Abstract
Drought causes substantial reductions in crop yields worldwide. Therefore, we set out to identify new chemical and genetic factors that regulate drought resistance in Arabidopsis thaliana. Karrikins (KARs) are a class of butenolide compounds found in smoke that promote seed germination, and have been reported to improve seedling vigor under stressful growth conditions. Here, we discovered that mutations in KARRIKIN INSENSITIVE2 (KAI2), encoding the proposed karrikin receptor, result in hypersensitivity to water deprivation. We performed transcriptomic, physiological and biochemical analyses of kai2 plants to understand the basis for KAI2-regulated drought resistance. We found that kai2 mutants have increased rates of water loss and drought-induced cell membrane damage, enlarged stomatal apertures, and higher cuticular permeability. In addition, kai2 plants have reduced anthocyanin biosynthesis during drought, and are hyposensitive to abscisic acid (ABA) in stomatal closure and cotyledon opening assays. We identified genes that are likely associated with the observed physiological and biochemical changes through a genome-wide transcriptome analysis of kai2 under both well-watered and dehydration conditions. These data provide evidence for crosstalk between ABA- and KAI2-dependent signaling pathways in regulating plant responses to drought. A comparison of the strigolactone receptor mutant d14 (DWARF14) to kai2 indicated that strigolactones also contributes to plant drought adaptation, although not by affecting cuticle development. Our findings suggest that chemical or genetic manipulation of KAI2 and D14 signaling may provide novel ways to improve drought resistance.
Author summary
MORE AXILLARY GROWTH2 (MAX2) is a central regulator of both strigolactone and karrikin (KAR) signaling pathways, which control many aspects of plant growth and development. MAX2 promotes plant resistance to drought. Application of a strigolactone analog enhances plant adaptation to drought, implying that strigolactone signaling regulates this trait; however, the potential contributions of the KAR signaling pathway to plant drought responses have not been explored. In this study, the functions of the KAR receptor KARRIKIN INSENSITIVE2 (KAI2) were analyzed to elucidate the role of KAI2-mediated signaling in plant drought adaptation. Our results indicate that the KAI2 signaling positively regulates drought resistance of Arabidopsis plants by both drought avoidance and drought tolerance strategies, such as through enhancing cuticle formation, stomatal closure, cell membrane integrity and anthocyanin biosynthesis. Our findings provide potential for manipulation of KAR- and/or strigolactone-mediated signaling pathways for improvement of drought resistance.
Introduction
Water deficit is a major constraint to crop productivity worldwide. As water resources are increasingly challenged by climate change and the demands of a growing human population, improvement of water use efficiency and development of drought-resistant crops will be critical innovations for agriculture [1]. Intensive efforts have been made toward discovering the hormones and genetic networks that control drought adaptation in plants, with the goal of developing chemical or genetic tools to manipulate drought resistance in the field. The most well-known drought-related phytohormone is abscisic acid (ABA), which accumulates in response to drought and other abiotic stresses. ABA triggers various physical and physiological mechanisms for plant protection, including stomatal closure, cuticle formation, and production of protective metabolites [2–4]. Another phytohormone, strigolactone (SL), was recently reported to promote drought resistance in several plant species, including Arabidopsis thaliana, Lotus japonicus, and Solanum lycopersicum, through both ABA-dependent and ABA-independent pathways [5–8]. SLs are perceived by the α/β-hydrolase protein DWARF14 (D14), which undergoes a conformational change after SL hydrolysis that promotes interactions with the F-box protein MORE AXILLARY GROWTH2 (MAX2)/DWARF3 (D3) and downstream effectors in the SUPPRESSOR OF MAX2 1 (SMAX1)-LIKE (SMXL)/DWARF53 (D53) family [9,10]. MAX2 functions as an adapter component of an Skp1–Cullin–F-box (SCF) E3 ubiquitin ligase complex that targets D53 in rice and its orthologs in Arabidopsis, SMXL6/7/8, for polyubiquitination and proteasomal degradation.
Intriguingly, MAX2 is not only a central regulator of SL signaling, but is also necessary for responses to karrikins (KARs) [9,11,12]. KARs are a class of butenolide compounds found in smoke that are thought to be ecologically important triggers of seed germination in the post-fire environment [13,14]. KAR treatment has additional effects on Arabidopsis growth that include inhibition of hypocotyl elongation, and promotion of cotyledon expansion and greening in seedlings [13]. The KAR signaling mechanism is likely to be very similar to that of SLs. In Arabidopsis, KAR perception requires KARRIKIN INSENSITIVE2 (KAI2)/HYPOSENSITIVE TO LIGHT (HTL), an ancient paralog of D14 [11]. Direct association of MAX2 and KAI2 has not been demonstrated, but residues that are required for D14 interaction with MAX2 are conserved in KAI2 proteins. Epistasis tests demonstrated that SMAX1 and SMXL2, which are in the same gene family as the SL pathway targets, act downstream of MAX2 as repressors of KAR responses; therefore, they are putative substrates of a KAR-activated KAI2-SCFMAX2 complex [15]. It must be noted that although Arabidopsis KAI2 can bind KARs in vitro and mediate growth responses to KAR treatments, KAR perception may not be its typical function. Instead, KARs may function as natural analogs of a hypothetical, endogenous KAI2 ligand (KL). Supporting this idea, KAI2 proteins with specialized ligand preferences have been identified in root parasitic plants; a KAR-responsive KAI2i paralog from Striga hermonthica only partially rescues Arabidopsis kai2 mutants, whereas a highly conserved KAI2c paralog from Pheliphanche aegyptiaca fully rescues kai2 but is not responsive to KAR1 [16,17]. Therefore, KARs may be useful as lead compounds to develop novel agonists of KAI2 signaling that improve crop performance.
Because MAX2 functions in two pathways, max2 phenotypes cannot be readily attributed to defects in the SL, KAR/KL, or both signaling mechanisms. Further complicating the matter, a commonly used SL analog known as GR24 is typically synthesized as a racemic mixture; one GR24 enantiomer has the stereochemical configuration of natural SLs and selectively activates D14, whereas the other has an unnatural configuration and can activate both KAI2 and D14 in Arabidopsis [18]. Two studies have shown that Arabidopsis max2 mutants have less drought-resistance than wild-type (WT), but it has been unclear whether this is due to a SL signaling defect because contradictory drought-sensitive phenotypes have been reported for SL-deficient mutants [5,6]. Spraying plants with rac-GR24 promotes survival after water deficit in both WT and SL-deficient max mutants [5], but this does not exclude the possibility that this is due, at least in part, to KAI2 activation. As KAR-containing smoke-water has been shown to increase germination, seedling vigor, and survival in crops grown under stressful environments, including high temperature and low osmotic potential conditions [14,19], we hypothesized that KAI2 signaling may contribute to MAX2-dependent drought resistance. Here we demonstrate a role for KAI2 in survival of Arabidopsis plants after water deficit, and identify physiological and molecular explanations for this function, suggesting the importance of an as-yet-unknown signal KL and KAI2-dependent signaling in plant drought resistance. Additionally, by analyzing the drought resistance of SL-specific d14 and kai2 receptor mutants we find that both pathways promote plant adaptation to water deficit.
Results
kai2 mutant plants are hypersensitive to drought
To investigate whether KAI2-mediated signaling contributes to drought resistance in Arabidopsis, we examined the effects of KAI2 loss-of-function on plant growth during water deficit using the gravimetric method [20]. We found that the biomass of kai2-2 mutant plants was higher than that of WT when they were grown under well-watered conditions (Fig 1A). However, the biomass reduction of kai2-2 mutants was higher than that of WT when they were grown under similar low soil water content after water withholding (Fig 1B and 1C). Additionally, when the kai2-2 and WT plants were grown under high density in the same pots, we observed that kai2-2 mutant plants displayed higher sensitivity to drought than WT (Fig 1D and 1E). These results together suggest that KAI2 signaling promotes adaptation to drought in Arabidopsis.
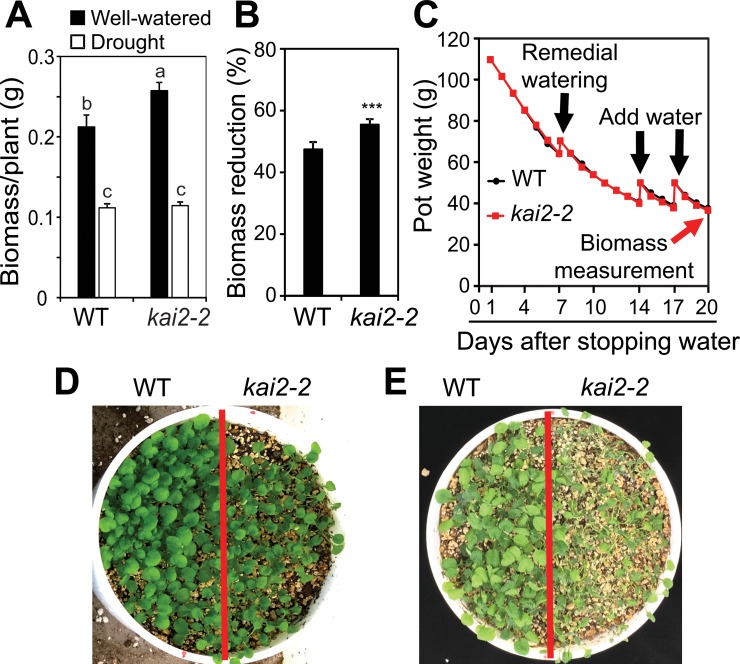
Fig 1. Effects of KAI2 on drought resistance.
(A) Biomass of kai2-2 and WT plants under drought stress and well-watered control (n = 9 biological replicates). The different letters above the error bars indicate significant differences (P < 0.05) in all combinations according to a Tukey's honest significant difference test. (B) Biomass reduction of kai2-2 and WT plants under drought stress relative to respective well-watered control. Data represent the means and standard errors (n = 9 biological replicates). Asterisks indicate significant differences as determined by a Student’s t-test, ***P < 0.001. (C) Averaged pot weights of kai2-2 and WT plants during drought stress (n = 9 biological replicates). Black arrows indicate when water was added to the root growth area in the soil. Red arrow indicates when biomass was measured. (D-E) WT and kai2-2 mutant plants were grown on water-saturated soil for 8 days. Watering was then stopped for 7 days (D) and 14 days (E).
