Abstract
Background
Recent studies have evaluated the associations between polymorphisms of the heat-shock protein 70 (HSP70) encoding genes and noise-induced hearing loss (NIHL). However, the conclusions of these studies are conflicting. The objective of this meta-analysis was to clarify the association between all known polymorphisms of HSP70 genetic loci and susceptibility to NIHL, based on existing reports.
Methods
We conducted a meta-analysis of the association between Hsp70 polymorphisms (rs1043618, rs1061581, rs2075800, rs2227956, and rs2763979) and NIHL risk in both Chinese and Caucasian males. All statistical analysis was done with was conducted using the “meta” package (version 4.6–0) of R version 3.3.2 and RStudio version 1.0.44. Online databases were searched for eligible case-control studies on February 13, 2017. The odds ratio (OR), 95% confidence interval (CI), and P value were calculated using Mantel-Haenszel statistics under a random- or fixed-effect model.
Results
A total of five studies, reported via four articles from online databases, were included in our meta-analysis. For rs1061581 (from three studies), a significant association was detected in the allele model, homozygote model, and dominant model (G versus A: OR (95% CI) = 1.32(1.05–1.67), GG versus AA: OR (95% CI) = 1.93(1.1–3.36), GG + AG versus AA: OR (95% CI) = 1.45(1.05–2.02)), but not in the heterozygote model or the recessive model. For rs1043618 (from five studies), rs2075800 (from two studies), rs2227956 (from four studies), rs2763979 (from two studies), no significant association was found for any genetic model. After subgroup analyses by ethnicity, significant associations were observed for the allele model, heterozygote model, and dominant model for rs1061581 and any genetic model for rs2227956 in Caucasians.
Conclusions
The rs1043618, rs2075800, and rs2763979 polymorphisms were not found to be associated with susceptibility to NIHL; however, the rs1061581 and rs2227956 polymorphisms were significantly associated with NIHL in Caucasian males.
Introduction
Noise-induced hearing loss (NIHL) is a preventable acquired hearing loss caused by industrial, military, and recreational noise exposure [1]. NIHL is considered a serious public health problem; WHO estimates show that 10% of individuals worldwide that are exposed to noise may develop NIHL [2]. Furthermore, the prevalence of NIHL was 24.4% among US adults aged 20–69 years, according to the 2011–2012 National Health and Nutrition Examination Survey [3]. In addition, before 2010, 20% or more of the population in Asian countries was reported to suffer from NIHL; this rate was as high as 89% among individuals in specific occupations [4]. NIHL susceptibility is influenced by numerous genetic and environmental factors; in particular, heat-shock protein 70 (HSP70) genes, which encode a group of proteins that protect cells from oxidative stress, are implicated. Other genes considered to influence NIHL susceptibility include inner ear potassium recycling pathway genes and monogenic deafness genes [5].
Heat shock proteins (HSPs) are a class of functionally related proteins whose expression is stimulated by physiological stress, ototoxic drugs, high temperature, and noise [6]. HSP70, which is highly conserved and is the most abundant chaperone, is present in various cellular compartments [7, 8]. The HSP70 gene family comprises HSP70-1, HSP70-2, and HSP70-hom [5]. Several studies have shown that the production of HSP70 in the inner ear protects hair cells by countering the ototoxic effects of cisplatin and aminoglycosides [9–11]. The influence of HSP70 genetic polymorphisms on NIHL susceptibility has been the focus of investigations for over ten years [12–14]. However, these studies have been unable to reach a consistent conclusion.
To the best of our knowledge, there has been no systematic review or meta-analysis of the association between HSP70 genetic variants and NIHL risk to date. The present report is the first meta-analysis, based on existing reports in the literature, of the relationship between all known HSP70 gene polymorphisms and susceptibility to NIHL in noise exposed workers.
Materials and methods
A review protocol was developed a priori and registered in the PROSPERO International prospective register of systematic reviews (http://www.crd.york.ac.uk/PROSPERO; registration number CRD42017062595), to provide full details of the methods used. Furthermore, the present meta-analysis was conducted in accordance with the standards of the Systematic Reviews of Genetic Association Studies [15] and the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) [16].
Literature search strategy
Relevant studies were identified by searching PubMed, EMBASE, Web of Science, China National Knowledge Infrastructure (CNKI), Wanfang Data, and SinoMed (Chinese biomedical literature service system). The search terms were: (“Hearing Loss, Noise-Induced” or “Acoustic Trauma”) and (“HSP70 Heat-Shock Proteins” or “HSP70”) in combination with (“Polymorphism, Single Nucleotide” or “gene” or “polymorphism” or “variants or alleles”). The full search strategy is available on PROSPERO (http://www.crd.york.ac.uk/PROSPERO/display_record.asp?ID=CRD42017062595). The last search was conducted on February 13, 2017.
Eligibility criteria
The following eligibility criteria were applied to select studies for inclusion in the meta-analysis: (1) articles evaluating the association of the HSP70 gene polymorphism with NIHL; (2) a clear definition of NIHL; (3) a case—control study; (4) published in either English or Chinese; and (5) sufficient published data for calculating odds ratios (ORs) with their 95% confidence intervals (CIs).
Exclusion criteria
The exclusion criteria were as follows: (1) duplicate publications (only the latest publication with the most complete or updated data was selected); (2) incomplete information; (3) insufficient data; (4) review articles or conference literatures.
Data extraction
Data were extracted by two reviewers (Song Lei and Liu Huang) independently according to the pre-specified data extraction form. The following information was extracted from each study: first author, population (country, ethnicity), source of controls, case/control sample size, genotype counts for cases and controls, and evidence of Hardy-Weinberg equilibrium (HWE). If the essential data were not reported, we attempted to contact the author of the relevant studies to obtain these. Differences, if any, were resolved by consensus after discussion.
Quality assessment for individual studies
The Newcastle-Ottawa Scale (NOS) was used to assess the methodological quality of the individual studies by the two reviewers (Song Lei and Yaqian Liu) [17]. Each study was evaluated and scored based on three criteria: selection (4 stars), comparability (2 stars), and exposure (3 stars). The NOS point ranged from 0 to 9 stars. Disagreement, if any, was resolved by discussion with a third reviewer (Lei Yang).
Data analysis
We first assessed the HWE in the control group using the chi-square test [18]. The P value, OR and corresponding 95% CI were calculated using Mantel–Haenszel statistics under the allele, homozygote, heterozygote, dominant, or recessive models. This differs from the published protocol as there is no explicit additive model; therefore, this was replaced with homozygote and heterozygote models. P values of less than 0.05 were considered to represent statistically significant associations between HSP70 gene polymorphisms and NIHL. Heterogeneity across individual studies was analyzed by the Cochran’s-Q statistic and the I2 statistic (P ≤ 0.10 and I2 ≥50% indicated the significance of heterogeneity)[19, 20]. A fixed-effect model was selected with no significant heterogeneity among studies. Otherwise, a random-effect model was used [19–22]. Subsequently, subgroup analyses were performed based on ethnicity to explore the sources of heterogeneity. The potential publication bias was evaluated by Begg’s funnel plot and Egger’s test [23]. All statistical analyses were conducted using the “meta” package (version 4.6–0) of R version 3.3.2 and RStudio version 1.0.44.
