ABSTRACT
Nucleic acid amplification tests (NAATs) are reliable tools for the detection of toxigenic Clostridium difficile from unformed (liquid or soft) stool samples. The objective of this study was to evaluate performance of the cobas Cdiff test on the cobas 4800 system using prospectively collected stool specimens from patients suspected of having C. difficile infection (CDI). The performance of the cobas Cdiff test was compared to the results of combined direct and broth-enriched toxigenic culture methods in a large, multicenter clinical trial. Additional discrepancy analysis was performed by using the Xpert C. difficile Epi test. Sample storage was evaluated by using contrived and fresh samples before and after storage at −20°C. Testing was performed on samples from 683 subjects (306 males and 377 females); 113 (16.5%) of 683 subjects were positive for toxigenic C. difficile by direct toxigenic culture, and 141 of 682 subjects were positive by using the combined direct and enriched toxigenic culture method (reference method), for a prevalence rate of 20.7%. The sensitivity and specificity of the cobas Cdiff test compared to the combined direct and enriched culture method were 92.9% (131/141; 95% confidence interval [CI], 87.4% to 96.1%) and 98.7% (534/541; 95% CI, 97.4% to 99.4%), respectively. Discrepancy analysis using results for retested samples from a second NAAT (Xpert C. difficile/Epi test; Cepheid, Sunnyvale, CA) found no false-negative and 4 false-positive cobas Cdiff test results. There was no difference in positive and negative results in comparisons of fresh and stored samples. These results support the use of the cobas Cdiff test as a robust aid in the diagnosis of CDI.
KEYWORDS: Clostridium difficile infection (CDI), health care-associated infection, nucleic acid amplification test, active surveillance testing (AST), toxin B (tcdB) gene
INTRODUCTION
Clostridium difficile is a Gram-positive, anaerobic, spore-forming bacillus identified as an etiological agent of antibiotic-associated diarrhea and pseudomembranous colitis in the late 1970s (1, 2). Historically, the incidence of C. difficile infection (CDI) ranged from 30 to 40 cases per 100,000 population in acute-care hospitals in the United States (3). The incidence rose to more than 140 cases per 100,000 population by 2013, with 60.7% clearly being health care associated (4). Currently, CDI is the most commonly diagnosed infectious cause of health care-associated diarrhea and is a significant cause of morbidity and mortality (5). Toxigenic strains of C. difficile typically produce two toxins, toxin A, which is an enterotoxin, and toxin B, which is a cytotoxin (6). A small percentage of strains produce only toxin B (7). There are no known naturally occurring toxin A-positive, toxin B-negative strains associated with clinical disease (8, 9). Diagnosis of CDI may be established by the presence of toxin B or toxins A and B in stool samples from patients with appropriate clinical symptoms, driving the need for accurate testing.
Early detection of CDI with the rapid implementation of contact precautions is considered critical for infection control to prevent transmission in the health care setting (10). There are various diagnostic approaches for CDI (11), and real-time PCR (quantitative PCR [qPCR]) is considered a reliable and rapid single test for the detection of C. difficile toxin (12–15). The purpose of this study was to compare the performance of the cobas Cdiff test to that of the direct and enriched toxigenic culture method for the detection of toxigenic C. difficile in stool samples.
(The data were presented in part at annual meetings of the American Society for Microbiology [16] and the Association for Molecular Pathology [17].)
RESULTS
Study population.
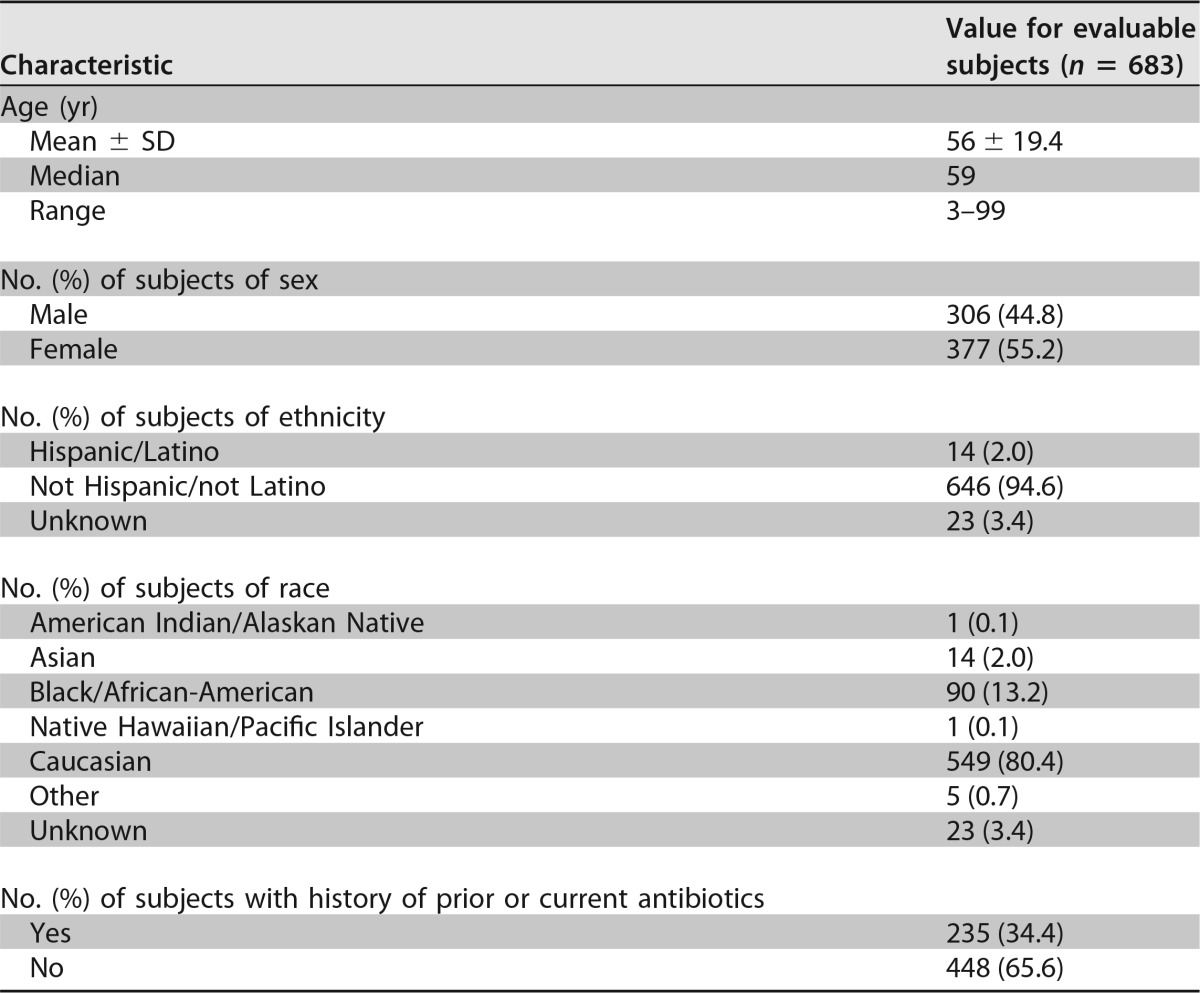
A clinical utility study was performed by using 683 specimens collected from 4 collection sites, and all specimens had valid test results with the cobas Cdiff test as well as direct culture from initial testing. One sample lacked a sufficient volume for repeat direct and enrichment culture methods and was not included in the final statistical analysis. Demographics and baseline characteristics of evaluable subjects are shown in Table 1. The age range of the patients was between 3 and 99 years, with a median age of 59 years. The gender distribution was 306 males (44.8%) and 377 females (55.2%).