Water loss, drought-induced cell membrane damage, and stomatal opening are increased in kai2 plants
We next investigated the physiological basis for drought sensitivity in kai2-2 mutant plants. Consistent with their reduced drought resistance, both kai2-2 and kai2-4 plants had lower relative water content (RWC) than WT on drying soil at similar soil moisture contents (Fig 2A and 2B, S1A and S1B Fig). A faster rate of water loss during drought might be attributable to several factors, such as increases in cell permeability and/or gas exchange through stomata. We used an electrolyte leakage assay to assess cell membrane integrity, and found that both kai2-2 and kai2-4 plants had higher electrolyte leakage than WT during water deficit (Fig 2C and S1C Fig). Notably, kai2-2 and kai2-4 plants had higher electrolyte leakage than WT even at time points when both genotypes had similar RWC (Fig 2D and S1D Fig). This finding implies that the degree of cell-membrane injury induced by water deprivation is more severe in kai2, contributing to its drought-susceptible phenotype.
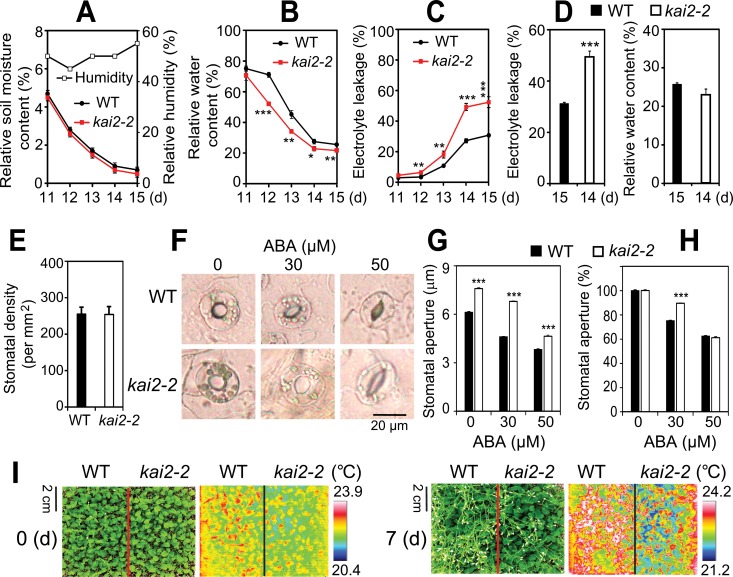
Fig 2. Drought-associated traits of kai2-2 leaves.
(A-C) kai2-2 and WT plants were grown and exposed to drought. At indicated time points, (A) soil relative moisture contents (n = 10) and relative humidity, (B) leaf relative water content (RWC) (n = 4 biological replicates), and (C) electrolyte leakage (n = 4 biological replicates) were determined. (D) Electrolyte leakage (Left) of kai2-2 and WT plants at a similar RWC (Right) during drought treatment (n = 4 biological replicates). Data represent the means and standard errors. (E) Average stomatal density of rosette leaves (abaxial side) from 14-day-old plate-grown kai2-2 and WT plants. Error bars represent standard errors (n = 15 leaves/15 independent plants/genotype). (F) Representative guard cells of rosette leaves from 21-day-old plate-grown kai2-2 and WT plants treated with 0, 30 and 50 μM ABA. (G-H) Average width of stomatal aperture of rosette leaves from 21-day-old WT and kai2-2 plants in the presence or absence of ABA. Aperture width are shown in micrometers (G) or in percentage of the average aperture width obtained from absence of ABA treatment (H). Error bars represent standard errors (n = 5 plants; for each plant the average of nine stomatal measurements from a single leaf was calculated). (I) Leaf surface temperatures of well-watered WT and kai2-2 plants before (0 d) and after a 7-d drought period (7 d). Plants were 21-d-old at the start of water withholding. Common optical camera (Left) and thermal imaging camera (Right) were used to take pictures at the same time. Asterisks indicate significant differences between the genotypes under well-watered control or drought conditions as determined by a Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.001.
We assessed whether differences in stomatal density or movement might also affect the rate of water loss in kai2-2 plants. Stomatal density was comparable between WT and kai2-2 rosette leaves (Fig 2E). However, kai2-2 had larger stomatal apertures than WT plants (Fig 2F and 2G). As ABA is induced during drought and promotes stomatal closure [21], we examined the responses of kai2-2 and WT stomata to ABA treatment. The stomatal apertures of kai2-2 plants were significantly larger than WT in the presence of 30 and 50 μM ABA (Fig 2F and 2G). Normalizing the ABA-treated data against the untreated apertures of WT and kai2-2 stomata indicated that kai2-2 stomata undergo a similar degree of closure as WT in the presence of 50 μM ABA, but are somewhat less sensitive to an intermediate (30 μM) ABA concentration (Fig 2H).
These results together suggested that kai2 plants may have increased transpiration rates during water deficit. We used leaf surface temperature measurements as a proxy to compare transpiration between kai2 and WT plants. We found that the leaf surfaces of both kai2-2 and kai2-4 plants were remarkably cooler than WT before and after 7 d of water withholding (Fig 2I and S1E Fig). Our findings collectively indicate that the differences in RWC due to stomatal water losses enhance the susceptibility of kai2 plants to drought.
KAI2 promotes ABA responses and ABA catabolism
The partial reduction in ABA-induced stomatal closure in the kai2 mutant raised the question of whether ABA responses are broadly impaired, which would likely impact drought adaptation [21]. To test this, we compared the inhibitory effects of ABA on cotyledon opening of kai2 and WT seedlings. While the cotyledon opening rates of kai2-2 and kai2-4 mutants were similar to WT in the absence of ABA, both kai2 alleles showed decreased sensitivity to ABA treatments (Fig 3A and S1F Fig). Thus, KAI2 positively regulates multiple ABA responses in Arabidopsis.
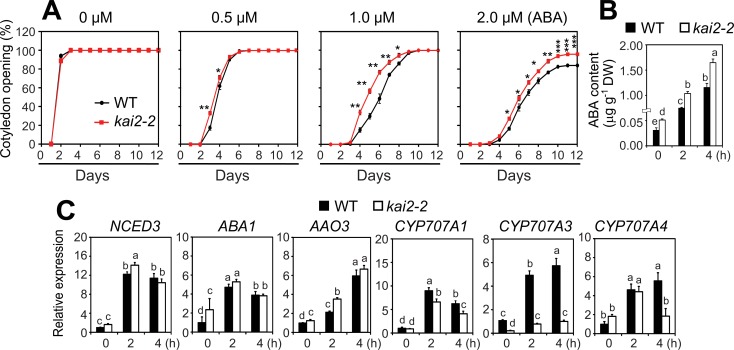
Fig 3. KAI2 effects on ABA responses and metabolism.
(A) Cotyledon opening percentage of kai2-2 and WT seedlings in the absence or presence of different concentrations of exogenous ABA. Data represent the means and standard deviation of 3 independent experiments (n = 50 seeds/genotype/experiment). Asterisks indicate significant differences as determined by a Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.001. (B) Endogenous ABA contents in leaves of 24-d-old kai2-2 and WT plants under normal and dehydration conditions. Data represent the means and standard errors (n = 5 plants). (C) Expression of genes involved in ABA biosynthesis and catabolism in leaves of 24-day-old kai2-2 and WT plants under normal and dehydration conditions. Relative expression levels were normalized to a value of 1 in the WT grown under normal conditions. Data represent the means and standard errors (n = 5 biological replicates). The different letters above the error bars indicate significant differences (P < 0.05) in all combinations according to a Tukey's honest significant difference test.
Next, we investigated whether KAI2 might also influence endogenous ABA levels in plants, especially under drought. We measured ABA content in the leaves of kai2-2 and WT plants during a 4 h time course of dehydration. Interestingly, kai2-2 leaves had significantly increased endogenous ABA contents than WT before and during dehydration (Fig 3B). To determine whether this was due to differential regulation of ABA metabolism-related genes, we analyzed the expression of several genes involved in ABA biosynthesis (NCED3, ABA1, and AAO3) and catabolism (CYP701A1, 3, and 4) with quantitative real-time RT-PCR (qRT-PCR). The most striking finding was that CYP707A3 transcripts were reduced several-fold in both well-watered and dehydrated kai2-2 plants in comparison to WT controls (Fig 3C). CYP707A3 encodes a major 8'-hydroxylase of ABA that is highly induced during dehydration [22]. Loss of CYP707A3 increases ABA content but, in contrast to kai2-2, causes ABA hypersensitivity [22]. Putatively, decreased sensitivity to endogenous ABA in kai2-2 might shift ABA homeostasis to higher levels through feedback reduction of ABA catabolism.
Comparative transcriptome analysis of KAI2 function during dehydration
To gain broader insights into how KAI2 signaling contributes to drought resistance, we performed transcriptomic profiling of kai2 and WT leaves undergoing dehydration. A microarray analysis was conducted as illustrated in Fig 4A and 4B. The microarray data can be accessed through accession number GSE90622, and the results of the transcriptome analysis are summarized in S1 Table. A preset criteria of |fold change ≥ 2| and false discovery rate-corrected P-values (i.e. q-values) <0.05 was used to identify differentially expressed genes (DEGs) in each comparison (Fig 4B and S2 Table). The microarray data was validated by expression analysis of several selected genes using qRT-PCR (S2 Fig).
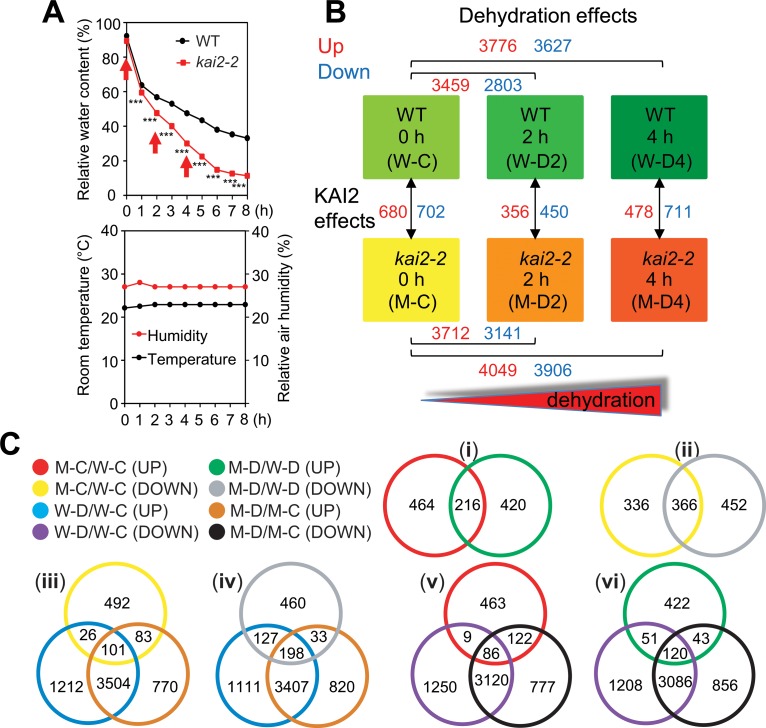
Fig 4. Transcriptome analysis of kai2-2 and WT plants under normal and dehydration conditions.
