Lower Middle America. Research permits were issued by J. Guevara (Oficina Ventanilla Unica, MINAE, Costa Rica), R. Manzanero (Belize Forest Department, to WW), Instituto Ecuatoriano Forestal y de Áreas Naturales y de Vida Silvestre (INEFAN, to WW), and Dirección de Licencias, Permisos y Trámites Ambientales, Ministerio de Medio Ambiente, Vivienda y Desarrollo Territorial de Colombia (Resolución No.1392 2008 to MMSC).
Data Availability
All relevant data are within the paper and its Supporting Information files. Accession numbers for gene sequences within GenBank are given in S1 Table.
Funding Statement
The research presented here was partially funded by a doctoral grant from CONICYT to MS-C (Proyecto Beca AT-24060101); a grant from Vicerrectoría de Investigación, UCR 741-A5-050 to MS; EU contracts TS3-CT91-0024 and IC18-CT96-OO32 and a Wellcome Trust Research Career Development Fellowship to WW; and by National Science Foundation grants DEB- 0416000 and 1638879 to CLP.
References
- 1.Davidse G, Sousa MS, Knapp S. Flora Mesoamericana, Volumen 1, Psilotaceae a Salviniaceae. Universidad Nacional Autónoma de México/Missouri Botanical Garden/The Natural History Museum, México, D.F; 1995. [Google Scholar]
- 2.Perlo BV. Birds of Mexico and Central America. Princeton University Press; 2006. [Google Scholar]
- 3.Wilson LD, Johnson JD. Distributional patterns of the herpetofauna of Mesoamerica, a biodiversity hotspot In: Wilson LD, Townsend JH, Johnson JD, editors. Conservation of Mesoamerican Amphibians and Reptiles. Utah: Eagle Mountain Publishing; 2010. pp. 30–235. [Google Scholar]
- 4.Graham A . In the Beginning: Early Events in the Development of Mesoamerica and the Lowland Maya Area In: Gómez–Pompa A, Allen MF, Fedick SL, Jiménez–Osorio JJ, editors. The Lowland Maya Area three millennia at the human-wildland human-wildland interface. New York: Food Products Press an Imprint of the Haworth Press; 2003. pp. 31–44. [Google Scholar]
- 5.Marshall JS. The geomorphology and physiographic provinces of Central America In: Bundschuh J, Alvarado G, editors. Central America: Geology, Resources, and Hazards. London: Taylor and Francis; 2007. pp. 75–122. [Google Scholar]
- 6.Leigh EG, O’Dea A, Vermeij GJ. Historical biogeography of the isthmus of Panama. Biol Rev. 2014; 148–172. [DOI] [PubMed] [Google Scholar]
- 7.Iturralde-Vinent MA. Meso-Cenozoic Caribbean paleogeography: implications for the historical biogeography of the region. Int Geol Rev. 2006; 48: 791–827. [Google Scholar]
- 8.O’Dea A, Jackson JBC, Fortunato H, Smith JT, D’Croz L, Johnson KG, et al. Environmental change preceded Caribbean extinction by 2 million years. PNAS. 2007; 104: 5501–5506. doi: 10.1073/pnas.0610947104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coates AG, Obando JA. The geological evolution of the Central American Isthmus In: Jackson JBC, Budd AF, Coates AG, editors. Evolution and Environment in Tropical América. Chicago: University of Chicago Press; 1996. pp. 21–56. [Google Scholar]
- 10.O’Dea A, Lessios HA, Coates AG, Eytan RI, Restrepo-Moreno SA, Cione AL, et al. Formation of the Isthmus of Panama. Science Advances. 2016; 2(8), e1600883 doi: 10.1126/sciadv.1600883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montes C, Cardona A, Jaramillo C, Pardo A, Silva JC, Valencia V, et al. Middle Miocene closure of Central American seaway. Science. 2015; 348: 226–229. doi: 10.1126/science.aaa2815 [DOI] [PubMed] [Google Scholar]
- 12.Savage JM. The enigma of the Central American herpetofauna: dispersals or vicariance?. Ann Mo Bot Gard. 1982; 69: 464–547. [Google Scholar]
- 13.Campbell JA. Distributional patterns of amphibians in Middle America In: Duellman WE, editor. Patterns of distribution of amphibians: a global perspective. USA: The Johns Hopkins University Press; 1999. pp. 111–157. [Google Scholar]
- 14.Rich PV, Rich TH. The Central American dispersal route: Biotic history and Paleogeography In: Janzen H, editor. Costa Rican Natural History. Chicago: University of Chicago Press; 1983. pp. 12–34. [Google Scholar]
- 15.Webb SD. Ecogeography and the Great American Interchange. Paleobiology. 1991; 17: 266–280. [Google Scholar]