TABLE 1.
Demographics and baseline characteristics of evaluable subjects
Comparison of the cobas Cdiff test with direct culture methods.
The performance of the cobas Cdiff test compared to that of the direct culture method is shown in Table 2. The overall percent agreement (OPA) was 95.3%. The three specimens with false-negative cobas Cdiff test results relative to the direct culture method were all negative by the Xpert Cdiff test. Of the 29 specimens with false-positive cobas Cdiff test results relative to direct culture, 15 were positive by the Xpert Cdiff test; 1 sample was not tested because of an insufficient specimen volume.
TABLE 2.
Comparison of the cobas Cdiff test with direct culturea
| cobas Cdiff test result | No. of specimens with result |
Total no. of specimens | |
|---|---|---|---|
| Positive by direct culture | Negative by direct culture | ||
| Positive | 110 | 29 | 139 |
| Negative | 3 | 541 | 544 |
| Total | 113 | 570 | 683 |
The positive percent agreement between the results of the cobas Cdiff test and direct culture was 97.3% (95% CI, 92.5% to 99.1%). The negative percent agreement between the results of the cobas Cdiff test and direct culture was 94.9% (95% CI, 92.8% to 96.4%). The overall percent agreement between the results of the cobas Cdiff test and direct culture was 95.3% (95% CI, 93.5% to 96.7%).
The clinical performance of the cobas Cdiff test compared with the combined results of initial and repeat direct and enriched toxigenic culture methods is shown in Table 3. These data demonstrate the benefit of the use of enrichment culture methods to enhance the recovery of C. difficile when cultures are used as a reference method. The sensitivity and specificity of the cobas Cdiff test were 92.9 and 98.7%, respectively; the positive predictive value (PPV) and negative predictive value (NPV) were 94.9 and 98.2%, respectively. In the discrepancy analysis, of the 7 specimens with false-positive cobas Cdiff test results relative to combined direct and enrichment culture methods, 3 were positive by the Xpert Cdiff test. The 10 specimens with false-negative cobas Cdiff test results were all negative by the Xpert C. difficile Epi test.
TABLE 3.
Comparison of the cobas Cdiff test with combined direct and enrichment cultureb
| cobas Cdiff test result | No. of speicmens with result |
Total no. of specimens | |
|---|---|---|---|
| Positive by combined direct and enrichment culture | Negative by combined direct and enrichment culture | ||
| Positive | 131 | 7 | 138 |
| Negative | 10 | 534 | 544 |
| Total | 141 | 541 | 682a |
One sample had an insufficient sample volume for repeat testing and is not included.
The sensitivity and specificity of the results of the cobas Cdiff test were 92.9% (131/141 samples; 95% CI, 87.4% to 96.1%) and 98.7% (534/541 samples; 95% CI, 97.4% to 99.4%), respectively. The positive and negative predictive values of the cobas Cdiff test were 94.9% (95% CI, 89.9% to 97.5%) and 98.2% (95% CI, 96.6% to 99.0%), respectively.
Impact of sample storage.
The results for 124 random samples returned to the reference laboratory for retesting had positive percent agreement, negative percent agreement, and overall percent agreement values between fresh and frozen stool specimens of 97.1% (33/34 samples), 97.8% (88/90), and 97.6% (121/124), respectively (chi-square statistic, 0.02; P = 0.89). For the 100 contrived specimens, the positive percent agreement between the cobas Cdiff test results before and those after one freeze-thaw cycle was 100.0% (100/100; 95% confidence interval [CI], 96.4 to 100.0%). There was no difference in the distributions of positive and negative cobas Cdiff test results when fresh and frozen specimens from the multicenter trial of 580 samples were compared (P = 1 by Fisher's exact test; P = 0.93 by the chi-square test likelihood ratio). Finally, there was 100% agreement between direct and repeat toxigenic culture results for 87 samples cultured fresh and following storage for at least 60 days at −20°C or colder (Kappa statistic, 1.0).
Analytical performance.
Analysis of the precision study data showed that at <1× the limit of detection (LOD), the positivity rate was 29%, and at 1× or 3× the LOD, it was 100%. The mean cycle threshold (CT) values were 38.5 at 1× the LOD and 37.5 at 3× the LOD, with standard deviations of 1.5% and 1.1%, respectively (18).
In the analysis of clinical reproducibility, the positive percent agreement values for results below the LOD, 1× the LOD, and 3× the LOD were 66.1% (95% CI, 58.7% to 73.0%), 100.0% (95% CI, 98.0% to 100.0%), and 100.0% (95% CI, 97.9% to 100.0%), respectively. The negative percent agreement was 100.0% (95% CI, 97.9% to 100.0%). The LOD for C. difficile strains ranged from 54 to 225 CFU per swab. The inclusivity study found that all 28 C. difficile strains were determined to be positive in ≥95% of replicates at densities ranging from 77.9 to 460 CFU/swab; 15 were determined to be positive 100% of the time (18).