(A) Relative water content (RWC) of leaves from 24-d-old well-watered kai2-2 and WT plants exposed to dehydration treatment. Data represent the means and standard errors (n = 5 plants). Asterisks indicate significant differences according to a Student’s t-test, ***P < 0.001. Rosette leaf samples collected in 3 biological repeats at 0, 2 and 4 h (arrows) were used for microarray analysis. Room temperature and relative room humidity were recorded during the dehydration period. (B) Diagrams illustrating experimental design and comparisons between the treatments. The number of differentially expressed genes (DEGs) identified from various comparisons are noted in red (upregulated relative to control) or blue (downregulated relative to control). Data were obtained from the microarray analysis of 3 biological repeats. (C) Venn diagram analysis showing the overlapping and non-overlapping DEGs among the comparisons. M-C/W-C, kai2-2 well-watered control 0 h vs. WT well-watered control 0 h; M-D2/W-D2, kai2-2 dehydrated 2 h vs. WT dehydrated 2 h; M-D4/W-D4, kai2-2 dehydrated 4 h vs. WT dehydrated 4 h; M-D/W-D, M-D2/W-D2 and/or M-D4/W-D4; W-D2/W-C, WT dehydrated 2 h vs. WT well-watered control 0 h; W-D4/W-C, WT dehydrated 4 h vs. WT well-watered control 0 h; W-D/W-C, W-D2/W-C and/or W-D4/W-C; M-D2/M-C, kai2-2 dehydrated 2 h vs. kai2-2 well-watered control 0 h; M-D4/M-C, kai2-2 dehydrated 4 h vs. kai2-2 well-watered control 0 h; M-D/M-C, M-D2/M-C and/or M-D4/M-C.
Under non-stressed, well-watered conditions, 680 transcripts were upregulated and 702 were downregulated in kai2-2, as indicated by the M-C/W-C (kai2-2 control/WT control) comparison (Fig 4B and S2A–S2K Table). We compared the DEGs in well-watered kai2-2 (M-C/W-C) to those we previously identified in well-watered max2-3 [5]. We found 66 downregulated genes and 56 upregulated genes in common between kai2-2 and max2-3 plants (S3 Table). The downregulated genes included several prominent KAR-induced transcripts that were identified in prior studies, such as DLK2 (At3g24420), BZS1/STH7 (At4g39070), KUF1 (At1g31350), GA3ox1, SMAX1 (At5g57710), SMXL2 (At4g30350), and At3g60290 encoding a 2OG-Fe(II) oxidoreductase [11,23–25]. Consistent with our qRT-PCR analysis (Fig 3C), CYP707A3 was also among this downregulated set of genes. Therefore, our analysis can provide reliable identification of genes that are regulated by KAI2.
We found that 216 (31.8%) of the upregulated transcripts and 366 (52.1%) of the downregulated transcripts in well-watered kai2-2 plants were differentially expressed in a similar manner after 2 h and/or 4 h of dehydration (see comparison to M-D/W-D; Fig 4C(i) to 4C(ii) and S4A and S4B Table). Another 420 and 452 unique genes were respectively upregulated or downregulated in kai2-2 relative to WT only after 2 h and/or 4 h dehydration, which may indicate genes whose regulation by KAI2 is only revealed under the drought condition (Fig 4C(i) and 4C(ii)). A high number of DEGs in kai2-2 plants were also dehydration-responsive. Specifically, 29.9% and 43.8% of the genes downregulated in kai2-2 under well-watered (M-C/W-C) or dehydration conditions (M-D/W-D), respectively, were induced by dehydration [Fig 4C(iii) and 4C(iv), and S4F and S4J Table]. Conversely, 31.9% to 33.7% of the genes upregulated in kai2-2 in this experiment were repressed by dehydration [Fig 4C(v) and 4C(vi), and S4O and S4S Table].
Transcriptome analysis of kai2 mutant reveals anthocyanin and cuticle defects
We hypothesized that some of these transcriptional perturbations might reveal altered biochemical or physiological responses that contribute to the reduced drought resistance of the kai2 plants. Therefore, we used MapMan to perform an in-depth survey of the functional categories of the DEGs identified in kai2-2 plants (S2–S5 Figs). We found two gene categories to be of particular interest.
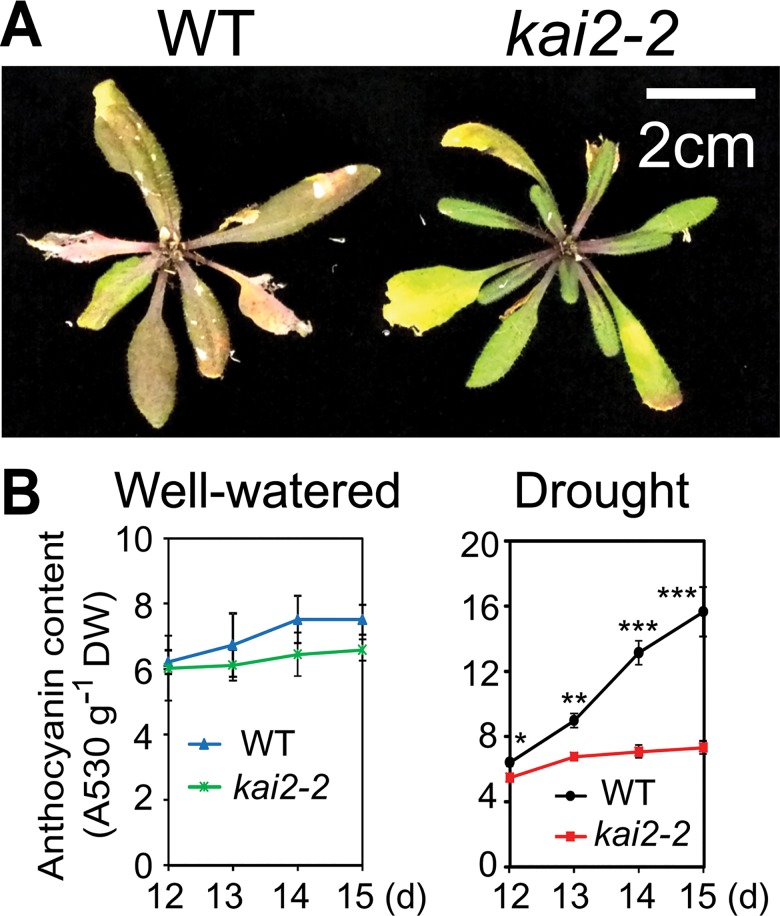
First, we noted that many dehydration-inducible genes that positively regulate or carry out the synthesis of anthocyanins/flavonoids, such as DFR, FLS1, F3’H, GL3, MYB75/PAP1 and MYB90/PAP2 [26], were downregulated in kai2-2 (S2–S4 Figs and S5A Table). By contrast, genes encoding negative regulators of anthocyanin biosynthesis, such as MYBL2, LBD37, and LBD39 [26], were dehydration-repressible and upregulated in kai2-2 under normal or dehydration conditions (S2–S4 Figs and S5A Table). Thus, we hypothesized that anthocyanin content might be lower in kai2 plants than WT, particularly during water deficit. Because anthocyanins are known to provide protection to plants against drought [27], reduced anthocyanin content might contribute to the drought susceptibility of kai2 plants. We observed that WT plants developed a darker leaf coloration than kai2-2 mutants at late developmental stages, especially after water was withheld for seed maturation (Fig 5A). We also compared the anthocyanin contents in kai2 and WT plants at various time points during a drought resistance assay. Anthocyanin levels were significantly lower in both kai2-2 and kai2-4 plants than WT under water deficit conditions (Fig 5B and S6A Fig).
Fig 5. Anthocyanin production in kai2-2 and WT plants.
(A) kai2-2 and WT plants were grown for 5 weeks, and watering was withheld for 10 days. Inflorescences were cut from representative plants before photographing. (B) Anthocyanin content in kai2-2 and WT plants under well-watered and drought conditions. Data represent the means and standard errors (n = 4 plants). Asterisks indicate significant differences between the genotypes under drought conditions as determined by a Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.001.
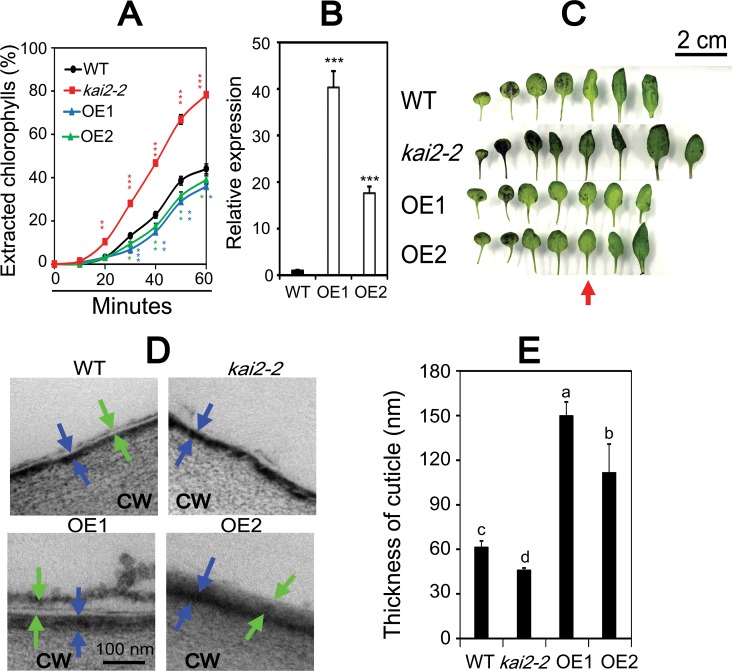
Second, many genes involved in cuticle formation, such as CER1, CER4, CYP96A15, MYB94, SHN1/WIN1, SHN2, SHN3 and ABCG13 [4,28], were found to be dehydration-inducible and downregulated in unstressed and/or dehydrated kai2-2 leaves (S2, S4, S5 Figs and S5B Table). We hypothesized that a cuticular defect could cause enhanced non-stomatal water loss that might explain the faster rate of RWC decline observed in drought-stressed kai2 plants. To examine this possibility, we carried out a chlorophyll (Chl) leaching assay of rosette leaves (Fig 6A). We found that Chl leached much faster from leaves of both kai2-2 and kai2-4 than that of WT (Fig 6A and S6B Fig), suggesting that kai2 mutants have higher cuticular water permeability than WT. In contrast, we noted lower Chl leaching rates from 35S:KAI2 overexpressor OE1 and OE2 plants than WT (Fig 6A and 6B), indicating that OE1 and OE2 plants have lower cuticular water permeability than WT. We also used toluidine blue (TB) staining to visualize potential defects in the leaf cuticle of kai2 mutants. More leaves, especially the older leaves, of kai2-2 and kai2-4 mutants were stained compared with WT, whereas reduced staining was observed in the leaves of OE1 and OE2 relative to that of WT (Fig 6C and S6C Fig), supporting the results of the Chl leaching assay (Fig 6A and S6B Fig). Furthermore, we examined the surface of the fifth leaves of kai2-2, OE1, OE2 and WT plants with transmission electron microscopy (TEM). A cuticle proper, which is a layer of lipid polymer filled with wax [29], was detected in leaves of WT, OE1 and OE2, but not in kai2-2 leaves (Fig 6D), revealing a significant structural defect that is likely to contribute to faster water loss in kai2 plants. Additionally, kai2-2 mutant plants were found to have thinner cuticles than WT, whereas OE1 and OE2 lines have thicker cuticles (Fig 6E).