- 16.Webb SD, Rancy A. Late Cenozoic evolution of the Neotropical mammal fauna In: Jackson JBC, Budd AF, Coates AG, editors. Evolution and Environmental in Tropical America. Chicago: University of Chicago Press; 1996. pp.335–358. [Google Scholar]
- 17.Bermingham E, Martin AP. Comparative mtDNA phylogeography of Neotropical freshwater fishes: testing shared history to infer the evolutionary landscape of lower Central America. Mol Ecol.1998; 7: 499–517. [DOI] [PubMed] [Google Scholar]
- 18.Smith BT, Klicka J. The profound influence of the Late Pliocene Panamanian uplift on the exchange, diversification, and distribution of New World birds. Ecography. 2010; 33: 333–342. [Google Scholar]
- 19.Daza JM, Castoe TA, Parkinson CL. Using regional comparative phylogeographic data from snake lineages to infer historical processes in Middle America. Ecography. 2010; 33: 343–354. [Google Scholar]
- 20.Bacon CD, Mora A, Wagner WL, Jaramillo CA. Testing geological models of evolution of the Isthmus of Panama in a phylogenetic framework. Bot J Linn Soc. 2013;171:287–300. [Google Scholar]
- 21.Gutiérrez-García TA, Vázquez-Domínguez E. Consensus between genes and stones in the biogeographic and evolutionary history of Central America. Quaternary Res. 2013; 79:311–324. [Google Scholar]
- 22.Bagley JC, Johnson JB. Phylogeography and biogeography of the lower Central American Neotropics: diversification between two continents and between two seas. Biol Rev. 2014; 89:767–790 doi: 10.1111/brv.12076 [DOI] [PubMed] [Google Scholar]
- 23.Castoe TA, Sasa M, Parkinson CL. Modeling nucleotide evolution at the mesoscale: The phylogeny of the Neotropical pitvipers of the Porthidium group (Viperidae: Crotalinae). Mol Phylogenet Evol. 2005; 37: 881–898. doi: 10.1016/j.ympev.2005.05.013 [DOI] [PubMed] [Google Scholar]
- 24.Castoe TA, Daza JM, Smith EN, Sasa M, Kuch U, Campbell JA, et al. Comparative phylogeography of pitvipers suggests a consensus of ancient Middle American highland biogeography. J Biogeogr. 2009;36: 88–103. [Google Scholar]
- 25.Bryson RW Jr., Murphy RW, Lathrop A, Lazcano D. Evolutionary drivers of phylogeographical diversity in the highlands of Mexico: A case study of the Crotalus triseriatus species group of montane rattlesnakes. J Biogeogr. 2011. 38; 697–710. [Google Scholar]
- 26.Parkinson CL. Molecular systematics and biogeographical history of pitvipers as determined by mitochondrial ribosomal DNA sequences. Copeia. 1999; 3:576–586. [Google Scholar]
- 27.Parkinson CL, Campbell JA, Chippindale PT. Multigene phylogenetic analyses of pitvipers; with comments on the biogeographical history of the group In: Schuett GW, Höggren M, Douglas ME. Greene HW, editors. Biology of the Vipers. USA: Eagle Mountain Publishing; 2002. a. pp. 93–110. [Google Scholar]
- 28.Parkinson CL, Zamudio KR, Greene HW. Phylogeography of the pitviper clade Agkistrodon: historical ecology, species status, and conservation of cantils. Mol Ecol. 2002b; 9: 411–420. [DOI] [PubMed] [Google Scholar]
- 29.Gutberlet RL Jr., Harvey MB. The evolution of New World venomous snakes In: Campbell JA, Lamar WW, editors. The Venomous Reptiles of the Western Hemisphere. New York: Cornell University Press; 2004. pp. 634–682. [Google Scholar]
- 30.Castoe TA, Parkinson CL. Bayesian mixed models and the phylogeny of pitvipers (Viperidae: Serpentes). Mol Phylogenet Evol. 2006; 39: 91–110. doi: 10.1016/j.ympev.2005.12.014 [DOI] [PubMed] [Google Scholar]
- 31.Murphy RW, Lathrop JFA, Feltham JV, Kovac V. Phylogeny of the rattlesnakes (Crotalus and Sistrurus) inferred from sequences of five mitochondrial DNA genes In: Schuett GW, Höggren M, Douglas ME, Greene HW, editors. Biology of the Vipers.USA: Eagle Mountain Publishing; 2002. pp. 69–92. [Google Scholar]
- 32.Wüster W, Ferguson JE, Quijada-Mascareñas JA, Pook CE, Salomão MG, Thorpe RS. Tracing an invasion: landbridges, refugia, and the phylogeography of the Neotropical rattlesnake (Serpentes: Viperidae: Crotalus durissus). Mol Ecol. 2005; 14: 1095–1108. doi: 10.1111/j.1365-294X.2005.02471.x [DOI] [PubMed] [Google Scholar]
- 33.Campbell JA, Lamar WW. The venomous Reptiles of the western Hemisphere Cornell University Press, Ithaca, NY; 2004. [Google Scholar]