Results
Selection and characteristics of studies
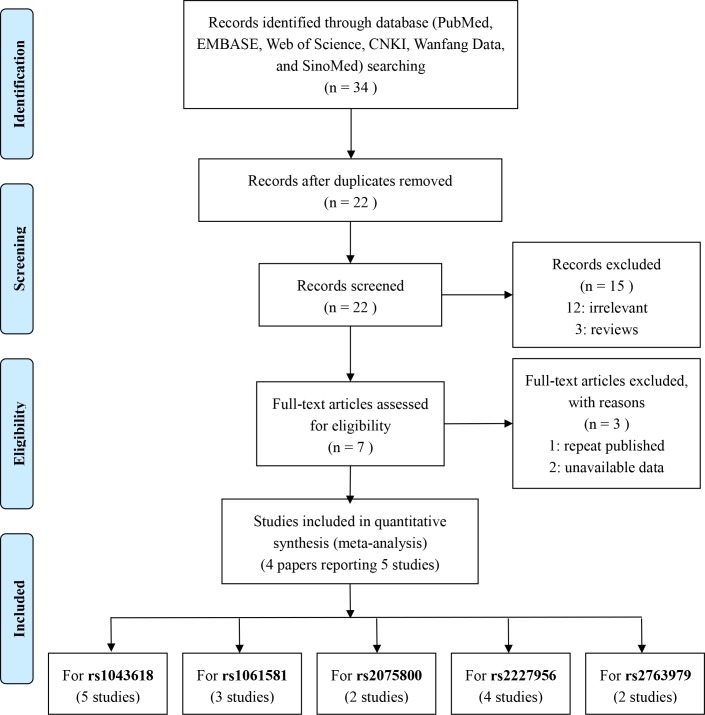
A total of 34 potential articles were retrieved through electronic databases, including EMBASE (n = 5), PubMed (n = 7), Web of Science (n = 8), SinoMed (n = 1), Wanfang (n = 7), and CNKI (n = 6), during initial searching. The study selection process is detailed in Fig 1 and S1 Table. After 12 duplicated articles were removed and 15 articles were excluded by screening the title and abstract, 12 articles were found to be unrelated and three articles were reviews [5, 24, 25]. The remaining seven articles were full-text-reviewed, and three were excluded: one [6] of these was a repeat publication and two [26, 27] did not contain sufficient data. Finally, five studies reported in four articles [12–14, 28] fulfilled the inclusion criteria and were included in the present meta-analysis (the article of Konings et al. reports two studies for Sweden and Poland).
Fig 1. Flow chart depicting the selection of eligible studies during the meta-analysis.
The selected study characteristics and data are listed in Table 1; there were two studies of Caucasian male populations, and one of these was hospital-based. The other studies were population-based and of Chinese populations including both males and females. In addition, among the five studies, five HSP70 gene loci polymorphisms associated with NIHL susceptibility were reported, including rs1043618 (615 cases and 925 controls, from five studies) [12–14, 28], rs1061581 (301 cases and 315 controls, from three studies) [13, 14], rs2075800 (303 cases and 608 controls, from two studies) [12, 28], rs2227956 (578 cases and 592 controls, from four studies) [12–14], and rs2763979 (313 cases and 608 controls, from two studies) [12, 28]. Various genotyping methods were utilized, including polymerase chain (PCR)-restriction fragment length polymorphism (RFLP), TaqMan, and SNPscan. One study [14] showed that the distribution of rs1061581 did not follow the HWE. Furthermore, the NOS scores of all studies ranged from 8 to 9 stars. This meta-analysis was carried out in accordance with the recommendations of the “Preferred Reporting Items for Systematic Reviews and Meta-Analyses” (PRISMA) statement, and Systematic Reviews of Genetic Association Studies (S1 File).
Table 1. Characteristics of studies included in the meta-analysis.
| Author | Year | Country | Ethnicity studied | Case | Control | P-HWE | Quality | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1043618 | GG | GC | CC | GG | GC | CC | |||||
| Li | 2017 | China | Chinese | 124 | 117 | 45 | 130 | 125 | 31 | 0.91 | 4/2/3 |
| Chang | 2011 | Taiwan | Chinese | 8 | 18 | 1 | 153 | 139 | 30 | 0.85 | 3/2/3 |
| Konings | 2009 | Sweden | Caucasian | 31 | 51 | 11 | 49 | 44 | 7 | 0.49 | 3/2/3 |
| Konings | 2009 | Poland | Caucasian | 44 | 58 | 14 | 46 | 58 | 12 | 0.31 | 3/2/3 |
| Yang | 2006 | China | Chinese | 37 | 43 | 13 | 35 | 48 | 18 | 0.83 | 4/1/3 |
| rs1061581 | AA | AG | GG | AA | AG | GG | |||||
| Konings | 2009 | Sweden | Caucasian | 24 | 55 | 13 | 44 | 45 | 11 | 0.92 | 3/2/3 |
| Konings | 2009 | Poland | Caucasian | 37 | 61 | 18 | 43 | 56 | 15 | 0.63 | 3/2/3 |
| Yang | 2006 | China | Chinese | 43 | 41 | 9 | 50 | 48 | 3 | 0.03 | 4/1/3 |
| rs2075800 | CC | TC | TT | CC | TC | TT | |||||
| Li | 2017 | China | Chinese | 128 | 128 | 30 | 112 | 132 | 42 | 0.76 | 4/2/3 |
| Chang | 2011 | Taiwan | Chinese | 10 | 15 | 2 | 113 | 166 | 43 | 0.14 | 3/2/3 |
| rs2227956 | TT | TC | CC | TT | TC | CC | |||||
| Li | 2017 | China | Chinese | 204 | 64 | 8 | 201 | 73 | 2 | 0.09 | 4/2/3 |
| Konings | 2009 | Sweden | Caucasian | 64 | 27 | 0 | 54 | 39 | 5 | 0.54 | 3/2/3 |
| Konings | 2009 | Poland | Caucasian | 95 | 22 | 1 | 81 | 32 | 4 | 0.70 | 3/2/3 |
| Yang | 2006 | China | Chinese | 58 | 34 | 1 | 67 | 32 | 2 | 0.41 | 4/1/3 |
| rs2763979 | CC | TC | TT | CC | TC | TT | |||||
| Li | 2017 | China | Chinese | 104 | 133 | 49 | 116 | 139 | 31 | 0.26 | 4/2/3 |
| Chang | 2011 | China | Chinese | 18 | 9 | 0 | 179 | 124 | 19 | 0.68 | 3/2/3 |
P-HWE: P-value for the Hardy-Weinberg equilibrium; Quality: Score of NOS scale.