Fig 6. Cuticle permeability of kai2-2, 35S:KAI2 transgenic lines OE1 and OE2, and WT plants.
(A) Chlorophyll leaching from rosette leaves of 28-day-old kai2-2, OE1, OE2 and WT plants at different time periods. Data represent the means and standard errors (n = 3 plants/genotype). (B) Fold-change of overexpression levels of KAI2 gene in leaves of 14-day-old OE1 and OE2 plants in comparison with WT (n = 5 biological replicates). Asterisks indicate significant differences between the WT and other genotypes under well-watered condition as determined by a Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.001. (C) Toluidine blue staining patterns of rosette leaves of 28-day-old kai2-2, OE1, OE2 and WT plants. Red arrow indicates the fifth leaves, which were used for transmission electron microscope (TEM) analysis.(D) TEM images of the surface of the fifth leaves (adaxial side) derived from kai2-2, OE1, OE2 and WT plants. CW, cell wall. Blue arrows indicate cuticular layer (electron-dense, darker-staining layer) and green arrows indicate wax-rich cuticle proper (electron-translucent layer). (E) Thickness of cuticle of the fifth leaves (adaxial side) derived from kai2-2, OE1, OE2 and WT plants. Data represent the means and standard errors (n = 3 biological replicates). Different letters above the error bars indicate significant differences (P < 0.05) among the genotypes according to a Tukey's honest significant difference test.
D14 also contributes to drought resistance
The role of SLs in drought resistance of Arabidopsis has been unclear. Bu et al. (2014) reported that max2 mutants have defects in drought survival, higher water loss from detached leaves, and decreased germinative greening under stress or ABA treatments, but SL-deficient max mutant plants resemble WT [6]. In contrast, Ha et al. (2014) showed reduced drought resistance and ABA hyposensitivity in max2 and SL-deficient max mutants [5]. Supporting this, SL-depleted tomato and L. japonicus have reduced resistance to drought and osmotic stress, respectively [7,8]. These conflicting observations may be reconcilable if SLs are only responsible for some of MAX2-dependent drought resistance.
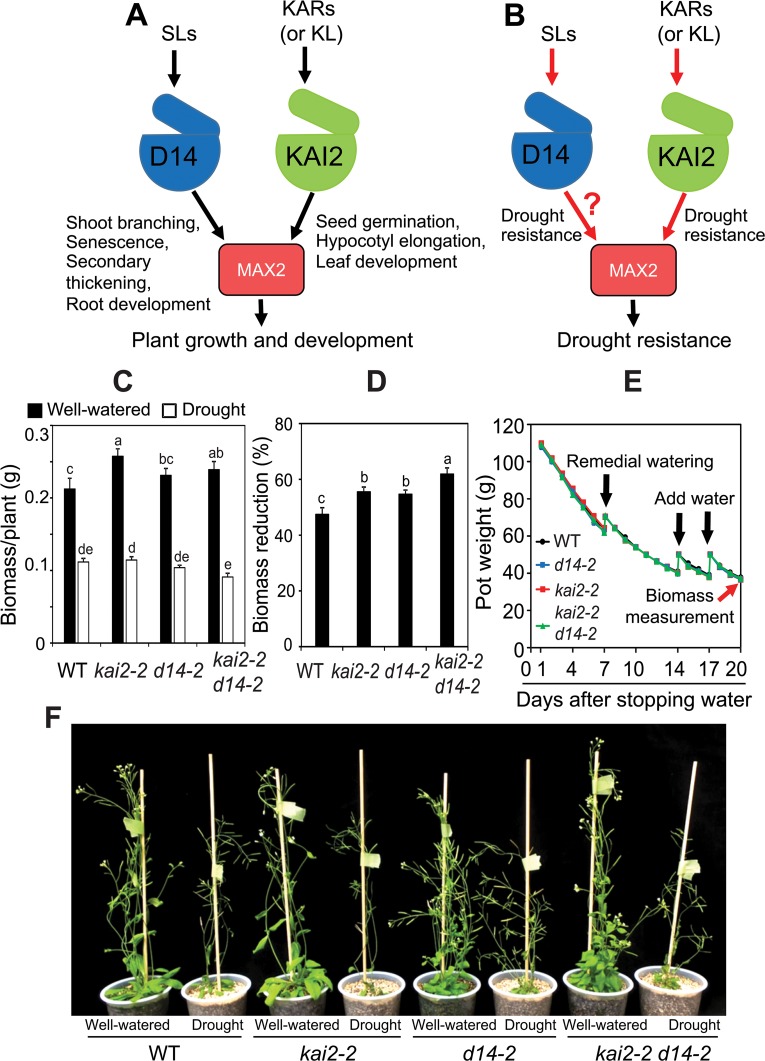
To date, all SCFMAX2-dependent SL responses in plant growth and development have been shown to be mediated by the SL receptor D14 (Fig 7A). Because our experiments showed that KAI2 is involved in drought resistance, it raised the question of whether D14 also participates in this response (Fig 7B). Thus, we analyzed drought resistance of the d14-2 allele [30]. We found that d14-2 had a higher reduction in biomass than WT in the gravimetric method-based drought-resistance assay (Fig 7C–7F). Therefore, D14 acts as a positive regulator of drought resistance. In addition, we noted that kai2-2 d14-2 double mutants were less able to accumulate biomass under water restrictions than either of the single mutants (Fig 7C–7F), suggesting that both D14- and KAI2-mediated signaling pathways may act together, perhaps through MAX2 [9,11,12], to enhance drought resistance in Arabidopsis. Interestingly, we did not observe cuticle defect in d14-2 leaves like in that of kai2-2 single and kai2-2 d14-2 double mutants (S7 Fig), suggesting that drought-sensitive phenotype of d14 plants is not associated with defect in cuticle development.
Fig 7. Models for functions of SLs and KARs/KAI2-ligand (KL) in plant growth, development and drought response.
(A) SL-regulation of shoot branching, senescence, secondary thickening and root development is mediated by SL-specific receptor D14. KAR-regulation (or hypothetical KL-regulation) of seed germination, hypocotyl elongation and leaf development is mediated by KAI2. MAX2 is the checkpoint for both SL and KAR/KL signaling pathways in plant growth and development. (B) SLs and KARs/KL regulate plant resistance to drought through D14-MAX2 and KAI2-MAX2 cascade, respectively. Question mark indicates the contribution of D14 to drought resistance was unknown until this study. (C) Biomass of kai2-2, d14-2, kai2-2 d14-2 and WT plants under well-watered control and drought stress (n = 9 biological replicates). (D) Biomass reduction of kai2-2, d14-2, kai2-2 d14-2 and WT plants under drought relative to respective well-watered control. Data represent the means and standard errors (n = 9 biological replicates). Different letters above the error bars indicate significant differences (P < 0.05) among the genotypes according to a Tukey's honest significant difference test. (E) Averaged pot weights of kai2-2, d14-2, kai2-2 d14-2 and WT plants during drought stress (n = 9 biological replicates). Black arrows indicate when water was added to the root growth area in the soil. Red arrow indicates when biomass was measured. (F) Plant phenotypes before harvest.
Discussion
Although KARs and SLs typically have distinct effects on plant growth and development, these signals are perceived by similar mechanisms that require MAX2 [12,31]. Several studies previously demonstrated that MAX2 and SLs have positive regulatory roles in plant adaptation to drought [5–8]; but possible contributions of KARs or KL had not been examined. In this study, we aimed to investigate the roles of KAI2-dependent signaling in plant response to drought using in-depth physiological, biochemical and molecular characterization of the loss-of-function kai2 mutants. We implicated KAI2-dependent signaling in plant response to drought as a positive regulator.
Our analyses of the kai2 mutants provided evidence that its reduced biomass under drought was associated with an inability to maintain high leaf RWC during water deficit (Fig 1B, Fig 2A and 2B, S1B Fig). A higher transpiration rate, which increases water loss, was suggested by remarkably lower leaf temperatures in kai2 plants than in WT under both well-watered and water-deficit conditions (Fig 2I and S1E Fig). We identified several ways in which KAI2-dependent signaling likely contributes to drought resistance. First, it is well-established that cell membrane stability, reflected by the levels of cellular electrolyte leakage, is a major factor contributing to the maintenance of water status in plants during water deficit [32,33]. Thus, the increase in electrolyte leakage linked with reduced RWC recorded in the kai2 mutants during drought (Fig 2A–2D and S1A–S1D Fig) suggested that kai2 suffered a severe stress-induced cell membrane damage. This might in part result from a decrease in reactive oxygen species (ROS)-scavenging antioxidants [33], as supported by the observed reduction in endogenous anthocyanin levels (Fig 5B and S6A Fig), ultimately leading to a higher rate of water loss.