- 34.Wüster W, Salomão MG, Quijada-Mascareñas JA, Thorpe RS, R.S.BBBSP. Origin and evolution of the South American pitviper fauna: evidence from mitochondrial DNA sequence data In: Schuett GW, Höggren M, Douglas ME, Greene HW, editors. Biology of the Vipers. USA: Eagle Mountain Publishing; 2002. a. pp. 111–128. [Google Scholar]
- 35.Jowett T. Preparation of nucleic acids In: Roberts DB, editor. Drosophila: a Practical Approach. Oxford: IRL Press; 1986. pp. 275–286. [Google Scholar]
- 36.Arévalo E, Davis SK, Sites JW Jr. Mitochondrial DNA sequence divergence and phylogenetic relationships among 8 chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in Central México. Syst Biol.1994; 43: 387–418. [Google Scholar]
- 37.Palumbi SR. Nucleic acids II: The polymerase chain reaction In: Hillis DM, Moritz C, Mable BK, editors. Molecular systematics. Sinauer Associates, Inc; 1996. pp. 205–247. [Google Scholar]
- 38.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser.1999; 41: 95–98. [Google Scholar]
- 39.Nicholas KB, Nicholas HB Jr., Deerfield DW. GeneDoc: analysis and visualization of genetic variation. Embnewnews. 1997; 4(14). [Google Scholar]
- 40.Triant DA, Dewoody JA. The occurrence, detection, and avoidance of mitochondrial DNA translocations in mammalian systematics and phylogeography. J Mammal. 2007; 88: 908–920. [Google Scholar]
- 41.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33:1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neath AA, Cavanaugh JE. The Bayesian information criterion: background, derivation, and applications. WIREs Comp Stat. 2012; 4: 199–203. [Google Scholar]
- 43.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford Univ. Press; 2000. [Google Scholar]
- 44.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1995; 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 45.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003; 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 46.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6. 2014. Available from http://beast.bio.ed.ac.uk/Tracer.
- 47.Zhang J, Kapli P, Pavlidis P, Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013; 29: 2869–2876. doi: 10.1093/bioinformatics/btt499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comp Biol. 2009; 5: 1–16, e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemey P, Rambaut A, Welch JJ, Suchard MA. Phylogeography takes a relaxed random walk in continuous space and time. Mol Biol Evol. 2010; 27: 1877–1885. doi: 10.1093/molbev/msq067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, et al. BEAST 2: A Software platform for Bayesian evolutionary analysis. Plos Comput Biol. 2014;10(4): e1003537 doi: 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol. 1998; 15: 1647–1657. [DOI] [PubMed] [Google Scholar]
- 52.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002; 51: 689–702. doi: 10.1080/10635150290102456 [DOI] [PubMed] [Google Scholar]
- 53.Parmley D, Holman JA.2007. Earliest fossil record of a pigmy rattlesnake (Viperidae: Sistrurus Garman). J Herpetol. 2007; 41: 141–144. [Google Scholar]
- 54.Holman JA. Fossil snakes of North America: origin, evolution, distribution, paleoecology Indiana Univ. Press; 2000. [Google Scholar]
- 55.Ho SYW, Phillips MJ. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Systematic Biol. 2009; 58:367–380. [DOI] [PubMed] [Google Scholar]
- 56.Castoe TA, Spencer CL, Parkinson CL. Phylogeographic Structure and Historical Demography of the Western Diamondback Rattlesnake (Crotalus atrox): a Perspective on North American Desert Biogeography. Mol Phylogenet Evol. 2007; 43:193:212. [DOI] [PubMed] [Google Scholar]
- 57.Carreño AL, Helenes J. Geology and ages of the islands In: Case TJ, Cody ML, Ezcurra E, editors. A new island biogeography of the Sea of Cortes. New York: Oxford University Press; 2002. pp. 14–40. [Google Scholar]
- 58.Hafner DJ, Riddle BR. Mammalian phylogeography and evolutionary history of northern Mexico’s deserts. In: Cartron JL, Ceballos G, Felger RS, editors. Biodiversity, ecosystems, and conservation in northern Mexico. Oxford; 2005. pp. 225–245.