Association between polymorphism rs1043618 and NIHL susceptibility
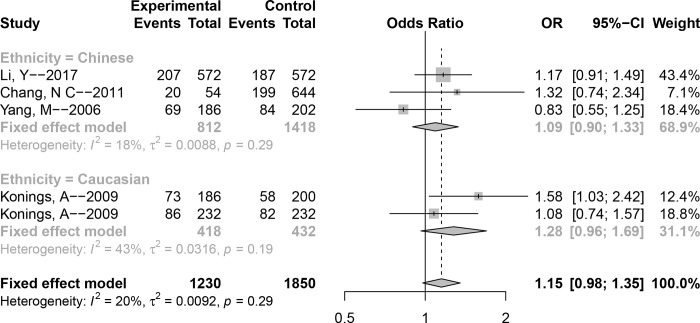
Next, the genetic association between polymorphism rs1043618 and susceptibility to NIHL was measured. The P value of Cochran’s-Q statistic for all genetic models was greater than 0.1, and I2 was less than 50%, suggesting that there was no obvious heterogeneity among the studies; therefore, the fixed-effect model was selected for pooling data. Overall, no significant association was found for any genetic model (C versus G: OR (95% CI) = 1.15 (0.98–1.35), GC versus GG: OR (95% CI) = 1.15 (0.91–1.45), CC versus GG: OR (95% CI) = 1.30 (0.91–1.86), CC + GC versus GG: OR (95% CI) = 1.19 (0.95–1.48), CC versus CG + GG: OR (95% CI) = 1.23 (0.88–1.72)). In addition, subgroup analyses by ethnicity showed no significant association in the Chinese population; this outcome was consistent with that observed in Caucasians. The main meta-analysis results are shown in detail in Fig 2 and Table 2.
Fig 2. Forest plot of the association between polymorphism rs1043618 and noise-induced hearing loss (NIHL) (Allele model: C versus G).
Table 2. Summary of pooled ORs in the meta-analysis.
| SNP | N | Allele model | Heterozygote model | Homozygote model | Dominant model | Recessive model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | I2 (%) | P-H | OR (95%CI) | I2 (%) | P-H | OR (95%CI) | I2 (%) | P-H | OR (95%CI) | I2 (%) | P-H | OR (95%CI) | I2 (%) | P-H | ||
| rs1043618 | C/G | GC/GG | CC/GG | CC + GC/GG | CC/CG + GG | |||||||||||
| Overall | 5 | 1.15(0.98–1.35) | 19.7 | 0.29 | 1.15(0.91–1.45) | 44 | 0.13 | 1.30(0.91–1.86) | 10.6 | 0.35 | 1.19(0.95–1.48) | 39.5 | 0.16 | 1.23(0.88–1.72) | 5.1 | 0.38 |
| Ethnicity | ||||||||||||||||
| Chinese | 3 | 1.09(0.90–1.33) | 18.2 | 0.29 | 1.06(0.80–1.42) | 54.5 | 0.11 | 1.18(0.77–1.82) | 29.4 | 0.24 | 1.10(0.84–1.44) | 42.6 | 0.17 | 1.16(0.78–1.73) | 45.4 | 0.16 |
| Caucasian | 2 | 1.28(0.96–1.69) | 42.9 | 0.19 | 1.35(0.90–1.45) | 44.7 | 0.18 | 1.63(0.84–3.18) | 4 | 0.31 | 1.40(0.95–2.06) | 52.2 | 0.15 | 1.40(0.75–2.63) | 0 | 0.54 |
| rs1061581 | G/A | AG/AA | GG/AA | GG+AG/AA | GG/AG + AA | |||||||||||
| Overall | 3 | 1.32(1.05–1.67) | 0 | 0.62 | 1.38(0.98–1.94) | 43.7 | 0.17 | 1.93(1.1–3.36) | 0 | 0.50 | 1.45(1.05–2.02) | 27.7 | 0.25 | 1.50(0.90–2.50) | 0 | 0.38 |
| Ethnicity | ||||||||||||||||
| Chinese | 1 | 1.27(0.82–1.97) | - | - | 0.99(0.55–1.78) | - | - | 3.49(0.89–13.71) | - | - | 1.14(0.65–2.00) | - | - | 3.50(0.92–13.35) | - | - |
| Caucasian | 2 | 1.34(1.02–1.77) | 0 | 0.33 | 1.64(1.07–2.49) | 41.9 | 0.19 | 1.68(0.91–3.11) | 0 | 0.49 | 1.65(1.10–2.47) | 41 | 0.19 | 1.26(0.72–2.21) | 0 | 0.87 |
| rs2075800 | T/C | CT/CC | TT/CC | TT + CT/CC | TT/CT + CC | |||||||||||
| 2 | 0.81(0.65–1.02) | 0 | 0.89 | 0.87(0.63–1.21) | 0 | 0.69 | 0.61(0.37–1.01) | 0 | 0.84 | 0.81(0.60–1.10) | 0 | 0.75 | 0.66(0.41–1.06) | 0 | 0.73 | |
| rs2227956 | C/T | TC/TT | CC/TT | CC + TC/TT | CC/TC + GG | |||||||||||
| Overall | 4 | 0.78(0.53–1.15) | 63.7 | 0.04 | 0.80(0.62–1.04) | 27.1 | 0.25 | 0.55(0.09–3.35) | 62.4 | 0.05 | 0.80(0.62–1.03) | 50.7 | 0.11 | 0.59(0.10–3.42) | 60.3 | 0.06 |
| Ethnicity | ||||||||||||||||
| Chinese | 2 | 1.06(0.75–1.47) | 0 | 0.86 | 0.96(0.69–1.33) | 0 | 0.33 | 1.91(0.31–11.84) | 41.3 | 0.19 | 1.01(0.74–1.39) | 0 | 0.52 | 1.85(0.26–12.88) | 47.7 | 0.17 |
| Caucasian | 2 | 0.54(0.37–0.78) | 0 | 0.90 | 0.59(0.38–0.90) | 0 | 0.99 | 0.15(0.03–0.86) | 0 | 0.58 | 0.53(0.35–0.81) | 0 | 0.91 | 0.17(0.03–0.99) | 0 | 0.60 |
| rs2763979 | T/C | CT/CC | TT/CC | TT + CT/CC | TT/CT + CC | |||||||||||
| 2 | 0.94(0.46–1.91) | 71.7 | 0.06 | 1.00(0.72–1.39) | 0 | 0.40 | 1.56(0.95–2.56) | 44.6 | 0.18 | 1.08(0.80–1.48) | 50 | 0.16 | 1.55(0.98–2.46) | 34.4 | 0.22 | |
ORs: odds ratio of the association between NIHL susceptibility and each genetic model for each single nucleotide polymorphisms (SNP); P-H: P-value for heterogeneity
Association between polymorphism rs1061581 and NIHL susceptibility
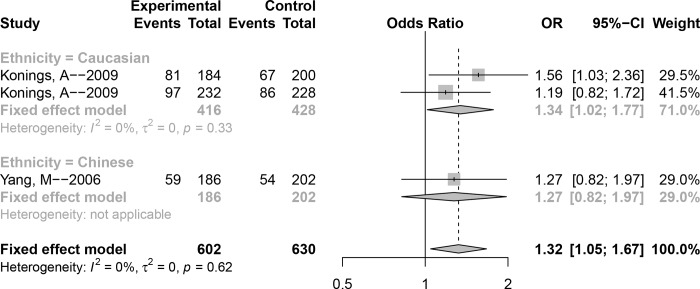
We additionally pooled analyses of polymorphism rs1061581 and NIHL; these are shown in Fig 3 and Table 2. The P value of Cochran’s-Q statistic of all genetic models were greater than 0.1 and I2 was less than 50%, indicating that there was no significant heterogeneity between the studies; therefore, the fixed-effect model was implemented. Overall, significant associations were detected in the allele model, homozygote model, and dominant model (G versus A: OR (95% CI) = 1.32 (1.05–1.67), GG versus AA: OR (95% CI) = 1.93 (1.1–3.36), GG + AG versus AA: OR (95% CI) = 1.45 (1.05–2.02)), but not in the heterozygote model or recessive model (AG versus AA: OR(95% CI) = 1.38 (0.98–1.94), GG versus AG + AA: OR (95% CI) = 1.50 (0.90–2.50)). Furthermore, subgroup analyses by ethnicity showed no significant association in the Chinese population; however, significant associations were observed for the allele model, heterozygote model, and dominant model in Caucasians.