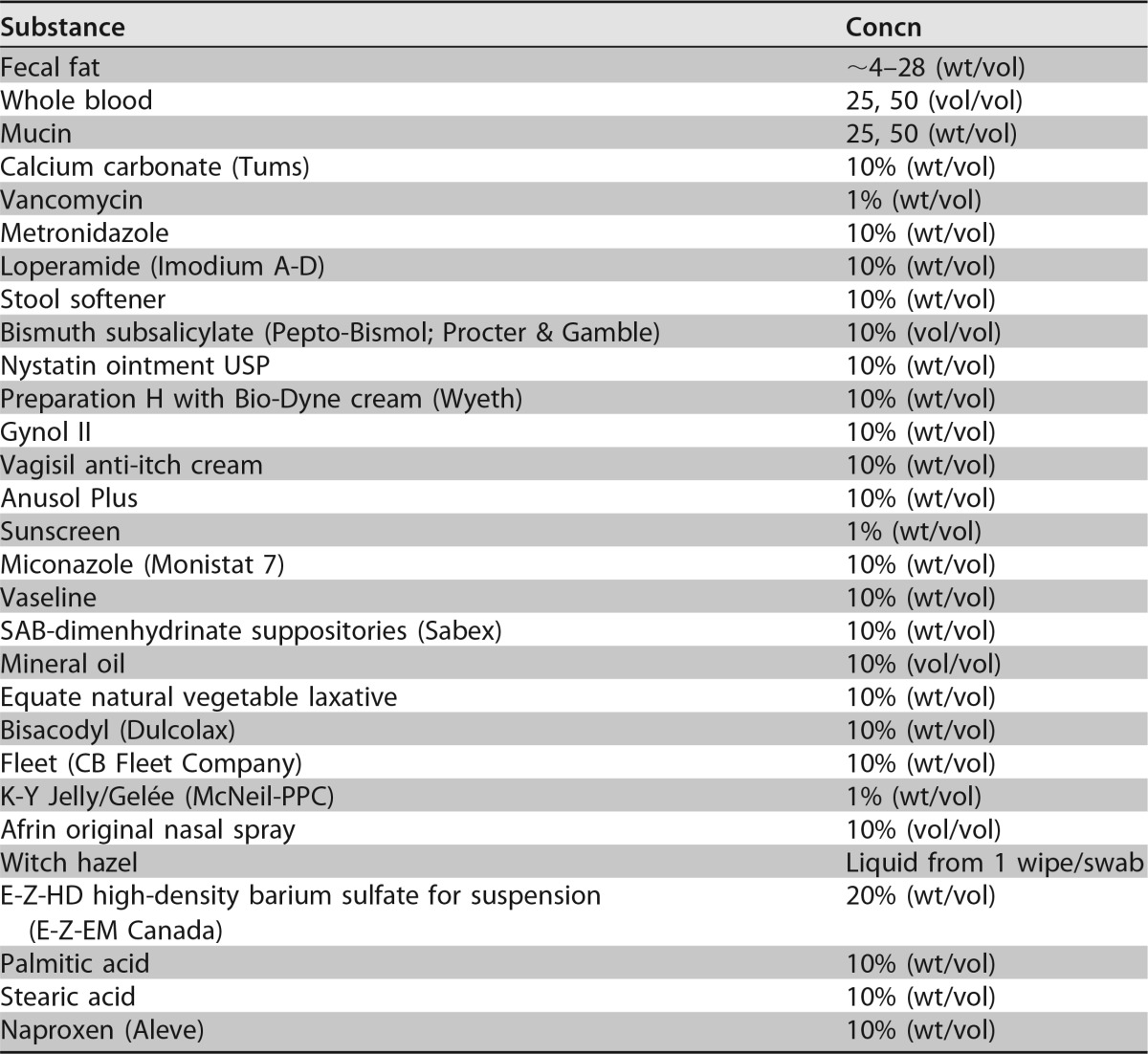
Analytical specificity testing found that that none of the organisms tested interfered with the detection of the intended C. difficile targets, and there were no false-positive results (19). The compounds listed in Table 4 were tested for interference. There was no interference found for any material other than mucin, which interfered with the detection of toxigenic C. difficile isolates at a 50% (vol/vol) concentration. For the cross-contamination analysis, in the nine checkerboard runs, 1 out of 423 negative samples produced a positive result, for a cross-contamination rate of ≤0.24%. None of the 282 negative samples from the carryover contamination runs produced a positive result (19).
TABLE 4.
Compounds tested for possible interference
Rate of invalid results.
Among 683 specimens tested for toxigenic C. difficile, the initial failure rate was 0% (0/683 specimens; 95% CI, 0% to 0.7%), and no retesting was needed, providing a final failure (invalid) rate of 0%. Thus, there were no invalid runs during this study.
Statistical opinion.
The performance of the cobas Cdiff test was comparable to that of toxigenic culture using unformed stool samples from subjects suspected of having CDI and met the criteria specified in the study protocol, assay product requirements, and clinical validation plan. The results support the intended use of the cobas Cdiff test as an aid in the diagnosis of CDI in humans in conjunction with clinical and epidemiological risk factors.
DISCUSSION
In our large, prospective, multisite clinical investigation, we demonstrated that the cobas Cdiff test is a robust test for the detection of toxigenic C. difficile. Our results extend those reported previously by Moure and colleagues, who found that the concordance between cobas Cdiff test results and glutamate dehydrogenase (GDH)/toxin gene screening was 97.6% (20). Also, we demonstrated the stability of sample storage, which will be useful for future test development of next-generation molecular and culture-based CDI diagnostics.
The genomics of C. difficile and the molecular structure of its toxin genes are well known (6, 9, 21, 22). Thus, the design of primers and probes for a robust real-time PCR assay is relatively straightforward. We demonstrated that the cobas Cdiff test had excellent analytical sensitivity, inclusivity, and reproducibility prior to beginning the clinical trial. We also demonstrated that there was minimal interference from other substances possibly found in the stool specimens of patients. Therefore, it was not surprising that the clinical performance of this test was robust. From the laboratory use perspective, we also demonstrated that the new assay is reliable in that the initial failure rate of the test was 0% (0 of 683 tests), which facilitates deployment in a diagnostic microbiology laboratory for clinical use.
For our literature review, we used PubMed.gov (MEDLINE) with the search terms Clostridium difficile diagnosis, Clostridium difficile genetics, and real-time PCR and the names of U.S. Food and Drug Administration (FDA)-cleared molecular tests that detect C. difficile to search the medical literature from 2005 through June 2017. We also searched Google using these terms as well as the FDA website (https://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm330711.htm) to determine which commercial tests were available as CDI assays. From this review, it is clear that there are potentially three main issues to consider when selecting a testing program for facilitating the diagnosis of CDI. One issue is how well the various molecular tests perform. The second issue is what the best testing approach is. The third issue, as an emerging approach to CDI control, is how the chosen test might perform in an admission active surveillance testing (AST) program to prevent the spread of C. difficile in acute-care hospitals.
The reported performances of the available molecular tests for the detection of toxigenic C. difficile were recently reviewed (11). Of the 11 tests reviewed, the assays that consistently showed >90% sensitivity and specificity were limited to the Cepheid Xpert C. difficile, Portrait Toxigenic C. difficile, and Illumigene C. difficile assays (22). In our large, multicenter trial, the cobas Cdiff test performed at least as well as the other assays when our results were compared to those results.
The question of which test or series of tests (e.g., algorithm) should a clinical laboratory implement is critical for this disease and very complex for the laboratory. A key argument has been that molecular qPCR tests are too sensitive and detect not only disease but also colonization. However, there is no evidence that disease is related to the level of toxin detected in stool (23). One of the frequently referenced clinical reports on this topic is that by Polage and colleagues, who concluded that PCR tests were too sensitive and detected an excess of colonized patients as well as those with disease (24). Unfortunately, those researchers did not use any clinical criteria or laboratory assessment of specimen quality to determine who should actually be evaluated (e.g., included in their data assessment were patients with <3 diarrheal stools per day who were not excluded from potential CDI cases) in the data that were critically analyzed (24). Thus, it is not surprising that those researchers found a high number of positive specimens from patients with no significant diarrhea, given that it is not unusual for hospitalized patients to be colonized with this organism (25). Our own data suggest that PCR-positive, direct stool toxin-negative patients have a significant risk for future complications if not treated (26). We found a significant positive effect of receiving treatment on reducing 90-day readmission for any reason, decreasing the inpatient length of stay (LOS) and total LOS (inpatient plus readmission LOS), and reduced charges in the enzyme-immunoassay-negative (EIA−)/PCR+ group versus the EIA+/PCR+ group, suggesting that at least some of these patients actually had clinically significant CDI. Importantly, a recent clinical effectiveness review (CER) from the Agency for Healthcare Research and Quality (AHRQ) found that nucleic acid amplification tests (NAATs) had high sensitivity and specificity for the laboratory detection of CDI with the highest-quality evidence (27). The lowest-quality evidence was the use of algorithms for this endeavor, and while the algorithms were specific, this approach lacked sensitivity (27). Clinical diagnosis is essential for the diagnosis of CDI (25), as is well stated in another recent review: “The diagnosis of CDI is primarily based on the clinical signs and symptoms and is only confirmed by laboratory testing” (11). Thus, in the appropriate clinical setting, the use of the most sensitive assay for the detection of toxigenic C. difficile in a patient's stool sample should remain the goal of the laboratory.