- 59.Roger AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992; 9:552–569. [DOI] [PubMed] [Google Scholar]
- 60.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010; 10: 564–567. doi: 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 61.Harpending HC, Sherry ST, Rogers AR, Stoneking M. Genetic structure of ancient human populations. Curr Anthr. 1993;34: 483–496. [Google Scholar]
- 62.Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Mol Biol Evol. 2006; 23: 1642. [DOI] [PubMed] [Google Scholar]
- 63.Fu YX, Li WH. Statistical tests of neutrality of mutations.Genetics.1993; 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simonsen KL, Churchill GA, Aquadro CF. Properties of statistical test of neutrality for DNA polymorphism data. Genetics. 1995; 141: 413–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wüster W, Thorpe RS, Salomão MG, Thomas L, Puorto G, Theakston RDG, et al. Origin and phylogenetic position of the Lesser Antillean species of Bothrops (Serpentes, Viperidae): biogeographical and medical implications. Bull Br Mus Nat Hist Zool. 2002b; 68: 101–106. [Google Scholar]
- 66.Fenwick AM, Gutberlet RL, Evans JA, Parkinson CL. Morphological and molecular evidence for phylogeny and classification of South American pitvipers, genera Bothrops, Bothriopsis, and Bothrocophias (Serpentes: Viperidae). Zool J Linn Soc-Lond. 2009; 156: 617–640. [Google Scholar]
- 67.Jadin RC, Gutberlet RL Jr., Smith EN. Phylogeny, evolutionary morphology, and hemipenis descriptions of the Middle American jumping pitvipers (Serpentes: Crotalinae: Atropoides). J Zoolog Syst Evol Res. 2010; 48: 360–365. [Google Scholar]
- 68.Carrasco PA, Mattoni CI, Leynaud GC, Scrocchi GJ. Morphology, phylogeny and taxonomy of South American bothropoid pitvipers (Serpentes, Viperidae). Zool Scr. 2012; 41:109–124. [Google Scholar]
- 69.Morrone J. Biogeograhical regionalisation of the Neotropical region. Zootaxa. 2014; 3782: 1–110. doi: 10.11646/zootaxa.3782.1.1 [DOI] [PubMed] [Google Scholar]
- 70.Gregory-Wodzicki KM. Uplift history of the central and northern Andes a review. Geol Soc Am Bull. 2000; 112: 1091–1105. [Google Scholar]
- 71.Collins LS, Coates AG, Berggren WA, Aubri MP, Zhang J. The late Miocene Panama Isthmian strait. Geology.1996; 24:687–690. [Google Scholar]
- 72.Schmidt DN. The closure history of the Central American seaway: evidence from isotopes and fossils to models and molecules In: Williams M, Haywood AM, Gregory FJ, Schmidt DN, editors. Deep-Time Perspectives on Climate Change: Marrying the Signal from Computer Models and Biological Proxies. London: The Geological Society, London; 2007. pp. 429–444. [Google Scholar]
- 73.Dowsett HJ, Cronin TM. High eustatic sea level during the middle Pliocene: Evidence from the southeastern U.S. Atlantic Coastal Plain. Geology.1990; 18:435–438. [Google Scholar]
- 74.Dowsett HJ, Chandler MA, Cronin TM, Dwyer GS. Middle Pliocene sea surface temperature variability. Paleoceanography. 2005. 20, PA2014, doi: 10.1029/2005PA001133 [Google Scholar]
- 75.Molnar P, Cane MA. El Niño’s tropical climate and teleconnections as a blueprint for pre-Ice Age climates. Paleoceanography. 2002; 17(2), 1021, doi: 10.1029/2001PA000663 [Google Scholar]
- 76.Nores M. The implications of Tertiary and Quaternary sea level rise events for avian distribution patterns in the lowlands of northern South America. Global Ecol Biogeogr. 2004; 13:149–161. [Google Scholar]