Fig 3. Forest plot of the association between polymorphism rs1061581 and noise-induced hearing loss (NIHL) (Allele model: G versus A).
Association between polymorphism rs2075800 and NIHL susceptibility
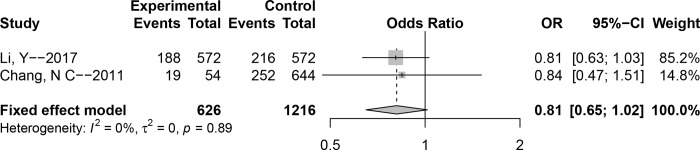
The association between polymorphism rs2075800 and the risk of NIHL was analyzed in two Chinese studies. The P value of Cochran’s-Q statistic for all genetic models was greater than 0.1, and that of I2 less than 50%, indicating that there was no significant heterogeneity between the studies; therefore, the fixed-effect model was applied. There were no notable associations for any of the genetic models (T versus C: OR (95% CI) = 0.81 (0.65–1.02), CT versus CC: OR (95% CI) = 0.87 (0.63–1.21), TT versus CC: OR (95% CI) = 0.61 (0.37–1.01), TT + CT versus CC: OR (95% CI) = 0.81 (0.60–1.10), TT versus CT + CC: OR (95%CI) = 0.66 (0.41–1.06)) (Fig 4 and Table 2).
Fig 4. Forest plot of association between polymorphism rs2075800 and noise-induced hearing loss (NIHL) (Allele model: T versus C).
Association between polymorphism rs2227956 and NIHL susceptibility
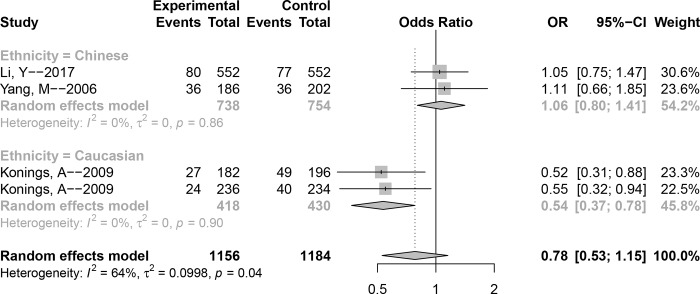
Data describing the association between polymorphism rs2227956 and the risk of NIHL are shown in Fig 5 and Table 2. The P value of Cochran’s-Q statistic of heterozygote model and dominant model was greater than 0.1, and that of I2 was less than 50%; therefore, the fixed-effect model was used. However, for the allele model (P = 0.04, I2 = 63.7%), homozygote model (P = 0.05, I2 = 62.4%), and recessive model (P = 0.06, I2 = 60.3%), the random-effect model was used. Overall, no significant association was found for any genetic model (C versus T: OR (95% CI) = 0.78 (0.53–1.15), TC versus TT: OR (95% CI) = 0.80 (0.62–1.04), CC versus TT: OR (95% CI) = 0.55 (0.09–3.35), CC + TC versus TT: OR (95% CI) = 0.80 (0.62–1.03), CC versus TC + GG: OR (95% CI) = 0.80 (0.35–1.80)). After subgroup analyses by ethnicity, the P value of Cochran’s-Q statistic of all models were greater than 0.1, and that of I2 was less than 50% in the Chinese and Caucasian populations. In addition, the association for all genetic models was significant in the Caucasian population but not in the Chinese.
Fig 5. Forest plot of the association between polymorphism rs2227956 and noise-induced hearing loss (NIHL) (Allele model: C versus T).
Association between polymorphism rs2763979 and NIHL susceptibility
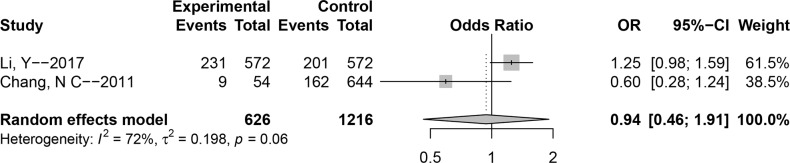
Besides above four genetic loci, Fig 6 and Table 2 show the pooled data of the association between polymorphism rs2763979 and NIHL susceptibility; the two studies of this polymorphism only focused on the Chinese population. The P value of Cochran’s-Q statistic was lower than 0.1, and that of I2 was more than 50% for the allele model (P = 0.06, I2 = 71.7%); therefore, we applied the random-effect model to this genetic model and the fixed-effect model to the other models. There were no notable associations for all genetic models (T versus C: OR (95% CI) = 0.94 (0.46–1.91), CT versus CC: OR (95% CI) = 1.00 (0.72–1.39), TT versus CC: OR (95% CI) = 1.56 (0.95–2.56), TT + CT versus CC: OR (95% CI) = 1.08 (0.80–1.48), TT versus CT + CC: OR (95% CI) = 1.55 (0.98–2.46)).
Fig 6. Forest plot of the association between polymorphism e rs2763979 and noise-induced hearing loss (NIHL) (allele model: T versus C).
Publication bias
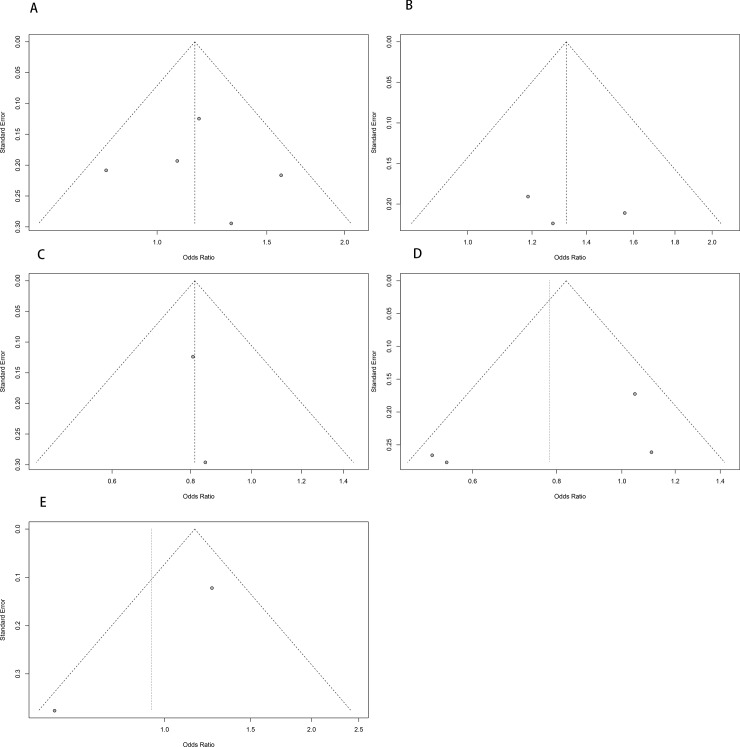
There was no publication bias with regards to the association between rs1043618, rs1061581, and rs2227956 polymorphisms and NIHL susceptibility, as identified using the Begg’s funnel plot or Egger’s regression test (Table 3). Funnel plots of the above three genetic loci in all genetic models were symmetrical (Fig 7). Owing to the limited number of studies of rs2075800 and rs2763979 included, funnel plot analysis and the Egger’s test were not carried out.