A novel question being asked of these tests is whether NAATs can be useful for the control of CDI in the acute-care setting by using them as screening tests (AST) at the time of admission for the detection of C. difficile carriers, who can then be subjected to contact precautions to prevent spread to other patients (10). A recent report illuminating the better understanding of CDI epidemiology indicates that asymptomatic carriers (e.g., colonized persons) are important sources of ongoing hospital transmission (28). Another investigation comparing the likely transmission of C. difficile from patients whose stool samples were positive by direct toxin testing as well as positive (by qPCR) for a toxin gene versus those that were positive only for the toxin gene found that symptomatic patients with qPCR-positive toxigenic C. difficile infection (without fecal toxin being detected) accounted for at least one-quarter of potential hospital transmission events (29). A third important study demonstrated that at the time of admission, patients acquire new organisms from their surrounding environment but that after the first night, they contribute bacteria (e.g., contaminate the room environment) until the microbiological representation of the patient and the room becomes quite homogeneous (30). This implies that if one is contemplating AST for pathogens that can spread within the hospital, a rapid (<24-h), sensitive test such as qPCR is needed. There has been one report that showed success in the use of the AST approach, where the investigators found an overall reduction in the number of patients who developed hospital-associated CDI from 416 patients (6.9 per 10,000 patient-days) during the control period to 38 patients (3.0 per 10,000 patient-days; 56.5% reduction) during the admission testing period (P < 0.001), suggesting that AST for toxigenic C. difficile is useful (31).
Our research had limitations in that there is no “perfect gold standard” for comparison with any new test developed to diagnose Clostridium difficile infection, but rather, they target toxigenic C. difficile. We used toxigenic culture as an accepted reference method for detecting toxigenic C. difficile (32), combined with broth enrichment to enhance sensitivity (14). We also demonstrated a potential cross-contamination rate of 0.24% (1 of 423 negative samples) when using high-signal-positive samples, demonstrating that cross-contamination is possible but that the risk is low. Thus, we believe that the clinical trial data are valid as a result of all the comparisons performed and that this study demonstrates the reliability of the cobas Cdiff test as a new automated test for the laboratory confirmation of CDI.
Future research will be useful in developing laboratory diagnostics that are able to separate colonization from disease attributed to C. difficile. This will be a challenge, since at this time, there is no acceptable, clear standard for definitively classifying who does and who does not have CDI. Also, defining the role of rapid molecular diagnostics in the prevention and control of health-care-onset CDI should be a priority, as this disease remains an increasing threat in our current environment. The best practice(s) to minimize this threat remains uncertain (10). Third, validation of assay performance using perirectal admission swabs for active surveillance testing to detect colonization is needed for this and other tests targeting toxigenic C. difficile.
In conclusion, we have demonstrated that the cobas Cdiff test is a reliable and robust automated assay for the detection of toxigenic C. difficile in stool samples from patients with diarrhea. It has entered the commercial testing market with other laboratory tests for confirming the diagnosis of CDI as a useful clinical diagnostic tool.
MATERIALS AND METHODS
Study population.
Stool samples were collected from 683 eligible patients over a 5-month period at 4 sites in the United States (NorthShore University HealthSystem, Evanston, IL; hospitals using TriCore Reference Laboratories located in Albuquerque, NM; Indiana University School of Medicine, Indianapolis, IN; and Thomas Jefferson University, Philadelphia, PA). Patients >12 months of age who were being tested for the diagnosis of CDI were identified and enrolled in the study. Each patient entering the study had an unformed stool sample of >5 ml submitted for testing. Patients were excluded from the study if they (i) submitted a formed stool sample, (ii) had been administered antibiotic therapy against C. difficile (e.g., oral and/or parenteral metronidazole, oral vancomycin, or fidaxomicin) during their present hospitalization, (iii) were previously enrolled in this study, (iv) were enrolled in any other study focused on the prevention or treatment of CDI within the previous 12 months, or (v) had a contraindication for the collection of stool samples according to institution policies and procedures.
Clinical trial design.
The clinical performance of the cobas Cdiff test was evaluated in a prospective, multisite investigation comparing the results of this test with those of toxigenic culture methods using remnant, deidentified, unformed stool samples from eligible subjects. Toxigenic culture was performed at a single site designated the reference laboratory (NorthShore University HealthSystem). The cobas Cdiff test was performed at the other three sites, according to the manufacturer's instructions.
This study was conducted in compliance with International Conference on Harmonization (ICH) guidelines, good clinical practice (GCP) guidelines, and regulations of the FDA. Approval by the institutional review boards at all four sites was obtained. This study was performed to obtain clearance of the cobas Cdiff test by the FDA.
Stool sample processing.
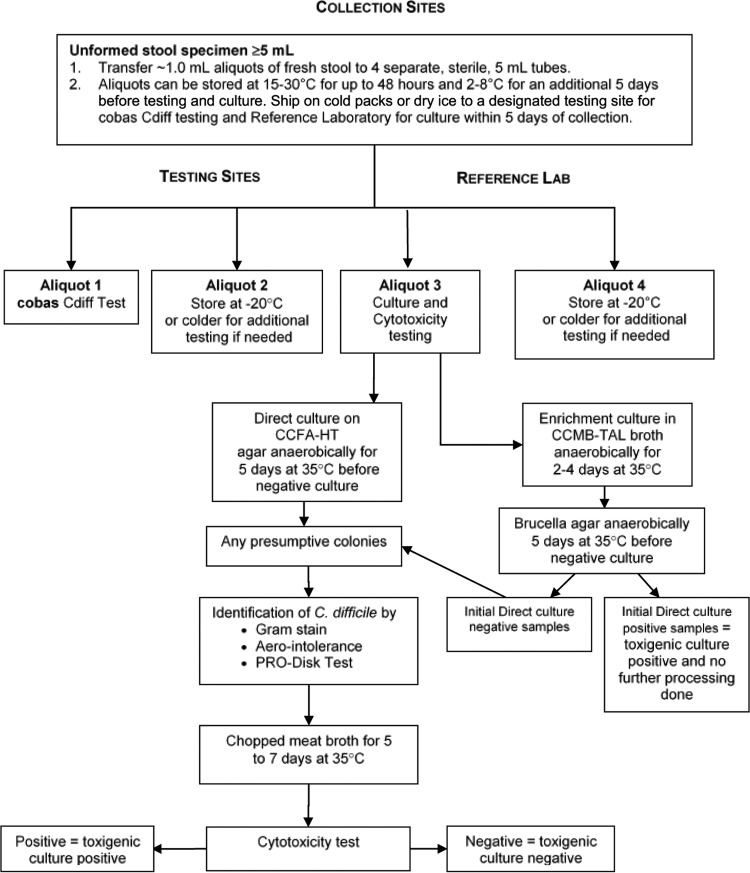
On the day of sample collection, fresh stool specimens were transported to the collection site laboratory at temperatures of between 2°C and 30°C. Upon receipt, fresh stool samples were processed within 48 h of collection. The accepted liquid and unformed samples of fresh stool were stirred to homogenize them, and aliquots were then prepared by transferring ∼1.0-ml portions into 4 separate, sterile, 5-ml cryogenic vials, as depicted in Fig. 1. Aliquots 1 and 2 were shipped on cold packs to one of the designated test sites for C. difficile PCR testing within 5 days of sample collection. Aliquots 3 and 4 were shipped on ice to the reference laboratory to arrive within the same time frame.