- 77.Hoorn C, Wesselingh FP, Ter Steege H, Bermudez MA, Mora A, Sevink J, et al. Amazonia through time: Uplift, climate change, landscape evolution and biodiversity. Science. 2010; 33:927–931. [DOI] [PubMed] [Google Scholar]
- 78.Kirby MX, MacFadden B. Was southern Central America an archipelago or a peninsula in the middle Miocene? A test using land-mammal body size. Palaeogeogr Palaeocl. 2005; 228:193–202. [Google Scholar]
- 79.Iturralde-Vinent MA, Macphee RDE. Paleogeography of the Caribbean region: Implications for Cenozoic biogeography. Bull Am Mus Nat Hist.1999; 238:1–95. [Google Scholar]
- 80.Brucks JT. Currents of the Caribbean and adjacent regions as deduced from drift-bottle studies. Bull. Mar. Sci. 1971; 21:455–465. [Google Scholar]
- 81.Fleming RF, Barron JA. Evidence of Pliocene Nothofagus in Antarctica from Pliocene marine sedimentary deposits (DSDP Site 274). Mar Micropaleontol.1996; 27: 227–236. [Google Scholar]
- 82.Keigwin LD Jr. Isotopic paleoceanography of the Caribbean and East Pacific: Role of Panama uplift in late Neogene time. Science. 1982; 217: 350–353. doi: 10.1126/science.217.4557.350 [DOI] [PubMed] [Google Scholar]
- 83.MacMillan I, Gans P, Alvarado G. Middle Miocene to present plate tectonic history of the southern Central American Volcanic Arc. Tectonophysics. 2004;392:325–348. [Google Scholar]
- 84.Crawford AJ, Bermingham E, Polanía C. The role of tropical dry forest as a long-term barrier to dispersal: a comparative phylogeographic analysis of dry forest tolerant and intolerant frogs. Mol Ecol. 2007; 16: 4789–4807. doi: 10.1111/j.1365-294X.2007.03524.x [DOI] [PubMed] [Google Scholar]
- 85.Wang IJ, Crawford AJ, Bermingham E. Phylogeography of the Pigmy Rain Frog (Pristimantis ridens) across the lowland wet forest of Ishmian Central America. Mol Phylogenet Evol. 2008; 47: 992–1004. doi: 10.1016/j.ympev.2008.02.021 [DOI] [PubMed] [Google Scholar]
- 86.Cheviron ZA, Hackett SJ, Capparella AP. Complex evolutionary history of a Neotropical lowland forest bird (Lepidothrix coronata) and its implications for historical hypotheses of the origin of Neotropical avian diversity. Mol Phylogenet Evol. 2005;36: 338–357. doi: 10.1016/j.ympev.2005.01.015 [DOI] [PubMed] [Google Scholar]
- 87.Daza JM, Smith EN, Páez VP, Parkinson CL. Complex evolution in the Neotropics: the origin and diversification of the widespread genus Leptodeira (Serpentes: Colubridae). Mol Phylogenet Evol. 2009; 53: 653–667. doi: 10.1016/j.ympev.2009.07.022 [DOI] [PubMed] [Google Scholar]
- 88.Savage JM. The Amphibians and Reptiles of Costa Rica: A Herpetofauna between Two continents, between Two Seas. University of Chicago, Press, Chicago, IL; 2002. [Google Scholar]
- 89.Vail PR, Hardenbol J. Sea level changes during the Tertiary. Oceanus. 1979; 22:71–79. [Google Scholar]
- 90.Excoffier L, Foll M, Petit RJ. Genetic consequences of range expansions. Annual Review of Ecology, Evolution, and Systematics, 2009; 40: 481–501. [Google Scholar]
- 91.Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. TREE. 2008; 23: 347–351. doi: 10.1016/j.tree.2008.04.004 [DOI] [PubMed] [Google Scholar]
- 92.Streicher JW, McEntee JP, Drzich LC, Card DC, Schield DR, Smart U, et al. Genetic surfing, not allopatric divergence, explains spatial sorting of mitochondrial haplotypes in venomous coralsnakes. Evolution. 2016; 70: 1435–1449. doi: 10.1111/evo.12967 [DOI] [PubMed] [Google Scholar]