Table 3. Begg’s funnel plot and Egger’s test of the meta-analysis.
| SNP | Allele model | Heterozygote model | Homozygote model | Dominant model | Recessive model | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Begg’s | Egger | Begg’s | Egger | Begg’s | Egger | Begg’s | Egger | Begg’s | Egger | |
| rs1043618 | 0.62 | 0.88 | 0.14 | 0.23 | 1.00 | 0.62 | 0.62 | 0.41 | 0.62 | 0.28 |
| rs1061581 | 0.60 | 0.71 | 0.60 | 0.33 | 0.12 | 0.21 | 0.60 | 0.32 | 0.12 | 0.10 |
| rs2075800 | - | - | - | - | - | - | - | - | - | - |
| rs2227956 | 0.17 | 0.32 | 0.50 | 0.70 | 0.17 | 0.07 | 0.50 | 0.50 | 0.17 | 0.06 |
| rs2763979 | - | - | - | - | - | - | - | - | - | - |
SNP: single nucleotide polymorphism.
Fig 7. Begg's funnel plot of the association between five single nucleotide polymorphisms (SNPs) and noise-induced hearing loss (NIHL) in the allele model.
A: for rs1043618; B: for rs1061581; C: for rs2075800; D: for rs2227956; E: for rs2763979.
Discussion
In the present meta-analysis, we evaluated the association between single nucleotide polymorphisms (SNPs) in the following HSP70 genes: HSP70-1 (rs1043618, 615 cases and 925 controls), HSP70-2 (rs1061581, 301 cases and 315 controls; rs2763979, 313 cases and 608 controls), HSP70-hom (rs2075800, 303 cases and 608 controls; rs2227956, 578 cases and 592 controls) and NIHL. Overall analysis indicated that there was no association between rs1043618, rs2075800, rs2227956, and rs2763979 and NIHL susceptibility for any of the genetic models. However, we identified a positive relationship between rs1061581 and NIHL in the allele model, homozygote model, and dominant model.
HSP70 is distributed in the inner ear, and has the potential to protect the hearing from noise, ototoxic drugs, or injury [29]. HSP70 overexpression can significantly protect the inner ear hair cells from cell death caused by aminoglycosides[11]. In addition, Geldanamycin, geranylgeranylacetone, and 17-Dimethylaminoethylamino-17-demethoxygeldanamycin can upregulate the expression of HSP70 to treat aminoglycoside-induced hearing loss[30–32]. Therefore, HSP70 gene polymorphisms might be associated with NIHL susceptibility. HSP70-1, HSP70-2, and HSP70-hom are located in the major histocompatibility complex (MHC) class III region of chromosome 6p21.3. Rs1043618 lies in the 5' untranslated region of HSP70-1, rs1061581 and rs2763979 are located in the 5' flanking region of HSP70-2; these loci are non-coding but perform a role in the control of gene expression. Rs2075800 and rs2227956 are located in exon 2 of HSP70-hom, the C-T of rs2075800 leads to a Glu-Lys substitution at position 602, and the A-G of rs2227956 leads to a Met-Thr substitution at position 493 (https://www.ncbi.nlm.nih.gov/snp/). All five SNPs of HSP70 genes have been explored in NIHL studies.
Two genome-wide association studies of NIHL-related SNPs in animals [33, 34] and one in humans [27] have been published; however, those studies did not examine the association between HSP70 genes and NIHL. This meta-analysis incorporates four articles that specifically investigate the association between HSP70 and NIHL; among these, only one [13] found positive results for a single SNP using crude data. However, NIHL is a complex disease that cannot be attributed to the influence of a single SNP, owing to the important role played by the interaction between genes, or between genes and the environment. Certain HSP70 haplotypes are associated with the development of NIHL [13, 14, 28]. These authors additionally found that age, smoking, drinking, cumulative noise exposure, noise exposure levels, and other factors influenced NIHL susceptibility. However, it is not possible to determine whether HSP70 is associated with NIHL based on small sample data.
To explore the heterogeneity among the studies, we conducted a subgroup analysis for rs1043618, rs1061581, and rs2227956. The results showed that the heterogeneity identified before subgroup analysis was reduced to an acceptable level after subgroup analysis by ethnicity, indicating that ethnicity is the main source of heterogeneity. For rs1043618, subgroup analysis did not reveal an association in either population. In addition, a significant association was observed between rs1061581 and NIHL in the allele model, heterozygote model, and dominant model in the Caucasian population, but not in the other two models. Furthermore, Caucasian individuals with the G allele were more susceptible to NIHL (allele model, G versus A: OR (95% CI) = 1.32(1.05–1.67)). Similarly, a significant association was detected between rs2227956 and NIHL in all genetic models in the Caucasian population; Caucasian individuals with the T allele were more susceptible to NIHL (allele model, C versus T: OR (95% CI) = 0.54(0.37–0.78)). However, no significant association was discovered in Chinese individuals for SNPs rs1061581 and rs2227956, suggesting that neither of these polymorphisms increase the risk of NIHL. The different results for the association between rs1061581 and rs2227956 and NIHL in the two different populations may be attributed to differences in ethnicity.
To the best of our knowledge, the present work is the first to evaluate the association between HSP70 gene polymorphisms and NIHL susceptibility via a meta-analysis. Our meta-analysis has several strengths. We did not limit the specific loci; all previously reported SNPs were included for analysis. According to the results of NOS, the methodological quality of the study was high. Moreover, we identified the main source of heterogeneity via subgroup analysis. Finally, no publication bias was identified by either Begg’s funnel plot or Egger’s regression test, except in the case of rs2075800 and rs2763979.
The present study also has several limitations. For instance, the number of included studies was very low for rs2075800 and rs2763979, which limited further analysis. Secondly, Yang’s study [14] showed that the distribution of rs1061581 did not follow the HWE; however, subgroup analysis was used separately. Thirdly, we were unable to extract sufficient adjustment data for certain factors, such as age and noise exposure level. Finally, as with three other meta-analysis studies [35–37] of the association between NIHL and other SNPs, our meta-analysis involved a small sample size, resulting in unstable pooled results.