FIG 1.
Processing of unformed stool specimens from subjects with suspected Clostridium difficile infection.
Toxigenic culture.
The toxigenic culture included initial and repeat direct and enrichment cultures of stool specimens followed by cytotoxicity testing. Direct culture was done by inoculating a portion of the sample into prereduced anaerobically sterilized (PRAS) selective medium and cycloserine-cefoxitin-fructose agar with horse blood and taurocholate (CCFA-HT; Anaerobe Systems, Morgan Hill, CA), followed by cytotoxicity testing on recovered C. difficile bacteria. Briefly, after incubation at 35°C, suspected colonies were identified as being C. difficile by Gram staining, aerotolerance testing, and testing using the Pro Disk test (Hardy Diagnostics, Santa Maria, CA) and then inoculated into anaerobic chopped-meat broth (Anaerobe Systems), which was incubated for 5 to 7 days at 35°C for cytotoxicity testing. The supernatant from the anaerobic chopped-meat broth was processed for the detection of C. difficile toxin B by cell culture cytotoxicity testing according to the manufacturer's instructions (C. Difficile Tox-B test; Techlab, Blacksburg, VA). An enriched toxigenic culture method was performed simultaneously using anaerobic cycloserine-cefoxitin-mannitol broth with taurocholate, lysozyme, and cysteine (CCMB-TAL; Anaerobe Systems), followed by subculturing on anaerobic Brucella agar plates (Anaerobe Systems), with identification and cytotoxicity testing of all recovered C. difficile isolates.
A specimen was considered positive for toxigenic C. difficile if a toxin-producing organism was recovered from a stool specimen in a direct or enriched toxigenic culture (any positive rule). If C. difficile was isolated from the direct culture and the isolate tested positive by a cell cytotoxicity assay, the enrichment culture was not further analyzed. By using aliquot 4 specimens that were frozen at −70°C for up to 5 months, 580 study samples were retested by direct and enriched culture methods. If a stool specimen was positive by the cobas Cdiff test but negative by the initial culture, a repeat culture was done (n = 29). Also, all other samples with a negative initial direct culture result (n = 541) had a repeat direct and enriched culture performed. Additionally, 10 specimens with a positive initial direct culture result were tested as positive controls. Specimens were classified as being negative for toxigenic C. difficile only if they tested negative by both initial as well repeat direct and enrichment culture methods.
Testing of patient samples on the cobas 4800 system.
After receipt of patient specimens in the laboratory, test orders were logged into the cobas 4800 system either manually, with a work list, or via a Laboratory Information System (LIS) interface and then tested. The cobas 4800 system is a walk-away platform to perform nucleic acid purification and real-time PCR (qPCR) set up in a 96-well plate, followed by manual sealing and transferring of the plate to the amplification and detection system (33). The system currently is available for methicillin-resistant Staphylococcus aureus (MRSA)-S. aureus (34), C. difficile (this study), herpes simplex virus 1 (HSV-1) and HSV-2 (35, 36), Chlamydia trachomatis-Neisseria gonorrhoeae (37–42), and high-risk human papillomavirus (43–45) testing and BRAF (46), epidermal growth factor receptor (EGFR) (47), and KRAS (48) mutation detection. The cobas MRSA/SA test, the cobas Cdiff test, and the cobas HSV 1 and 2 test can be performed simultaneously in a single testing run. This system uses the cobas x 480 instrument, which includes an automated pipetter to extract, purify, and prepare target nucleic acid for qPCR (e.g., automated specimen preparation). Bacterial lysis of the stool specimen is achieved with proteinase K, detergent, and a chaotropic agent. The purified DNA is bound to magnetic glass particles (MGPs), washed, and eluted. The cobas x 480 instrument next transfers the working master mix reagent and moves the specimen to the qPCR microwell plate. The plate is then transferred to the cobas z 480 instrument where the amplification and detection of target DNA and controls occur by using specific primers and probes. Data analysis and report generation are performed with cobas 4800 software. The cobas Cdiff test uses positive and negative controls (cobas 4800 Cdiff Controls and Cofactor kit) and an internal control (cobas 4800 system internal control kit). One set of positive and negative controls is included in each run. Valid results must be obtained for both the positive and negative controls for the cobas 4800 software to display a reportable result.
Discrepancy analysis.
Discrepancy analysis using the Xpert C. difficile/Epi test (Cepheid, Sunnyvale, CA) was performed on all samples with discordant results between the cobas Cdiff test and the combined direct and enrichment culture method. Eight concordant positive and eight concordant negative samples were also tested as controls. The results of the Xpert C. difficile/Epi test determined the final reference result for the discrepancy analysis.
Testing of fresh and archived (frozen) stool specimens.
To evaluate the effect of specimen storage on the results of toxigenic culture and the cobas Cdiff test, aliquots from contrived specimens and unformed stool samples prospectively collected from patients suspected of having CDI were tested fresh and following storage at −20°C or colder. Briefly, four studies were performed; specifically, (i) toxigenic culture was performed on fresh samples at the reference laboratory before the samples were shipped frozen on dry ice to Roche Molecular Systems, Inc. (Pleasanton, CA), for PCR, and following PCR, a subset was returned for a repeat culture; (ii) a panel of 100 contrived specimens was prepared by using fresh negative stool specimens spiked with toxigenic C. difficile at various densities (2.5-, 5.0-, 10-, and 20-fold-higher densities than the assay limit of detection) and tested fresh plus following storage for 32 days at −20°C ± 5°C; (iii) the assay also was run on the 580 prospectively collected samples, both fresh and following storage at −70°C for up to 5 months; and (iv) a repeat toxigenic culture was performed on a subset of clinical samples (15 culture-positive and 72 culture-negative samples) stored at −20°C or colder for ≥60 days.
Analytical performance.
The technical performance verification (TPV) study was conducted by using a panel composed of C. difficile cultures diluted into a negative stool matrix in cobas PCR medium to a density below the LOD, near the LOD, and above the LOD as well as a negative level composed of only the stool suspension in cobas PCR medium. We used three unique lots of cobas Cdiff test reagents and three instruments for a total of 36 runs over 12 days.