Second, a reduction in ABA-regulated stomatal closure in kai2 plants, which may contribute to increased water loss (Fig 2F–2H), might enhance drought susceptibility. The impaired stomatal movement in kai2 mutants was in good agreement with the strong downregulation of ABCG40/AT1G15520 and ABCG22/AT5G06530 in both unstressed and stressed kai2 plants (12.1- and 17.7-fold for ABCG40, and 3.92- and 1.79-fold for ABCG22, respectively, in normal and dehydrated kai2 vs WT) (S1 Table). ABCG40 is a key ABA transporter, and ABCG22 putatively has a similar function; these genes were shown to regulate stomatal closure and drought resistance in Arabidopsis [34,35]. The abcg40 mutant has inefficient ABA-mediated stomatal closure [34], whereas abcg22 shows a defect in stomatal closing that is perhaps dependent on ABA signaling [35]. Both abcg22 and abcg40 mutants exhibited drought-susceptible phenotypes, further supporting the link [34,35]. Notably, previous studies reported that the drought-susceptible max2 plants also have impaired ABA-mediated stomatal closure [5,6], and exhibit downregulation of ABCG22 and ABCG40 under both normal and dehydration conditions [5]. Furthermore, the impairment of ABA-mediated stomatal closure and increased cotyledon opening rates of kai2 mutants treated with exogenous ABA (Figs 2F–2H and 3A, S1F Fig) indicate hyposensitive responses to ABA. This finding suggests that crosstalk between ABA- and KAI2-dependent signaling pathways may influence plant adaptation to drought. We noted an increase in ABA content in kai2 leaves during dehydration (Fig 3B), which might be attributed to the downregulation of the key ABA catabolic enzyme CYP707A3 (Fig 3C and S2 Fig). This result suggests that kai2 mutants may produce higher levels of ABA to compensate for its reduced ability to respond to ABA; such a feedback mechanism may attenuate the severity of the kai2 drought-resistant phenotype.
Third, non-stomatal water evaporation associated with higher cuticular permeability is likely to increase the rate of RWC decline of kai2 plants under drought [28]. Unlike in leaves of WT, in leaves of kai2 plants we did not detect a cuticle proper (Fig 6D). Additionally, we identified thinner cuticles in leaves of kai2 than that of WT by using TEM analysis (Fig 6D and 6E), and observed higher permeability through a Chl leaching assay and TB staining (Fig 6A and 6C, S6B–S6C Fig). At a molecular level, our comparative transcriptome analysis suggested that the altered structure of the kai2 cuticle may be due to downregulation of several genes involved in the biosynthesis and transport of wax, such as CER1, CYP96A15, WSD1, MYB94, MYB16, SHN1, SHN2 and SHN3 (S3–S6 Figs) [4,36]. Some of the KAI2-regulated cuticle formation-related genes, such as the MYB16 and MYB94 transcription factors, are also controlled by ABA [4]. Similar to our observations, the drought-sensitive max2 mutant plants were found to have a defect in cuticular architecture [6], which might be attributed to downregulation of genes involved in cuticle formation as well (S5B Table) [5]. Additionally, loss of KAI2 also downregulated the expression of cutin biosynthesis-related genes (S5 Fig). For example, CED1/At1g64670 [37,38], which minimizes water loss through not only by enhancing cuticle structure but also by mediating ABA and osmotic stress signaling [39], was downregulated in kai2 versus WT after 4 h dehydration (S5 Fig). These findings support the positive role of KAI2 in mediating drought resistance, and the link among cuticle formation, KAI2 signaling, ABA signaling and osmotic stress responses in plants.
Fourth, the decline in anthocyanin production in kai2 plants during drought may contribute to its enhanced drought sensitivity. This finding is supported by a number of studies that have found a positive correlation between drought resistance and anthocyanin levels in Arabidopsis as the ROS-scavenging antioxidant function of anthocyanins can protect cells from drought [27,40]. The reduced anthocyanin accumulation in kai2 plants (Fig 5B and S6A Fig) may be explained by transcriptional misregulation of the anthocyanin biosynthetic pathway (S2–S4 Figs). In agreement with this idea, several anthocyanin/flavonoid biosynthesis-related genes were found to be upregulated in KAR1-treated Arabidopsis seeds, and downregulated in kai2 seedlings grown under different light conditions, leading to reduced accumulation of anthocyanin pigments [24,41]. Similarly, many anthocyanin biosynthesis-related genes were shown to be downregulated in max2 plants under dehydration conditions (S5A Table) [5]. Furthermore, a quantitative proteomic analysis of max2 seedlings identified a set of proteins involved in flavonoid biosynthesis that have reduced abundance relative to WT; a subset of these proteins are induced by rac-GR24 treatment [42]. Both purified enantiomers of GR24 were able to stimulate flavonol production, and importantly, rac-GR24 enhanced flavonol accumulation in both d14 and kai2 mutants [42]. Therefore, there is ample evidence to support regulation of anthocyanin and flavonoid syntheses by both MAX2-dependent D14 and KAI2 signaling pathways. Thus, we propose that accumulation of anthocyanin under drought might be an important aspect of stress resistance conferred by KAI2. Altogether these observations support the involvement of a KAR/KL-KAI2-SCFMAX2 signaling cascade in regulating plant drought adaptation through controlling cell membrane stability, stomatal movement, cuticle development and anthocyanin production.
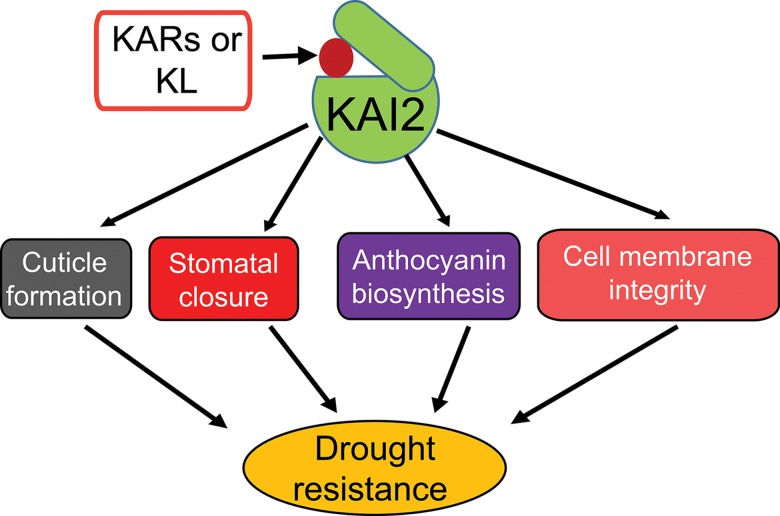
In summary, our results demonstrate that KAI2-mediated signaling positively regulates drought resistance in Arabidopsis. KAI2 is activated through the binding of KARs (or as-yet-unknown KL), resulting in the regulation of downstream genes involved in cuticle formation, stomatal closure, anthocyanin biosynthesis and membrane integrity. These adjustments may collectively contribute to plant adaptation to drought (Fig 8). Importantly, our results suggest that genetic manipulation to enhance KAI2 and D14 signaling pathways, either alone or together, is a promising avenue for the improvement of crop productivity in arid lands.
Fig 8. Model illustrating functions of KAI2 in plant resistance to drought.
Karrikins (KARs) or a putative, endogenous KAI2 ligand (KL) activate KAI2 signaling, which promotes plant resistance to drought through several biochemical and physiological processes.
Materials and methods
Plant materials
Arabidopsis thaliana Columbia ecotype (Col-0) was used as WT in all experiments. The kai2-2 (SGT6839) and kai2-4 (GT6185) alleles were obtained from the Arabidopsis Biological Resource Center [11,43]. The kai2-2 and kai2-4 mutants were originally in the Ler background and were backcrossed with Col-0 six times [43]. The d14-2 mutant was obtained from the TILLING project (http://tilling.fhcrc.org/) after backcrossing twice with Col-0 [30]. The kai2-2 d14-2 double mutant was made by crossing the d14-2 and kai2-2 mutants. To make KAI2-overexpressing plants, the KAI2 cDNA was amplified by PCR (primers listed in S6 Table), and cloned into the pGWB2 expression vector under the control of the CaMV35S promoter [44]. The resulting 35S:KAI2 plasmid was introduced into WT (Col-0) plants using Agrobacterium tumefaciens-mediated transformation method [45]. Two homozygous 35S:KAI2 transgenic lines OE1 and OE2 with a single transgene copy obtained by selection for three consecutive generations were used in the study.
Drought resistance assayes
Drought-responsive phenotypes were examined using the gravimetric method described by Harb and Pereira [20]. Briefly, 2-week-old plants grown on germination medium (GM) were transferred to plastic pots (7×7 cm in diameter and height) containing 28.7 g dry soil (Dio Propagation Mix No.2 for Professional; Dio Chemicals Ltd.). Plants were then grown on pots saturated with water for 10 days before they were exposed to drought stress. After watering was stopped for 7 days, remedial watering of several pots was conducted to ensure that all pots had almost the same amount of soil water content. Seven days after remedial watering, each pot received a suitable amount of water to reach the weight of 60 g. The pots were then dried for 3 days; thereafter, they received a suitable volume of water to again reach the weight of 60 g. This process was repeated two times so that each pot had similar soil water content during drying. The weight of the pots was measured every day during the experiment. Twenty days after drought stress, whole aerial parts of plants were cut and packed in paper bags. The well-watered plants were also harvested at the same time. The bags were then oven-dried at 65°C for two days, and the dry weight (biomass) of aerial parts of each plant was measured. The biomass reduction was calculated using the following equation:
In addition, we also adapted the method of Bu et al. (2014) [6] to compare the drought resistance of the kai2 mutant and WT plants grown in high density. Seeds of mutant and WT plants (50 seeds/each/pot) were sown directly side-by-side in the same pots (7×7 cm in diameter and height) containing soil (Dio Propagation Mix No.2 for Professional; Dio Chemicals Ltd.) saturated with water. Eight days after sowing, water was withheld from plants for 14 days. Photographs were taken at days 7th (15-day-old plants) and 14th (22-day-old plants) during the drought assay.