Conclusion
In summary, our meta-analysis comprehensively and systematically evaluated the association between HSP70 gene polymorphisms and NIHL susceptibility. The results suggested that rs1043618, rs2075800, and rs2763979 polymorphisms are not associated with susceptibility to NIHL, while rs1061581 and rs2227956 polymorphisms are significantly associated with NIHL in Caucasians males. Given the limited sample size, the conclusions of this study should be treated with caution, and large sample studies are necessary in the future. These results will be useful for genetic testing of NIHL susceptibility in Caucasian males. In addition, NIHL susceptible populations can be identified by genetic testing and the corresponding measures can be taken to protect their hearing.
Supporting information
(ZIP)
Forest plot (Figure A—Figure T). (ZIP). Figure A in S2 File: Forest plot of association between the rs1043618 polymorphism and NIHL (Heterozygote model: GC versus GG). Figure B in S2 File: Forest plot of association between the rs1043618 polymorphism and NIHL (Homozygote model: CC versus GG). Figure C in S2 File: Forest plot of association between the rs1043618 polymorphism and NIHL (Dominant model: CC+GC versus GG). Figure D in S2 File: Forest plot of association between the rs1043618 polymorphism and NIHL (Recessive model: CC versus CG+GG). Figure E in S2 File: Forest plot of association between the rs1061581 polymorphism and NIHL (Heterozygote model: AG versus AA). Figure F in S2 File: Forest plot of association between the rs1061581 polymorphism and NIHL (Homozygote model: GG versus AA). Figure G in S2 File: Forest plot of association between the rs1061581 polymorphism and NIHL (Dominant model: GG+AG versus AA). Figure H in S2 File: Forest plot of association between the rs1061581 polymorphism and NIHL (Recessive model: GG versus AG+AA). Figure I in S2 File: Forest plot of association between the rs2075800 polymorphism and NIHL (Heterozygote model: CT versus CC). Figure J in S2 File: Forest plot of association between the rs2075800 polymorphism and NIHL (Homozygote model: TT versus CC). Figure K in S2 File: Forest plot of association between the rs2075800 polymorphism and NIHL (Dominant model: TT+CT versus CC). Figure L in S2 File: Forest plot of association between the rs2075800 polymorphism and NIHL (Recessive model: TT versus CT+CC). Figure M in S2 File: Forest plot of association between the rs2227956 polymorphism and NIHL (Heterozygote model: TC versus TT). Figure N in S2 File: Forest plot of association between the rs2227956 polymorphism and NIHL (Homozygote model: CC versus TT). Figure O in S2 File: Forest plot of association between the rs2227956 polymorphism and NIHL (Dominant model: CC+TC versus TT). Figure P in S2 File: Forest plot of association between the rs2227956 polymorphism and NIHL (Recessive model: CC versus TC+GG). Figure Q in S2 File: Forest plot of association between the rs2763979 polymorphism and NIHL (Heterozygote model: CT versus CC). Figure R in S2 File: Forest plot of association between the rs2763979 polymorphism and NIHL (Homozygote model: TT versus CC). Figure S in S2 File: Forest plot of association between the rs2763979 polymorphism and NIHL (Dominant model: TT+CT versus CC). Figure T in S2 File: Forest plot of association between the rs2763979 polymorphism and NIHL (Recessive model: TT versus CT+CC).
(ZIP)
Begg’s funnel plot (Figure A—Figure T). (ZIP). Figure A in S3 File: Begg’s funnel plot of association between the rs1043618 polymorphism and NIHL (Heterozygote model: GC versus GG). Figure B in S3 File: Begg’s funnel plot of association between the rs1043618 polymorphism and NIHL (Homozygote model: CC versus GG). Figure C in S3 File: Begg’s funnel plot of association between the rs1043618 polymorphism and NIHL (Dominant model: CC+GC versus GG). Figure D in S3 File: Begg’s funnel plot of association between the rs1043618 polymorphism and NIHL (Recessive model: CC versus CG+GG). Figure E in S3 File: Begg’s funnel plot of association between the rs1061581 polymorphism and NIHL (Heterozygote model: AG versus AA). Figure F in S3 File: Begg’s funnel plot of association between the rs1061581 polymorphism and NIHL (Homozygote model: GG versus AA). Figure G in S3 File: Begg’s funnel plot of association between the rs1061581 polymorphism and NIHL (Dominant model: GG+AG versus AA). Figure H in S3 File: Begg’s funnel plot of association between the rs1061581 polymorphism and NIHL (Recessive model: GG versus AG+AA). Figure I in S3 File: Begg’s funnel plot of association between the rs2075800 polymorphism and NIHL (Heterozygote model: CT versus CC). Figure J in S3 File: Begg’s funnel plot of association between the rs2075800 polymorphism and NIHL (Homozygote model: TT versus CC). Figure K in S3 File: Begg’s funnel plot of association between the rs2075800 polymorphism and NIHL (Dominant model: TT+CT versus CC). Figure L in S3 File: Begg’s funnel plot of association between the rs2075800 polymorphism and NIHL (Recessive model: TT versus CT+CC). Figure M in S3 File: Begg’s funnel plot of association between the rs2227956 polymorphism and NIHL (Heterozygote model: TC versus TT). Figure N in S3 File: Begg’s funnel plot of association between the rs2227956 polymorphism and NIHL (Homozygote model: CC versus TT). Figure O in S3 File: Begg’s funnel plot of association between the rs2227956 polymorphism and NIHL (Dominant model: CC+TC versus TT). Figure P in S3 File: Begg’s funnel plot of association between the rs2227956 polymorphism and NIHL (Recessive model: CC versus TC+GG). Figure Q in S3 File: Begg’s funnel plot of association between the rs2763979 polymorphism and NIHL (Heterozygote model: CT versus CC). Figure R in S3 File: Begg’s funnel plot of association between the rs2763979 polymorphism and NIHL (Homozygote model: TT versus CC). Figure S in S3 File: Begg’s funnel plot of association between the rs2763979 polymorphism and NIHL (Dominant model: TT+CT versus CC). Figure T in S3 File: Begg’s funnel plot of association between the rs2763979 polymorphism and NIHL (Recessive model: TT versus CT+CC).