The reproducibility of the results of the cobas 4800 system was established by a multisite investigation using simulated clinical samples evaluated across lot, site/instrument, operator, and day and within a run. Reproducibility test panels consisted of four specimens, with three replicates each, using various densities of C. difficile strain ATCC 43255 (negative, below the LOD, 1× the LOD, and 3× the LOD) in a pooled, C. difficile-negative, unformed stool matrix in cobas PCR medium. These specimens were tested at three sites by two operators/day (six operators total) for 5 days per lot over two lots for an overall total of 712 valid tests. We modeled the target CT value as the dependent variable using the random effects lot, site, operator, and day.
The LOD was determined by analyzing quantified cultures of seven toxigenic C. difficile strains diluted into a pooled negative stool specimen matrix in cobas PCR medium. All densities were analyzed by using three unique reagent lots. At least 21 replicates per reagent lot were tested at each density. To assess inclusivity, the cobas Cdiff test was evaluated with 28 additional strains of toxigenic C. difficile tested at a minimum of three densities and with 40 replicates per density level. The lowest density that had a ≥95% hit (positive-detection) rate was considered the LOD for each strain.
Analytical specificity was evaluated with a panel that consisted of 103 bacteria, fungi, and viruses as well as one human tissue cell line (18). In addition, 28 Clostridium genus organisms, including nontoxigenic C. difficile, were tested (18). Bacteria were quantified as CFU per milliliter, human cells were quantified as cells per milliliter, and viruses were quantified as PFU per milliliter. The only exceptions were for the following microorganisms: Chlamydia trachomatis was quantified as elementary bodies (EB) per milliliter, and cytomegalovirus, human echovirus, and human enterovirus were quantified as IU per milliliter. Bacteria and human cells were spiked to 1 × 106 CFU/ml and 1 × 106 cells/ml, respectively, and all viruses were spiked to 1 × 105 PFU/ml, except for adenovirus type 40, cytomegalovirus (human herpesvirus 5 [HHV5]), and human rotavirus, which were spiked to lower densities due to stock density limitations. Testing was performed with the organisms alone or with two C. difficile isolates present individually at 3× the LOD.
Interference was evaluated for 26 medications, fecal fat, whole blood, and mucin. The effects of the potentially interfering substances were evaluated in the presence and absence of two toxigenic C. difficile isolates spiked to ∼3× the LOD of the cobas Cdiff test (54 CFU/ml and 113 CFU/ml for the two strains).
Potential cross-contamination was assessed by testing a series of high-positive toxigenic C. difficile samples and negative samples in a checkerboard configuration on the cobas 4800 system. High-positive samples were prepared by spiking a pooled negative stool matrix sufficient to generate a CT value (i.e., the number of PCR cycles until the PCR growth curve was classified as positive) of the 95th percentile of the clinical specimen population. A total of 94 samples were included per run, with nine runs being performed, using three cobas 4800 systems with alternating checkerboard configurations. A total of three runs of negative samples following these checkerboard runs were performed to assess carryover contamination.
Statistical analysis.
SAS/STAT software (SAS, Cary, NC) was used to perform all analyses. Statistical analyses of clinical samples were chosen based on statistical guidance from the FDA (49, 50) and in accordance with the guidelines of Clinical and Laboratory Standards Institute method EP12-A2 for evaluating qualitative test performance (19). For the clinical reproducibility study, 95% two-sided exact binomial confidence intervals were used. In the studies designed to examine the difference between results from fresh and those from frozen samples, statistical analysis was done by using two-sided Fisher's exact test and the likelihood ratio of the chi-square test.
ACKNOWLEDGMENTS
We sincerely appreciate the contributions of Yosh Ohhashi and all the laboratory personnel from the study testing sites to this project.
This work was supported by Roche Molecular Diagnostics, Pleasanton, CA.
For potential conflicts of interest, L.R.P. has received speaking honoraria from Becton Dickinson, Cepheid, Roche, and CareFusion. L.R.P. has received research funding from Becton Dickinson, Cepheid, Nanosphere, 3M, GeneWEAVE, and Roche. S.A.Y. has received research funding from Roche. He has also consulted for Roche and Quidel, Inc. T.E.D. has received research funding from BioFire, Becton Dickinson, Cepheid, GenMark, Hologic, Luminex, Nanosphere, and Roche. Z.-X.W. has received research funding from Roche and has consulted for Roche. J.D., C.N., O.L., J.C.O., and M.A.L. are employees of Roche Molecular Diagnostics.
L.R.P. had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. L.R.P., O.L., and M.A.L. were responsible for the study concept and design. L.R.P., S.A.Y., T.E.D., Z.-X.W., and M.A.L. performed acquisition, analysis, or interpretation of data. L.R.P., O.L., and M.A.L. drafted the manuscript. L.R.P., S.A.Y., T.E.D., Z.-X.Y., J.D., C.N., O.L., J.C.O., and M.A.L. performed critical revision of the manuscript for important intellectual content. J.D. performed statistical analysis. C.N., O.L., J.C.O., and M.A.L. provided administrative, technical, or material support. L.R.P., O.L., and M.A.L. supervised the study.
REFERENCES
- 1.Bartlett JG, Chang TW, Moon N, Onderdonk AB. 1978. Antibiotic-induced lethal enterocolitis in hamsters: studies with eleven agents and evidence to support the pathogenic role of toxin-producing clostridia. Am J Vet Res 39:1525–1530. [PubMed] [Google Scholar]
- 2.Larson HE, Price AB, Honour P, Borriello SP. 1978. Clostridium difficile and the aetiology of pseudomembranous colitis. Lancet i:1063–1066. doi: 10.1016/S0140-6736(78)90912-1. [DOI] [PubMed] [Google Scholar]
- 3.McDonald LC, Owings M, Jernigan DB. 2006. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996-2003. Emerg Infect Dis 12:409–415. doi: 10.3201/eid1205.051064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2013. Annual report for the Emerging Infections Program for Clostridium difficile infection. https://www.cdc.gov/hai/eip/pdf/cdiff/2013-annual-report.pdf. Accessed 8 May 2017.