RWC and electrolyte leakage of plants exposed to drought stress
RWC and electrolyte leakage of the detached aerial parts of different genotypes exposed to drought stress were determined according to Nishiyama et al. (2011) [46]. Briefly, two-week-old mutants and WT plants grown on GM were transferred side-by-side (30 plants for each phenotype) to trays (21×30×5cm in width, length and height) containing soil (Dio Propagation Mix No.2 for Professional; Dio Chemicals Ltd.) saturated with water. After one week of transfer, water irrigation was stopped to induce drought stress. During the assay, relative soil moisture content of both sides of the trays was followed using a HydroSense soil moisture probe (Campbell Scientific Inc.) as previously described [46]. Shoots of well-watered and drought-stressed plants (n = 4/genotype) were harvested every day from day 11 to day 15 for determination of RWC using the procedure adapted from Barrs and Weatherley [47]. Briefly, detached shoot samples were weighed to determine the weight (W) of each individual, and the samples were then placed into 50-mL tubes containing 40 mL of deionized water for 3-hour rehydration to full turgidity under room temperature. After rehydration, water was gently removed from shoot samples by filter paper, and the turgid weight (TW) of each sample was determined. Subsequently, the shoot samples were put into paper bags, oven-dried (65°C) for 48 h, and then dry weight (DW) of each oven-dried sample was measured. RWC was calculated by using the following equation: RWC (%) = [(W-DW)/(TW-DW)] × 100. Detached aerial parts of different genotypes (n = 4/genotype) exposed to drought stress as described above were also harvested for determination of electrolyte leakage as previously described by Nishiyama et al. (2011) [46].
Stomatal aperture and density
Stomatal aperture and stomatal density were measured as previously described [5], with a slight modification for stomatal aperture closure assay. Epidermal peels from leaves of 21-day-old plants grown on GM plates were incubated in a solution containing 0.2 mM CaCl2, 10 mM KCl, and 10 mM Mes·KOH (pH 6.15) under white light (300 μmol·m−2·s−1) for 12 h. Thereafter, the samples were incubated in the same buffer solution containing different concentrations of ABA for 1 h before the stomatal aperture was measured.
Leaf surface temperature and quantification of ABA
Surface temperature of leaves was determined by using a thermal video system (TVS-8500; Nippon Avionics) [48]. Quantification of ABA in leaves of 24-day-old WT and kai2-2 plants grown on soil, which were detached and exposed to dehydration according to published method [5], was performed as previously described [49].
Cotyledon opening assay
Cotyledon opening assay was used as a means to assess the ABA sensitivity of various genotypes. Seeds were sowed on GM containing 1% sucrose and various concentrations of ABA, and opened cotyledons were counted according to published method [46].
Anthocyanin content
WT and mutant plants were grown on the same tray and subjected to a drought treatment as described above. At indicated time points, aerial parts (without inflorescence) of stressed and well-watered plants were separately collected, and frozen dry weight and anthocyanin content were measured [50].
Chl leaching assay and TB staining
Chlorophyll leaching assays were performed as described previously [6]. Briefly, the aerial parts (without inflorescence) of 4-week-old plants were incubated on ice for 30 min, and then immersed in 40 mL 80% ethanol (v/v) at room temperature. Solution samples (100 μL) were taken every 10 min after immersion to quantify the amount of chlorophyll content. TB staining was used to detect cuticular defects on leaves [51]. Aerial parts (without inflorescence) of 4-week-old plants were submerged into a solution of 0.05% (w/v) TB for 2 h. Plants were then gently transferred to water and softly shaken to remove excessive TB. Rosette leaves were separated and placed on dry paper for taking photos.
TEM analysis
For cuticle observation, the fifth rosette leaves from 4-week-old kai2-2, d14-2, kai2-2 d14-2, 35S:KAI2 (OE1, OE2), and WT plants were analyzed as described previously [52], with a slight modification. Briefly, the top part of the leaves (5 mm) was cut into 1 × 3 mm rectangles and fixed with 4% paraformaldehyde and 2% glutaraldehyde in 50 mM sodium cacodylate buffer (pH 7.4) overnight at 4°C. The ultrathin sections were observed with a JEOL JEM-1400 TEM at 80 kV. Detailed observation and analysis using TEM were performed according to Toyooka et al. (2000) [53]. The thickness of the cuticle was measured using ImageJ software (https://imagej.nih.gov/ij/index.html).
Dehydration treatment of soil-grown seedlings and transcriptome analysis
Aerial rossette leaves of 24-day-old WT and kai2-2 plants grown on soil under well-watered conditions were detached and exposed to dehydration for the indicated time periods for determination of RWC and sample collection as previously described [5]. RWC of dehydrated shoot samples was determined following the same method adapted from Barrs and Weatherley [47] described above. Leaves of WT and kai2-2 plants treated by dehydration for 0, 2 and 4 h were collected in 3 biological repeats for transcriptome analysis using the Arabidopsis Oligo 44K DNA microarray (Version 4.0, Agilent Technology) [54]. The criteria of |fold-change ≥ 2| and a false discovery rate corrected p-value (q-value) <0.05 were used in identifying the DEGs [55]. The raw microarray data and detailed protocol were deposited in the Gene Expression Omnibus database (GSE90622). When necessary, MapMan (http://mapman.gabipd.org), or Arabidopsis eFP browser (http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi) were used to analyze the data.
qRT-PCR analysis
Total RNA was extracted using TRIzol Reagent (Invitrogen). Previously described procedures were used for cDNA synthesis and qRT-PCR analysis [56], in which UBQ10 was used as a reference gene. Primer pairs used in qRT-PCR are listed in S6 Table.
Supporting information
(A-C) kai2-4 and WT plants were grown and exposed to drought. At indicated time points, (A) soil relative moisture contents (n = 10) and relative humidity, (B) leaf relative water content (RWC) (n = 4 biological replicates), and (C) electrolyte leakage (n = 4 biological replicates) were determined. (D) Electrolyte leakage (Left) of kai2-4 and WT plants at a similar RWC (Right) during drought treatment (n = 4 biological replicates). (E) Leaf surface temperature of well-watered (21-day-old) kai2-4 and WT plants. Thermal imaging camera (Top) and common optical camera (Bottom) were used to take pictures at the same time. (F) Cotyledon opening percentage of kai2-4 and WT seeds in the absence or presence of different concentrations of exogenous ABA. Data represent the means and standard errors of 3 independent experiments (n = 50 seeds/genotype/experiment). Asterisks indicate significant differences as determined by a Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
(A) Heatmap presentation indicates the fold-changes in expression of representative genes derived from microarray analysis. (B) Heatmap presentation indicates the fold-changes in expression of representative genes using qRT-PCR. Expression data were obtained from microarray analysis or qRT-PCR of 24-day-old Arabidopsis rosette leaf samples that were collected from 3 independent plants for microarray analysis (n = 3). UBQ10 was used as reference gene in qRT-PCR analysis. Relative expression levels are indicated by intensities of colors expressed in fold-change with saturation at 6. Red and blue colors indicate up- and downregulation, respectively. Note that not all data points shown in (a) passed the q-value < 0.05. M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h.
(TIF)
Heatmap presentation indicates the fold-changes in gene expression derived from microarray data. Relative expression levels are indicated by intensities of colors expressed in fold-change with saturation at 6. Red and blue colors indicate up- and downregulation, respectively. Note that not all data points shown passed the q-value < 0.05. M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h.
(TIF)
(A) Downregulated genes in M-C/W-C and/or M-D/W-D. (B) Upregulated genes in M-C/W-C and/or M-D/W-D. Green and red colors show down- and upregulation, respectively. Colored bars in each panel indicate fold-changes in gene expression. M-C/W-C, kai2-2 well-watered control versus WT well-watered control; M-D/W-D represents M-D2/W-D2 and/or M-D4/W-D4; M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h. For repetitive genes, their highest fold-change was used in the analysis.
(TIF)
Heatmap presentation indicates the fold-changes in gene expression derived from microarray data. Relative expression levels are indicated by intensities of colors expressed in fold-change with saturation at 6. Red and blue colors indicate up- and downregulation, respectively. Note that not all data points shown passed the q-value < 0.05. M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h.
(TIF)
(A) Anthocyanin content in kai2-4 and WT plants under drought conditions. Data represent the means and standard errors (n = 4 plants). (B) Chlorophyll leaching from rosette leaves of 28-day-old kai2-4 and WT plants at different time periods. Data represent the means and standard errors (n = 3 plants/genotype). (C) Toluidine blue staining patterns of rosette leaves of 28-day-old kai2-4 and WT plants. Asterisks indicate significant differences as determined by a Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
(A) Thickness of cuticle of the fifth leaves (adaxial side) derived from kai2-2, d14-2, kai2-2 d14-2 and WT plants. Blue arrows indicate cuticular layer (electron-dense, darker-staining layer) and green arrows indicate wax-rich cuticle proper (electron-translucent layer). (B) Transmission electron microscope images of the surface of the fifth leaves (adaxial side) derived from kai2-2, d14-2, kai2-2 d14-2 and WT plants. CW, cell wall. Data represent the means and standard errors (n = 3 biological replicates). Different letters above the error bars indicate significant differences (P < 0.05) among the genotypes according to a Tukey's honest significant difference test.
(TIF)
(XLS)
(A) List of upregulated genes in the M-C/W-C comparison. (B) List of upregulated genes in the M-D2/W-D2 comparison. (C) List of upregulated genes in the M-D4/W-D4 comparison. (D) List of upregulated genes in the M-D/W-D (e.g. in M-D2/W-C and/or M-D4/W-C) comparison. (E) List of upregulated genes in the W-D2/W-C comparison. (F) List of upregulated genes in the W-D4/W-C comparison. (G) List of upregulated genes in the W-D/W-C (e.g. in W-D2/W-C and/or W-D4/W-C) comparison. (H) List of upregulated genes in the M-D2/M-C comparison. (I) List of upregulated genes in the M-D4/M-C comparison. (J) List of upregulated genes in the M-D/M-C (e.g. in M-D2/M-C and/or M-D4/M-C) comparison. (K) List of downregulated genes in the M-C/W-C comparison. (L) List of downregulated genes in the M-D2/W-D2 comparison. (M) List of downregulated genes in the M-D4/W-D4 comparison. (N) List of downregulated genes in the M-D/W-D (e.g. in M-D2/W-D2 and/or M-D4/W-D4) comparison. (O) List of downregulated genes in the W-D2/W-C comparison. (P) List of downregulated genes in the W-D4/W-C comparison. (Q) List of downregulated genes in the W-D/W-C (e.g. in W-D2/W-C and/or W-D4/W-C) comparison. (R) List of downregulated genes in the M-D2/M-C comparison. (S) List of downregulated genes in the M-D4/M-C comparison. (T) List of downregulated genes in the M-D/M-C (e.g. in M-D2/M-C and/or M-D4/M-C) comparison. M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h.