(ZIP)
(XLSX)
Acknowledgments
We would like to thank Shugang Li, Jiaming Liu, and Ping Yang who have contributed to this paper. At the same time, we would like to thank Editage [http://www.editage.cn] for English language editing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Zhejiang Key Research and Development Program (grant number 2015C03050), the Major Science and Technology Innovation Project of Hangzhou in China (grant number 20152013A01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sha SH, Schacht J. Emerging therapeutic interventions against noise-induced hearing loss. Expert Opin Investig Drugs. 2017;26(1):85–96. doi: 10.1080/13543784.2017.1269171 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basner M, Babisch W, Davis A, Brink M, Clark C, Janssen S, et al. Auditory and non-auditory effects of noise on health. Lancet. 2014;383(9925):1325–32. doi: 10.1016/S0140-6736(13)61613-X ; PubMed Central PMCID: PMCPMC3988259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll YI, Eichwald J, Scinicariello F, Hoffman HJ, Deitchman S, Radke MS, et al. Vital Signs: Noise-Induced Hearing Loss Among Adults—United States 2011–2012. MMWR Morb Mortal Wkly Rep. 2017;66(5):139–44. doi: 10.15585/mmwr.mm6605e3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuente A, Hickson L. Noise-induced hearing loss in Asia. Int J Audiol. 2011;50 Suppl 1:S3–10. doi: 10.3109/14992027.2010.540584 . [DOI] [PubMed] [Google Scholar]
- 5.Sliwinska-Kowalska M, Pawelczyk M. Contribution of genetic factors to noise-induced hearing loss: a human studies review. Mutat Res. 2013;752(1):61–5. doi: 10.1016/j.mrrev.2012.11.001 . [DOI] [PubMed] [Google Scholar]
- 6.Li Y. The relationship between occupational noise-induced hearing loss and single nucleotide polymorphisms of gene HSP70 [Master]: Zhengzhou University; 2015. [Google Scholar]
- 7.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14(10):630–42. doi: 10.1038/nrm3658 ; PubMed Central PMCID: PMCPMC4340576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mashaghi A, Kramer G, Lamb DC, Mayer MP, Tans SJ. Chaperone action at the single-molecule level. Chem Rev. 2014;114(1):660–76. doi: 10.1021/cr400326k . [DOI] [PubMed] [Google Scholar]
- 9.May LA, Kramarenko II, Brandon CS, Voelkel-Johnson C, Roy S, Truong K, et al. Inner ear supporting cells protect hair cells by secreting HSP70. J Clin Invest. 2013;123(8):3577–87. doi: 10.1172/JCI68480 ; PubMed Central PMCID: PMCPMC3967657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Berrocal JR, Nevado J, Gonzalez-Garcia JA, Sanchez-Rodriguez C, Sanz R, Trinidad A, et al. Heat shock protein 70 and cellular disturbances in cochlear cisplatin ototoxicity model. J Laryngol Otol. 2010;124(6):599–609. doi: 10.1017/S0022215110000496 . [DOI] [PubMed] [Google Scholar]
- 11.Taleb M, Brandon CS, Lee FS, Harris KC, Dillmann WH, Cunningham LL. Hsp70 inhibits aminoglycoside-induced hearing loss and cochlear hair cell death. Cell Stress Chaperones. 2009;14(4):427–37. doi: 10.1007/s12192-008-0097-2 ; PubMed Central PMCID: PMCPMC2728278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Yu S, Gu G, Chen G, Zheng Y, Jiao J, et al. Polymorphisms of heat shock protein 70 genes (HSPA1A, HSPA1B and HSPA1L) and susceptibility of noise-induced hearing loss in a Chinese population: A case-control study. PLoS One. 2017;12(2):e0171722 doi: 10.1371/journal.pone.0171722 ; PubMed Central PMCID: PMCPMC5300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konings A, Van Laer L, Michel S, Pawelczyk M, Carlsson PI, Bondeson ML, et al. Variations in HSP70 genes associated with noise-induced hearing loss in two independent populations. European Journal of Human Genetics. 2009;17(3):329–35. doi: 10.1038/ejhg.2008.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, Tan H, Yang Q, Wang F, Yao H, Wei Q, et al. Association of hsp70 polymorphisms with risk of noise-induced hearing loss in Chinese automobile workers. Cell Stress and Chaperones. 2006;11(3):233–9. doi: 10.1379/CSC-192R.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sagoo GS, Little J, Higgins JP. Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Med. 2009;6(3):e28 doi: 10.1371/journal.pmed.1000028 ; PubMed Central PMCID: PMCPMC2650724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–9, W64. . [DOI] [PubMed] [Google Scholar]
- 17.Wells GA, O’Connell D, Peterson J, Welch V, Shea B, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta- analyses. www.ohrica/programs/clinical_epidemiology/oxfordasp (accessed on March 10, 2017).
- 18.Schaid DJ, Jacobsen SJ. Biased tests of association: comparisons of allele frequencies when departing from Hardy-Weinberg proportions. Am J Epidemiol. 1999;149(8):706–11. . [DOI] [PubMed] [Google Scholar]
- 19.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60. doi: 10.1136/bmj.327.7414.557 ; PubMed Central PMCID: PMCPMC192859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zintzaras E, Ioannidis JP. Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol. 2005;28(2):123–37. doi: 10.1002/gepi.20048 . [DOI] [PubMed] [Google Scholar]
- 21.Thakkinstian A, McElduff P, D'Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24(9):1291–306. doi: 10.1002/sim.2010 . [DOI] [PubMed] [Google Scholar]
- 22.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58. doi: 10.1002/sim.1186 . [DOI] [PubMed] [Google Scholar]
- 23.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34. ; PubMed Central PMCID: PMCPMC2127453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clifford RE, Hoffer M, Rogers R. The Genomic Basis of Noise-induced Hearing Loss: A Literature Review Organized by Cellular Pathways. Otol Neurotol. 2016;37(8):e309–16. doi: 10.1097/MAO.0000000000001073 . [DOI] [PubMed] [Google Scholar]
- 25.Hu L, Deng J. Progression of noise-induced hearing loss associated SNP in human Occupation and Health. 2014;(18):2664–9. [Google Scholar]
- 26.Yang M, Tan H, Zheng J, Wang F, Jiang C, He M, et al. Relationship between hOGG1 and PMCA2 Polymorphisms and Noise Induced Hearing Loss Susceptibility in Chinese Han Population. The 4th National Symposium on Environmental and Occupational Medicine 2005: 11.