- 5.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voth DE, Ballard JD. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 18:247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson S, Sambol SP, Brazier JS, Delmee M, Avesani V, Merrigan MM, Gerding DN. 2003. International typing study of toxin A-negative, toxin B-positive Clostridium difficile variants. J Clin Microbiol 41:1543–1547. doi: 10.1128/JCM.41.4.1543-1547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawa D. 1 June 2011. Molecular detection of toxigenic C. difficile: toxin A or B gene? Med Lab Observer https://www.mlo-online.com/molecular-detection-of-toxigenic-c-difficile-toxin-a-or-b-gene.php#respond Accessed 5 May 2017. [PubMed]
- 9.Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald LC, Kutty PK. 25 July 2017, posting date Clostridium difficile infection: prevention and control. Topic 2687, p 1–12. UpToDate, version 26.0. Wolters Kluwer, Philadelphia, PA: https://www.uptodate.com/contents/2687. [Google Scholar]
- 11.Martínez-Meléndez A, Camacho-Ortiz A, Morfin-Otero R, Maldonado-Garza HJ, Villarreal-Treviño L, Garza-González E. 2017. Current knowledge on the laboratory diagnosis of Clostridium difficile infection. World J Gastroenterol 23:1552–1567. doi: 10.3748/wjg.v23.i9.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshpande A, Pasupuleti V, Rolston DD, Jain A, Deshpande N, Pant C, Hernandez AV. 2011. Diagnostic accuracy of real-time polymerase chain reaction in detection of Clostridium difficile in the stool samples of patients with suspected Clostridium difficile infection: a meta-analysis. Clin Infect Dis 53:e81–e90. doi: 10.1093/cid/cir505. [DOI] [PubMed] [Google Scholar]
- 13.Kufelnicka AM, Kirn TJ. 2011. Effective utilization of evolving methods for the laboratory diagnosis of Clostridium difficile infection. Clin Infect Dis 52:1451–1457. doi: 10.1093/cid/cir201. [DOI] [PubMed] [Google Scholar]
- 14.Peterson LR, Mehta MS, Patel PA, Hacek DM, Harazin M, Nagwekar PP, Thomson RB Jr, Robicsek A. 2011. Laboratory testing for Clostridium difficile infection: light at the end of the tunnel. Am J Clin Pathol 136:372–380. doi: 10.1309/AJCPTP5XKRSNXVIL. [DOI] [PubMed] [Google Scholar]
- 15.Tenover FC, Baron EJ, Peterson LR, Persing DH. 2011. Laboratory diagnosis of Clostridium difficile infection: can molecular amplification methods move us out of uncertainty? J Mol Diagn 13:573–582. doi: 10.1016/j.jmoldx.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterson L, Young S, Davis T, Wang Z-X, Duncan J, Noutsios C, Ohhashi Y, Liesenfeld O, Osiecki J, Lewinski M. 2015. Evaluation of stool specimens with the cobas C. difficile test performed on the cobas 4800 system for the detection of Clostridium difficile toxin B compared with toxigenic culture, abstr 2420. Abstr 115th Gen Meet Am Soc Microbiol, New Orleans, LA, 30 May to 2 June 2015. [Google Scholar]
- 17.Peterson LR, Young S, Davis T Jr, Wang ZX, Duncan J, Noutsios C, Ohhashi Y, Liesenfeld O, Osiecki J, Lewinski MA. 2015. Detection of toxigenic Clostridium difficile from fresh and archived (frozen) stool specimens by toxigenic culture and the cobas Cdiff test performed on the cobas 4800 system, abstr ID69. Abstr 2015 Annu Meet Assoc Mol Pathol, Austin, TX, 5 to 7 November 2015. [Google Scholar]
- 18.US Food and Drug Administration. 2014. 510(k) substantial equivalence determination decision summary. US Food and Drug Administration, Washington, DC: https://www.accessdata.fda.gov/cdrh_docs/reviews/K142422.pdf Accessed 25 August 2017. [Google Scholar]
- 19.CLSI. 2008. EP12-A2: user protocol for evaluation of qualitative test performance; approved guideline. CLSI, Wayne PA. [Google Scholar]
- 20.Moure R, Cañizares Á, Muíño M, Lobato M, Fernández A, Rodríguez M, Gude MJ, Tomás M, Bou G. 2016. Use of the cobas 4800 system for the rapid detection of toxigenic Clostridium difficile and methicillin-resistant Staphylococcus aureus. J Microbiol Methods 120:50–52. doi: 10.1016/j.mimet.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Knight DR, Elliott B, Chang BJ, Perkins TT, Riley TV. 2015. Diversity and evolution in the genome of Clostridium difficile. Clin Microbiol Rev 28:721–741. doi: 10.1128/CMR.00127-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott B, Androga GO, Knight DR, Riley TV. 2017. Clostridium difficile infection: evolution, phylogeny and molecular epidemiology. Infect Genet Evol 49:1–11. doi: 10.1016/j.meegid.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 23.Anikst VE, Gaur RL, Schroeder LF, Banaei N. 2016. Organism burden, toxin concentration, and lactoferrin concentration do not distinguish between clinically significant and nonsignificant diarrhea in patients with Clostridium difficile. Diagn Microbiol Infect Dis 84:343–346. doi: 10.1016/j.diagmicrobio.2015.11.022. [DOI] [PubMed] [Google Scholar]
- 24.Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, Nguyen HH, Huang B, Tang YW, Lee LW, Kim K, Taylor S, Romano PS, Panacek EA, Goodell PB, Solnick JV, Cohen SH. 2015. Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern Med 175:1792–1801. doi: 10.1001/jamainternmed.2015.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curry SR. 2017. Clostridium difficile. Clin Lab Med 37:341–369. doi: 10.1016/j.cll.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Smith B, Schaefer E, Liu E, Schoonmaker M, Ridgway J, Robicsek A, Peterson L. 2015. The significance and utility of a highly sensitive PCR test for the diagnosis of Clostridium difficile infection (CDI). Open Forum Infect Dis 2(Suppl 1):949. doi: 10.1093/ofid/ofv133.665. [DOI] [Google Scholar]
- 27.Butler M, Olson A, Drekonja D, Shaukat A, Schwehr N, Shippee N, Wilt TJ. 2016. Early diagnosis, prevention, and treatment of Clostridium difficile: update. Comparative effectiveness review no. 172. (Prepared by the Minnesota Evidence-Based Practice Center under contract no. 290-2012-00016-I.) AHRQ publication no. 16-EHC012-EF. Agency for Healthcare Research and Quality, Rockville, MD: https://www.ncbi.nlm.nih.gov/books/NBK361173/pdf/Bookshelf_NBK361173.pdf. Accessed 6 June 2017. [PubMed] [Google Scholar]
- 28.Caroff DA, Yokoe DS, Klompas M. 17 May 2017. Evolving insights into the epidemiology and control of Clostridium difficile in hospitals. Clin Infect Dis doi: 10.1093/cid/cix456. [DOI] [PubMed] [Google Scholar]