(XLS)
(A) List of upregulated genes. (B) List of downregulated genes.
(XLS)
(A) Genes upregulated in M-C/W-C and M-D/W-D. (B) Genes downregulated in M-C/W-C and M-D/W-D. (C) Genes upregulated in W-D/W-C and M-D/M-C. (D) Genes downregulated in M-C/W-C and upregulated in W-D/W-C. (E) Genes downregulated in M-C/W-C and upregulated in M-D/M-C. (F) Genes downregulated in M-C/W-C and upregulated in W-D/W-C and/or M-D/M-C. (G) Genes downregulated in M-C/W-C and upregulated in both W-D/W-C and M-D/M-C. (H) Genes downregulated in M-D/W-D and upregulated in W-D/W-C. (I) Genes downregulated in M-D/W-D and upregulated in M-D/M-C. (J) Genes downregulated in M-D/W-D and upregulated in W-D/W-C and/or M-D/M-C. (K) Genes downregulated in M-D/W-D and upregulated in both W-D/W-C and M-D/M-C. (L) Genes downregulated in W-D/W-C and M-D/M-C. (M) Genes upregulated in M-C/W-C and downregulated in W-D/W-C. (N) Genes upregulated in M-C/W-C and downregulated in M-D/M-C. (O) Genes upregulated in M-C/W-C and downregulated in W-D/W-C and/or M-D/M-C. (P) Genes upregulated in M-C/W-C and downregulated in both W-D/W-C and M-D/M-C. (Q) Genes upregulated in M-D/W-D and downregulated in W-D/W-C. (R) Genes upregulated in M-D/W-D and downregulated in M-D/M-C. (S) Genes upregulated in M-D/W-D and downregulated in W-D/W-C and/or M-D/M-C. (T) Genes upregulated in M-D/W-D and downregulated in both W-D/W-C and M-D/M-C. M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h.
(XLS)
(A) List of DEGs related to anthocyanin biosynthesis. (B) List of DEGs related to cuticle formation.
(XLS)
(XLS)
Acknowledgments
We thank Y. Kanno and M. Wakazaki and H. Sakamoto for excellent assistance in ABA measurements, TEM analysis and stomatal movement assay, respectively.
Data Availability
The raw microarray data are available from the Gene Expression Omnibus database, accession number GSE90622 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90622). All other data are within the paper and its Supporting Information files.
Funding Statement
This project was supported in part by the Japan Society for the Promotion of Science (#17K07459 to LSPT), the Japan Science and Technology Agency (JST), Core Research for Evolutionary Science and Technology (CREST to MoS), and the National Science Foundation (IOS-1737153 to DCN). The ABA measurements were supported by the Japan Advanced Plant Science Research Network. MGM acknowledges the postdoc fellowship from the Japan Society for the Promotion of Science. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jogaiah S, Govind SR, Tran LS (2013) Systems biology-based approaches toward understanding drought tolerance in food crops. Crit Rev Biotechnol 33: 23–39. doi: 10.3109/07388551.2012.659174 [DOI] [PubMed] [Google Scholar]
- 2.Daszkowska-Golec A, Szarejko I (2013) Open or close the gate-stomata action under the control of phytohormones in drought stress conditions. Front Plant Sci 4: 138 doi: 10.3389/fpls.2013.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 5: 170 doi: 10.3389/fpls.2014.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui F, Brosche M, Lehtonen MT, Amiryousefi A, Xu E, et al. (2016) Dissecting abscisic acid signaling pathways involved in cuticle formation. Mol Plant 9: 926–938. doi: 10.1016/j.molp.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 5.Ha CV, Leyva-Gonzalez MA, Osakabe Y, Tran UT, Nishiyama R, et al. (2014) Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc Natl Acad Sci U S A 111: 851–856. doi: 10.1073/pnas.1322135111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bu Q, Lv T, Shen H, Luong P, Wang J, et al. (2014) Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol 164: 424–439. doi: 10.1104/pp.113.226837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, He H, Vitali M, Visentin I, Charnikhova T, et al. (2015) Osmotic stress represses strigolactone biosynthesis in Lotus japonicus roots: exploring the interaction between strigolactones and ABA under abiotic stress. Planta 241: 1435–1451. doi: 10.1007/s00425-015-2266-8 [DOI] [PubMed] [Google Scholar]
- 8.Visentin I, Vitali M, Ferrero M, Zhang Y, Ruyter-Spira C, et al. (2016) Low levels of strigolactones in roots as a component of the systemic signal of drought stress in tomato. New Phytol 212: 954–963. doi: 10.1111/nph.14190 [DOI] [PubMed] [Google Scholar]
- 9.Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, et al. (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci U S A 108: 8897–8902. doi: 10.1073/pnas.1100987108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao R, Ming Z, Yan L, Li S, Wang F, et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536: 469–473. doi: 10.1038/nature19073 [DOI] [PubMed] [Google Scholar]
- 11.Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YKM, et al. (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295. doi: 10.1242/dev.074567 [DOI] [PubMed] [Google Scholar]
- 12.Li W, Tran LS (2015) Are karrikins involved in plant abiotic stress responses? Trends Plant Sci 20: 535–538. doi: 10.1016/j.tplants.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 13.Smith SM, Li J (2014) Signalling and responses to strigolactones and karrikins. Curr Opin Plant Biol 21: 23–29. doi: 10.1016/j.pbi.2014.06.003 [DOI] [PubMed] [Google Scholar]
- 14.Ghebrehiwot HM, Kulkarni MG, Kirkman KP, Van Staden J (2008) Smoke-water and a smoke-isolated butenolide improve germination and seedling vigour of Eragrostis tef (Zucc.) trotter under high temperature and low osmotic potential. J Agron Crop Sci 194: 270–277. [Google Scholar]
- 15.Stanga JP, Morffy N, Nelson DC (2016) Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 243: 1397–1406. doi: 10.1007/s00425-015-2458-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, et al. (2015) PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349: 540–543. doi: 10.1126/science.aab1140 [DOI] [PubMed] [Google Scholar]
- 17.Conn CE, Nelson DC (2016) Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front Plant Sci 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scaffidi A, Waters MT, Sun YMK, Skelton BW, Dixon KW, et al. (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol 165: 1221–1232. doi: 10.1104/pp.114.240036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jamil M, Kanwal M, Aslam MM, Khan SU, Malook I, et al. (2014) Effect of plant-derived smoke priming on physiological and biochemical characteristics of rice under salt stress condition. Aust J Crop Sci 8: 159–170. [Google Scholar]
- 20.Harb A, Pereira A (2011) Screening Arabidopsis genotypes for drought stress resistance. Methods Mol Biol 678: 191–198. doi: 10.1007/978-1-60761-682-5_14 [DOI] [PubMed] [Google Scholar]
- 21.Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS (2014) ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol 202: 35–49. doi: 10.1111/nph.12613 [DOI] [PubMed] [Google Scholar]
- 22.Umezawa T, Okamoto M, Kushiro T, Nambara E, Oono Y, et al. (2006) CYP707A3, a major ABA 8'-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J 46: 171–182. doi: 10.1111/j.1365-313X.2006.02683.x [DOI] [PubMed] [Google Scholar]
- 23.Nelson DC, Riseborough JA, Flematti GR, Stevens J, Ghisalberti EL, et al. (2009) Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol 149: 863–873. doi: 10.1104/pp.108.131516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson DC, Flematti GR, Riseborough JA, Ghisalberti EL, Dixon KW, et al. (2010) Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc Natl Acad Sci U S A 107: 7095–7100. doi: 10.1073/pnas.0911635107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanga JP, Smith SM, Briggs WR, Nelson DC (2013) SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol 163: 318–330. doi: 10.1104/pp.113.221259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo N, Cheng F, Wu J, Liu B, Zheng S, et al. (2014) Anthocyanin biosynthetic genes in Brassica rapa. BMC Genomics 15: 426 doi: 10.1186/1471-2164-15-426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, et al. (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77: 367–379. doi: 10.1111/tpj.12388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, et al. (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480. doi: 10.1105/tpc.104.022897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeats TH, Rose JK (2013) The formation and function of plant cuticles. Plant Physiol 163: 5–20. doi: 10.1104/pp.113.222737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seto Y, Sado A, Asami K, Hanada A, Umehara M, et al. (2014) Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc Natl Acad Sci U S A 111: 1640–1645. doi: 10.1073/pnas.1314805111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morffy N, Faure L, Nelson DC (2016) Smoke and hormone mirrors: action and evolution of karrikin and strigolactone signaling. Trends Genet 32: 176–188. doi: 10.1016/j.tig.2016.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishiyama R, Watanabe Y, Leyva-Gonzalez MA, Ha CV, Fujita Y, et al. (2013) Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc Natl Acad Sci U S A 110: 4840–4845. doi: 10.1073/pnas.1302265110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demidchik V, Straltsova D, Medvedev SS, Pozhvanov GA, Sokolik A, et al. (2014) Stress-induced electrolyte leakage: the role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. J Exp Bot 65: 1259–1270. doi: 10.1093/jxb/eru004 [DOI] [PubMed] [Google Scholar]
- 34.Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, et al. (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci U S A 107: 2355–2360. doi: 10.1073/pnas.0909222107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuromori T, Sugimoto E, Shinozaki K (2011) Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J 67: 885–894. doi: 10.1111/j.1365-313X.2011.04641.x [DOI] [PubMed] [Google Scholar]
- 36.Greer S, Wen M, Bird D, Wu XM, Samuels L, et al. (2007) The cytochrome p450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol 145: 653–667. doi: 10.1104/pp.107.107300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakobson L, Lindgren LO, Verdier G, Laanemets K, Brosche M, et al. (2016) BODYGUARD is required for the biosynthesis of cutin in Arabidopsis. New Phytologist 211: 614–626. doi: 10.1111/nph.13924 [DOI] [PubMed] [Google Scholar]
- 38.Kurdyukov S, Faust A, Nawrath C, Bar S, Voisin D, et al. (2006) The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339. doi: 10.1105/tpc.105.036079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang ZY, Xiong LM, Li WB, Zhu JK, Zhu JH (2011) The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell 23: 1971–1984. doi: 10.1105/tpc.110.081943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen KH, Van Ha C, Nishiyama R, Watanabe Y, Leyva-Gonzalez MA, et al. (2016) Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc Natl Acad Sci U S A 113: 3090–3095. doi: 10.1073/pnas.1600399113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun XD, Ni M (2011) HYPOSENSITIVE TO LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation. Mol Plant 4: 116–126. doi: 10.1093/mp/ssq055 [DOI] [PubMed] [Google Scholar]