- 27.Grondin Y, Bortoni ME, Sepulveda R, Ghelfi E, Bartos A, Cotanche D, et al. Genetic Polymorphisms Associated with Hearing Threshold Shift in Subjects during First Encounter with Occupational Impulse Noise. PLoS One. 2015;10(6):e0130827 Epub 2015/06/30. doi: 10.1371/journal.pone.0130827 ; PubMed Central PMCID: PMCPMC4488244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang NC, Ho CK, Lin HY, Yu ML, Chien CY, Ho KY. Association of polymorphisms of heat shock protein 70 with susceptibility to noise-induced hearing loss in the Taiwanese population. Audiology & neuro-otology. 2011;16(3):168–74. doi: 10.1159/000317119 [DOI] [PubMed] [Google Scholar]
- 29.Neely JG, Thompson AM, Gower DJ. Detection and localization of heat shock protein 70 in the normal guinea pig cochlea. Hear Res. 1991;52(2):403–6. . [DOI] [PubMed] [Google Scholar]
- 30.Yu Y, Szczepek AJ, Haupt H, Mazurek B. Geldanamycin induces production of heat shock protein 70 and partially attenuates ototoxicity caused by gentamicin in the organ of Corti explants. J Biomed Sci. 2009;16:79 doi: 10.1186/1423-0127-16-79 ; PubMed Central PMCID: PMCPMC2746196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sone M, Hayashi H, Yamamoto H, Hoshino T, Mizushima T, Nakashima T. Upregulation of HSP by geranylgeranylacetone protects the cochlear lateral wall from endotoxin-induced inflammation. Hear Res. 2005;204(1–2):140–6. doi: 10.1016/j.heares.2005.01.012 . [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Yu Y, Chu H, Bing D, Wang S, Zhou L, et al. 17-DMAG induces Hsp70 and protects the auditory hair cells from kanamycin ototoxicity in vitro. Neurosci Lett. 2015;588:72–7. doi: 10.1016/j.neulet.2014.12.060 . [DOI] [PubMed] [Google Scholar]
- 33.Lavinsky J, Crow AL, Pan C, Wang J, Aaron KA, Ho MK, et al. Genome-wide association study identifies nox3 as a critical gene for susceptibility to noise-induced hearing loss. PLoS genetics. 2015;11(4):e1005094 Epub 2015/04/17. doi: 10.1371/journal.pgen.1005094 ; PubMed Central PMCID: PMCPMC4399881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White CH, Ohmen JD, Sheth S, Zebboudj AF, McHugh RK, Hoffman LF, et al. Genome-wide screening for genetic loci associated with noise-induced hearing loss. Mammalian genome: official journal of the International Mammalian Genome Society. 2009;20(4):207–13. Epub 2009/04/02. doi: 10.1007/s00335-009-9178-5 ; PubMed Central PMCID: PMCPMC2823581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Li J, Peng K, Fu ZY, Tang J, Yang MJ, et al. Association of the C47T polymorphism in superoxide dismutase gene 2 with noise-induced hearing loss: a meta-analysis. Braz J Otorhinolaryngol. 2017;83(1):80–7. doi: 10.1016/j.bjorl.2016.01.008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu JN, Wu SS, He CH, Zhang CY, Mu HX, Ma WS, et al. Association between CDH23 gene polymorphisms and susceptibility to noise-induced hearing loss in the Chinese population: a meta-analysis. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 2016;34(12):920–3. doi: 10.3760/cma.j.issn.1001-9391.2016.12.009 . [DOI] [PubMed] [Google Scholar]
- 37.Zhou S, Wang R, Zhou J, Liu S, Zhou B, Cao L. Association of GSTM1 and GSTT1 polymorphisms with noise-induced hearing loss: a meta-analysis. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 2014;32(2):123–5. . [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(ZIP)
Forest plot (Figure A—Figure T). (ZIP). Figure A in S2 File: Forest plot of association between the rs1043618 polymorphism and NIHL (Heterozygote model: GC versus GG). Figure B in S2 File: Forest plot of association between the rs1043618 polymorphism and NIHL (Homozygote model: CC versus GG). Figure C in S2 File: Forest plot of association between the rs1043618 polymorphism and NIHL (Dominant model: CC+GC versus GG). Figure D in S2 File: Forest plot of association between the rs1043618 polymorphism and NIHL (Recessive model: CC versus CG+GG). Figure E in S2 File: Forest plot of association between the rs1061581 polymorphism and NIHL (Heterozygote model: AG versus AA). Figure F in S2 File: Forest plot of association between the rs1061581 polymorphism and NIHL (Homozygote model: GG versus AA). Figure G in S2 File: Forest plot of association between the rs1061581 polymorphism and NIHL (Dominant model: GG+AG versus AA). Figure H in S2 File: Forest plot of association between the rs1061581 polymorphism and NIHL (Recessive model: GG versus AG+AA). Figure I in S2 File: Forest plot of association between the rs2075800 polymorphism and NIHL (Heterozygote model: CT versus CC). Figure J in S2 File: Forest plot of association between the rs2075800 polymorphism and NIHL (Homozygote model: TT versus CC). Figure K in S2 File: Forest plot of association between the rs2075800 polymorphism and NIHL (Dominant model: TT+CT versus CC). Figure L in S2 File: Forest plot of association between the rs2075800 polymorphism and NIHL (Recessive model: TT versus CT+CC). Figure M in S2 File: Forest plot of association between the rs2227956 polymorphism and NIHL (Heterozygote model: TC versus TT). Figure N in S2 File: Forest plot of association between the rs2227956 polymorphism and NIHL (Homozygote model: CC versus TT). Figure O in S2 File: Forest plot of association between the rs2227956 polymorphism and NIHL (Dominant model: CC+TC versus TT). Figure P in S2 File: Forest plot of association between the rs2227956 polymorphism and NIHL (Recessive model: CC versus TC+GG). Figure Q in S2 File: Forest plot of association between the rs2763979 polymorphism and NIHL (Heterozygote model: CT versus CC). Figure R in S2 File: Forest plot of association between the rs2763979 polymorphism and NIHL (Homozygote model: TT versus CC). Figure S in S2 File: Forest plot of association between the rs2763979 polymorphism and NIHL (Dominant model: TT+CT versus CC). Figure T in S2 File: Forest plot of association between the rs2763979 polymorphism and NIHL (Recessive model: TT versus CT+CC).
(ZIP)
Begg’s funnel plot (Figure A—Figure T). (ZIP). Figure A in S3 File: Begg’s funnel plot of association between the rs1043618 polymorphism and NIHL (Heterozygote model: GC versus GG). Figure B in S3 File: Begg’s funnel plot of association between the rs1043618 polymorphism and NIHL (Homozygote model: CC versus GG). Figure C in S3 File: Begg’s funnel plot of association between the rs1043618 polymorphism and NIHL (Dominant model: CC+GC versus GG). Figure D in S3 File: Begg’s funnel plot of association between the rs1043618 polymorphism and NIHL (Recessive model: CC versus CG+GG). Figure E in S3 File: Begg’s funnel plot of association between the rs1061581 polymorphism and NIHL (Heterozygote model: AG versus AA). Figure F in S3 File: Begg’s funnel plot of association between the rs1061581 polymorphism and NIHL (Homozygote model: GG versus AA). Figure G in S3 File: Begg’s funnel plot of association between the rs1061581 polymorphism and NIHL (Dominant model: GG+AG versus AA). Figure H in S3 File: Begg’s funnel plot of association between the rs1061581 polymorphism and NIHL (Recessive model: GG versus AG+AA). Figure I in S3 File: Begg’s funnel plot of association between the rs2075800 polymorphism and NIHL (Heterozygote model: CT versus CC). Figure J in S3 File: Begg’s funnel plot of association between the rs2075800 polymorphism and NIHL (Homozygote model: TT versus CC). Figure K in S3 File: Begg’s funnel plot of association between the rs2075800 polymorphism and NIHL (Dominant model: TT+CT versus CC). Figure L in S3 File: Begg’s funnel plot of association between the rs2075800 polymorphism and NIHL (Recessive model: TT versus CT+CC). Figure M in S3 File: Begg’s funnel plot of association between the rs2227956 polymorphism and NIHL (Heterozygote model: TC versus TT). Figure N in S3 File: Begg’s funnel plot of association between the rs2227956 polymorphism and NIHL (Homozygote model: CC versus TT). Figure O in S3 File: Begg’s funnel plot of association between the rs2227956 polymorphism and NIHL (Dominant model: CC+TC versus TT). Figure P in S3 File: Begg’s funnel plot of association between the rs2227956 polymorphism and NIHL (Recessive model: CC versus TC+GG). Figure Q in S3 File: Begg’s funnel plot of association between the rs2763979 polymorphism and NIHL (Heterozygote model: CT versus CC). Figure R in S3 File: Begg’s funnel plot of association between the rs2763979 polymorphism and NIHL (Homozygote model: TT versus CC). Figure S in S3 File: Begg’s funnel plot of association between the rs2763979 polymorphism and NIHL (Dominant model: TT+CT versus CC). Figure T in S3 File: Begg’s funnel plot of association between the rs2763979 polymorphism and NIHL (Recessive model: TT versus CT+CC).
(ZIP)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.