- 93.Clark RW, Brown WS, Stechert R, Zamudio K. Integrating individual behaviour and landscape genetics: the population structure of timber rattlesnake hibernacula. Molecular Ecology. 2008; 17: 719–730. doi: 10.1111/j.1365-294X.2007.03594.x [DOI] [PubMed] [Google Scholar]
- 94.Brito JC, Álvares FJ. Patterns of road mortality in Vipera latastei and V. seoanei from northern Portugal. Amphibian-Reptilia 2004; 25: 459–465. [Google Scholar]
- 95.Petit RJ, Excoffier L. Gene flow and species delimitation. Trends in Ecology & Evolution. 2009; 24: 386–393. [DOI] [PubMed] [Google Scholar]
- 96.Bryson JRW, De Oca ANM, Jaeger JR, Riddle BR. Elucidation of cryptic diversity in a widespread Nearctic treefrog reveals episodes of mitochondrial gene capture as frogs diversified across a dynamic landscape. Evolution, 2010; 64: 2315–2330. doi: 10.1111/j.1558-5646.2010.01014.x [DOI] [PubMed] [Google Scholar]
- 97.Bryson JRW, Pastorini J, Burbrink FT, Forstner MR. A phylogeny of the Lampropeltis mexicana complex (Serpentes: Colubridae) based on mitochondrial DNA sequences suggests evidence for species-level polyphyly within Lampropeltis. Molecular phylogenetics and evolution, 2007; 43:674–684. doi: 10.1016/j.ympev.2006.11.025 [DOI] [PubMed] [Google Scholar]
- 98.Ruane S, Bryson JRW, Pyron RA, Burbrink FT. Coalescent species delimitation in milksnakes (genus Lampropeltis) and impacts on phylogenetic comparative analyses. Systematic Biology, 2014; 63: 231–250. doi: 10.1093/sysbio/syt099 [DOI] [PubMed] [Google Scholar]
- 99.Burnham RJ, Graham A. The History of Neotropical vegetation: new developments and status. Ann Mo Bot Gard. 1999. 86; 546–589. [Google Scholar]
- 100.Morrone JJ. Presentación sintética de un nuevo esquema biogeográfico de América Latina y el Caribe. Red Iberoamericana de Biogeografía y Entomología Sistemática. Monografías Tercer Milenio. Vol. 2, SEA, Zaragoza, España;2002.
- 101.Candenillas-Ordinola RE. Diversidad, ecología y análisis biogeográfico de los murciélagos del Parque Nacional Cerros de Amotape,Tumbes Perú. M.Sc. Thesis. Universidad Nacional Mayor de San Marcos, Perú. 2010. Available from: http://cybertesis.unmsm.edu.pe/handle/cybertesis/2121.
- 102.Linares-Palomino R, Kvist LP, Aguirre-Mendoza Z, Gonzales-Inca C. Diversity and endemism of woody plant species in the Equatorial Pacific seasonally dry forests. Biodivers Conserv. 2010; 19:169–185. [Google Scholar]
- 103.Sasa M, Wasko DK, Lamar WW. Natural history of the terciopelo Bothrops asper (Serpentes: Viperidae) in Costa Rica. Toxicon. 2009; 54: 904–922. doi: 10.1016/j.toxicon.2009.06.024 [DOI] [PubMed] [Google Scholar]
- 104.Sasa M. Morphological variation in the lancehead pitviper Bothrops asper (Garman) (Serpentes: Viperidae) from Middle America. Rev Biol Trop. 2002; 50: 259–271. [PubMed] [Google Scholar]
- 105.Saldarriaga MM, Sasa M, Pardo R, Mendez MA. Phenotypic differences in a cryptic predator: factors influencing morphological variation in the terciopelo Bothrops asper (Garman, 1884; Serpentes:Viperidae). Toxicon. 2009; 54: 923–937. doi: 10.1016/j.toxicon.2009.05.031 [DOI] [PubMed] [Google Scholar]
- 106.Lomonte B, Carmona E. Individual expression patterns of myotoxin isoforms in the venom of the snake Bothrops asper. Comp Biochem Phys B. 1992; 102: 325–329. [DOI] [PubMed] [Google Scholar]
- 107.Alape-Girón A, Flores-Díaz M, Sanz L, Madrigal M, Escolano J, Sasa M, et al. Studies on the venom proteome of Bothrops asper: perspectives and applications.Toxicon. 2009; 54:938–948. doi: 10.1016/j.toxicon.2009.06.011 [DOI] [PubMed] [Google Scholar]