- 29.Mawer DPC, Eyre DW, Griffiths D, Fawley WN, Martin JSH, Quan TP, Peto TEA, Crook DW, Walker AS, Wilcox MH. 2017. Contribution to Clostridium difficile transmission of symptomatic patients with toxigenic strains who are fecal toxin negative. Clin Infect Dis 64:1163–1170. doi: 10.1093/cid/cix079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lax S, Sangwan N, Smith D, Larsen P, Handley KM, Richardson M, Guyton K, Krezalek M, Shogan BD, Defazio J, Flemming I, Shakhsheer B, Weber S, Landon E, Garcia-Houchins S, Siegel J, Alverdy J, Knight R, Stephens B, Gilbert JA. 2017. Bacterial colonization and succession in a newly opened hospital. Sci Transl Med 9:eaah6500. doi: 10.1126/scitranslmed.aah6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longtin Y, Paquet-Bolduc B, Gilca R, Garenc C, Fortin E, Longtin J, Trottier S, Gervais P, Roussy JF, Lévesque S, Ben-David D, Cloutier I, Loo VG. 2016. Effect of detecting and isolating Clostridium difficile carriers at hospital admission on the incidence of C. difficile infections: a quasi-experimental controlled study. JAMA Intern Med 176:796–804. doi: 10.1001/jamainternmed.2016.0177. [DOI] [PubMed] [Google Scholar]
- 32.Burnham C-AD, Carroll KC. 2013. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev 26:604–630. doi: 10.1128/CMR.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Der Pol B. 2013. Cobas 4800: a fully automated system for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae. Expert Rev Mol Diagn 13:131–140. doi: 10.1586/erm.12.141. [DOI] [PubMed] [Google Scholar]
- 34.Peterson LR, Woods CW, Davis TE Jr, Wang Z-X, Young SA, Osiecki JC, Lewinski MA, Liesenfeld O. 13 July 2017. Performance of the cobas MRSA/SA test for simultaneous detection of methicillin-susceptible and methicillin-resistant Staphylococcus aureus from nasal swabs. Am J Clin Pathol doi: 10.1093/ajcp/aqx040. [DOI] [PubMed] [Google Scholar]
- 35.Binnicker MJ, Espy MJ, Duresko B, Irish C, Mandrekar J. 2017. Automated processing, extraction and detection of herpes simplex virus types 1 and 2: a comparative evaluation of three commercial platforms using clinical specimens. J Clin Virol 89:30–33. doi: 10.1016/j.jcv.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Van Der Pol B. 2016. Type-specific detection of herpes simplex virus type 1 and type 2 using the cobas HSV 1 and 2 test on the cobas 4800 platform. Expert Rev Mol Diagn 16:1145–1154. doi: 10.1080/14737159.2016.1243473. [DOI] [PubMed] [Google Scholar]
- 37.Chernesky MA, Jang D, Gilchrist J, Smieja M, Arias M, Hatchette T, Poirier A, Mayne D, Ratnam S. 2017. Comparison of cobas 4800, m2000, Viper XTR, and Infinity 80 automated instruments when processing urine specimens for the diagnosis of Chlamydia trachomatis and Neisseria gonorrhoeae. Sex Transm Dis 44(3):161–165. [DOI] [PubMed] [Google Scholar]
- 38.Peuchant O, de Diego S, Le Roy C, Frantz-Blancpain S, Hocké C, Bébéar C, de Barbeyrac B. 2015. Comparison of three real-time PCR assays for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae in young pregnant women. Diagn Microbiol Infect Dis 83:335–337. doi: 10.1016/j.diagmicrobio.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Bromhead C, Liyanarachchy N, Mayes J, Upton A, Balm M. 2015. Supplementary testing is not required in the cobas 4800 CT/NG test for Neisseria gonorrhoeae weak-positive urogenital samples. J Clin Microbiol 53:327–328. doi: 10.1128/JCM.01976-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ratnam S, Jang D, Gilchrist J, Smieja M, Poirier A, Hatchette T, Flandin JF, Chernesky M. 2014. Workflow and maintenance characteristics of five automated laboratory instruments for the diagnosis of sexually transmitted infections. J Clin Microbiol 52:2299–2304. doi: 10.1128/JCM.03549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chernesky M, Jang D, Gilchrist J, Hatchette T, Poirier A, Flandin JF, Smieja M, Ratnam S. 2014. Head-to-head comparison of second-generation nucleic acid amplification tests for detection of Chlamydia trachomatis and Neisseria gonorrhoeae on urine samples from female subjects and self-collected vaginal swabs. J Clin Microbiol 52:2305–2310. doi: 10.1128/JCM.03552-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geelen TH, Rossen JW, Beerens AM, Poort L, Morré SA, Ritmeester WS, van Kruchten HE, van de Pas MM, Savelkoul PH. 2013. Performance of cobas 4800 and m2000 real-time assays for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in rectal and self-collected vaginal specimen. Diagn Microbiol Infect Dis 77:101–105. doi: 10.1016/j.diagmicrobio.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 43.Pettus JR, Wilson TL, Steinmetz HB, Lefferts JA, Tafe LJ. 2016. Utility of the Roche Cobas 4800 for detection of high-risk human papillomavirus in formalin-fixed paraffin-embedded oropharyngeal squamous cell carcinoma. Exp Mol Pathol 102:47–49. doi: 10.1016/j.yexmp.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Lee DH, Hwang NR, Lim MC, Yoo CW, Joo J, Kim JY, Park SY, Hwang SH. 2016. Comparison of the performance of Anyplex II HPV HR, the Cobas 4800 human papillomavirus test and Hybrid Capture 2. Ann Clin Biochem 53:561–567. doi: 10.1177/0004563215614036. [DOI] [PubMed] [Google Scholar]
- 45.Álvarez-Argüelles ME, de Oña-Navarro M, Rojo-Alba S, Torrens-Muns M, Junquera-Llaneza ML, Antonio-Boga J, Pérez-Castro S, Melón-García S. 2015. Quantification of human papilloma virus (HPV) DNA using the Cobas 4800 system in women with and without pathological alterations attributable to the virus. J Virol Methods 222:95–102. doi: 10.1016/j.jviromet.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 46.Mourah S, Denis MG, Narducci FE, Solassol J, Merlin JL, Sabourin JC, Scoazec JY, Ouafik L, Emile JF, Heller R, Souvignet C, Bergougnoux L, Merlio JP. 2015. Detection of BRAF V600 mutations in melanoma: evaluation of concordance between the Cobas 4800 BRAF V600 mutation test and the methods used in French National Cancer Institute (INCa) platforms in a real-life setting. PLoS One 10:e0120232. doi: 10.1371/journal.pone.0120232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ardakani NM, Giardina T, Grieu-Iacopetta F, Tesfai Y, Carrello A, Taylor J, Robinson C, Spagnolo D, Amanuel B. 2016. Detection of epidermal growth factor receptor mutations in lung adenocarcinoma: comparing Cobas 4800 EGFR assay with Sanger bidirectional sequencing. Clin Lung Cancer 17:e113–e119. doi: 10.1016/j.cllc.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 48.Harlé A, Busser B, Rouyer M, Harter V, Genin P, Leroux A, Merlin JL. 2013. Comparison of COBAS 4800 KRAS, TaqMan PCR and high resolution melting PCR assays for the detection of KRAS somatic mutations in formalin-fixed paraffin embedded colorectal carcinomas. Virchows Arch 462:329–335. doi: 10.1007/s00428-013-1380-x. [DOI] [PubMed] [Google Scholar]
- 49.US Food and Drug Administration. 2007. Statistical guidance on reporting results from studies evaluating diagnostic tests. US Food and Drug Administration, Washington, DC: https://www.fda.gov/RegulatoryInformation/Guidances/ucm071148.htm. Accessed 6 June 2017. [Google Scholar]
- 50.US Food and Drug Administration. 2015. Guideline for industry and Food and Drug Administration staff. Class II special controls guideline document: toxin gene amplification assays for the detection of Clostridium difficile. US Food and Drug Administration, Washington, DC. [Google Scholar]