- 42.Walton A, Stes E, Goeminne G, Braem L, Vuylsteke M, et al. (2016) The response of the root proteome to the synthetic strigolactone GR24 in Arabidopsis. Mol Cell Proteomics 15: 2744–2755. doi: 10.1074/mcp.M115.050062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umehara M, Cao M, Akiyama K, Akatsu T, Seto Y, et al. (2015) Structural requirements of strigolactones for shoot branching inhibition in rice and Arabidopsis. Plant Cell Physiol 56: 1059–1072. doi: 10.1093/pcp/pcv028 [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, et al. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41. doi: 10.1263/jbb.104.34 [DOI] [PubMed] [Google Scholar]
- 45.Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 46.Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, et al. (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183. doi: 10.1105/tpc.111.087395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrs HDW, P.E. (1962) A re-examination of the relative turgidity technique for estimating water deficit in leaves. Aust J Biol Sci 15: 413–428. [Google Scholar]
- 48.Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, et al. (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci U S A 109: 9653–9658. doi: 10.1073/pnas.1203567109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanno Y, Oikawa T, Chiba Y, Ishimaru Y, Shimizu T, et al. (2016) AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun 7: 13245 doi: 10.1038/ncomms13245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ito S, Nozoye T, Sasaki E, Imai M, Shiwa Y, et al. (2015) Strigolactone regulates anthocyanin accumulation, acid phosphatases production and plant growth under low phosphate condition in Arabidopsis. PLoS One 10: e0119724 doi: 10.1371/journal.pone.0119724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y (2004) A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J 37: 139–146. [DOI] [PubMed] [Google Scholar]
- 52.Ukitsu H, Kuromori T, Toyooka K, Goto Y, Matsuoka K, et al. (2007) Cytological and biochemical analysis of COF1, an Arabidopsis mutant of an ABC transporter gene. Plant Cell Physiol 48: 1524–1533. doi: 10.1093/pcp/pcm139 [DOI] [PubMed] [Google Scholar]
- 53.Toyooka K, Okamoto T, Minamikawa T (2000) Mass transport of proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicle is involved in protein mobilization in germinating seeds. J Cell Biol 148: 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishiyama R, Le DT, Watanabe Y, Matsui A, Tanaka M, et al. (2012) Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS One 7: e32124 doi: 10.1371/journal.pone.0032124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.To TK, Nakaminami K, Kim JM, Morosawa T, Ishida J, et al. (2011) Arabidopsis HDA6 is required for freezing tolerance. Biochem Biophys Res Commun 406: 414–419. doi: 10.1016/j.bbrc.2011.02.058 [DOI] [PubMed] [Google Scholar]
- 56.Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, et al. (2011) Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res 18: 17–29. doi: 10.1093/dnares/dsq032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A-C) kai2-4 and WT plants were grown and exposed to drought. At indicated time points, (A) soil relative moisture contents (n = 10) and relative humidity, (B) leaf relative water content (RWC) (n = 4 biological replicates), and (C) electrolyte leakage (n = 4 biological replicates) were determined. (D) Electrolyte leakage (Left) of kai2-4 and WT plants at a similar RWC (Right) during drought treatment (n = 4 biological replicates). (E) Leaf surface temperature of well-watered (21-day-old) kai2-4 and WT plants. Thermal imaging camera (Top) and common optical camera (Bottom) were used to take pictures at the same time. (F) Cotyledon opening percentage of kai2-4 and WT seeds in the absence or presence of different concentrations of exogenous ABA. Data represent the means and standard errors of 3 independent experiments (n = 50 seeds/genotype/experiment). Asterisks indicate significant differences as determined by a Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
(A) Heatmap presentation indicates the fold-changes in expression of representative genes derived from microarray analysis. (B) Heatmap presentation indicates the fold-changes in expression of representative genes using qRT-PCR. Expression data were obtained from microarray analysis or qRT-PCR of 24-day-old Arabidopsis rosette leaf samples that were collected from 3 independent plants for microarray analysis (n = 3). UBQ10 was used as reference gene in qRT-PCR analysis. Relative expression levels are indicated by intensities of colors expressed in fold-change with saturation at 6. Red and blue colors indicate up- and downregulation, respectively. Note that not all data points shown in (a) passed the q-value < 0.05. M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h.
(TIF)
Heatmap presentation indicates the fold-changes in gene expression derived from microarray data. Relative expression levels are indicated by intensities of colors expressed in fold-change with saturation at 6. Red and blue colors indicate up- and downregulation, respectively. Note that not all data points shown passed the q-value < 0.05. M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h.
(TIF)
(A) Downregulated genes in M-C/W-C and/or M-D/W-D. (B) Upregulated genes in M-C/W-C and/or M-D/W-D. Green and red colors show down- and upregulation, respectively. Colored bars in each panel indicate fold-changes in gene expression. M-C/W-C, kai2-2 well-watered control versus WT well-watered control; M-D/W-D represents M-D2/W-D2 and/or M-D4/W-D4; M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h. For repetitive genes, their highest fold-change was used in the analysis.
(TIF)
Heatmap presentation indicates the fold-changes in gene expression derived from microarray data. Relative expression levels are indicated by intensities of colors expressed in fold-change with saturation at 6. Red and blue colors indicate up- and downregulation, respectively. Note that not all data points shown passed the q-value < 0.05. M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h.
(TIF)
(A) Anthocyanin content in kai2-4 and WT plants under drought conditions. Data represent the means and standard errors (n = 4 plants). (B) Chlorophyll leaching from rosette leaves of 28-day-old kai2-4 and WT plants at different time periods. Data represent the means and standard errors (n = 3 plants/genotype). (C) Toluidine blue staining patterns of rosette leaves of 28-day-old kai2-4 and WT plants. Asterisks indicate significant differences as determined by a Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
(A) Thickness of cuticle of the fifth leaves (adaxial side) derived from kai2-2, d14-2, kai2-2 d14-2 and WT plants. Blue arrows indicate cuticular layer (electron-dense, darker-staining layer) and green arrows indicate wax-rich cuticle proper (electron-translucent layer). (B) Transmission electron microscope images of the surface of the fifth leaves (adaxial side) derived from kai2-2, d14-2, kai2-2 d14-2 and WT plants. CW, cell wall. Data represent the means and standard errors (n = 3 biological replicates). Different letters above the error bars indicate significant differences (P < 0.05) among the genotypes according to a Tukey's honest significant difference test.
(TIF)
(XLS)
(A) List of upregulated genes in the M-C/W-C comparison. (B) List of upregulated genes in the M-D2/W-D2 comparison. (C) List of upregulated genes in the M-D4/W-D4 comparison. (D) List of upregulated genes in the M-D/W-D (e.g. in M-D2/W-C and/or M-D4/W-C) comparison. (E) List of upregulated genes in the W-D2/W-C comparison. (F) List of upregulated genes in the W-D4/W-C comparison. (G) List of upregulated genes in the W-D/W-C (e.g. in W-D2/W-C and/or W-D4/W-C) comparison. (H) List of upregulated genes in the M-D2/M-C comparison. (I) List of upregulated genes in the M-D4/M-C comparison. (J) List of upregulated genes in the M-D/M-C (e.g. in M-D2/M-C and/or M-D4/M-C) comparison. (K) List of downregulated genes in the M-C/W-C comparison. (L) List of downregulated genes in the M-D2/W-D2 comparison. (M) List of downregulated genes in the M-D4/W-D4 comparison. (N) List of downregulated genes in the M-D/W-D (e.g. in M-D2/W-D2 and/or M-D4/W-D4) comparison. (O) List of downregulated genes in the W-D2/W-C comparison. (P) List of downregulated genes in the W-D4/W-C comparison. (Q) List of downregulated genes in the W-D/W-C (e.g. in W-D2/W-C and/or W-D4/W-C) comparison. (R) List of downregulated genes in the M-D2/M-C comparison. (S) List of downregulated genes in the M-D4/M-C comparison. (T) List of downregulated genes in the M-D/M-C (e.g. in M-D2/M-C and/or M-D4/M-C) comparison. M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h.
(XLS)
(A) List of upregulated genes. (B) List of downregulated genes.
(XLS)
(A) Genes upregulated in M-C/W-C and M-D/W-D. (B) Genes downregulated in M-C/W-C and M-D/W-D. (C) Genes upregulated in W-D/W-C and M-D/M-C. (D) Genes downregulated in M-C/W-C and upregulated in W-D/W-C. (E) Genes downregulated in M-C/W-C and upregulated in M-D/M-C. (F) Genes downregulated in M-C/W-C and upregulated in W-D/W-C and/or M-D/M-C. (G) Genes downregulated in M-C/W-C and upregulated in both W-D/W-C and M-D/M-C. (H) Genes downregulated in M-D/W-D and upregulated in W-D/W-C. (I) Genes downregulated in M-D/W-D and upregulated in M-D/M-C. (J) Genes downregulated in M-D/W-D and upregulated in W-D/W-C and/or M-D/M-C. (K) Genes downregulated in M-D/W-D and upregulated in both W-D/W-C and M-D/M-C. (L) Genes downregulated in W-D/W-C and M-D/M-C. (M) Genes upregulated in M-C/W-C and downregulated in W-D/W-C. (N) Genes upregulated in M-C/W-C and downregulated in M-D/M-C. (O) Genes upregulated in M-C/W-C and downregulated in W-D/W-C and/or M-D/M-C. (P) Genes upregulated in M-C/W-C and downregulated in both W-D/W-C and M-D/M-C. (Q) Genes upregulated in M-D/W-D and downregulated in W-D/W-C. (R) Genes upregulated in M-D/W-D and downregulated in M-D/M-C. (S) Genes upregulated in M-D/W-D and downregulated in W-D/W-C and/or M-D/M-C. (T) Genes upregulated in M-D/W-D and downregulated in both W-D/W-C and M-D/M-C. M-C, kai2-2 well-watered control; M-D2, kai2-2 dehydrated 2 h; M-D4, kai2-2 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h.
(XLS)
(A) List of DEGs related to anthocyanin biosynthesis. (B) List of DEGs related to cuticle formation.
(XLS)
(XLS)
Data Availability Statement
The raw microarray data are available from the Gene Expression Omnibus database, accession number GSE90622 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90622). All other data are within the paper and its Supporting Information files.