- 108.Segura A, Herrera M, Villalta M, Vargas M, Uscanga-Reynell A, Ponce de León-Rosales S, et al. Venom of Bothrops asper from Mexico and Costa Rica: Intraspecific variation and cross-neutralization by antivenoms. Toxicon. 2012; 59: 158–162. doi: 10.1016/j.toxicon.2011.11.005 [DOI] [PubMed] [Google Scholar]
- 109.Gutiérrez JM, Chaves F, Bolaños R. Estudio comparativo de venenos de ejemplares recién nacidos y adultos de Bothrops asper. Rev Biol Trop. 1980;28: 341–351. [PubMed] [Google Scholar]
- 110.Laines J, Segura A, Villalta M, Herrera M, Vargas M, Alvarez G, et al. Toxicity of Bothrops sp snake venoms from Ecuador and preclinical assessment of the neutralizing efficancy of a polyespecific antivenom from Costa Rica. Toxicon. 2014; 88: 34–37. doi: 10.1016/j.toxicon.2014.06.008 [DOI] [PubMed] [Google Scholar]
- 111.Saravia P, Rojas E, Escalante T, Arce V, Chaves E, Velásquez R, et al. The venoms of Bothrops asper from Guatemala: toxic activities and neutralization by antivenoms. Toxicon. 2001; 39: 401–405. [DOI] [PubMed] [Google Scholar]
- 112.Bolaños R, Cerdas L. Producción y control de sueros antiofídicos en Costa Rica. B Ofic Sanit Panam. 1980; 88: 189–196. [PubMed] [Google Scholar]
- 113.Gutiérrez JM, Chaves F, Bolaños R, Cerdas L, Rojas E, Arroyo O, et al. Neutralización de los efectos locales del veneno de Bothrops asper por un antiveneno polivalente. Toxicon.1981; 19: 493–500. [DOI] [PubMed] [Google Scholar]
- 114.Aragón F, Gubensek F. Bothrops asper venom from the Atlantic and Pacific zones of Costa Rica. Toxicon. 1981; 19:797–805. [DOI] [PubMed] [Google Scholar]
- 115.Zamudio KR, Greene HW. Phylogeography of the bushmaster (Lachesis muta: Viperidae): implications for Neotropical biogeography, systematics, and conservation. Biol J Linn Soc. 1997; 62: 421–442. [Google Scholar]
- 116.Wüster W, Crookes S, Ineich I, Mané Y, Pook CE, Trape JF, et al. The phylogeny of cobras inferred from mitochondrial DNA sequences: Evolution of venom spitting and the phylogeography of the African spitting cobras (Serpentes: Elapidae: Naja nigricollis complex). Mol Phylogenet Evol. 2007; 45: 437–453. doi: 10.1016/j.ympev.2007.07.021 [DOI] [PubMed] [Google Scholar]
- 117.Eberhad JR, Bermingham E. Phylogeny and Biogeography of the Amazona ochrocephala (Aves: Psitacidae) complex. The Auk. 2004; 121: 318–332. [Google Scholar]
- 118.Matthee CA, Burzla VJD, Taylor JF, Davis SK. Mining the mammalian genome for artiodactyl systematics. Systematic Biol. 2001; 50: 367–390. [PubMed] [Google Scholar]
- 119.Willows-Munro S, Robinson TJ, Matthee CA.2005. Utility of nuclear DNA intron markers at lower taxonomic levels: Phylogenetic resolution among nine Tragelaphus spp. Mol Phylogenet Evol. 2005; 35: 624–636. doi: 10.1016/j.ympev.2005.01.018 [DOI] [PubMed] [Google Scholar]
- 120.Sanders KL, Lee MS, Leys R, Foster R, Keogh JS. Molecular phylogeny and divergence dates for Australasian elapids and sea snakes (hydrophiinae): evidence from seven genes for rapid evolutionary radiations. J Evol Biol. 2008; 21: 682–95. doi: 10.1111/j.1420-9101.2008.01525.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accession Numbers With Asterisks Are Sequences Obtained Of GeneBank.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Accession numbers for gene sequences within GenBank are given in S1 Table.